We narrowed to 7,918 results for: chloramphenicol
-
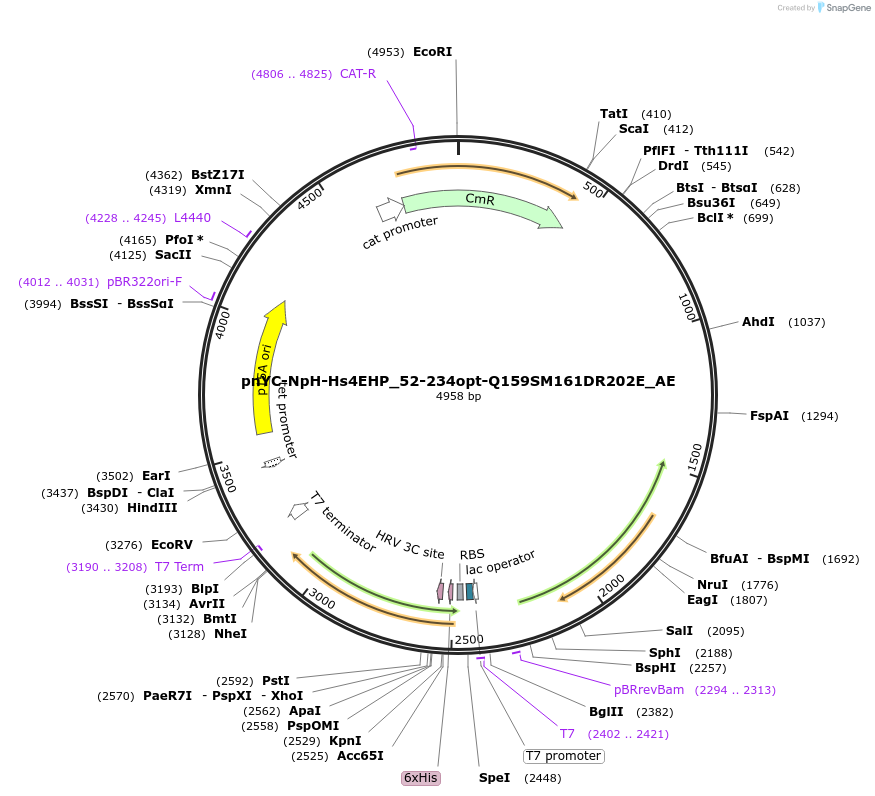
Plasmid#148567PurposeBacterial Expression of Hs4EHP_52-234opt-Q159SM161DR202EDepositorInsertHs4EHP_52-234opt-Q159SM161DR202E (EIF4E2 Human)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
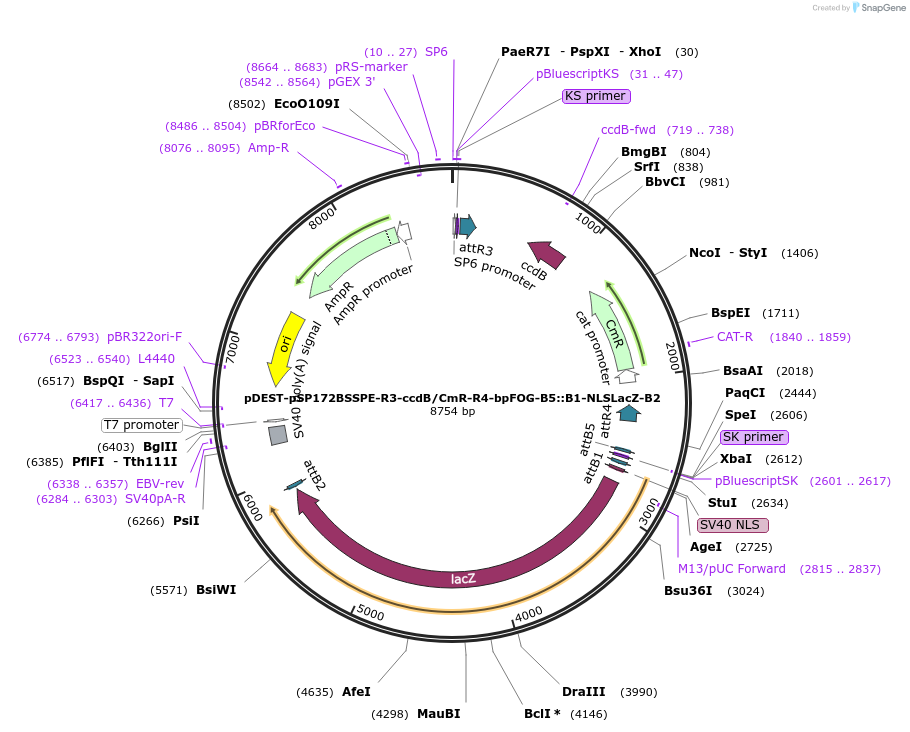
pDEST-pSP172BSSPE-R3-ccdB/CmR-R4-bpFOG-B5::B1-NLSLacZ-B2
Plasmid#186414PurposeAttR3/R4 destination vector for ascidian electroporation with AttB1/B2 recombined NLSLacZ under control of AttB4/B5-recombined pFOG ectodermal regulatory sequence.DepositorInsertlacZ (lacZ E. coli)
UseGateway destination vectorTagsNuclear localization signalPromoterBasal promoter of the FOG geneAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
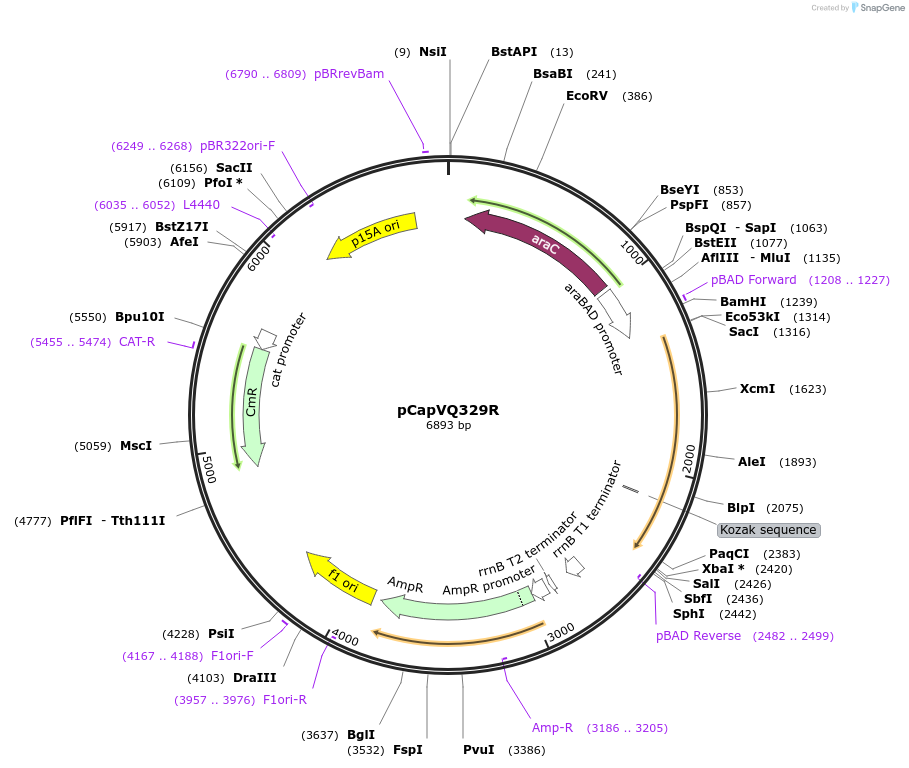
pCapVQ329R
Plasmid#183029Purposethe patatin-like phosphodiesterase CapV variant CapVQ329R cloned in the L-arabinose inducible vector pBAD28DepositorInsertpatatin-like phospholipase variant CapVQ329R
TagsNoExpressionBacterialMutationglutamine 329 changed to argininePromoterpBAD promoterAvailable SinceMay 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
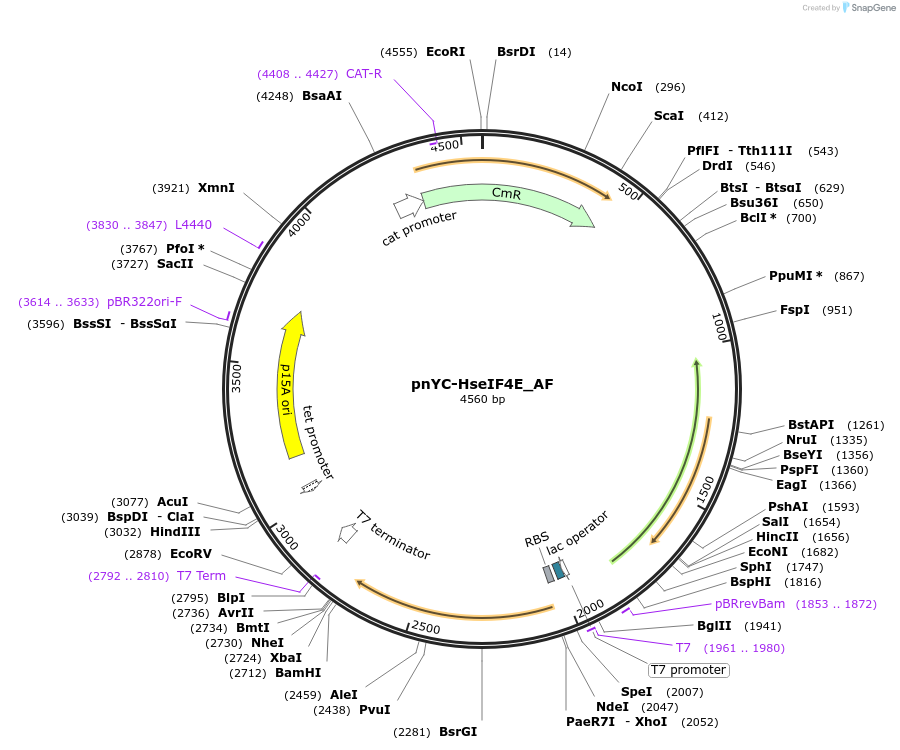
pnYC-HseIF4E_AF
Plasmid#148669PurposeBacterial Expression of HseIF4EDepositorInsertHseIF4E (EIF4E Human)
ExpressionBacterialMutationone silent mutation compared to the sequence of i…Available SinceMarch 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
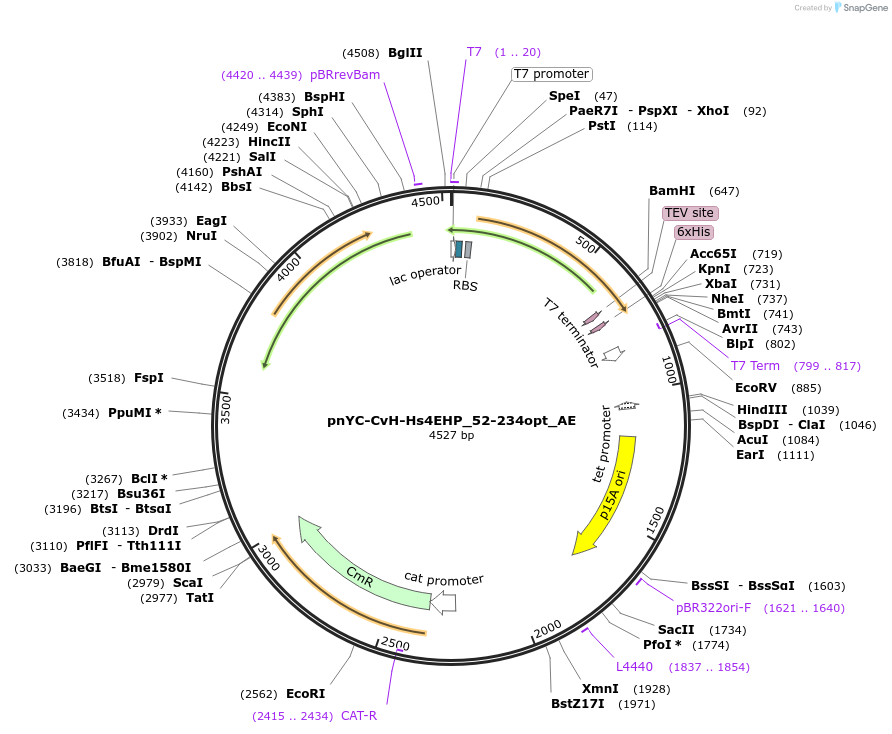
pnYC-CvH-Hs4EHP_52-234opt_AE
Plasmid#148564PurposeBacterial Expression of Hs4EHP_52-234optDepositorInsertHs4EHP_52-234opt (EIF4E2 Human)
ExpressionBacterialAvailable SinceMarch 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
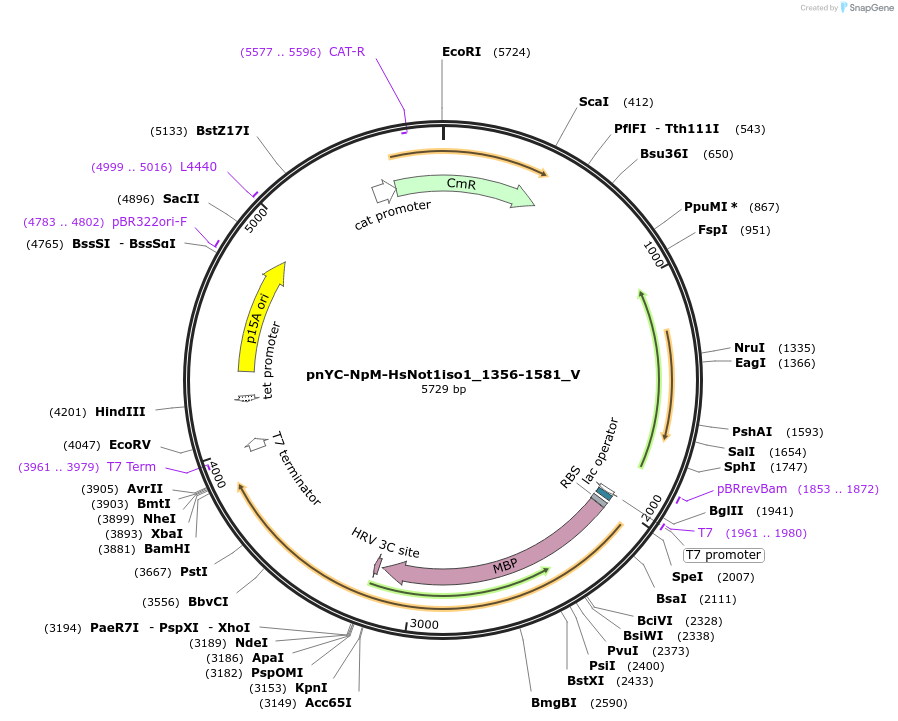
pnYC-NpM-HsNot1iso1_1356-1581_V
Plasmid#147779PurposeBacterial Expression of HsNot1iso1_1356-1581DepositorInsertHsNot1iso1_1356-1581 (CNOT1 Human)
ExpressionBacterialAvailable SinceMarch 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
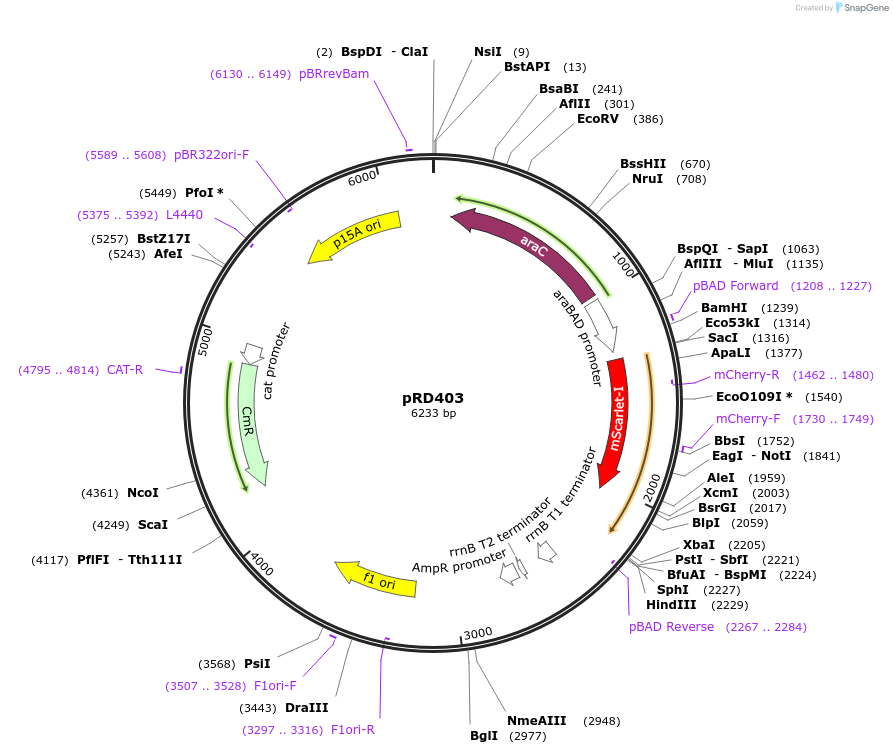
pRD403
Plasmid#163108PurposeExpression of mScarlet-I_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInsertmScarlet-I_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
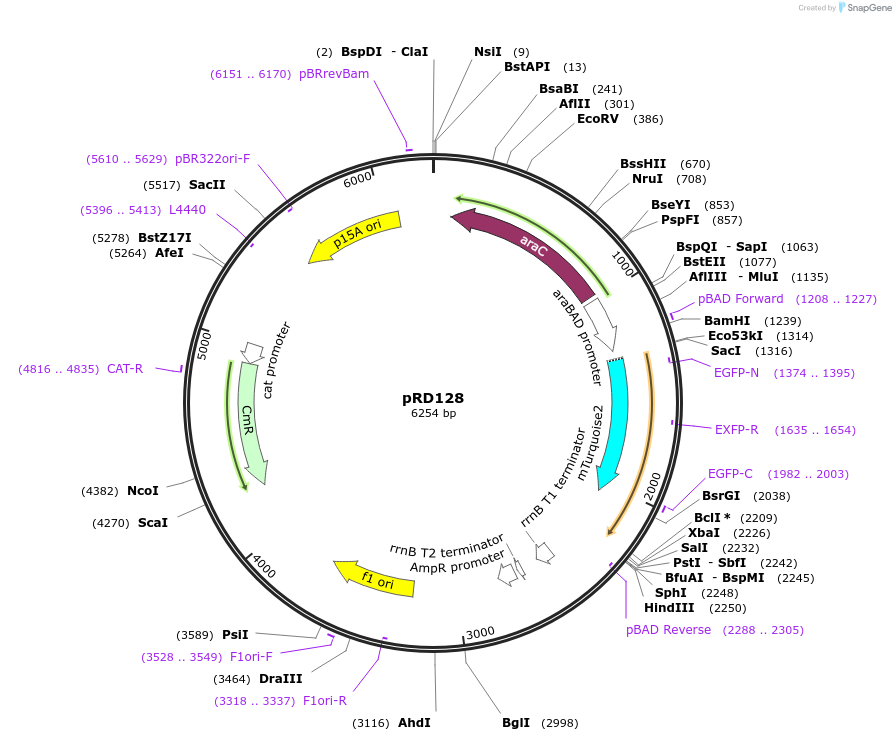
pRD128
Plasmid#163097PurposeExpression of mTurquoise2_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInsertmTurquoise2_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
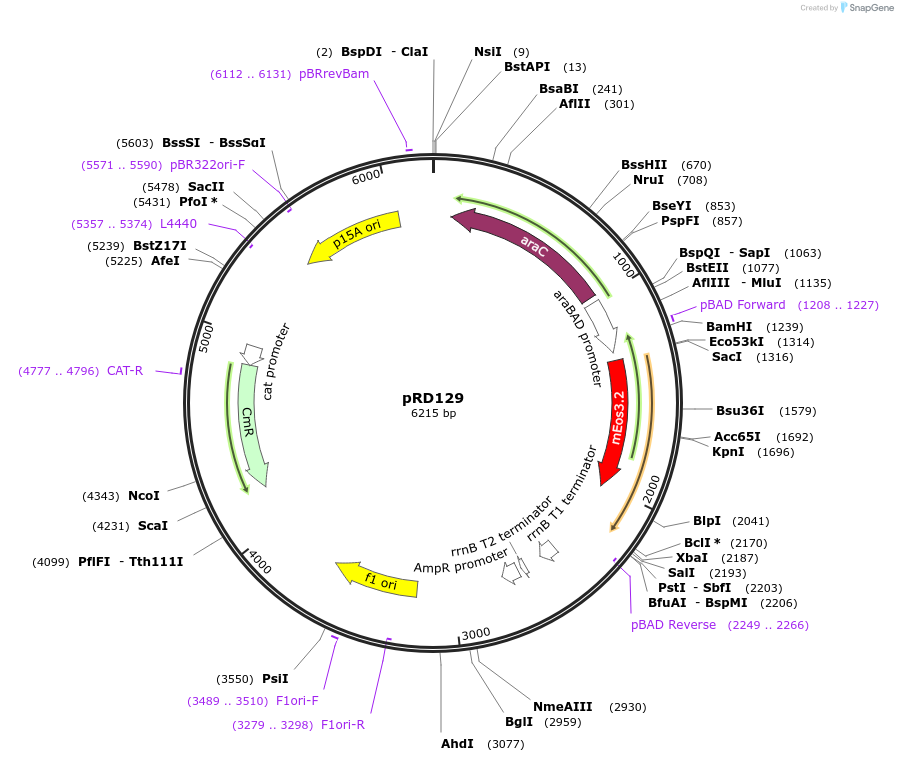
pRD129
Plasmid#163098PurposeExpression of mEos3.2_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInsertmEos3.2_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
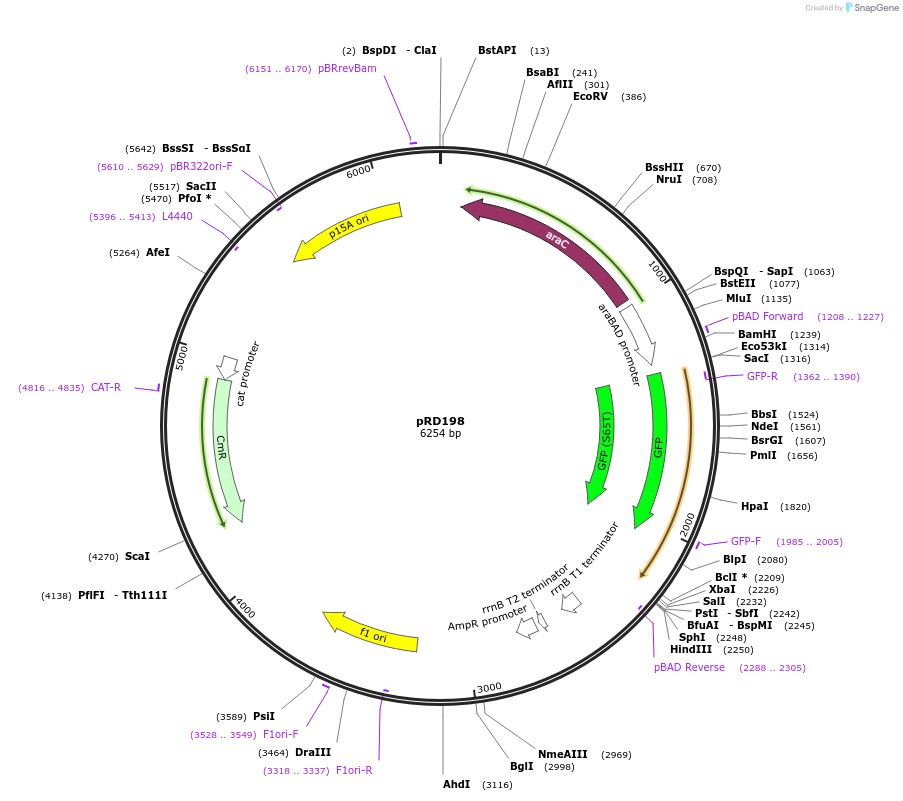
pRD198
Plasmid#163101PurposeExpression of mEGFP_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInsertmEGFP_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
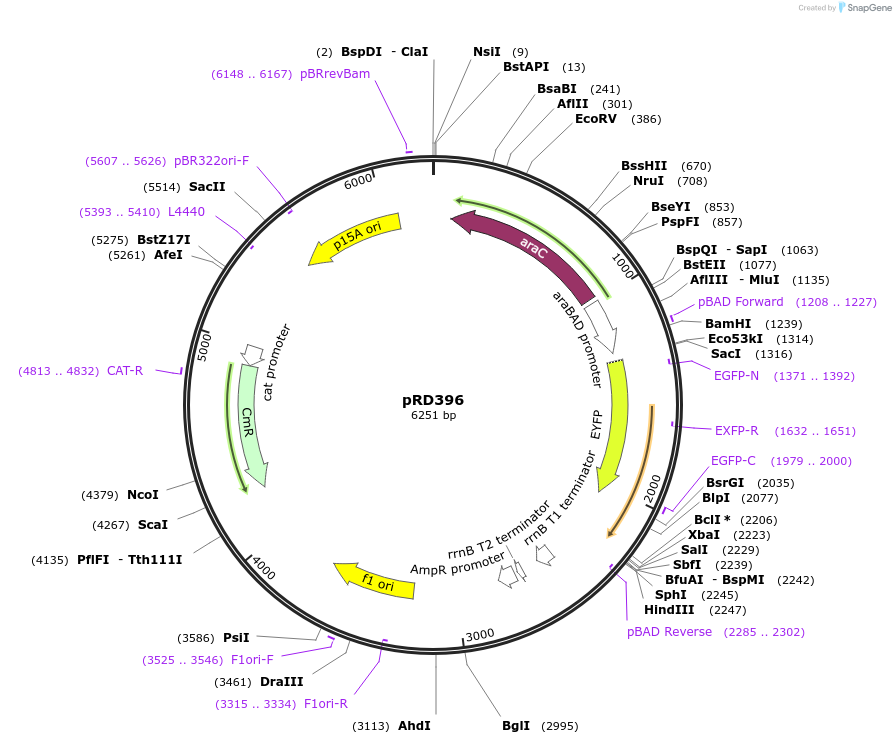
pRD396
Plasmid#163103PurposeExpression of eYFP_H-NSdbd in Escherichia coli (and, potentially, other bacteria)DepositorInserteYFP_H-NSdbd
ExpressionBacterialPromoteraraBAD promoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
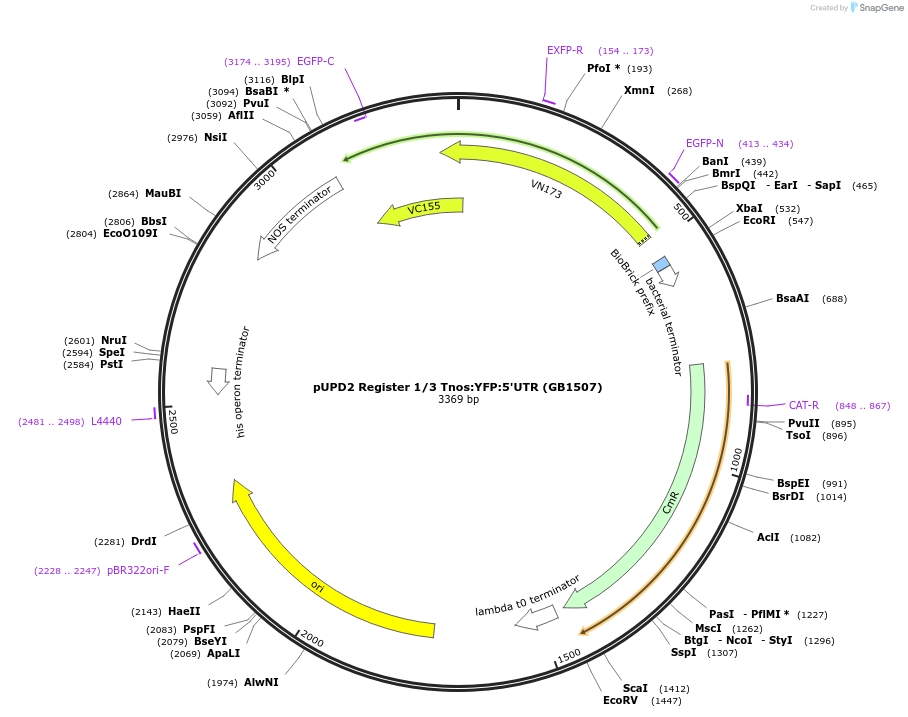
pUPD2 Register 1/3 Tnos:YFP:5'UTR (GB1507)
Plasmid#160583PurposePart 1/3 for assembly of site-specific recombination based Registers. Composed of the reversed sequences of the TNos terminator,YFP CDS and 5' UTR of 35S promoter (TNos:YFP:5'UTR)DepositorInsertYFP
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceAug. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
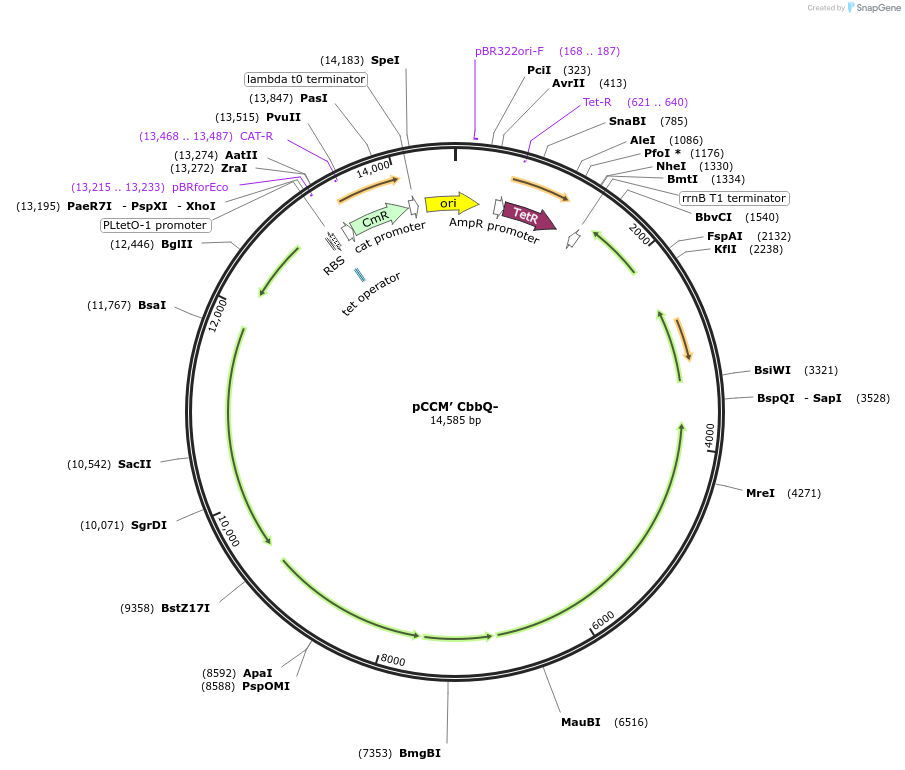
pCCM’ CbbQ-
Plasmid#162711PurposeSecond CCM operon with inactive CbbQ (K46A, E107Q) rubisco activaseDepositorInsertSecond CCM operon cloned from H. neapolitanus
ExpressionBacterialMutationInactivation of CbbQ subunit of CbbOQ rubisco act…PromoterPLtet0-1 promoterAvailable SinceFeb. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
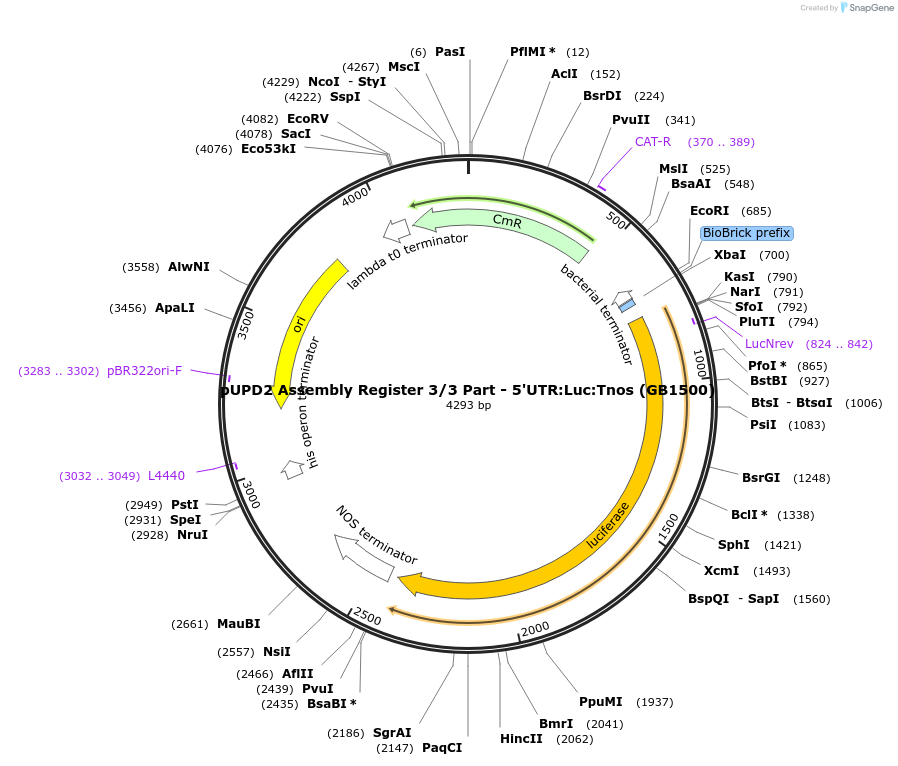
pUPD2 Assembly Register 3/3 Part - 5'UTR:Luc:Tnos (GB1500)
Plasmid#160581PurposePart 3/3 for assembly of site-specific recombination based Registers. Composed of the 5'UTR sequence of 35S promoter, coding sequence of Luc and NOS terminatorDepositorInsertLuc
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJan. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
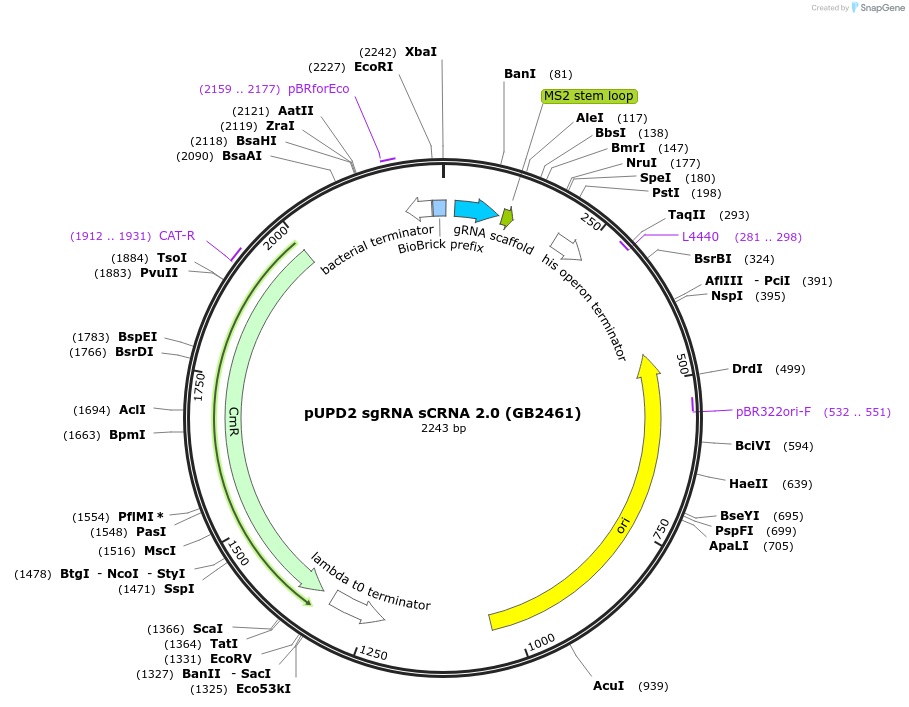
pUPD2 sgRNA sCRNA 2.0 (GB2461)
Plasmid#160593PurposeVersion of the native Cas9-sgRNA with one native WT aptamer sequence and F6 aptamer sequence recognized for Ms2 coat protein, in 3'.DepositorInsertsgRNA sCRNA 2.0
UseCRISPRMutationBsaI and BsmBI sites removedAvailable SinceJan. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
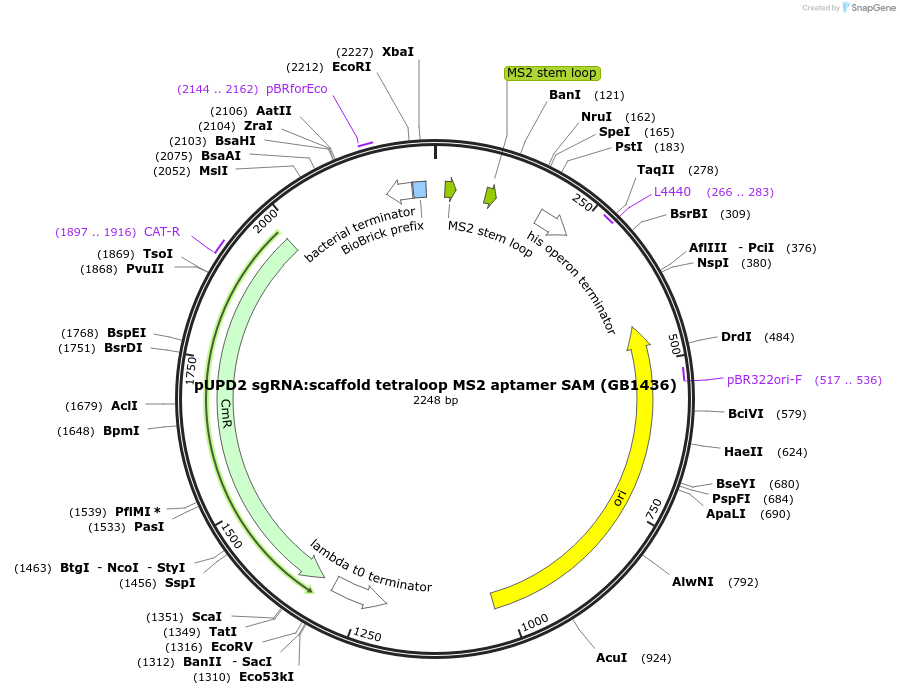
pUPD2 sgRNA:scaffold tetraloop MS2 aptamer SAM (GB1436)
Plasmid#160570PurposeA version of scaffold sgRNA with the sequence of MS2 aptamer inside the tetraloop of the scaffoldDepositorInsertsgRNA:scaffold tetraloop MS2 aptamer SAM
UseCRISPRMutationBsaI and BsmBI sites removedAvailable SinceDec. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
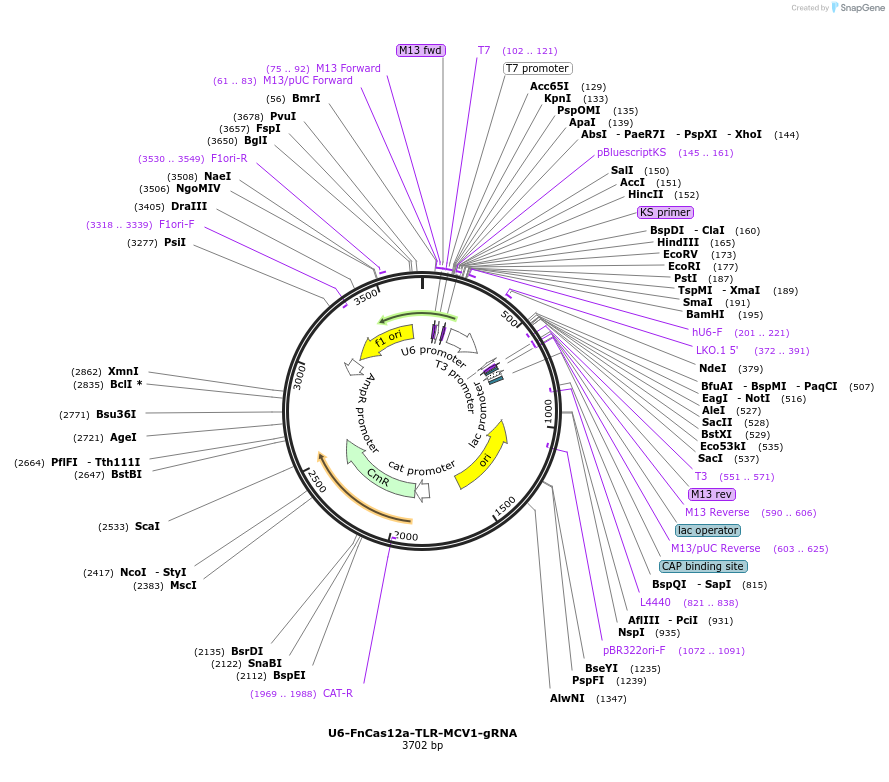
U6-FnCas12a-TLR-MCV1-gRNA
Plasmid#117412PurposeFnCas12a-gRNA targeting TLR2.0DepositorInsertFnCas12a gRNA targeting TLR 2.0
PromoterU6Available SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
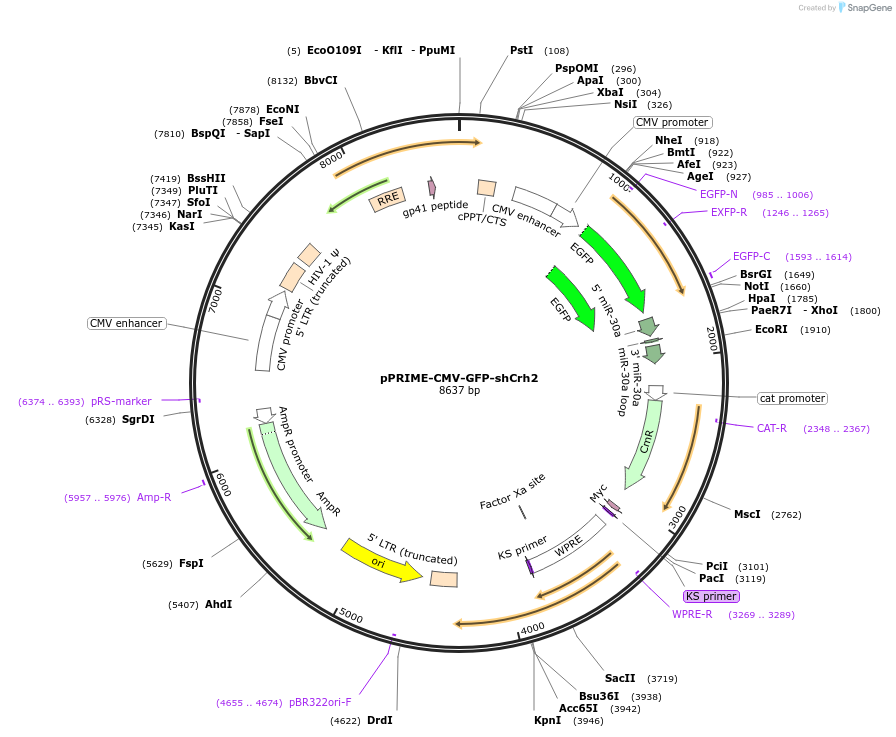
pPRIME-CMV-GFP-shCrh2
Plasmid#132710PurposeEncodes short hairpin RNA (shRNA) #2 that targets the 3’-untranslated region of the rat Crh geneDepositorAvailable SinceOct. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
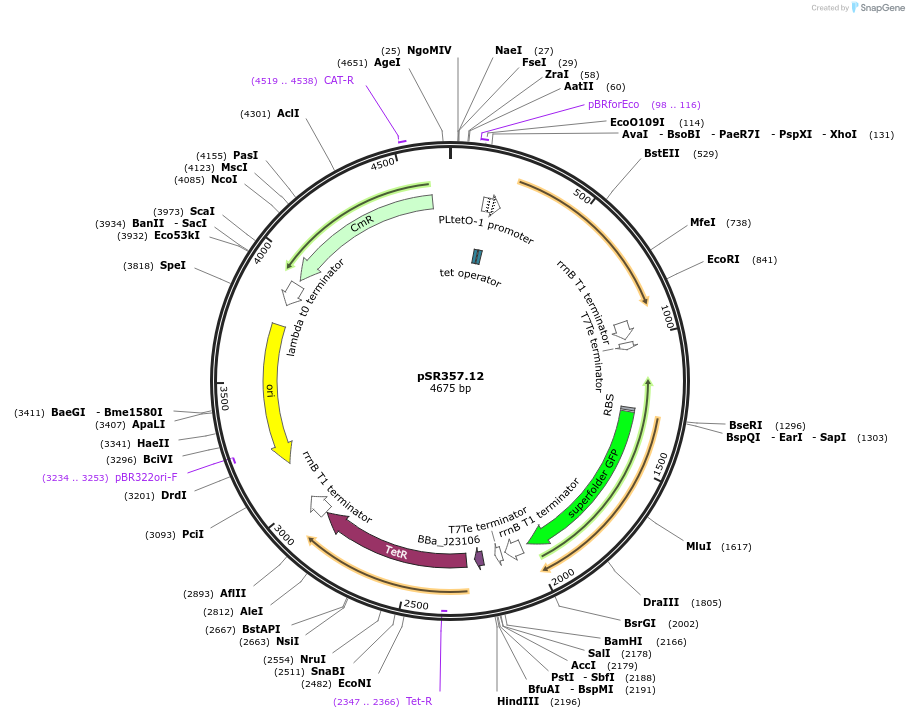
pSR357.12
Plasmid#125107PurposeExpression of narL(1-134aa)-ydfI(129-213aa) chimera under PLtetO-1, output Promoter PydfJ115DepositorInsertsnarL(REC)-ydfI(DBD)134
sfgfp
tetR
UseSynthetic BiologyExpressionBacterialMutationA84T - see depositor comments below and Q183K-see…Available SinceJune 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
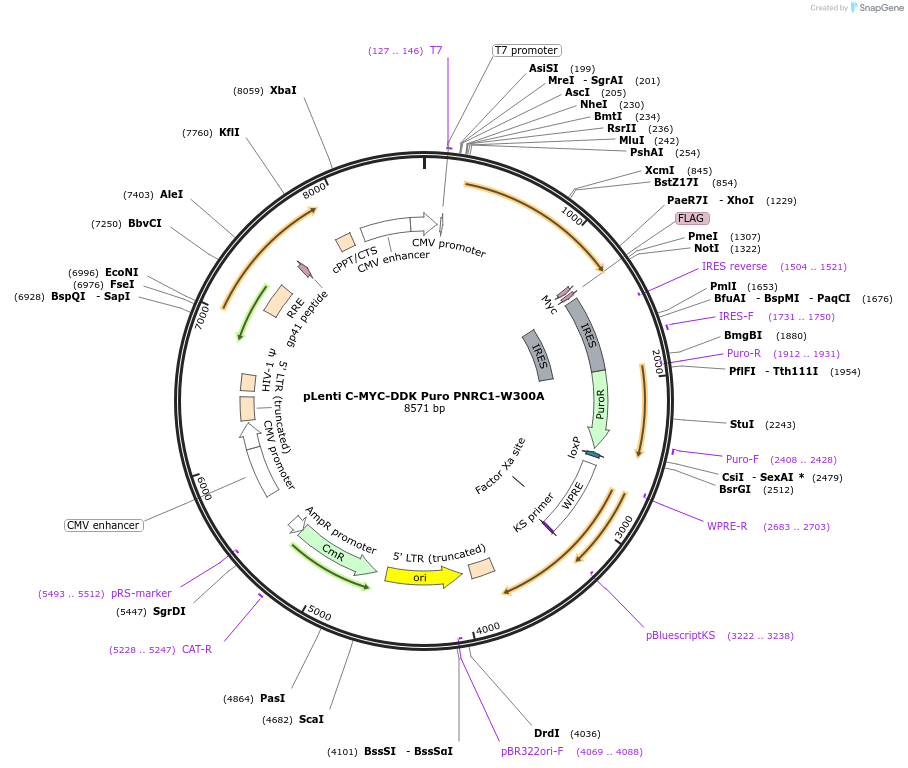
pLenti C-MYC-DDK Puro PNRC1-W300A
Plasmid#123301PurposeMammalian expression of PNRC1-W300A mutant C-MYC-FLAGDepositorAvailable SinceMarch 26, 2019AvailabilityAcademic Institutions and Nonprofits only