We narrowed to 7,456 results for: PROC
-
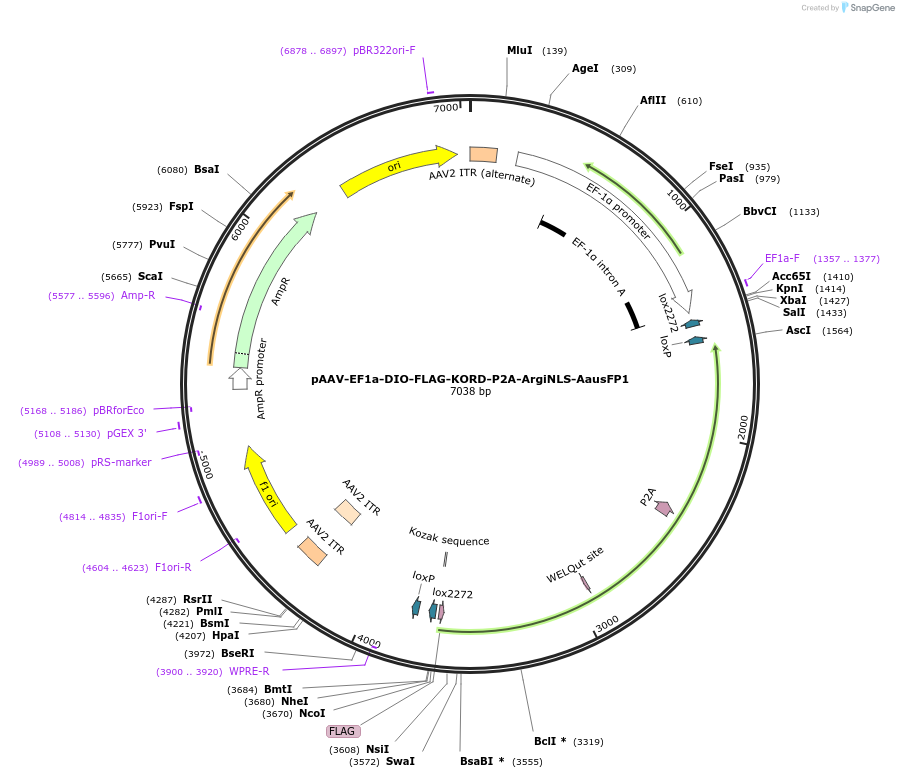
Plasmid#220612PurposeCre-dependent co-expression of FLAG-tagged inhibitory KORD DREADD receptor, and a single-cell discriminating version of AausFP1 as a reporter.DepositorInsertFLAG-KORD-P2A-ArgiNLS-AausFP1
UseAAV and Cre/LoxTagsFLAG; ArgiNLSExpressionMammalianPromoterEF1aAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
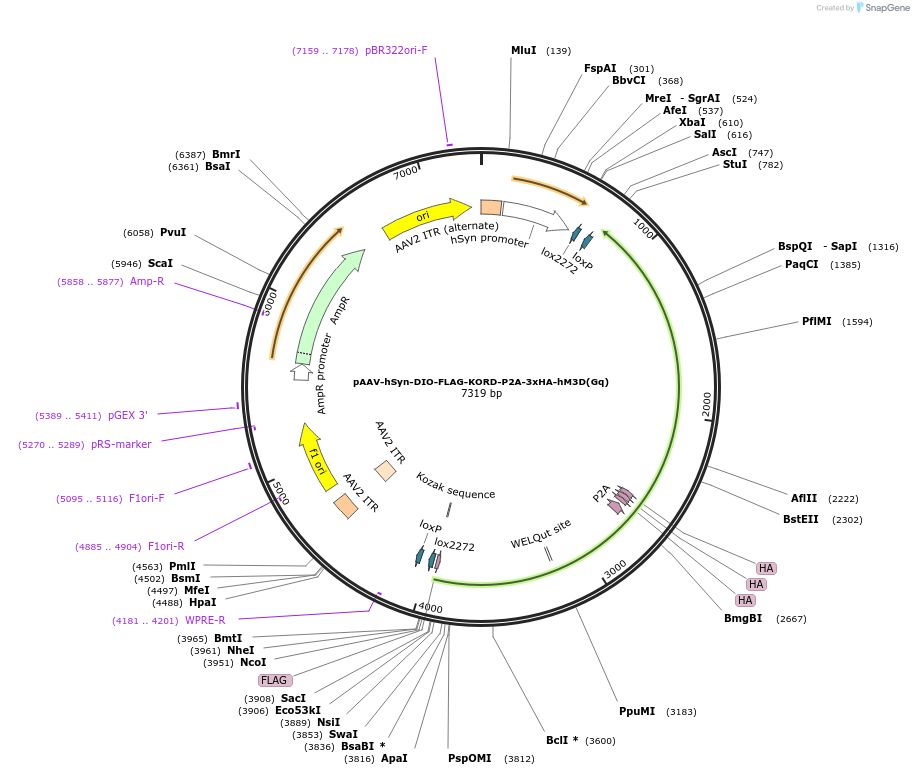
pAAV-hSyn-DIO-FLAG-KORD-P2A-3xHA-hM3D(Gq)
Plasmid#220611PurposeNeuron-specific, Cre-dependent co-expression of FLAG-tagged inhibitory KORD DREADD receptor, and 3x HA-tagged excitatory hM3D(Gq) DREADD receptor.DepositorInsertFLAG-KORD-P2A-3x HA-hM3D(Gq)
UseAAV and Cre/LoxTagsFLAG; ArgiNLSExpressionMammalianPromoterhSynAvailable SinceAug. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
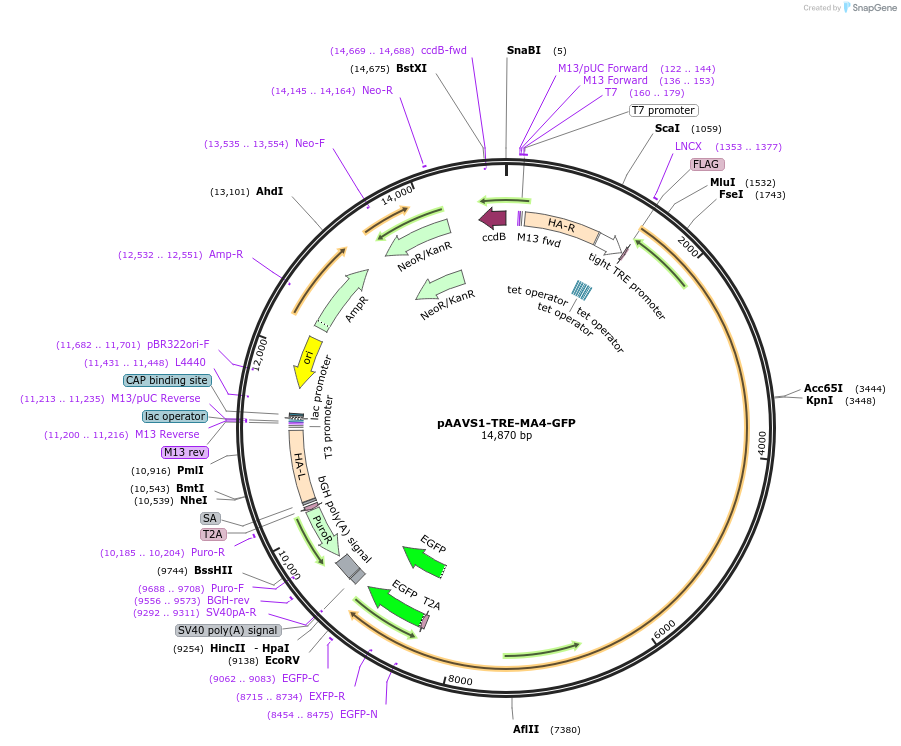
pAAVS1-TRE-MA4-GFP
Plasmid#114380PurposeDoxycycline inducible MLL-AF4 and GFP expression. The construct has been used with CAG-rtTA for making engraftable iPSC derived HSCs.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
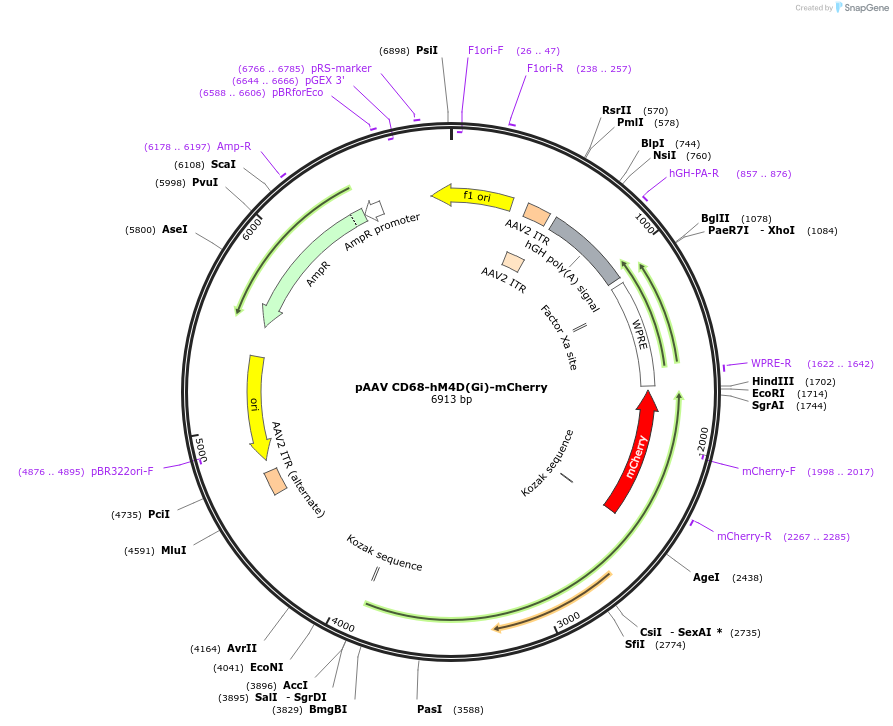
pAAV CD68-hM4D(Gi)-mCherry
Plasmid#75033PurposeExpression of hM4D(Gi) using the CD68 promoter.DepositorHas ServiceAAV1 and AAV9InserthM4D(Gi)-mCherry
UseAAVExpressionMammalianPromoterCD68Available SinceOct. 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
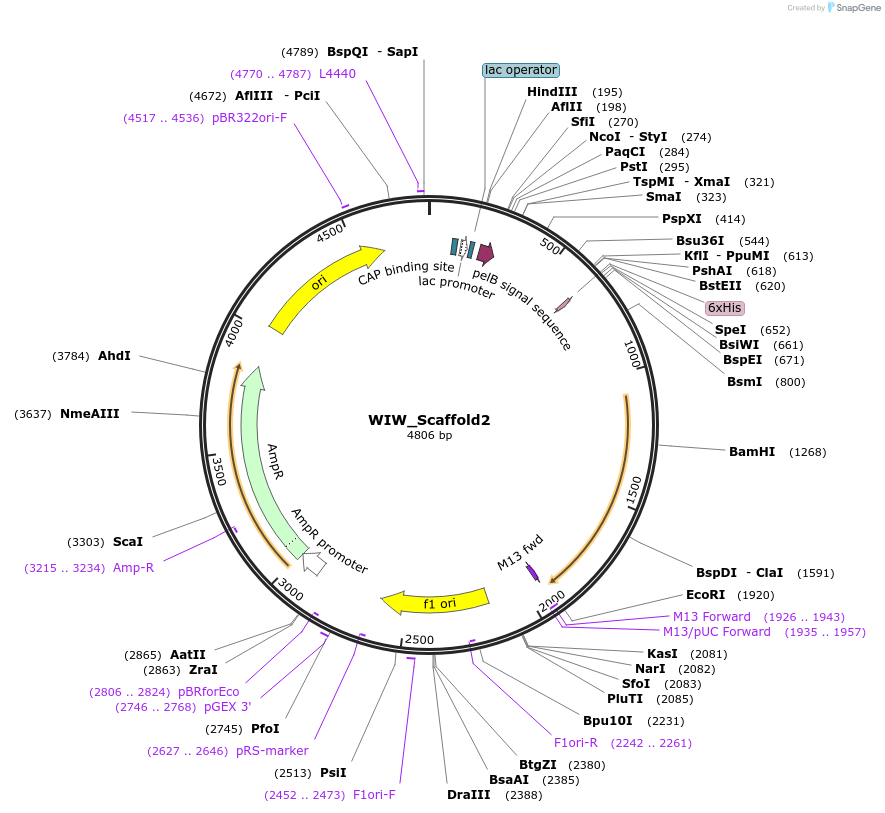
WIW_Scaffold2
Plasmid#205574PurposeNanobody that inhibits seeding of tau fibrilsDepositorInsertWIW_Scaffold2
Tags6X HisExpressionBacterialAvailable SinceDec. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
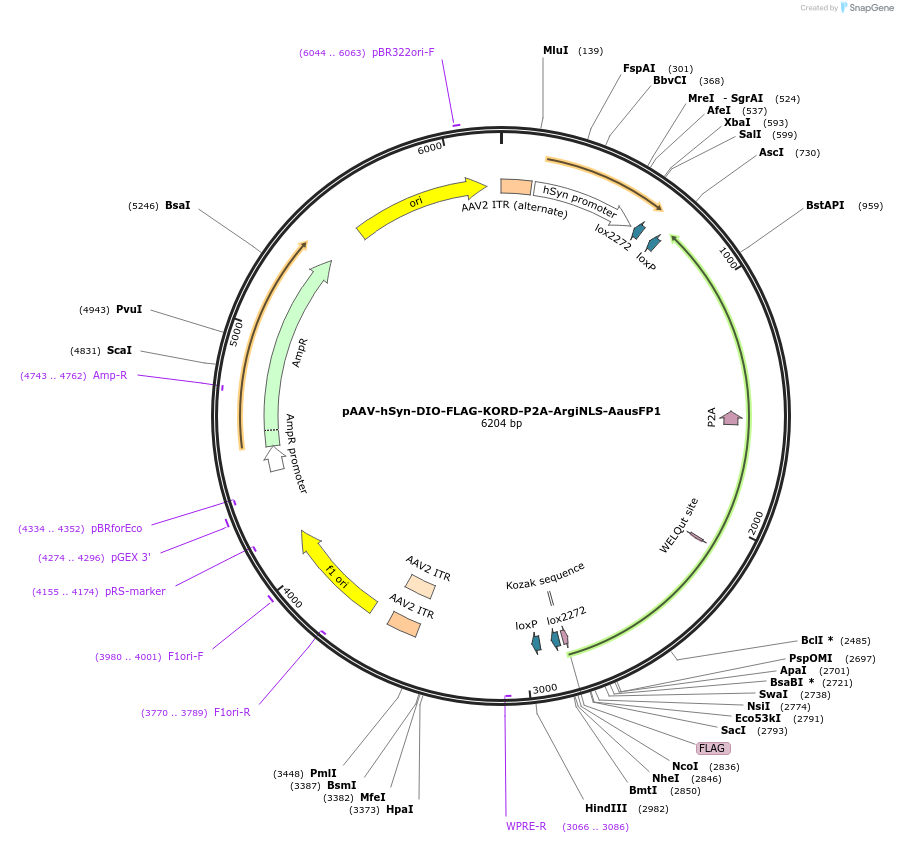
pAAV-hSyn-DIO-FLAG-KORD-P2A-ArgiNLS-AausFP1
Plasmid#220610PurposeNeuron-specific, Cre-dependent co-expression of FLAG-tagged inhibitory KORD DREADD receptor, and a single-cell discriminating version of AausFP1 as a reporterDepositorInsertFLAG-KORD-P2A-ArgiNLS-AausFP1
UseAAV and Cre/LoxTagsFLAG; ArgiNLSExpressionMammalianPromoterhSynAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
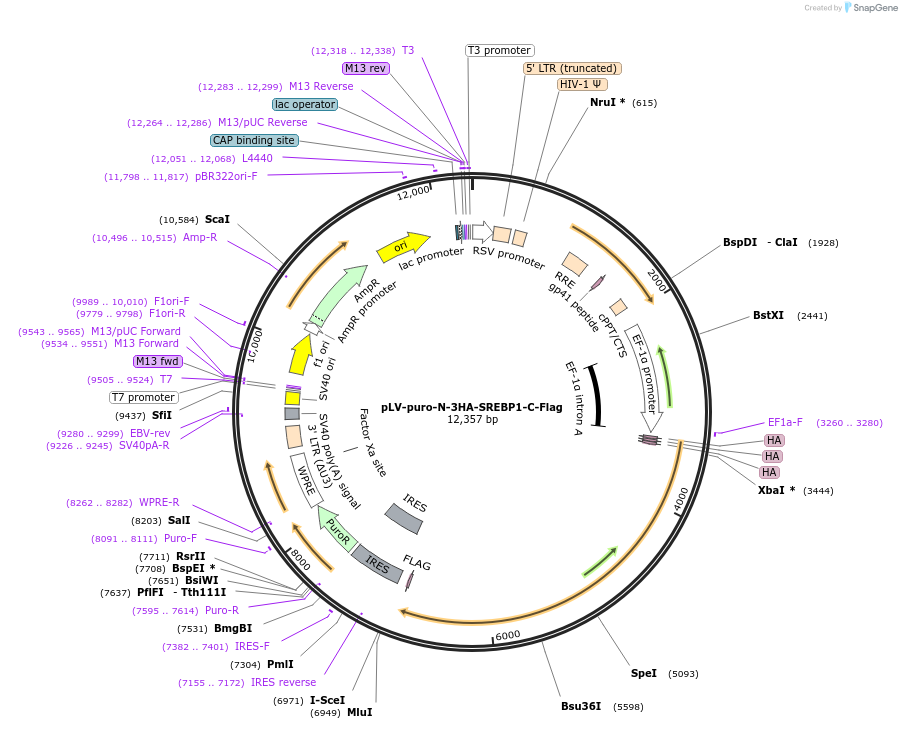
pLV-puro-N-3HA-SREBP1-C-Flag
Plasmid#134286PurposeLentivector encoding 3XHA (N-term) and Flag (C-term)-tagged SREBP1DepositorInsertSREBP1 (Srebf1 Mouse)
UseLentiviralTags3x HA and FlagExpressionMammalianMutationmouse SREBP1 isoform a precursorPromoterEF1aAvailable SinceMarch 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
pAAV-EF1a-DIO-FLAG-hM4D(Gi)-P2A-ArgiNLS-AausFP1
Plasmid#220613PurposeCre-dependent co-expression of FLAG-tagged inhibitory hM4D(Gi) DREADD receptor, and a single-cell discriminating version of AausFP1 as a reporter.DepositorInsertFLAG-hM4D(Gi)-P2A-ArgiNLS-AausFP1
UseAAV and Cre/LoxTagsFLAG; ArgiNLSExpressionMammalianPromoterEF1aAvailable SinceDec. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
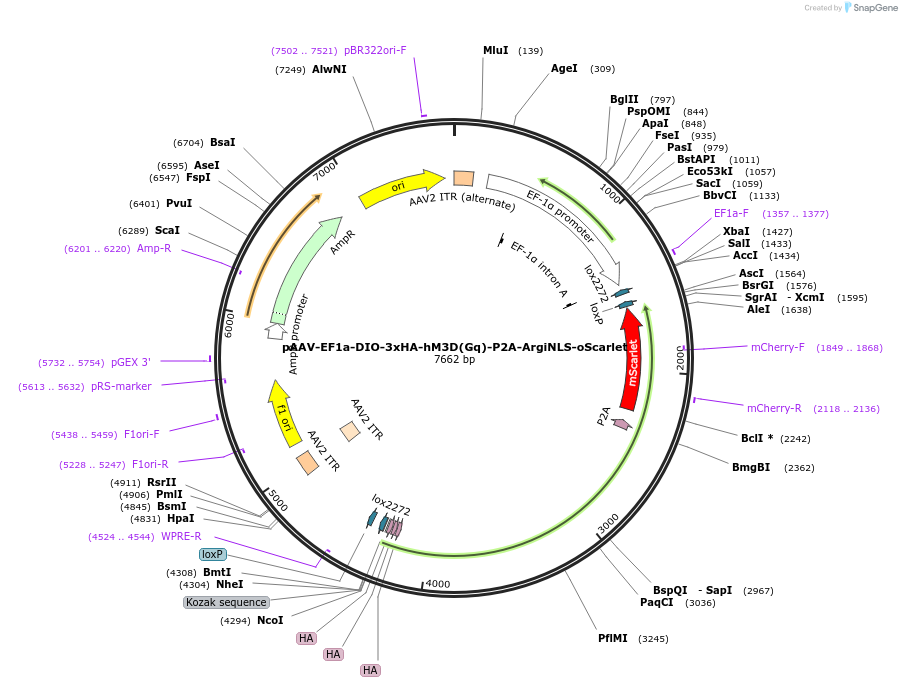
pAAV-EF1a-DIO-3xHA-hM3D(Gq)-P2A-ArgiNLS-oScarlet
Plasmid#220614PurposeCre-dependent co-expression of 3x HA-tagged excitatory hM3D(Gq) DREADD receptor, and a single-cell discriminating version of oScarlet as a reporter.DepositorInsert3x HA-hM3D(Gq)-P2A-AgiNLS-oScarlet
UseAAV and Cre/LoxTags3x HA; ArgiNLSExpressionMammalianPromoterEF1aAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
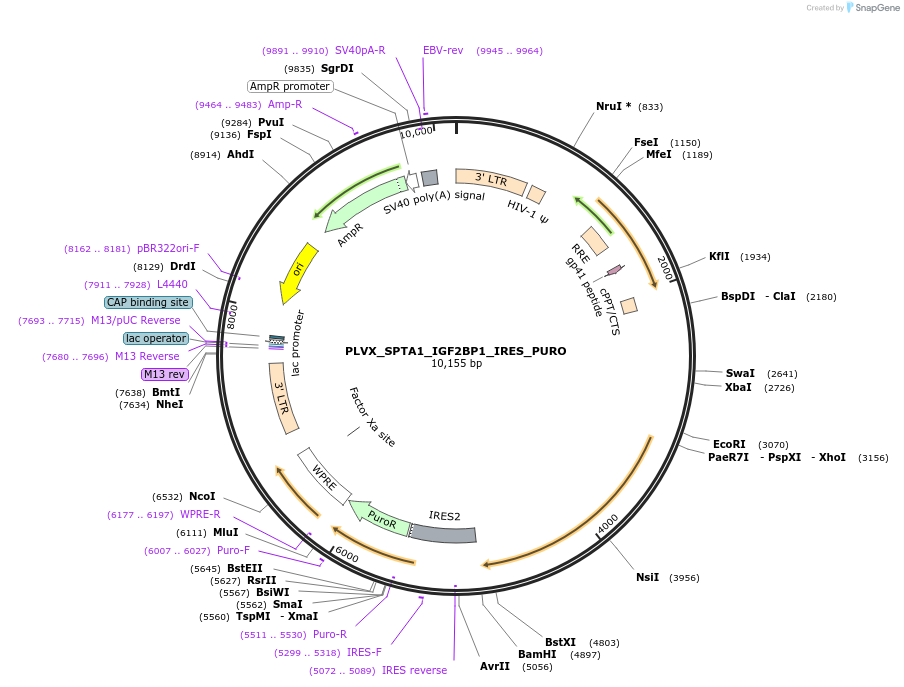
PLVX_SPTA1_IGF2BP1_IRES_PURO
Plasmid#100016PurposeTo over-express IGF2BP1 in Adult Erythroblasts using tissue specific promoter with IRES for puromycin selection.DepositorInsertInsulin like growth factor 2 mRNA binding protein 1 (IGF2BP1 Human)
UseLentiviralExpressionMammalianPromoterSPTA1 erythroid specific promoterAvailable SinceOct. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
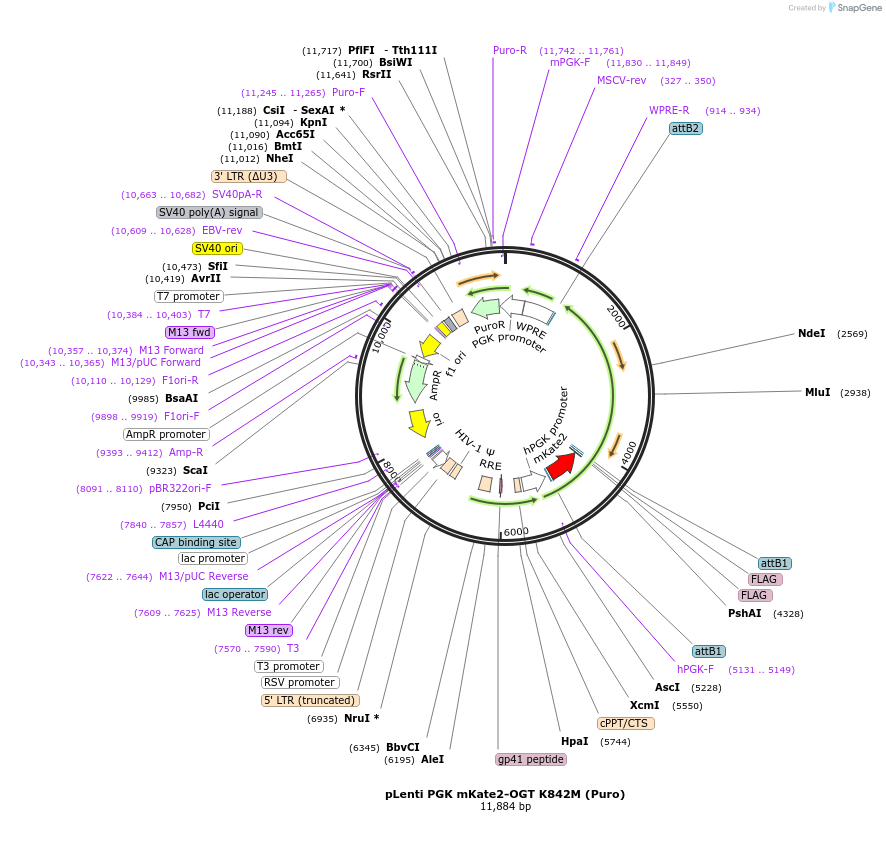
pLenti PGK mKate2-OGT K842M (Puro)
Plasmid#154291PurposeLentiviral vector encoding full length mKate2-2xFLAG-OGT (full length human O-GlcNAc transferase, K842M variant, catalytic dead)DepositorInsertO-GlcNAc Transferase (OGT Human)
UseLentiviralTags2x FLAG and mKate2ExpressionMammalianMutationK842M mutation (Catalytic Inactive)PromoterPGKAvailable SinceDec. 30, 2020AvailabilityAcademic Institutions and Nonprofits only -
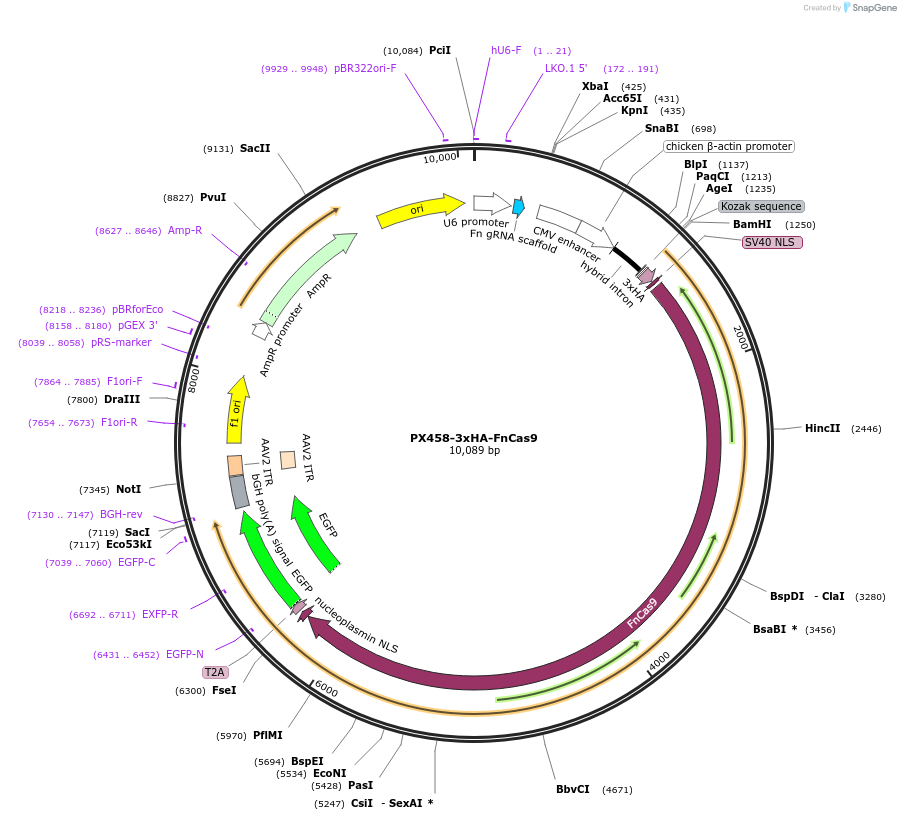
PX458-3xHA-FnCas9
Plasmid#130969Purpose3xHA tagged Cas9 from F. novicida with T2A-EGFP and cloning backbone for sgRNADepositorInsertFnCas9
UseCRISPRTags3xHA-NLS and NLS-T2A-EGFPExpressionMammalianPromoterCbhAvailable SinceNov. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
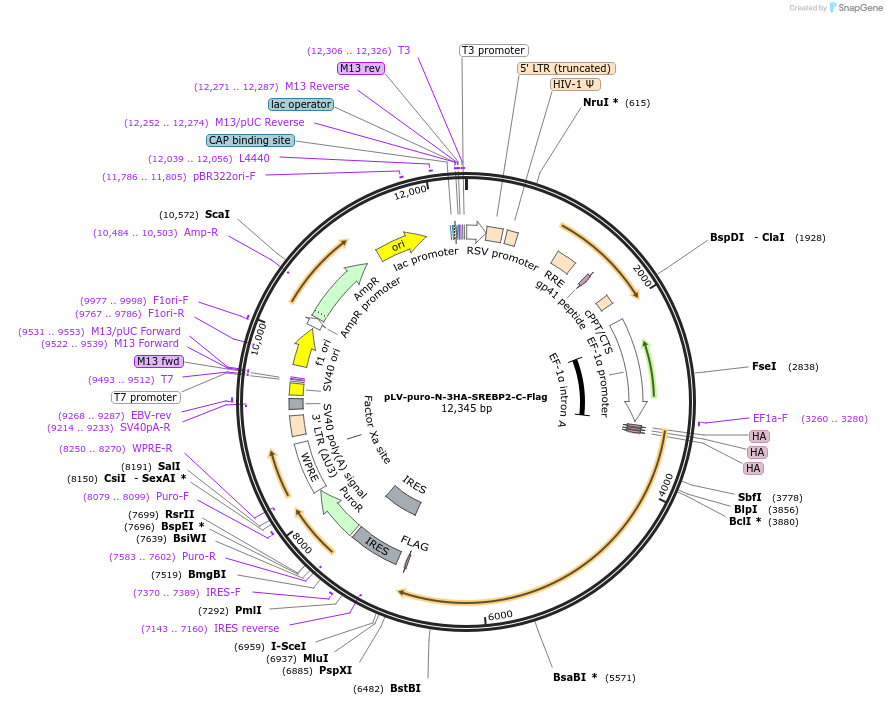
pLV-puro-N-3HA-SREBP2-C-Flag
Plasmid#134287PurposeLentivector encoding 3XHA (N-term) and Flag (C-term)-tagged SREBP2DepositorInsertSREBP2 (Srebf2 Mouse)
UseLentiviralTags3x HA and FlagExpressionMammalianMutationmouse SREBP2 precursorPromoterCMVAvailable SinceMarch 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
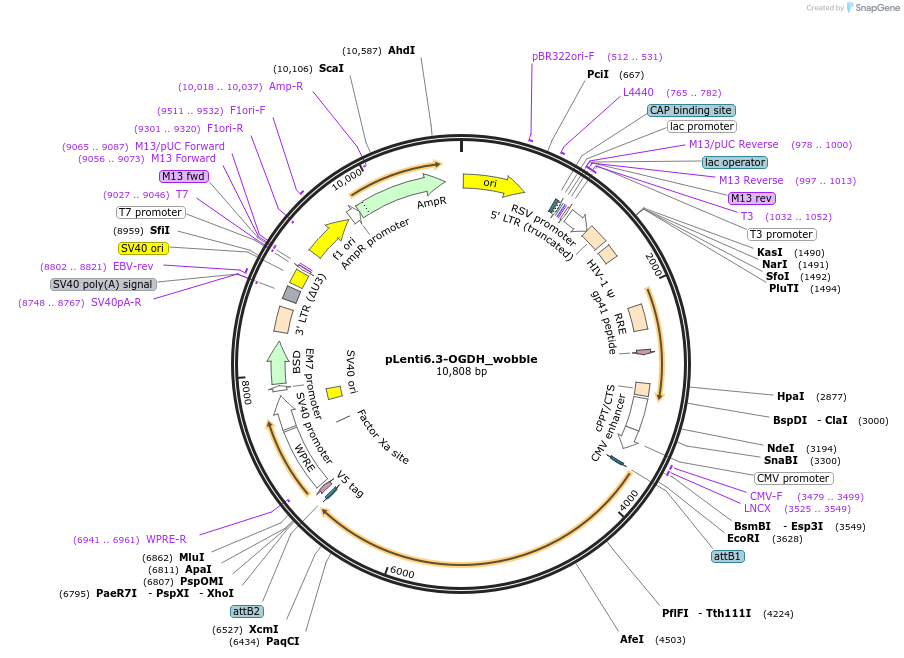
pLenti6.3-OGDH_wobble
Plasmid#110938PurposeOGDH ORF containing silent wobble mutations insensitive to corresponding shRNAsDepositorInsertOGDH wobble cDNA (OGDH Human)
UseLentiviralMutation15 silent wobble mutations corresponding to three…PromoterSV40Available SinceJune 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
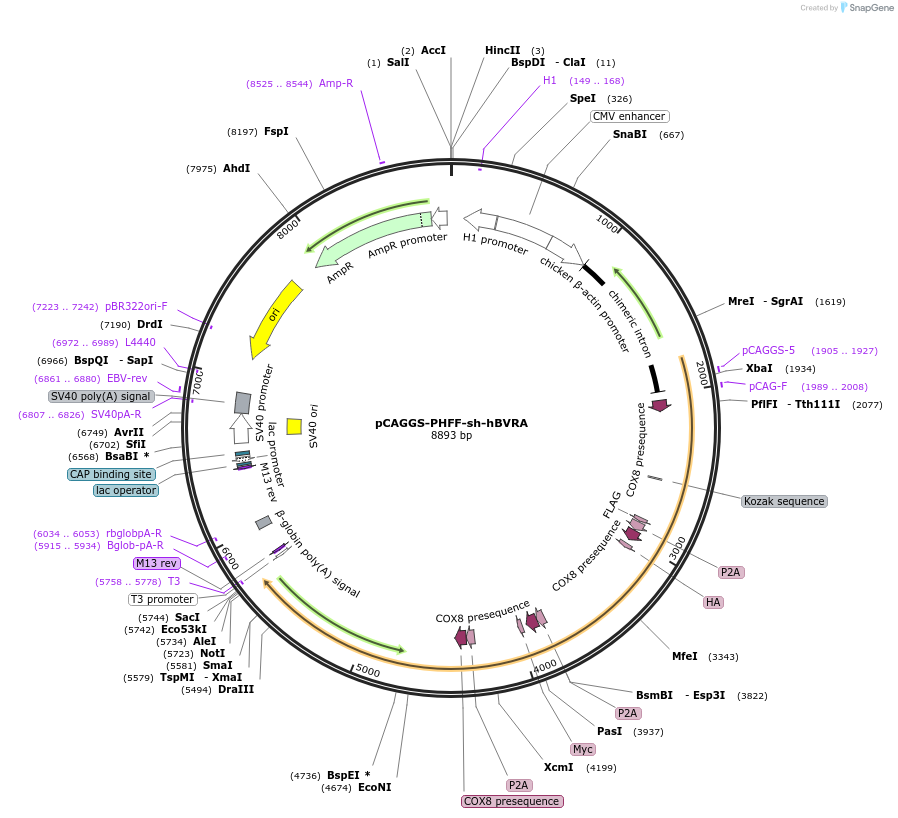
pCAGGS-PHFF-sh-hBVRA
Plasmid#100285PurposeExpression vector for phycocyanobilin (PCB) synthesis and shRNA for human BVRA gene in mammalian cellsDepositorInsertsMTS-PcyA-FLAG-P2A-MTS-HA-HO1-P2A-MTS-Myc-Fd-P2A-MTS-Fnr-T7 (synthetic genes)
shRNA for human BVRA
TagsFLAG, T7 and Mitochondria-targeting sequence (MTS…ExpressionMammalianMutationAll genes are codon-optimized for expression in h…PromoterCAG promoter and H1 promoterAvailable SinceSept. 20, 2017AvailabilityAcademic Institutions and Nonprofits only -
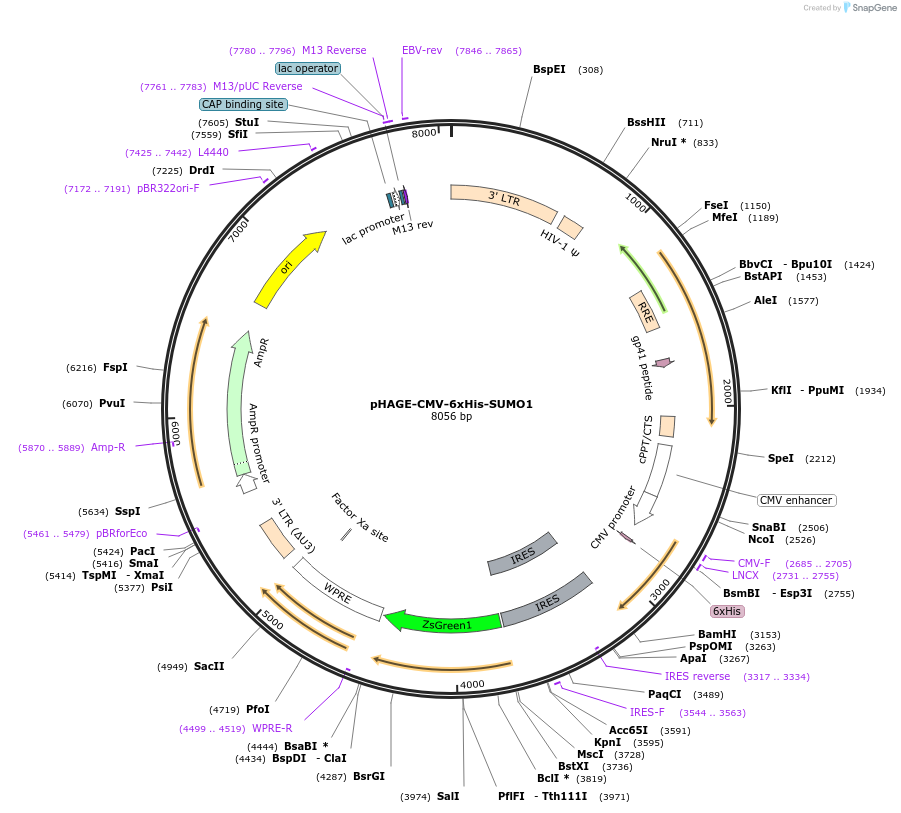
pHAGE-CMV-6xHis-SUMO1
Plasmid#164939PurposeExpresses 6xHis tagged human Small Ubiquitin Like Modifier 1 (SUMO1) wild type cDNADepositorAvailable SinceMay 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
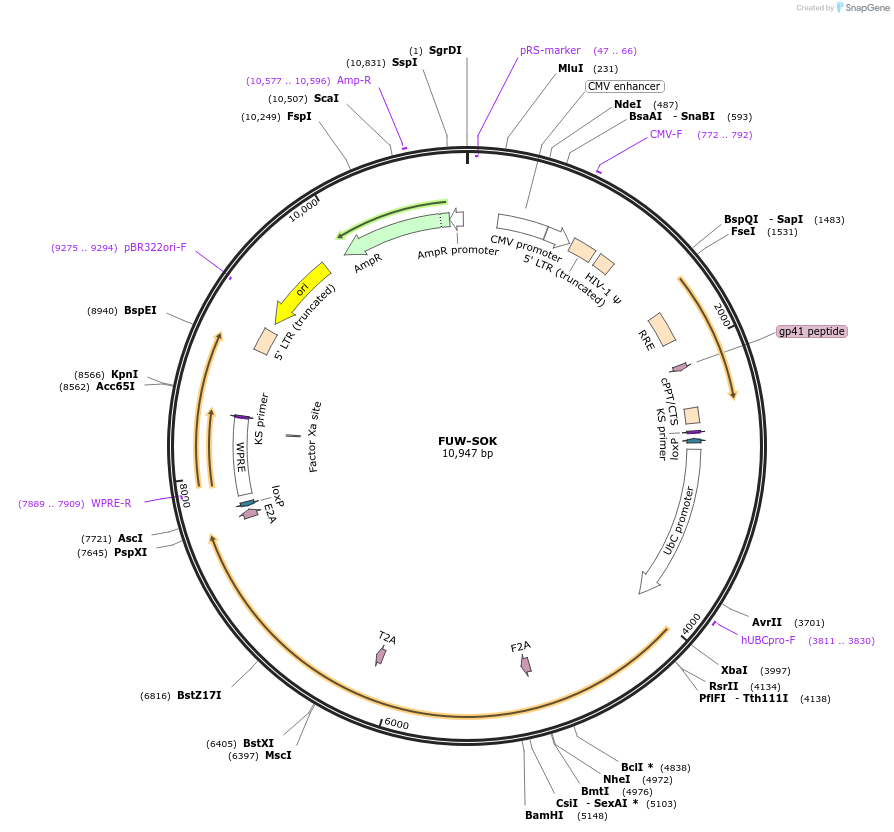
FUW-SOK
Plasmid#20327DepositorAvailable SinceFeb. 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
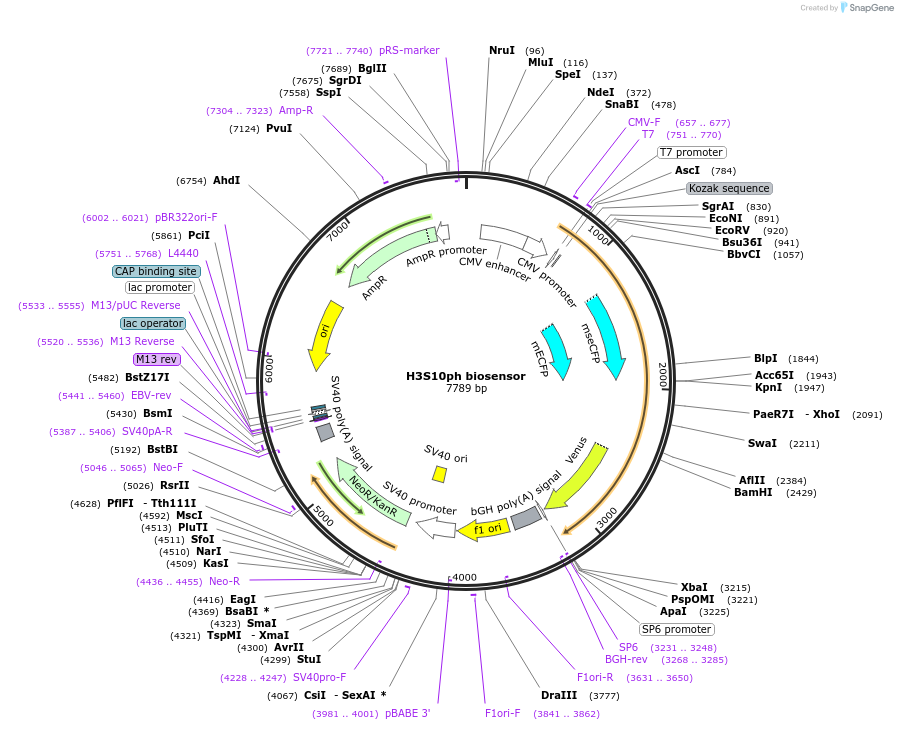
H3S10ph biosensor
Plasmid#120809PurposeFRET biosensor. To monitor histone H3 Serine 10 phosphorylation in mammalian cells by FRETDepositorInsertMouse histone H3-CFP-FHA2-histone H3 peptide (1-14aa)-YFP
ExpressionMammalianMutationThreonine 3, Threonine 6, and Threonine 11 are mu…PromoterCMVAvailable SinceFeb. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
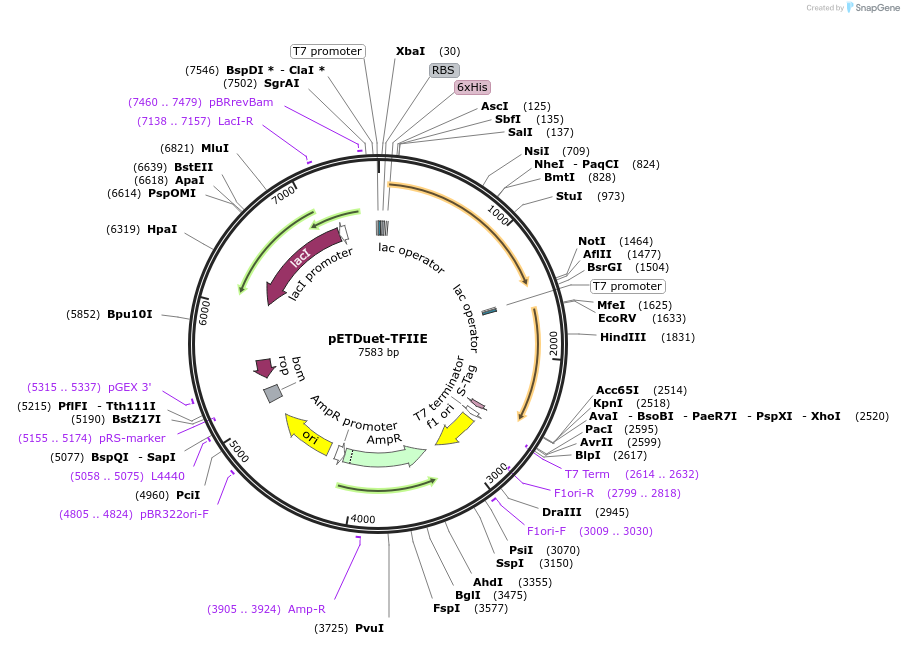
pETDuet-TFIIE
Plasmid#171083PurposeCo-expresses human TFIIE. The resulting plasmid was used to generate a single expression construct encoding 2 subunits, with His-tag on TF2E1.Originally from MN Gonzalez, was submitted with permissionDepositorTagsHisExpressionBacterialPromoterT7 promoterAvailable SinceJuly 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
hV1aR-GFP
Plasmid#67846Purposemammalian expression of human V1a receptor with GFP tagDepositorAvailable SinceOct. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
hV1b-GFP-1
Plasmid#67847Purposemammalian expression of human V1b receptor with GFP tagDepositorAvailable SinceOct. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
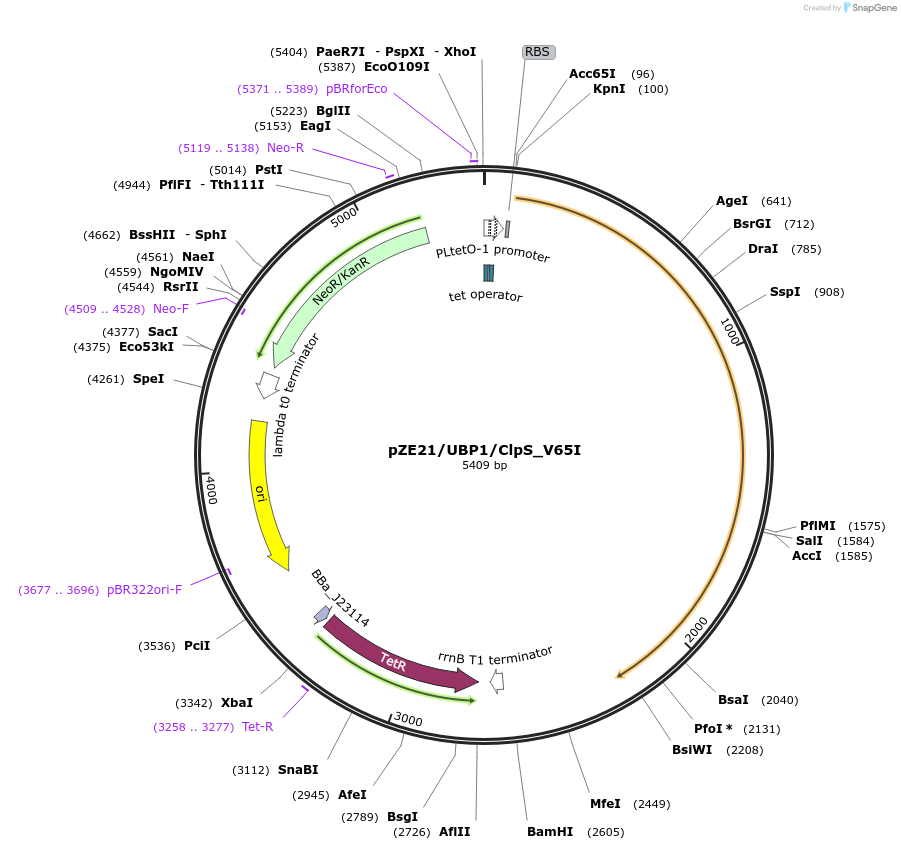
pZE21/UBP1/ClpS_V65I
Plasmid#98567PurposeExpresses truncated codon-opt S. cerevisiae UBP1 and the V65I engineered variant of E. coli ClpS in an artificial operonDepositorInsertsTruncated ubiquitin cleavase, codon optimized
Mutated ATP-dependent Clp protease adapter protein
ExpressionBacterialMutationContains the V65I mutation that increases discrim…Available SinceFeb. 7, 2018AvailabilityAcademic Institutions and Nonprofits only -
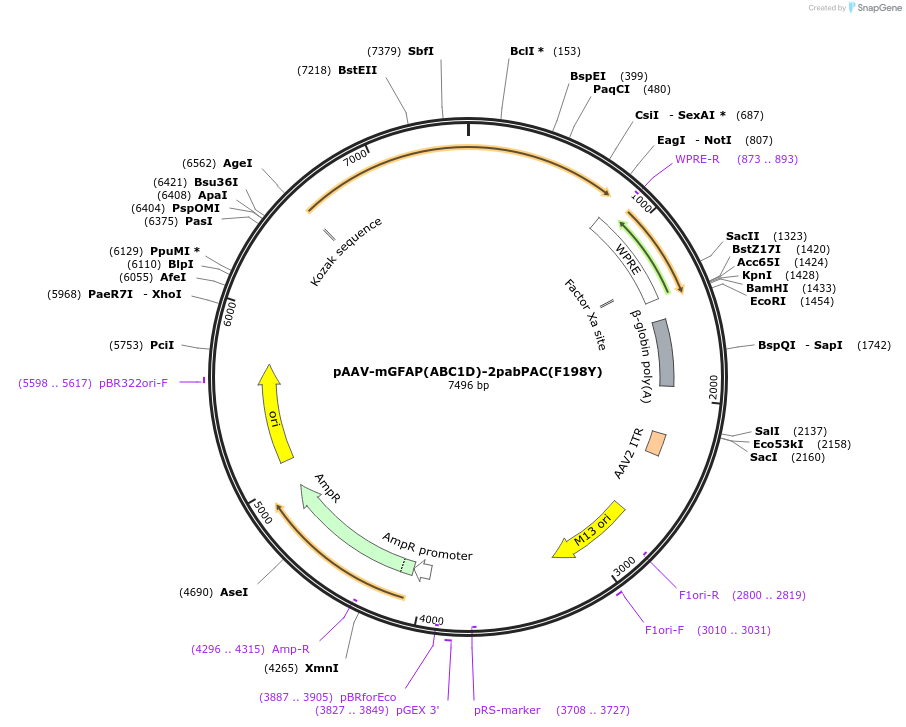
pAAV-mGFAP(ABC1D)-2pabPAC(F198Y)
Plasmid#234548Purposeto express a version with reduced constitutive activity (thanks to point mutation F198Y) of the optogenetic tool 2pabPAC, which increases cAMP levels when stimulated by blue light, in astrocytesDepositorInsert2pabPAC(F198Y)
UseAAVMutationF198YPromotermGFAP(ABC1D)Available SinceJuly 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
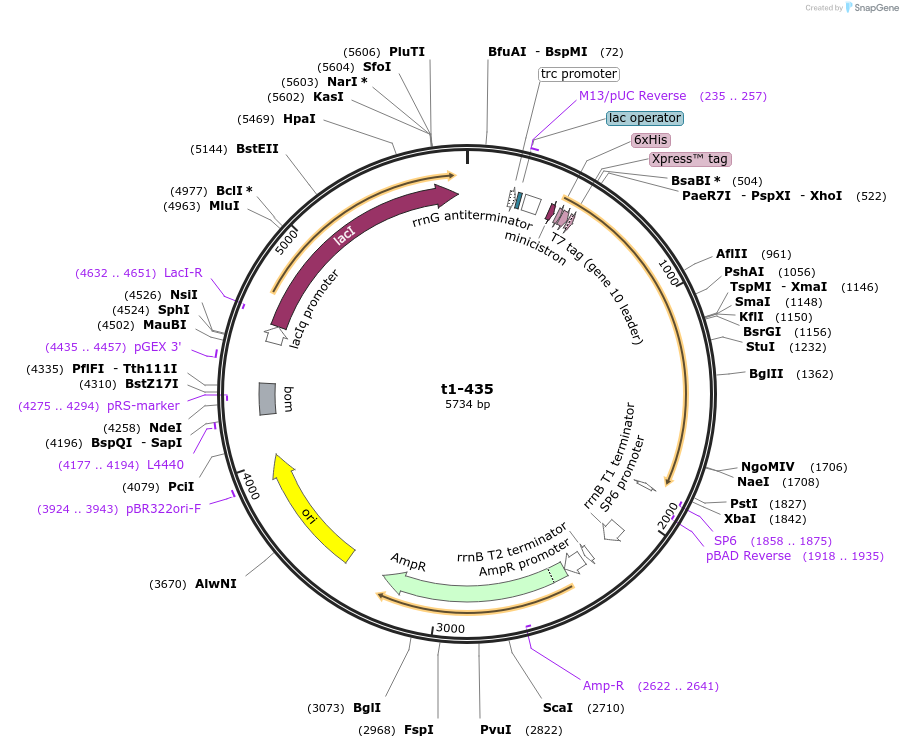
t1-435
Plasmid#166578PurposeFor expression of human talin head (residues 1-435) in E. coli. N-terminal His6-tag, Xpress-epitope (DLYDDDDK) and enterokinase cleavage site for tag removal.DepositorInsertTln1 Head (1-435) (TLN1 Human)
TagsHis6-tag, Xpress-epitope (DLYDDDDK) and enterokin…ExpressionBacterialMutationresidues 1-435 onlyAvailable SinceApril 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
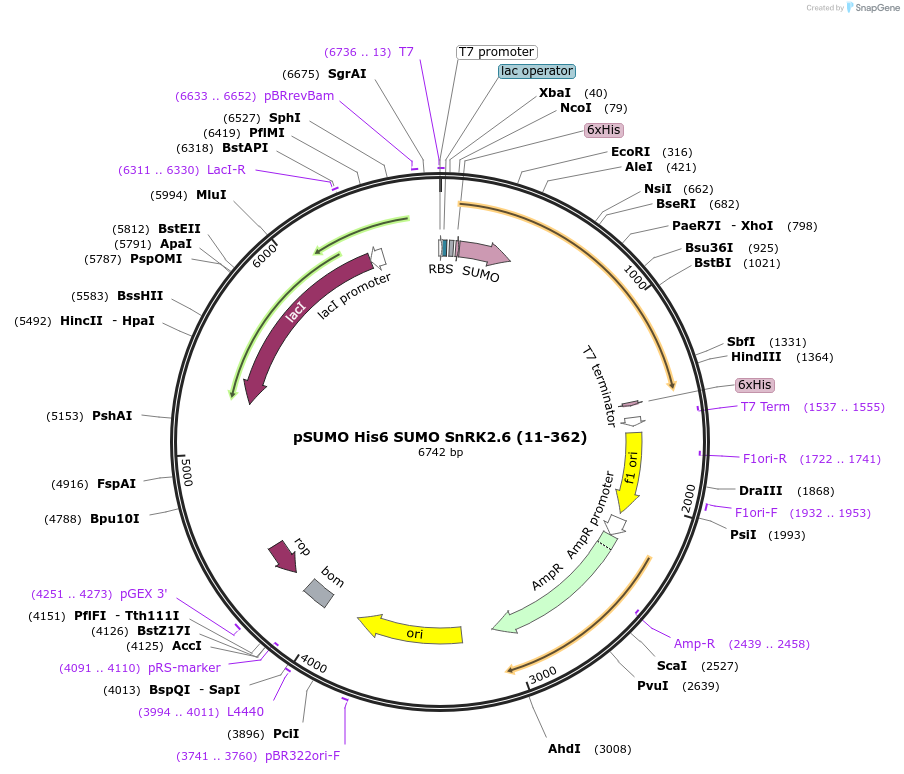
pSUMO His6 SUMO SnRK2.6 (11-362)
Plasmid#177847PurposeBacterial Expression of SnRK2.6DepositorAvailable SinceDec. 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCEP4-FLAG-TAPBPR-TM-VC
Plasmid#135499PurposeMammalian expression of VC-fused and FLAG-tagged TAPBPR with MHC TMDepositorInsertTAPBPR (TAPBPL Human)
TagsLuminal/Extracellular FLAG epitope tag, Signal pe…ExpressionMammalianMutationswitched transmembrane domain with that of MHC-IPromoterCMVAvailable SinceFeb. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
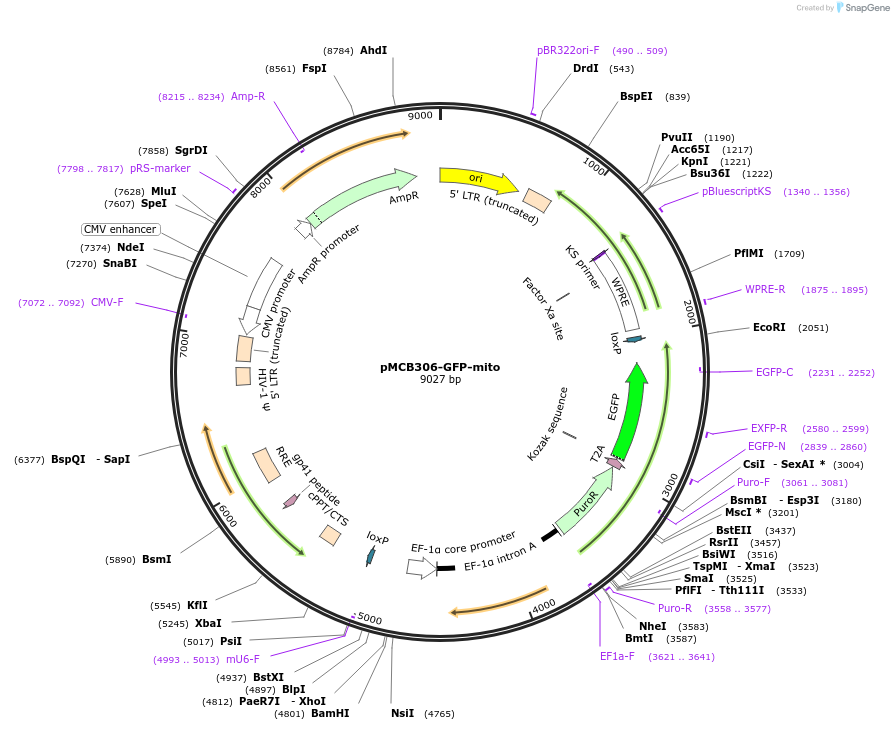
pMCB306-GFP-mito
Plasmid#229232PurposeExpresses GFP localized to mitochondriaDepositorAvailable SinceFeb. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
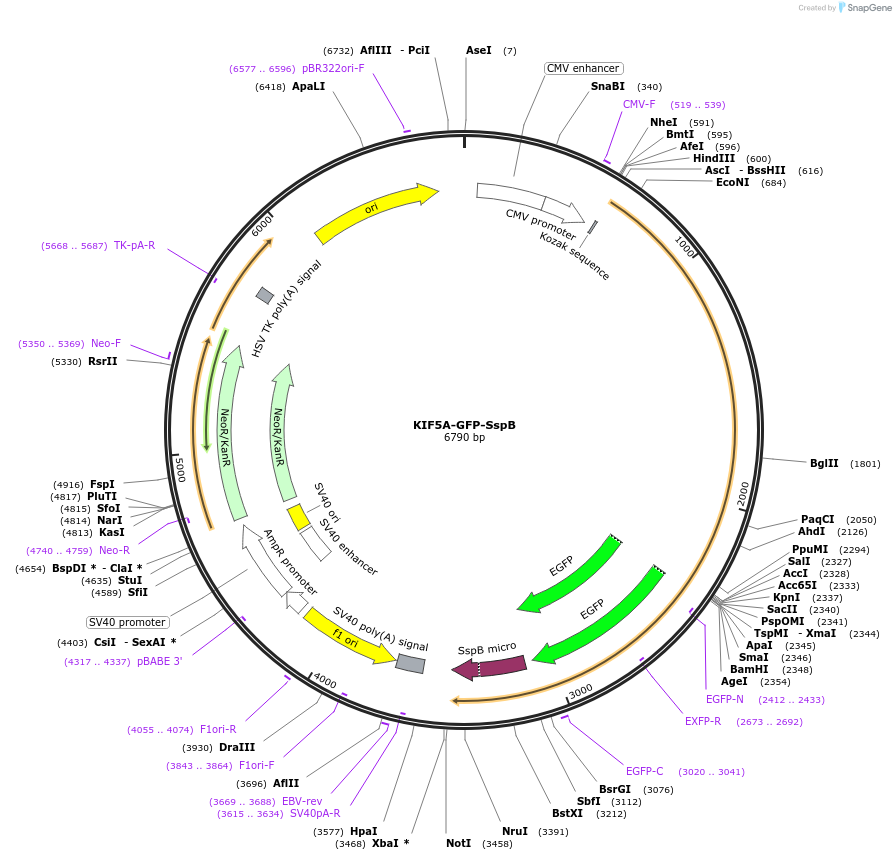
KIF5A-GFP-SspB
Plasmid#214401PurposeExpresses fusion of Kinesin heavy chain 5A (1-572) with GFP and SspB (micro)DepositorAvailable SinceMarch 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
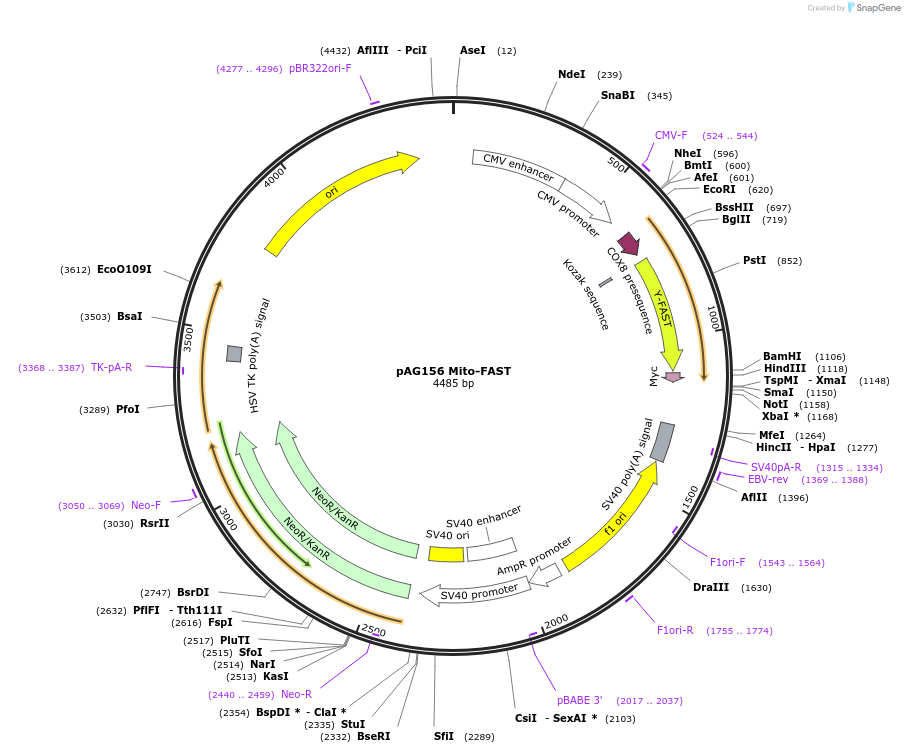
pAG156 Mito-FAST
Plasmid#130723PurposeExpresses FAST (also called YFAST) fused to a mitochondria targeting sequence (mito) in mammalian cellsDepositorInsertMito-FAST
TagsMyc-tagExpressionMammalianPromoterCMVAvailable SinceSept. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
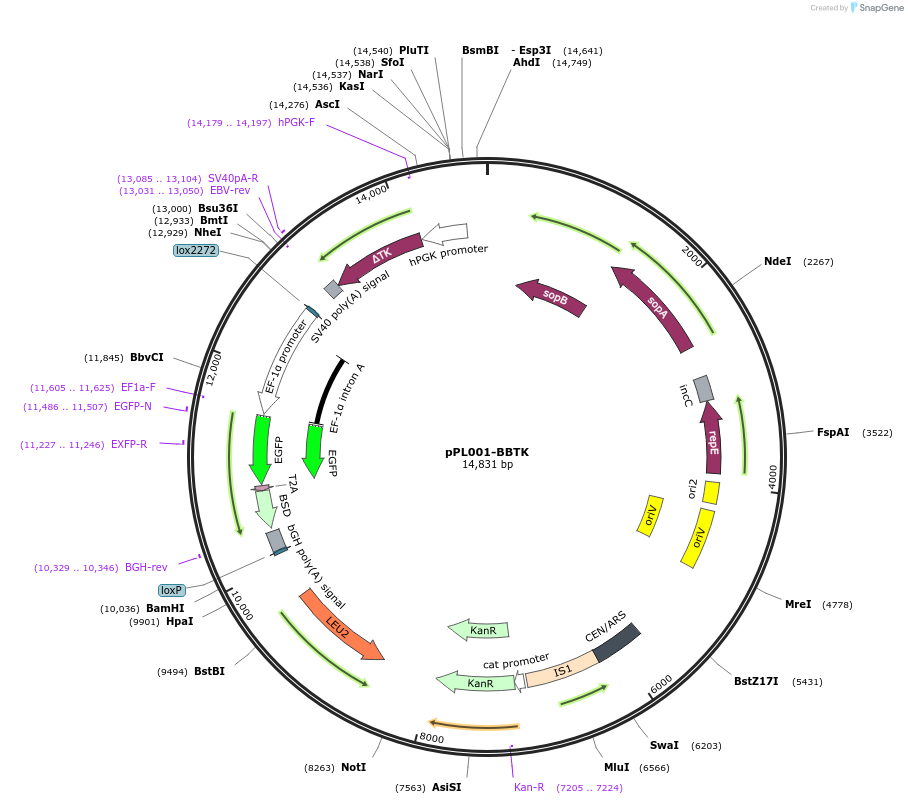
pPL001-BBTK
Plasmid#184751PurposeBig-IN positive control payload encoding EF1a-driven GFP-T2A-BSD. Harbors a backbone counterselectable TK cassette.DepositorInsertpEF1a-eGFP-T2A-BSD-bGHpA
UseCre/Lox, Mouse Targeting, and Synthetic Biology ;…ExpressionMammalianPromoterEF1aAvailable SinceJan. 17, 2023AvailabilityAcademic Institutions and Nonprofits only