We narrowed to 2,840 results for: fas
-
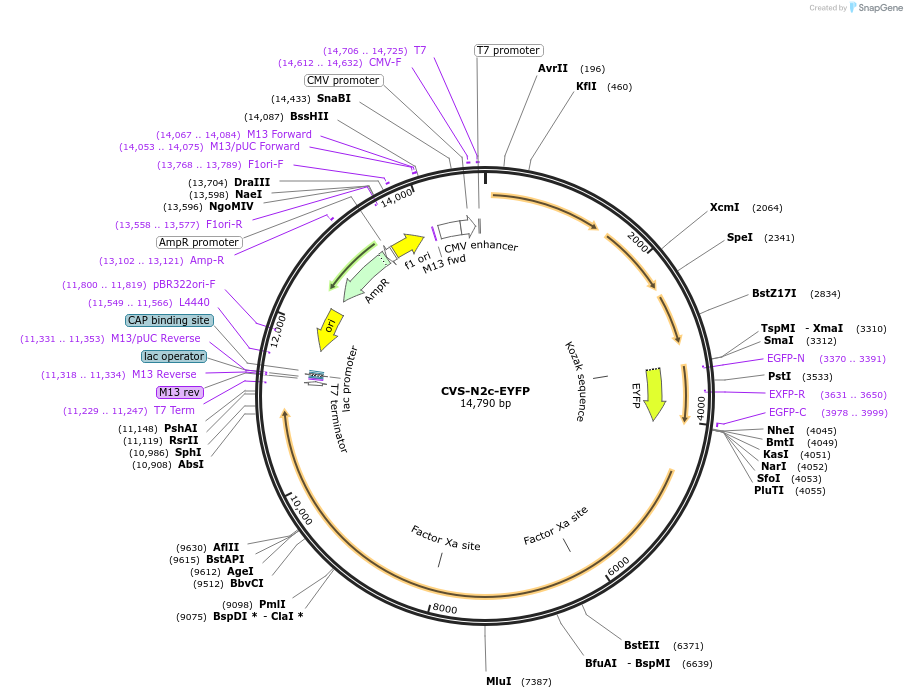
Plasmid#172383PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertEYFP
UseNeurotropic virusAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
CVS-N2c-mCitrine
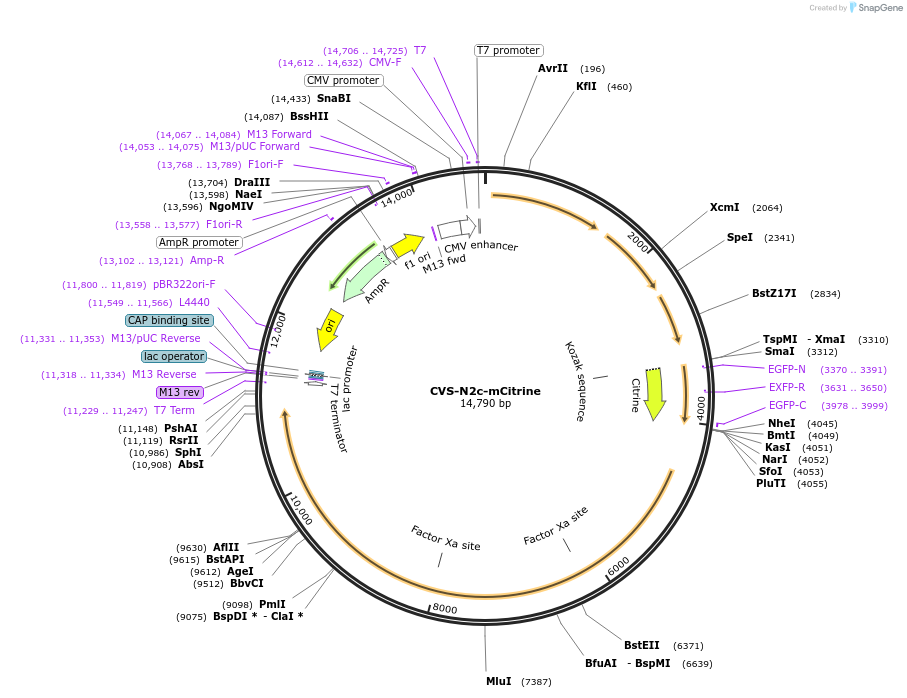
Plasmid#172384PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertmCitrine
UseNeurotropic virusAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
CVS-N2c-EGFP-ChIEF
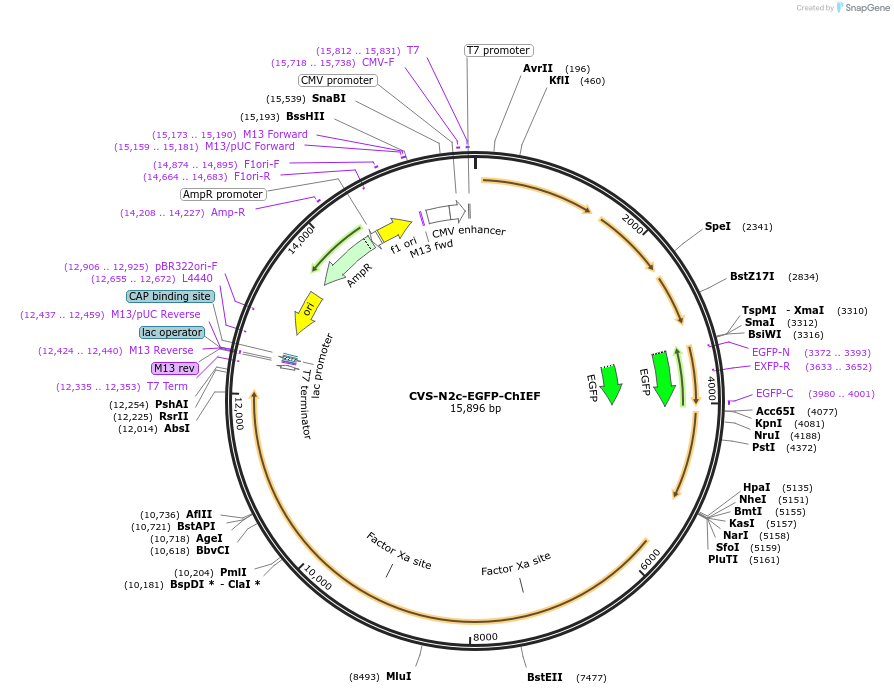
Plasmid#172371PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertEGFP + ChIEF
UseNeurotropic virusAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
pSV human p55 alpha (HA tag)
Plasmid#11498DepositorAvailable SinceMarch 3, 2006AvailabilityAcademic Institutions and Nonprofits only -
CVS-N2c-nl.EGFP-FlpO
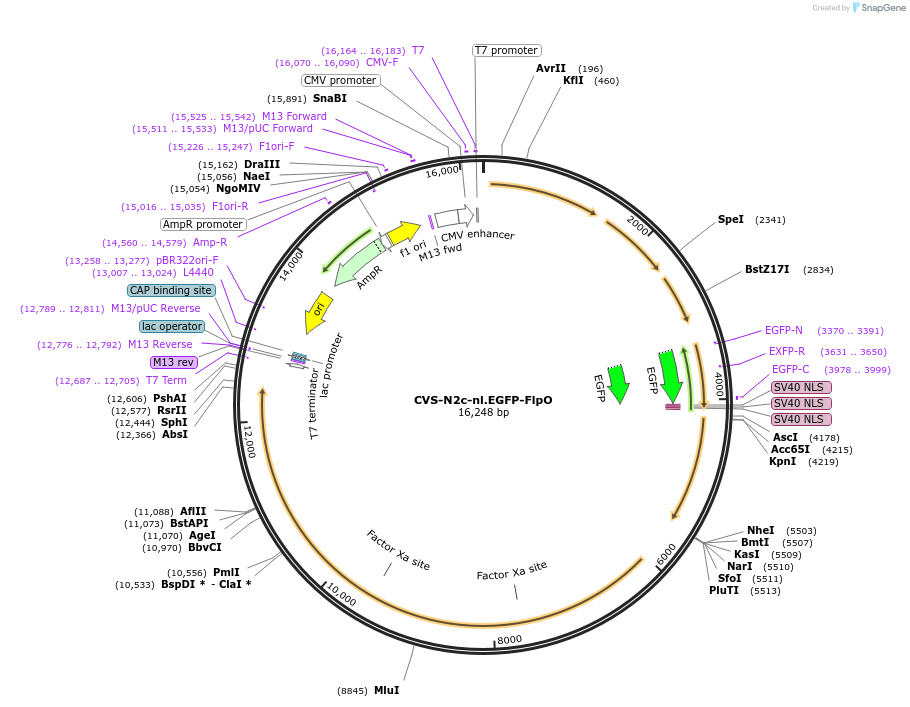
Plasmid#172379PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertnuclear-localized EGFP + FlpO
UseNeurotropic virusAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
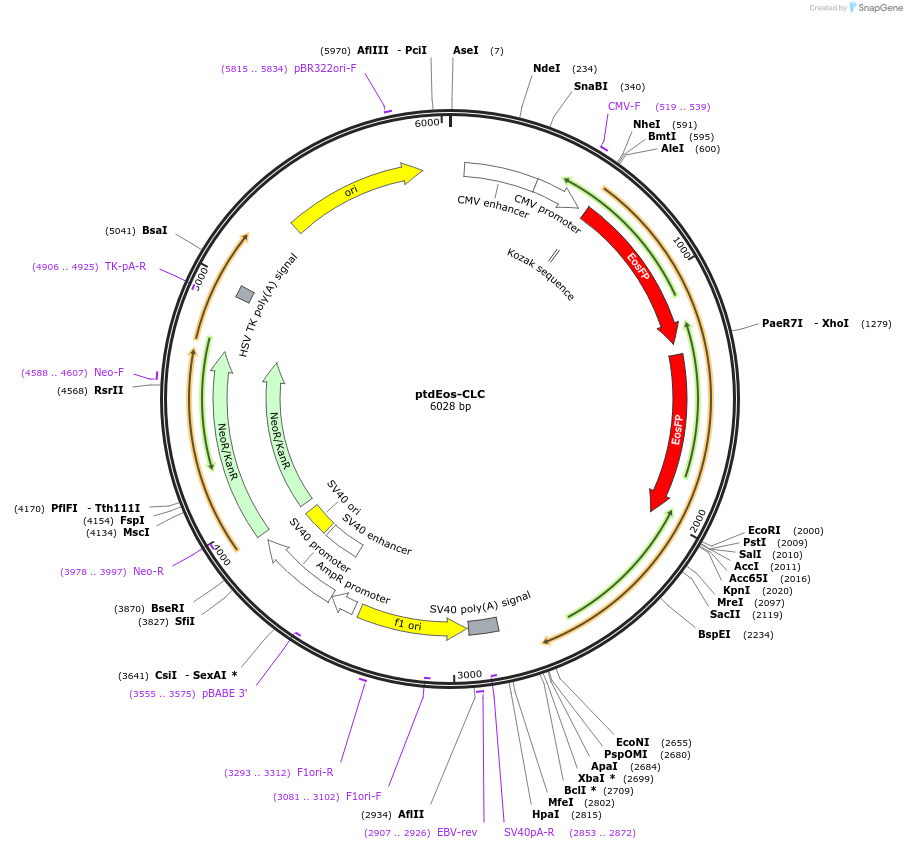
ptdEos-CLC
Plasmid#38009DepositorInsertsExpressionBacterial and MammalianPromoterCMVAvailable SinceJan. 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
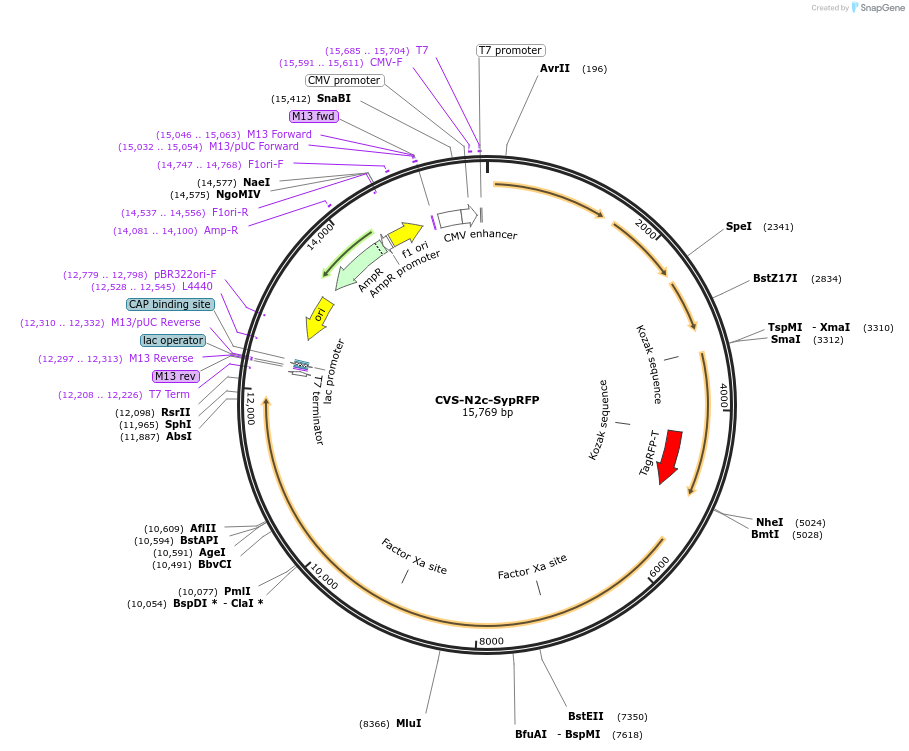
CVS-N2c-SypRFP
Plasmid#172381PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertSynaptophysin1-RFP fusion protein
UseNeurotropic virusAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
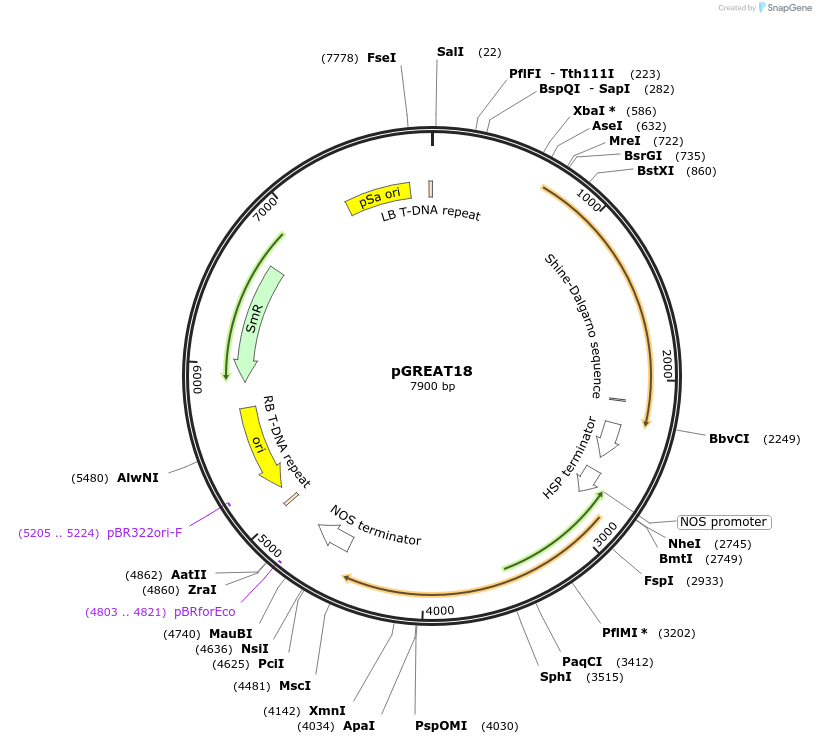
pGREAT18
Plasmid#170906PurposeGoldenBraid Double Transcriptional Unit - pAtUBQ10_E-Luc_AtHSP18.2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpAtUBQ10Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
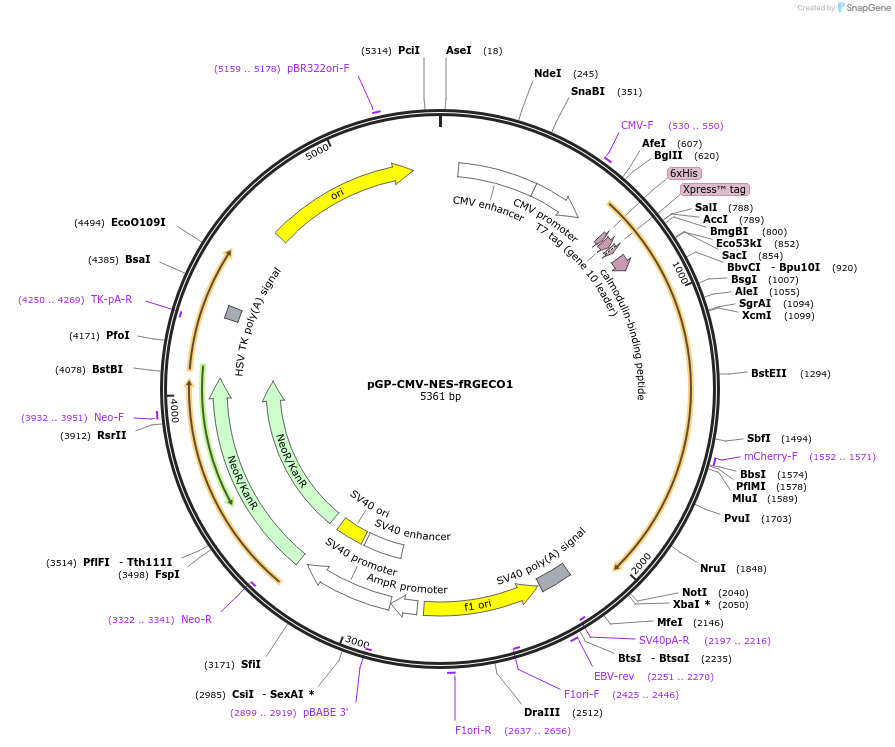
pGP-CMV-NES-fRGECO1
Plasmid#125247PurposeExpresses red fluorescent calcium sensor in mammalian cellsDepositorInsertfRGECO2
TagsNES signalExpressionMammalianAvailable SinceNov. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
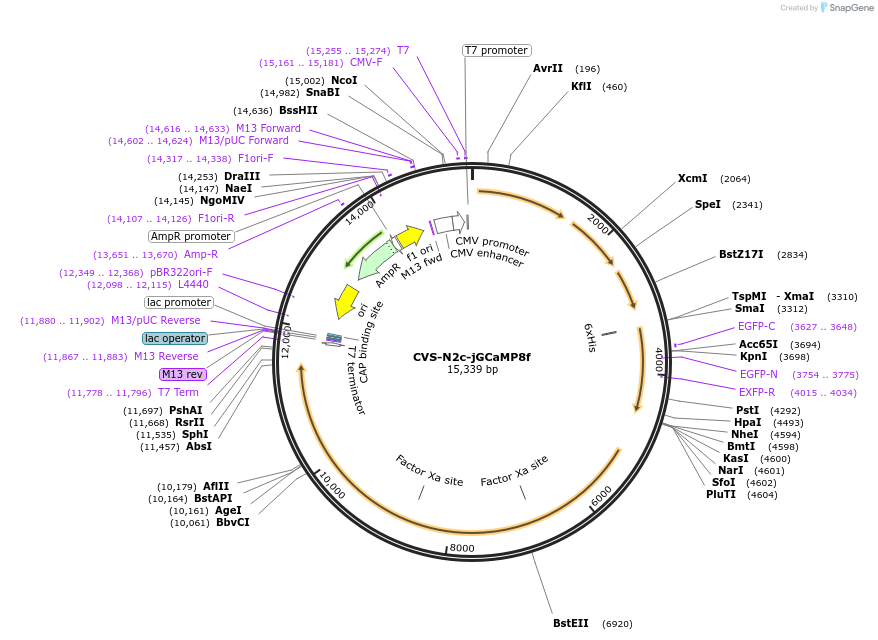
CVS-N2c-jGCaMP8f
Plasmid#172387PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertGCaMP8f
UseNeurotropic virusAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
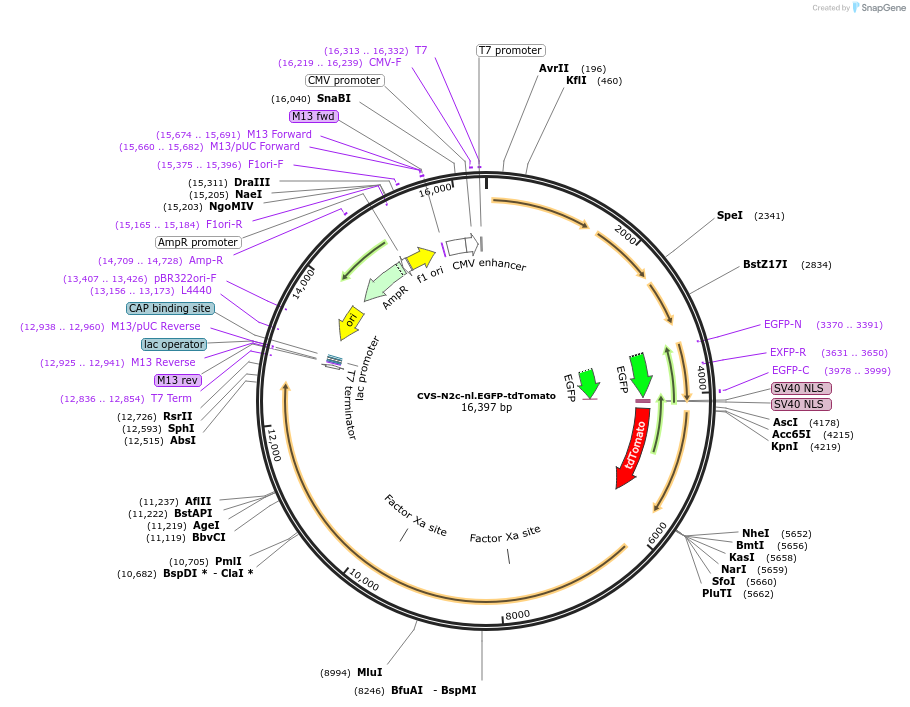
CVS-N2c-nl.EGFP-tdTomato
Plasmid#172382PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertnuclear-localized EGFP + tdTomato
UseNeurotropic virusAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
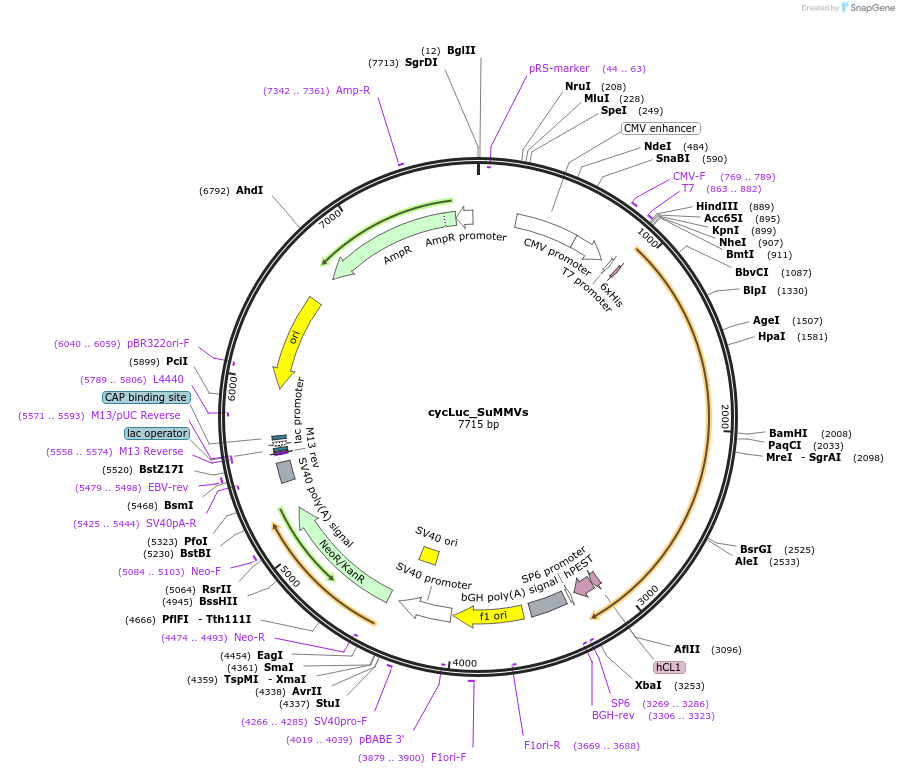
cycLuc_SuMMVs
Plasmid#119210PurposeCircularly permutated firefly luciferase with SuMMVs cleavage site (Luciferase reporter for detection of proteolytic activity)DepositorInsertcyclic luciferase with SuMMVp cleavage site
UseLuciferaseTagsAU1 tagExpressionMammalianPromoterCMVAvailable SinceFeb. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
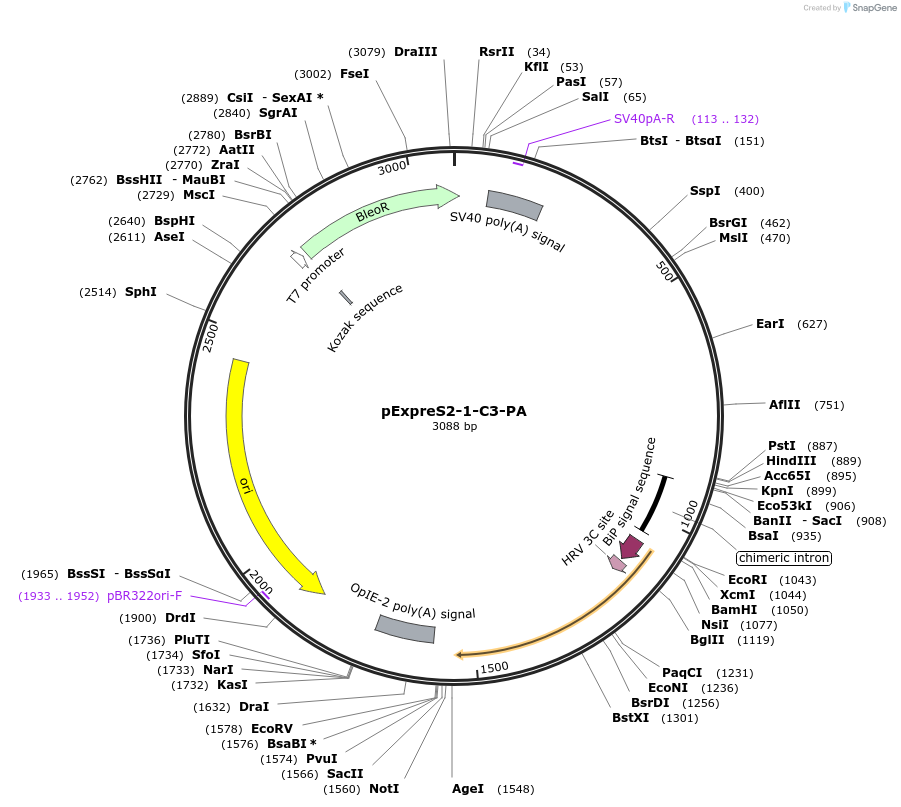
pExpreS2-1-C3-PA
Plasmid#175446PurposeHiFi-compatible destination vector for secreted overexpression of 3C-cleavable, C-terminal Protein A tagged proteins with ExpreS2 PlatformDepositorTypeEmpty backboneTags3C-Protein A and BiP Secretion PeptideExpressionInsectPromoterFused Actin-HSP70Available SinceMarch 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
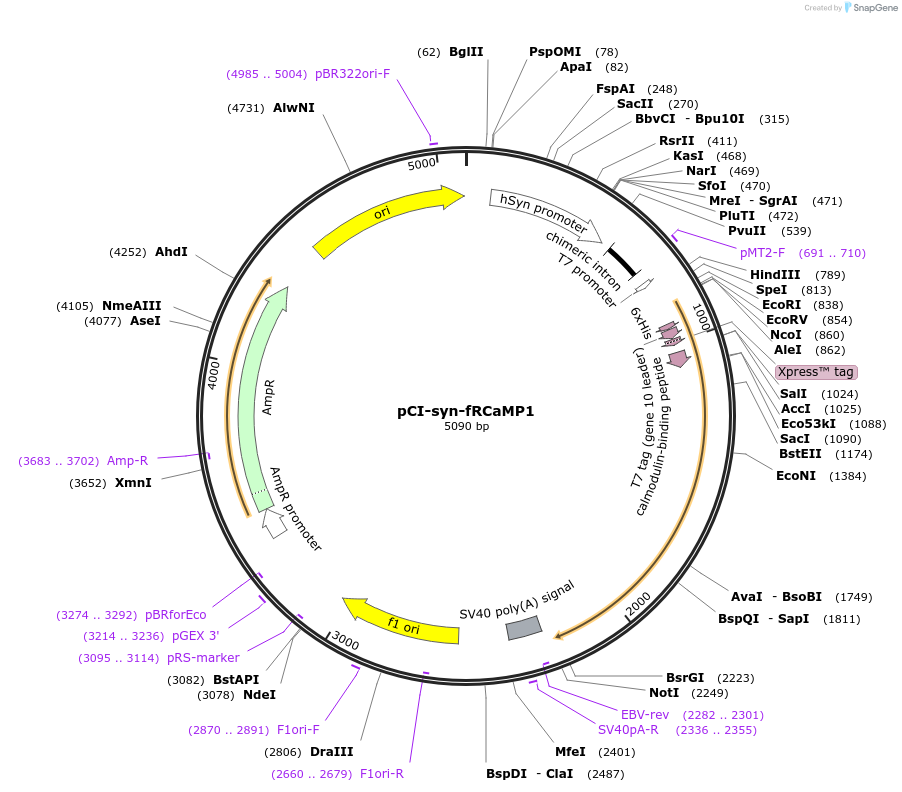
pCI-syn-fRCaMP1
Plasmid#125245PurposeExpresses red fluorescent calcium sensor in hippocampal slicesDepositorInsertfRCaMP1
ExpressionMammalianMutationW44YPromotersynapsinAvailable SinceNov. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
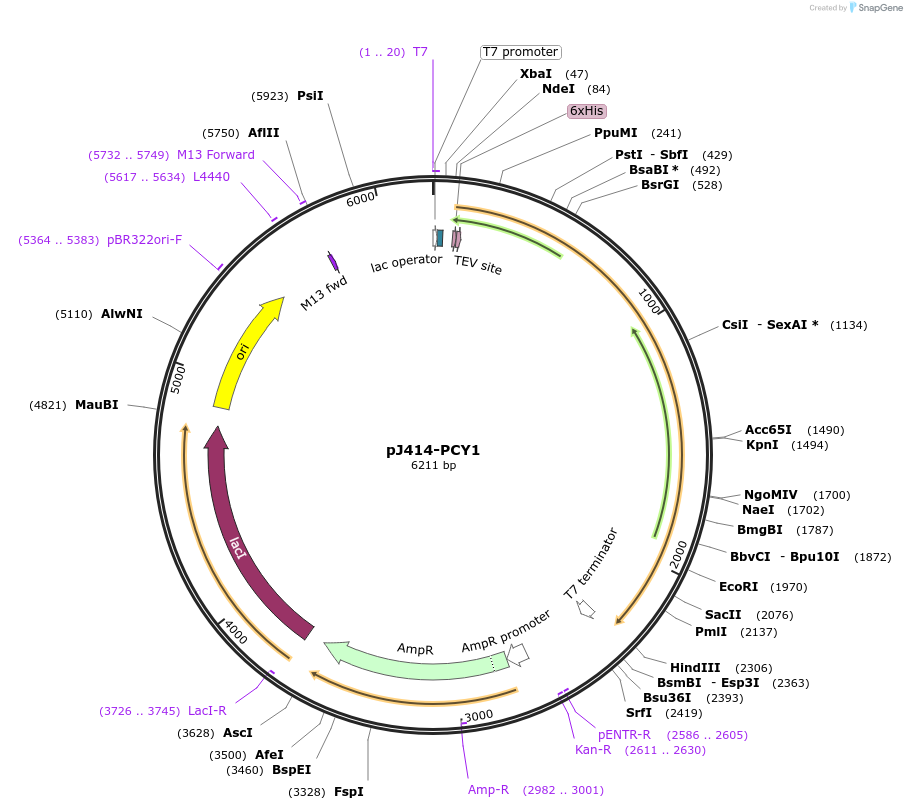
pJ414-PCY1
Plasmid#98150PurposeExpresses PCY1 from Saponaria vaccaria possessing a N-terminal His6-tag (TEV cleavable) in E. coli.DepositorInsertPeptide cyclase 1
TagsHis6-TEVExpressionBacterialMutationWTPromoterT7Available SinceMarch 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
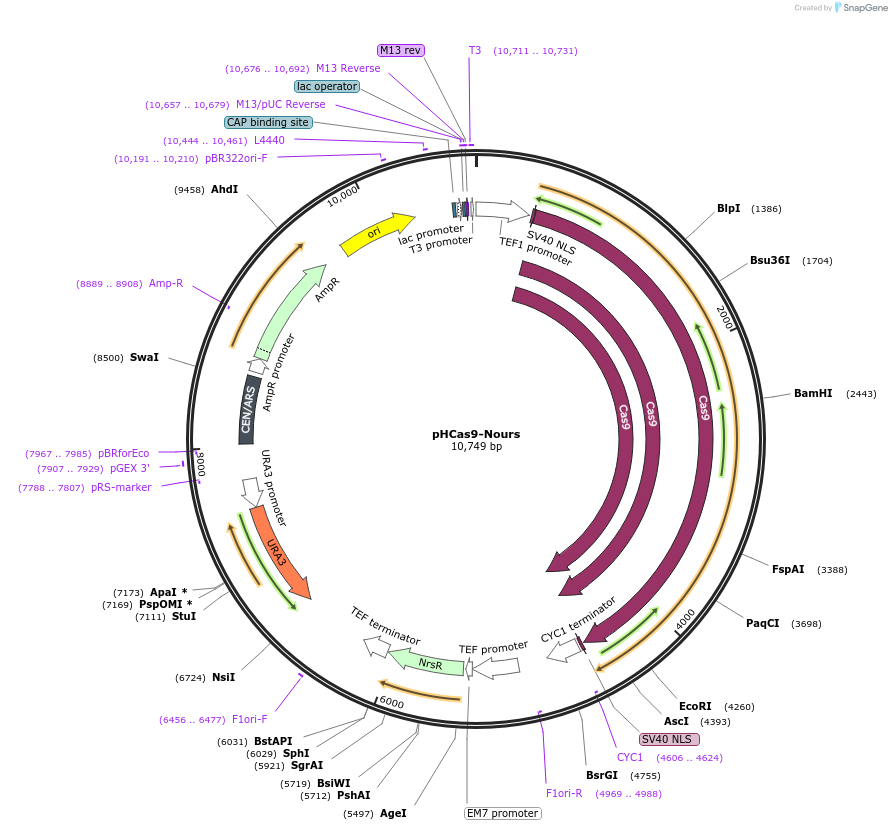
pHCas9-Nours
Plasmid#107733PurposeEscherichia coli- S. cerevisiae shuttle plasmid harbors a Cas9 gene, a natMX6 and URA3 selection markers used in S. cerevisiaeDepositorInsertCas9
UseCRISPRTagsSV40 NLSExpressionYeastMutationHuman optimizedPromoterTEF1Available SinceJan. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSR-siKlf5
Plasmid#41740DepositorInsertKlf5 (Klf5 Mouse)
UseRNAiAvailable SinceJan. 18, 2013AvailabilityAcademic Institutions and Nonprofits only -
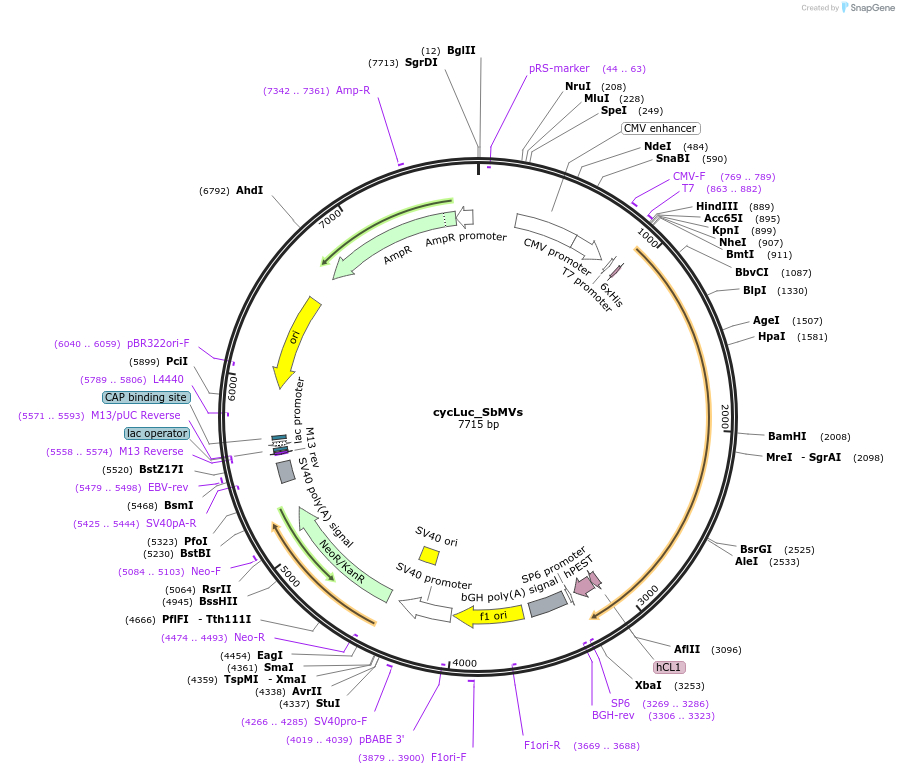
cycLuc_SbMVs
Plasmid#119209PurposeCircularly permutated firefly luciferase with SbMV cleavage site (Luciferase reporter for detection of proteolytic activity)DepositorInsertcyclic luciferase with SbMVp cleavage site
UseLuciferaseTagsAU1 tagExpressionMammalianPromoterCMVAvailable SinceFeb. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
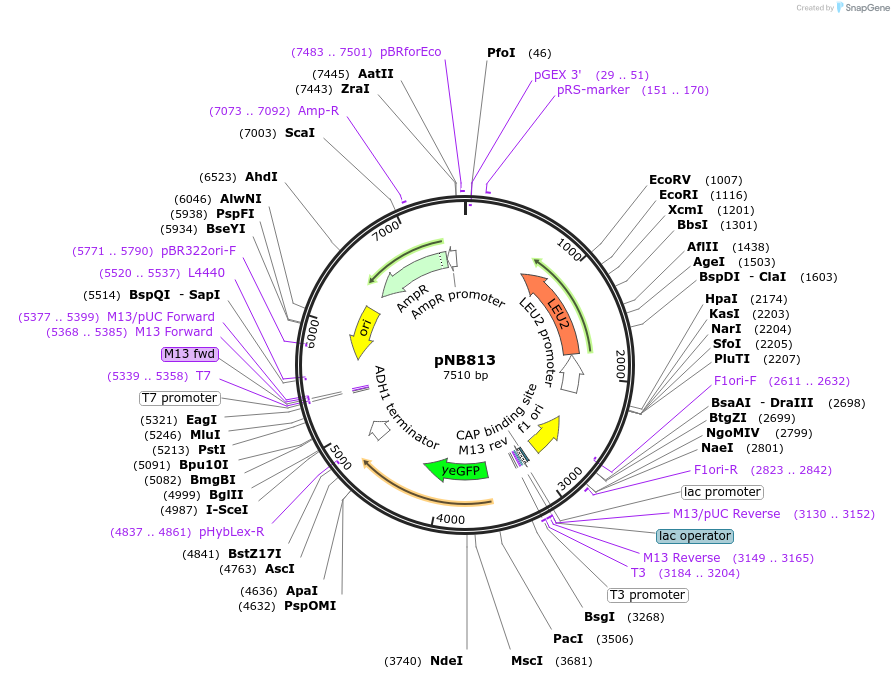
pNB813
Plasmid#60161PurposeYeast codon-optimized yellow fluorescent protein (yEVenus) with PEST degron driven by SIC1 promoter.DepositorInsertyEVenus-PEST
TagsPEST degronExpressionYeastPromoterSIC1prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
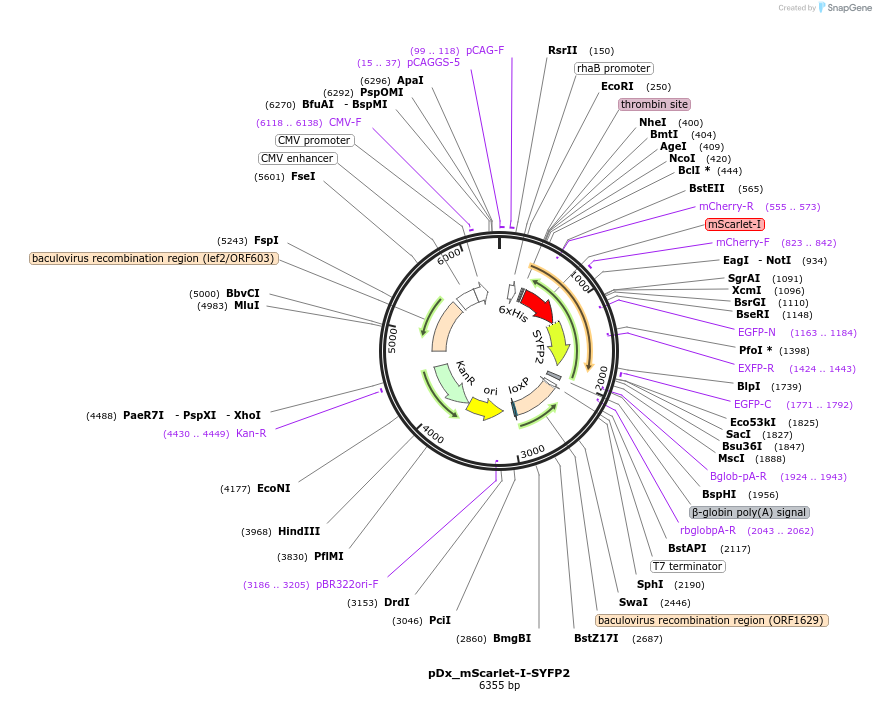
pDx_mScarlet-I-SYFP2
Plasmid#189761PurposeDual expression vector for bacteria and mammalian cells producing mScarlet-I-SYFP2 fusion protein for FRET analysisDepositorInsertmScarlet-I-SYFP2
Tags6xHisExpressionBacterial and MammalianPromoterCMV & RhaAvailable SinceMarch 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
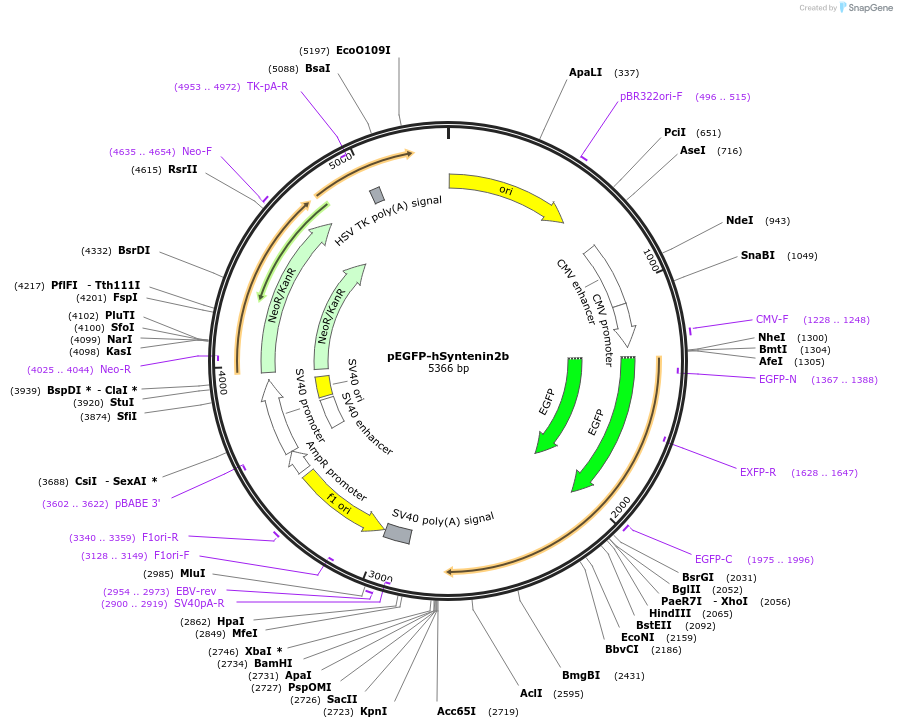
pEGFP-hSyntenin2b
Plasmid#89437Purposemammalian expression of EGFP tagged human Syntenin2bDepositorAvailable SinceJuly 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
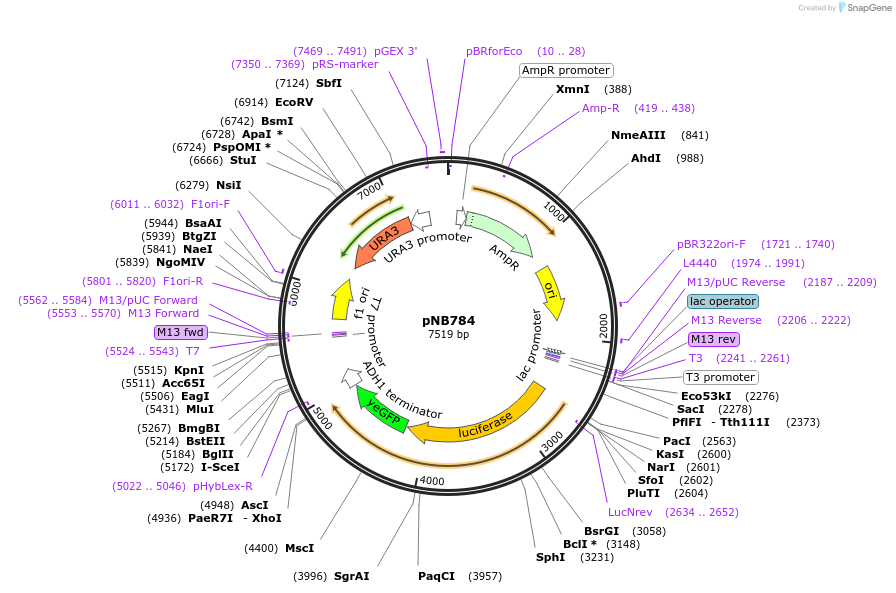
pNB784
Plasmid#60163PurposeFirefly luciferase (Promega) yeast codon-optimized yellow fluorescent protein (yEVenus) fusion driven by SIC1 promoter.DepositorInsertFLuc-yEVenus
ExpressionYeastMutationFLuc has A4V & S504G (no functional effect)PromoterSIC1prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
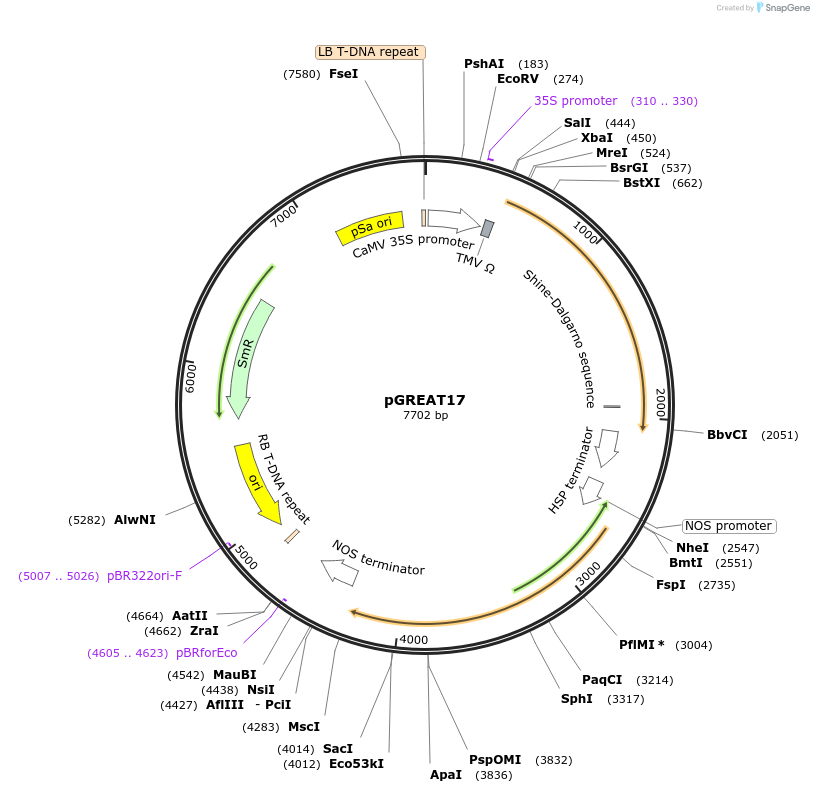
pGREAT17
Plasmid#170905PurposeGoldenBraid Double Transcriptional Unit - p35SoTMV_E-Luc_AtHSP18.2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…Promoterp35SoTMVAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
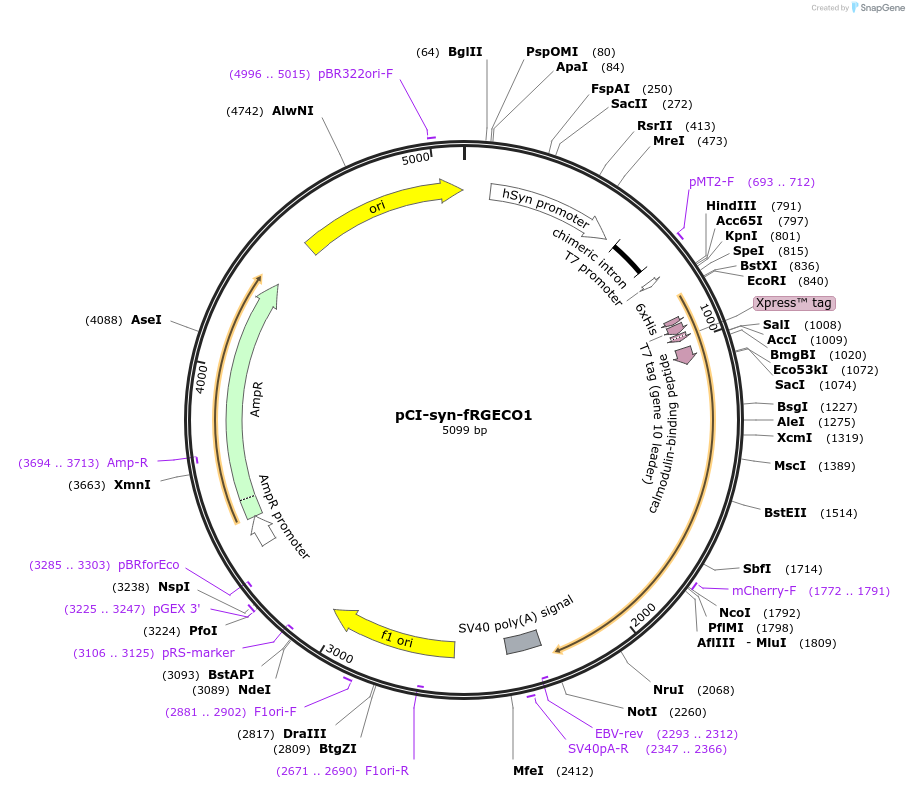
pCI-syn-fRGECO1
Plasmid#125243PurposeExpresses red fluorescent calcium sensor in hippocampal slicesDepositorInsertfRGECO1
ExpressionMammalianMutationD323APromotersynapsinAvailable SinceNov. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
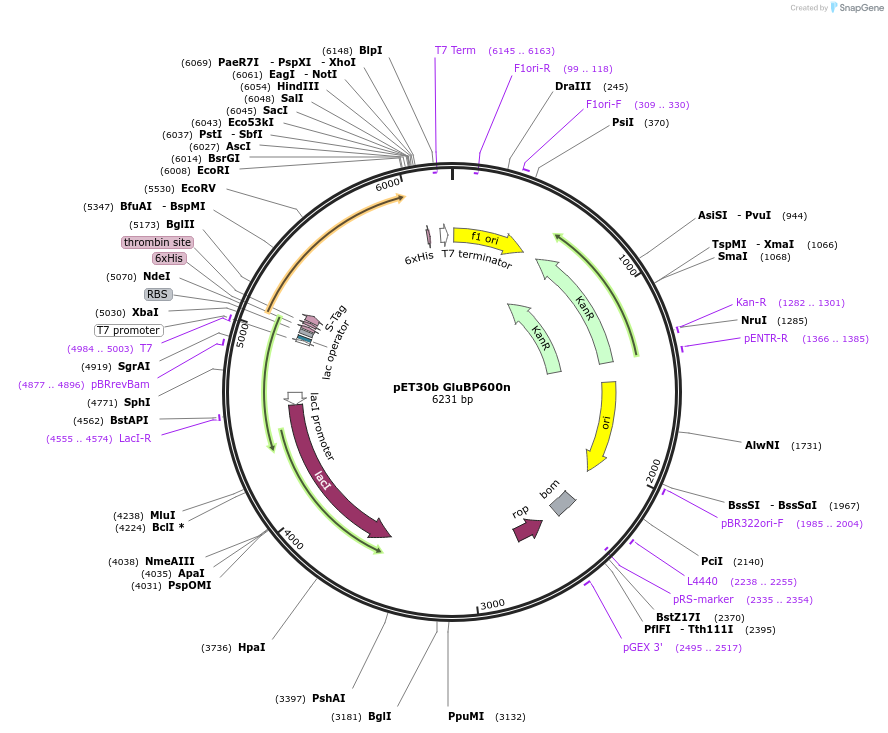
pET30b GluBP600n
Plasmid#119835PurposeProduction of recombinant GluBP600n high affinity protein in E. coliDepositorInsertGluBP600n
TagsHisExpressionBacterialPromoterT7Available SinceJan. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
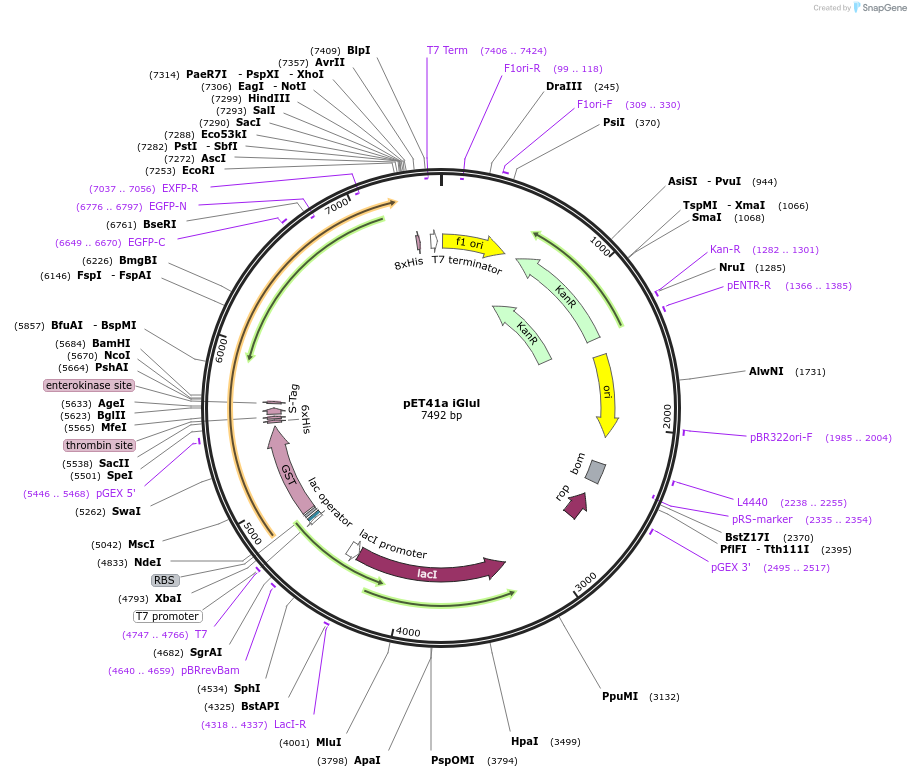
pET41a iGlul
Plasmid#119832PurposeProduction of recombinant iGluSnFR T92A low affinity variant, a single-wavelength glutamate indicator, in E. coliDepositorInsertiGlul
TagsGSTExpressionBacterialMutationT92APromoterT7Available SinceJan. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
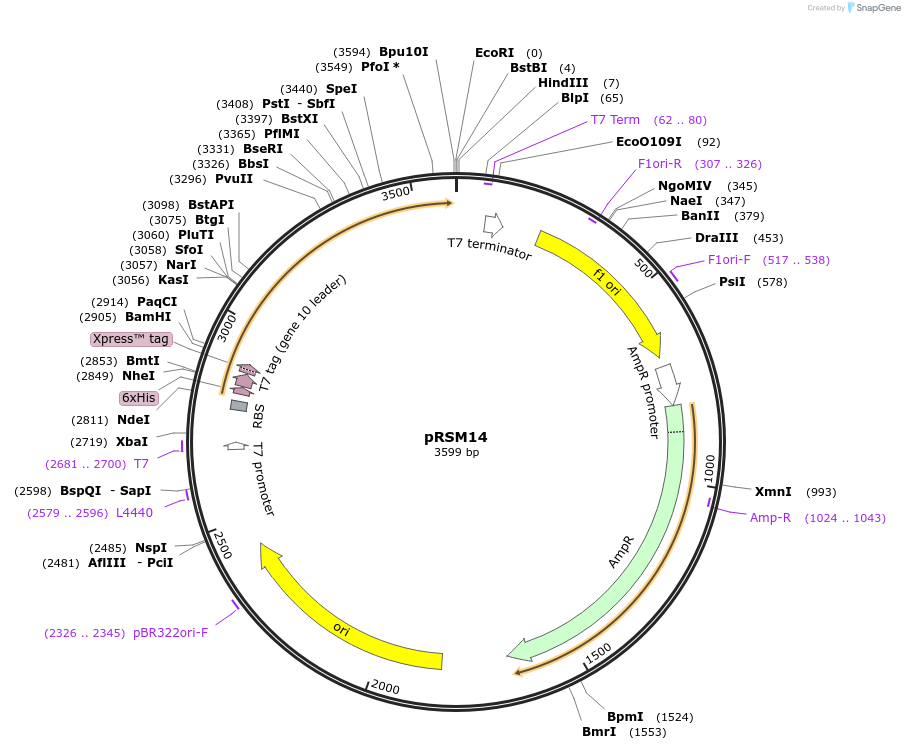
pRSM14
Plasmid#101645PurposeEncodes the fluorescent protein efasCFP for constitutive expression in E. coli.DepositorInsertefasCFP
Tags6XHisExpressionBacterialAvailable SinceNov. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
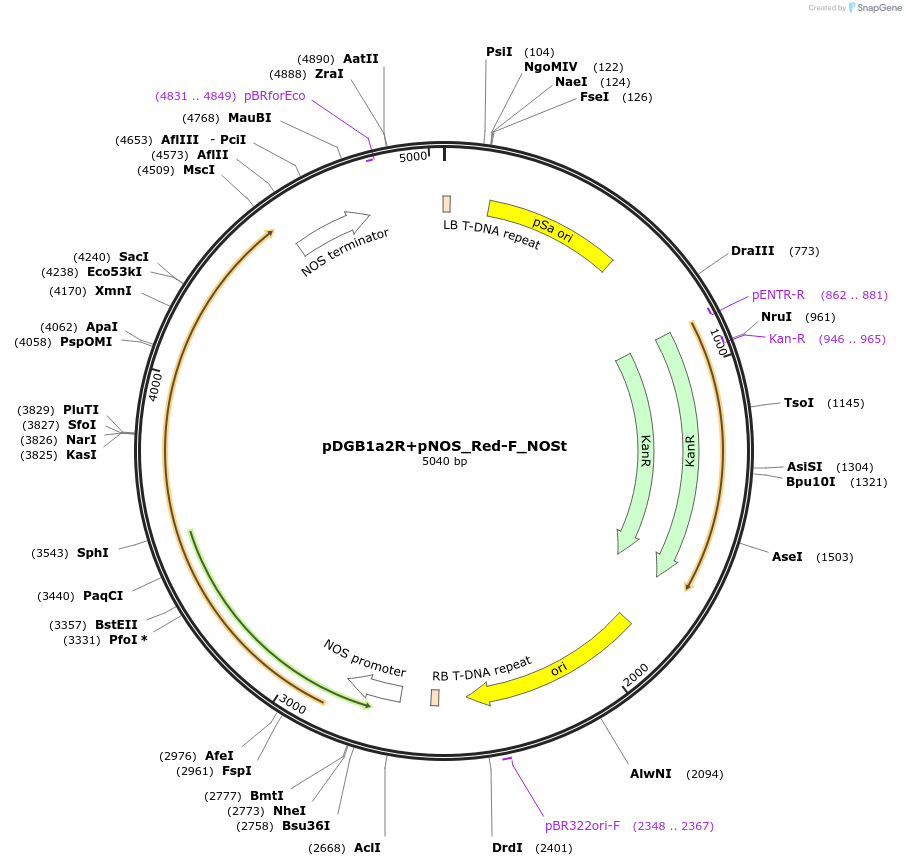
pDGB1a2R+pNOS_Red-F_NOSt
Plasmid#170888PurposeGoldenBraid Transcriptional Unit - pNOS_Red-F_NOSt Reverse orientationDepositorInsertRed-F
UseLuciferase and Synthetic BiologyExpressionPlantMutationp.S286Y Red mutantPromoterpNOSAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
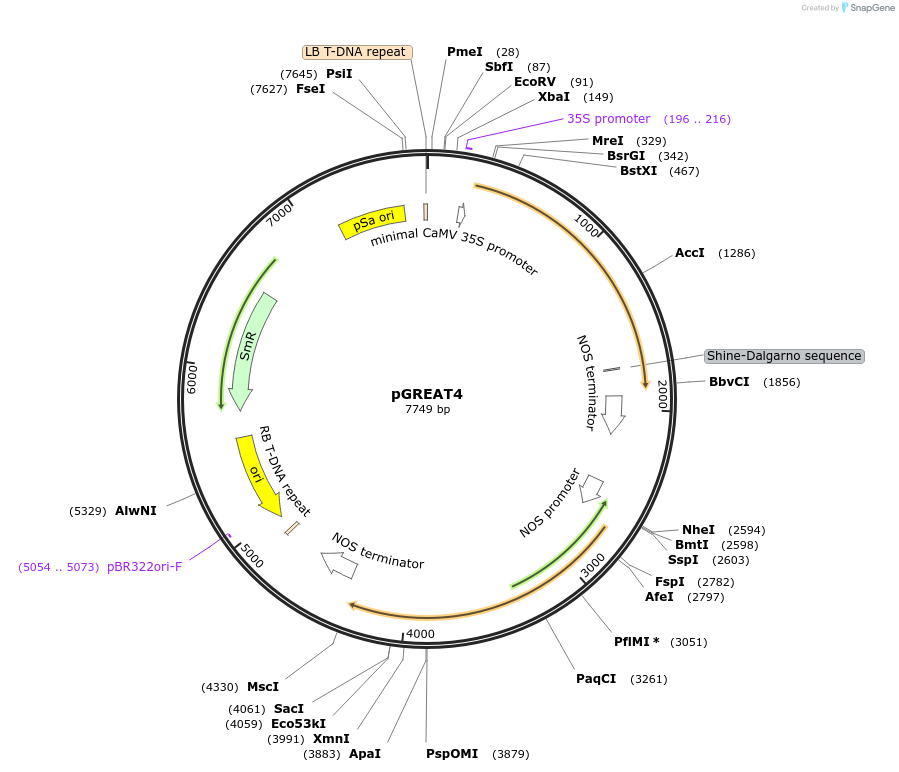
pGREAT4
Plasmid#170892PurposeGoldenBraid Double Transcriptional Unit - pG10-90_E-Luc_NOSt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpG10-90Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
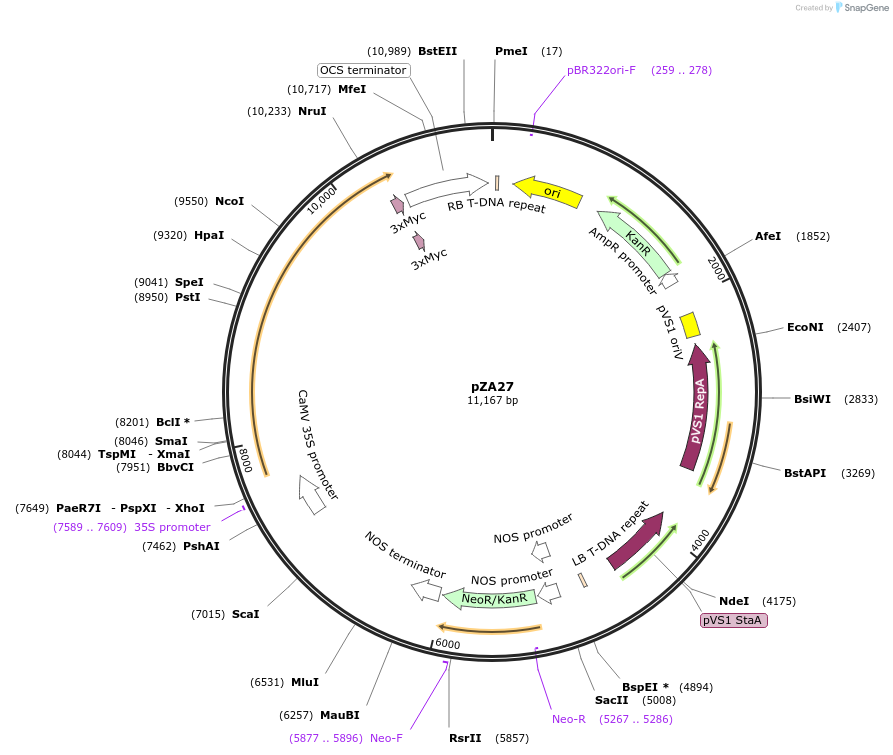
pZA27
Plasmid#158505PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 L17E-4xMyc. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
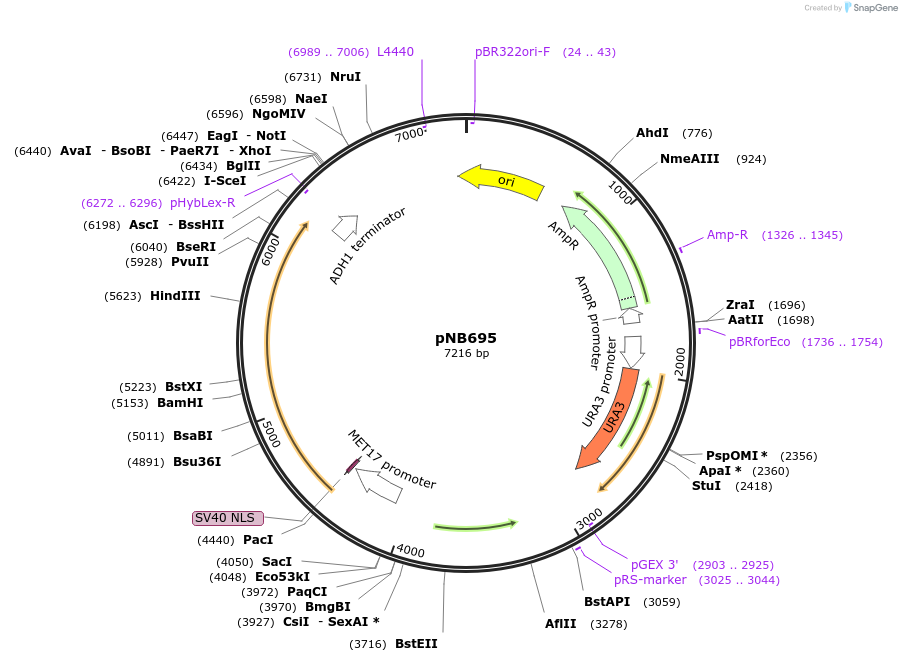
pNB695
Plasmid#60144PurposeDisintegrator plasmid encoding MET17 promoter driven green beetle luciferase with nuclear localization signal (NLS) and ClpXP degron tag (ssrA).DepositorInsertGrLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterMET17prAvailable SinceDec. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
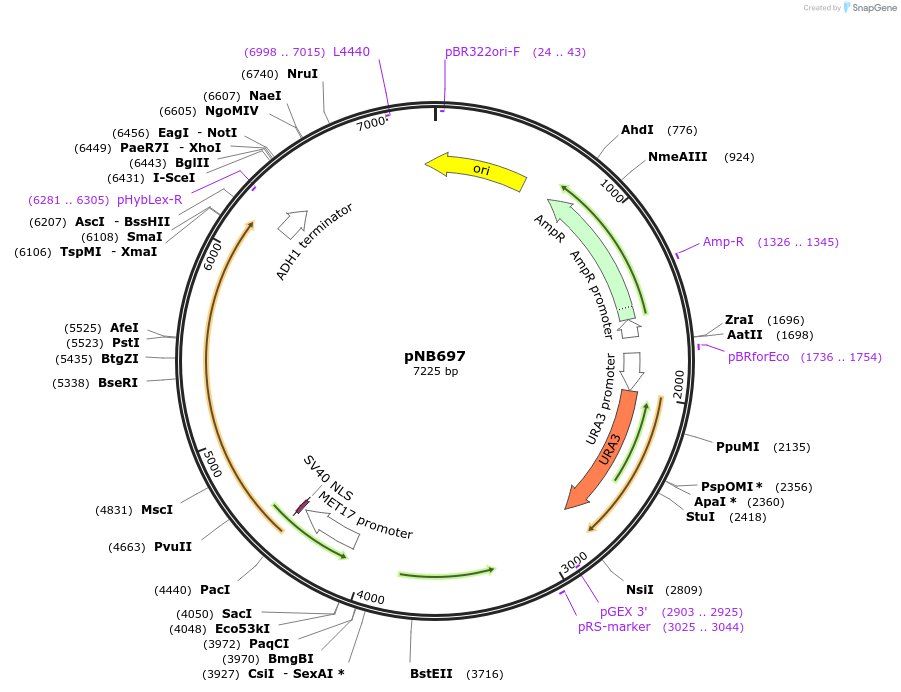
pNB697
Plasmid#60146PurposeDisintegrator plasmid encoding MET17 promoter driven red beetle luciferase with nuclear localization signal (NLS) and ClpXP degron tag (ssrA).DepositorInsertRdLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterMET17prAvailable SinceDec. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
TAL3439
Plasmid#47158DepositorInsertZebrafishCommunity-fastkd3-right (im:7147223 Zebrafish)
UseT7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceAug. 13, 2013AvailabilityAcademic Institutions and Nonprofits only -
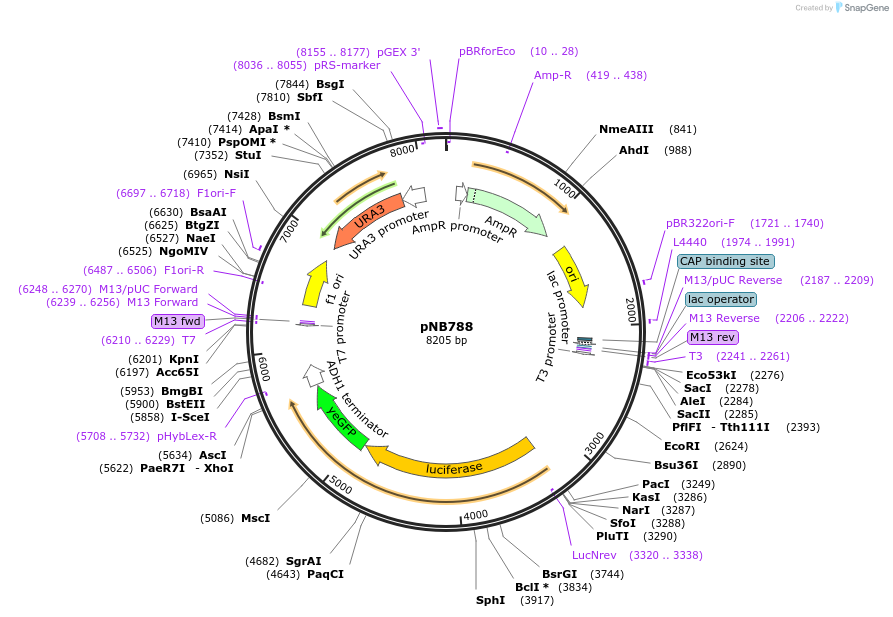
pNB788
Plasmid#60165PurposeFirefly luciferase (Promega) yeast codon-optimized yellow fluorescent protein (yEVenus) fusion driven by RNR1 promoter.DepositorInsertFLuc-yEVenus
ExpressionYeastMutationFLuc has A4V & S504G (no functional effect)PromoterRNR1prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
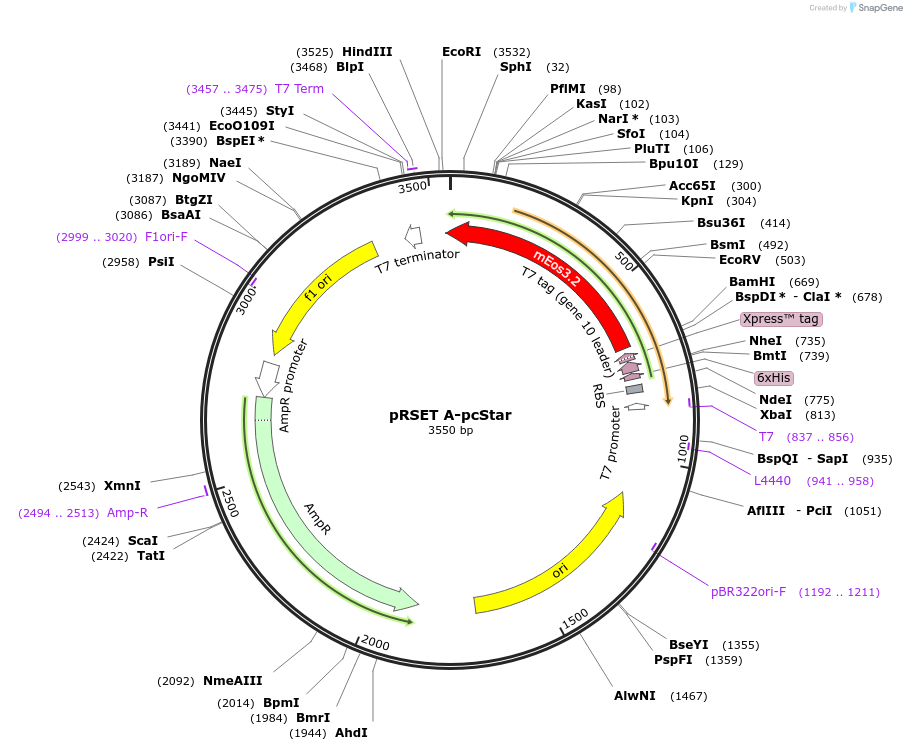
pRSET A-pcStar
Plasmid#226339PurposeBacterial expression of RSFP pcStar; photoconvertible red fluorescent protein with high brightness, high photoconversion contrast, and early detectable fluorescenceDepositorInsertpcStar
TagsHisExpressionBacterialPromoterT7Available SinceJune 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
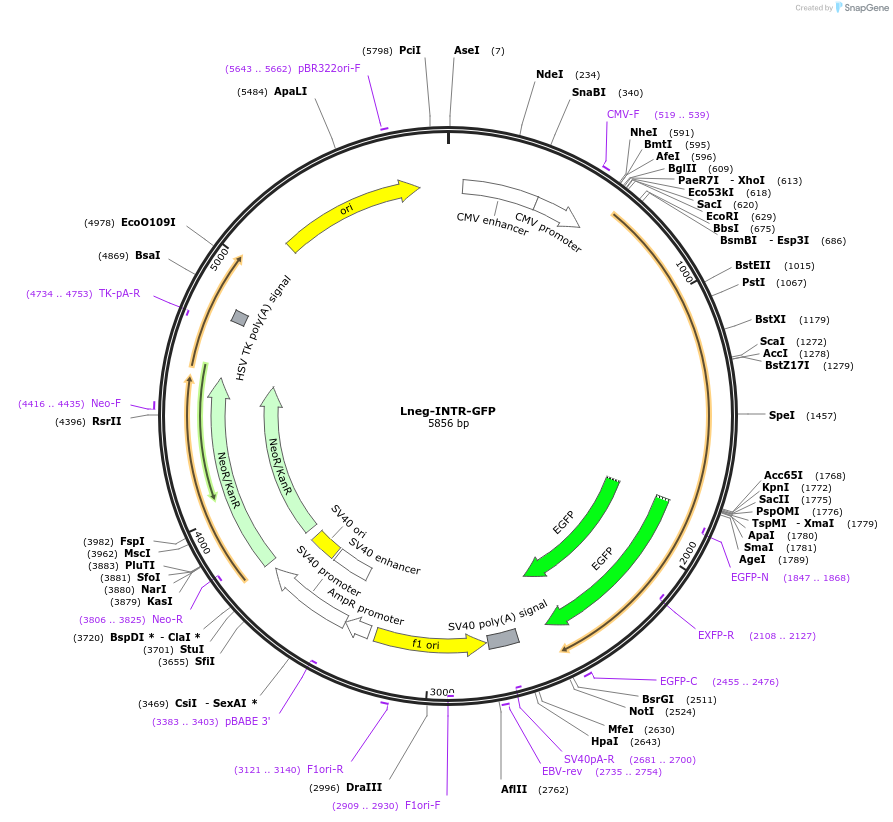
Lneg-INTR-GFP
Plasmid#208902Purposeencodes the inotocin receptor (oxytocin/vasopressin-like) from the ant Lasius neglectus, tagged with GFPDepositorInsertINTR
TagsEGFPExpressionMammalianAvailable SinceApril 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
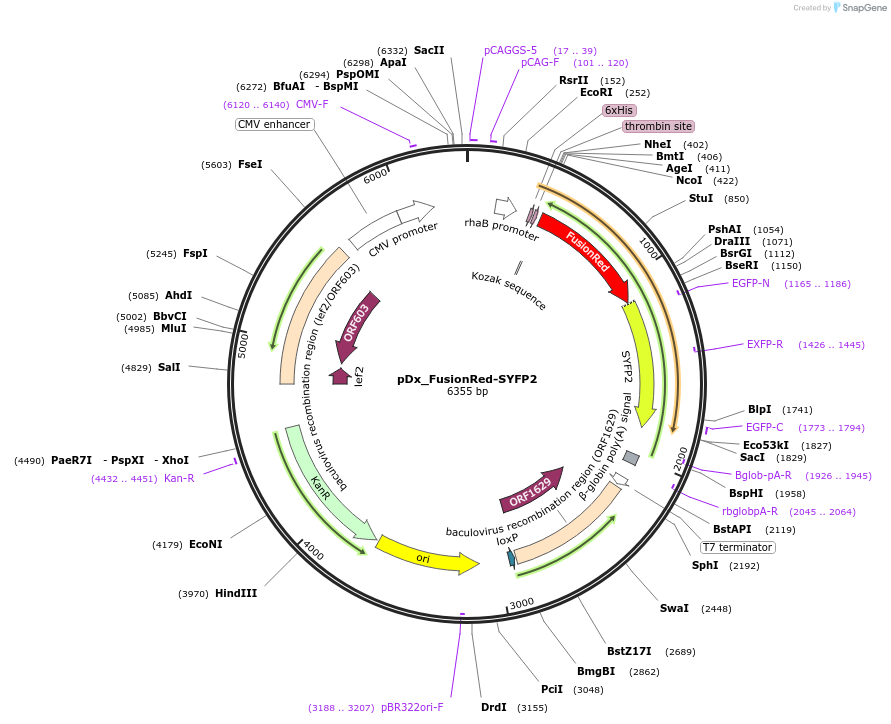
pDx_FusionRed-SYFP2
Plasmid#189760PurposeDual expression vector for bacteria and mammalian cells producing FusionRed-SYFP2 fusion protein for FRET analysisDepositorInsertFusionRed-SYFP2
Tags6xHisExpressionBacterial and MammalianPromoterCMV & RhaAvailable SinceMarch 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
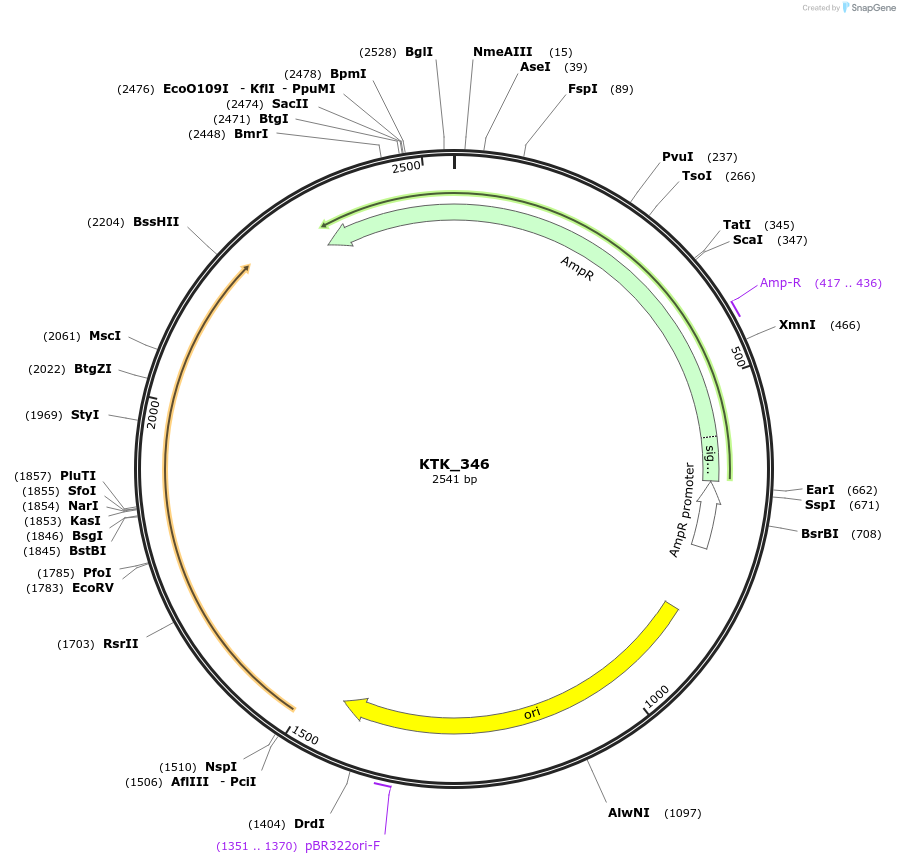
KTK_346
Plasmid#180549PurposeEntry vector containing gfasPurpleDepositorInsertgfasPurple
ExpressionBacterialAvailable SinceFeb. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
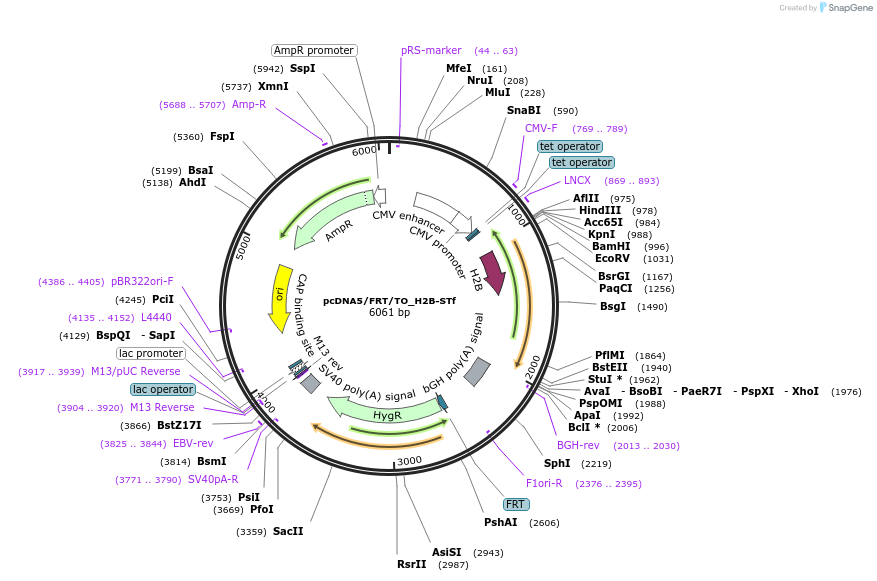
pcDNA5/FRT/TO_H2B-STf
Plasmid#181998PurposeCMV overexpression of SNAP-tag fast fusion to H2B for nuclear localization in mammalian cell linesDepositorInsertH2B-STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
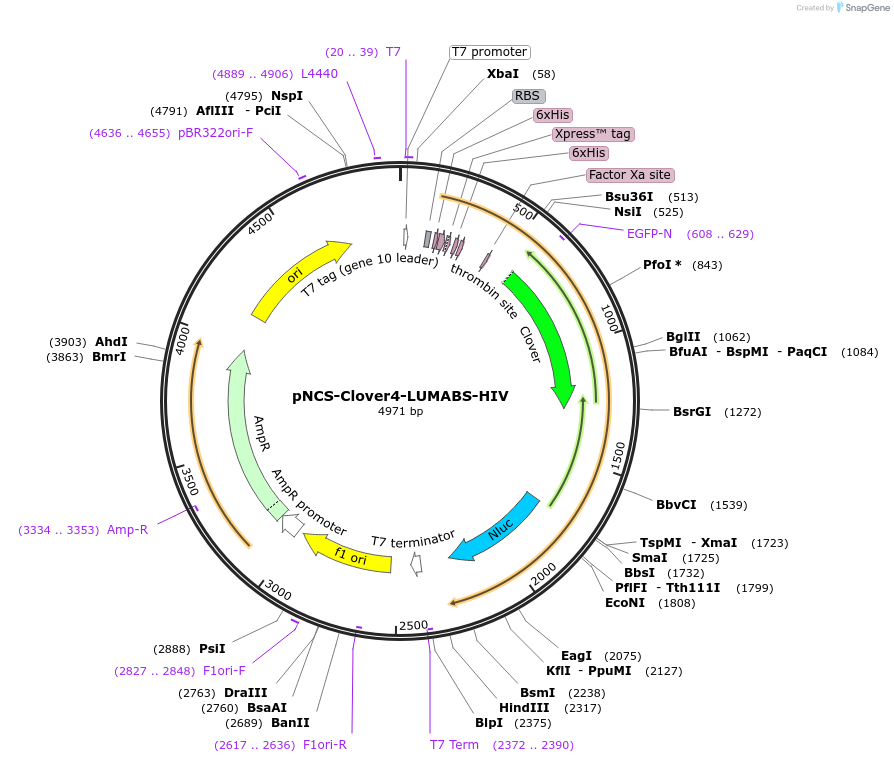
pNCS-Clover4-LUMABS-HIV
Plasmid#183485PurposeTo express the Clover4-LUMABS-HIV sensor for Acquired Immunodeficiency Virus antibody dection.DepositorInsertClover4-LUMABS-HIV
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only