We narrowed to 9,891 results for: Coli
-
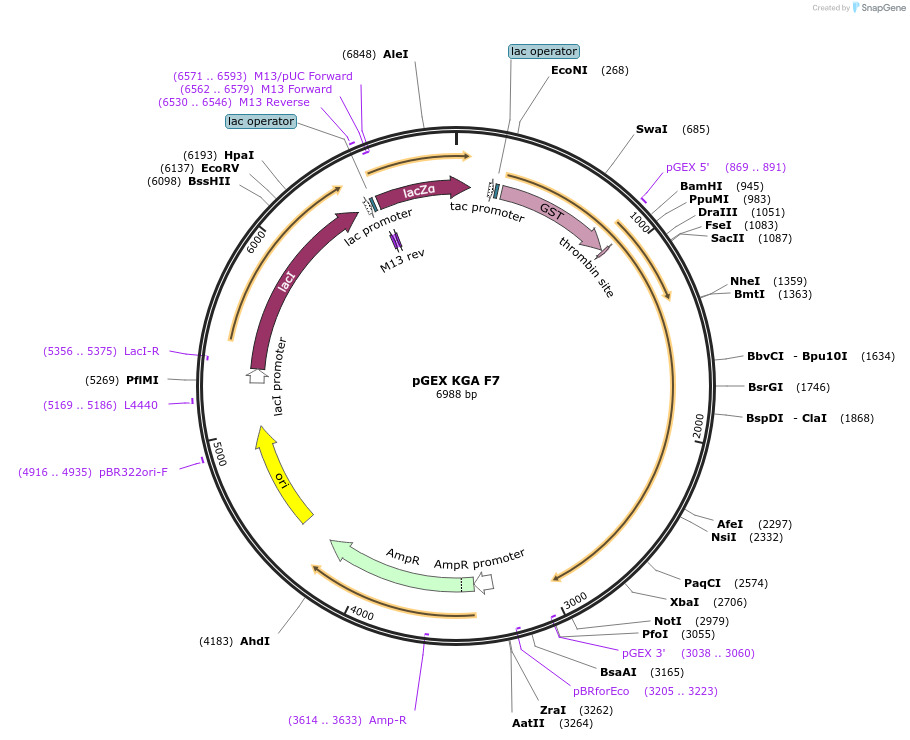
Plasmid#227822PurposeTo express in E. coli KGA F7 with 33 residues mutated to alanine at the interface for interacting with PDZD8DepositorInsertKGA isoform of F7 (GLS Human)
TagsGSTExpressionBacterialMutationP137, L139, E140, L142, Y145, G150, Q151, E152, K…Available SinceJune 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
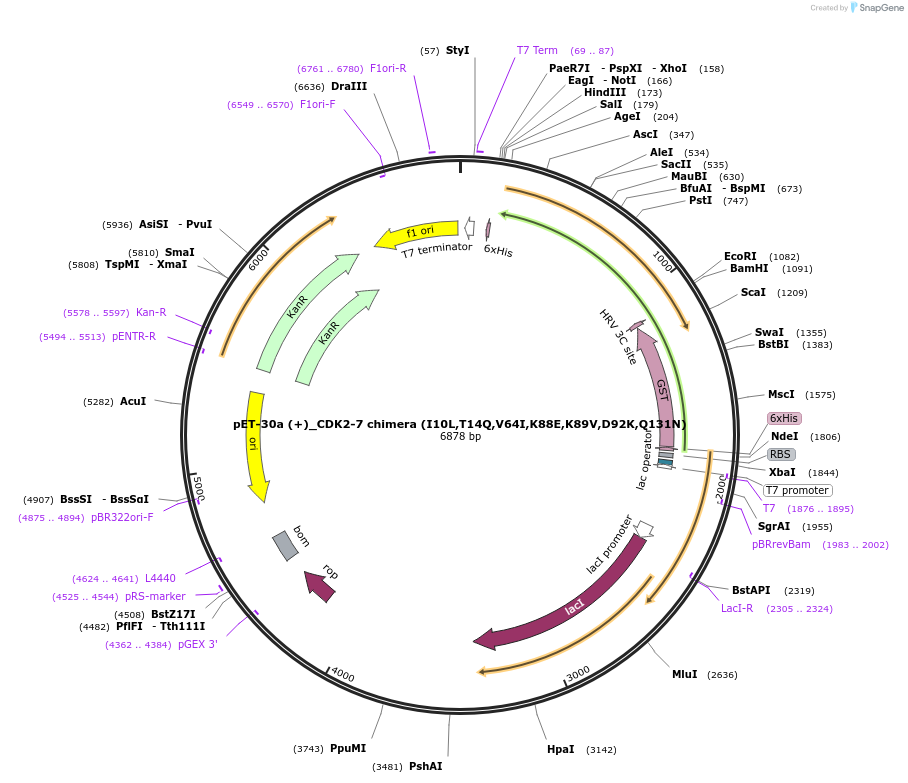
pET-30a (+)_CDK2-7 chimera (I10L,T14Q,V64I,K88E,K89V,D92K,Q131N)
Plasmid#217014PurposeExpression of CDK2-7 chimera in E.coliDepositorTagsC3 protease cleavable GST tagExpressionBacterialMutationCDK2 (I10L,T14Q,V64I,K88E,K89V,D92K,Q131N)Available SinceJuly 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
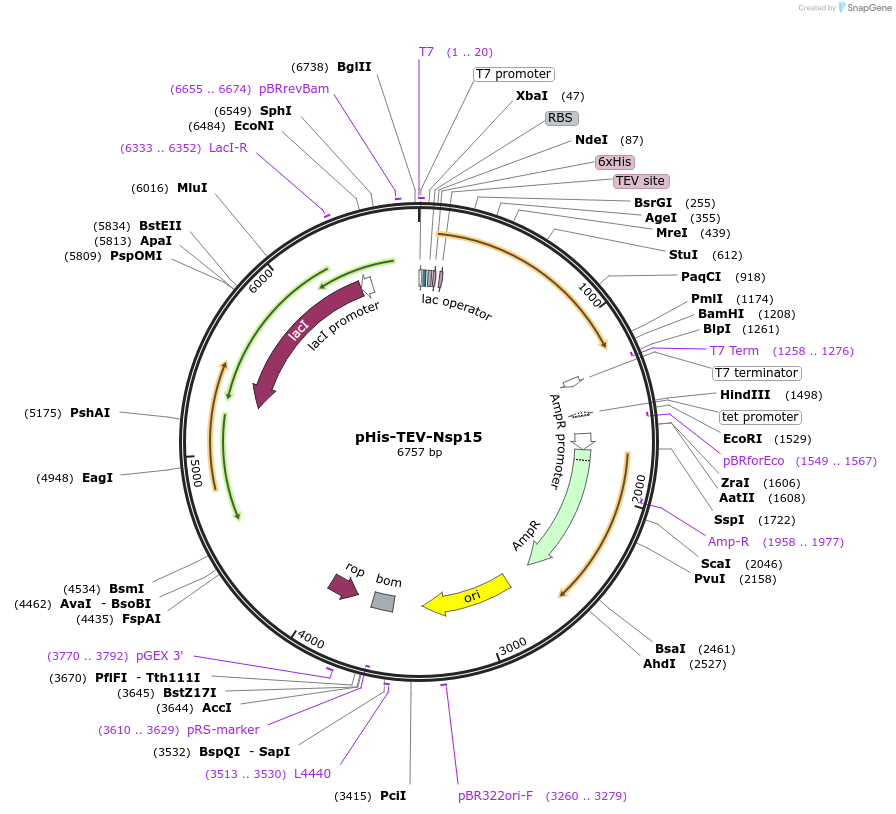
pHis-TEV-Nsp15
Plasmid#187656PurposeE. coli plasmid (T7lac promoter) for full-length, wild-type SARS CoV-2 Nsp15 (uridine-specific nidoviral endoribonuclease, expression-optimized gene) with N-terminal TEV protease cleavable His6-tagDepositorInsertNsp15 (N Severe acute respiratory syndrome coronavirus 2)
TagsTEV protease cleavable His6-tagExpressionBacterialMutationnonePromoterT7lacAvailable SinceMarch 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
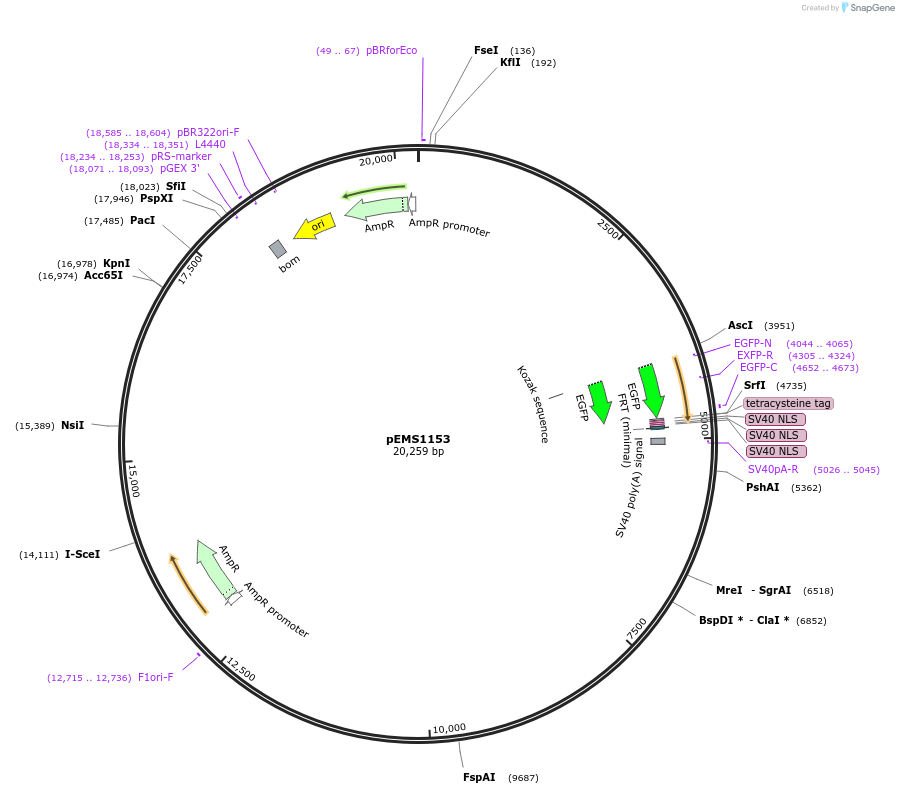
pEMS1153
Plasmid#29068PurposeExpression of the reporter gene under the control of the MiniPromoter in this plasmid has not been testedDepositorAvailable SinceJan. 9, 2012AvailabilityAcademic Institutions and Nonprofits only -
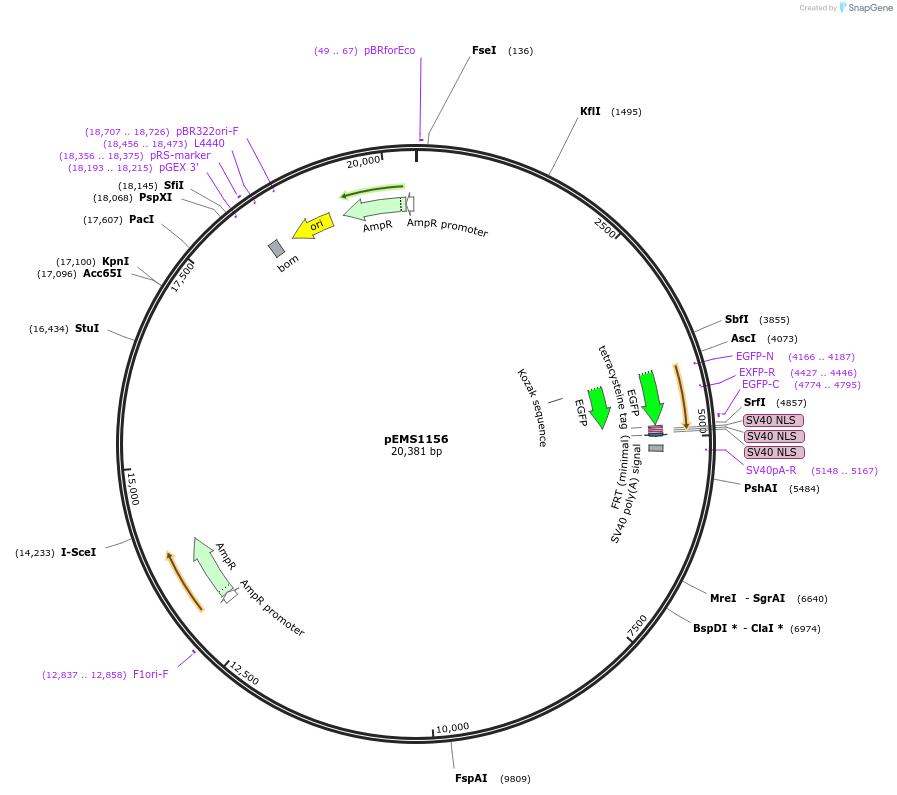
pEMS1156
Plasmid#29071PurposeExpression of the eGFP reporter gene was not detected in the brain and eye.DepositorAvailable SinceJan. 25, 2012AvailabilityAcademic Institutions and Nonprofits only -
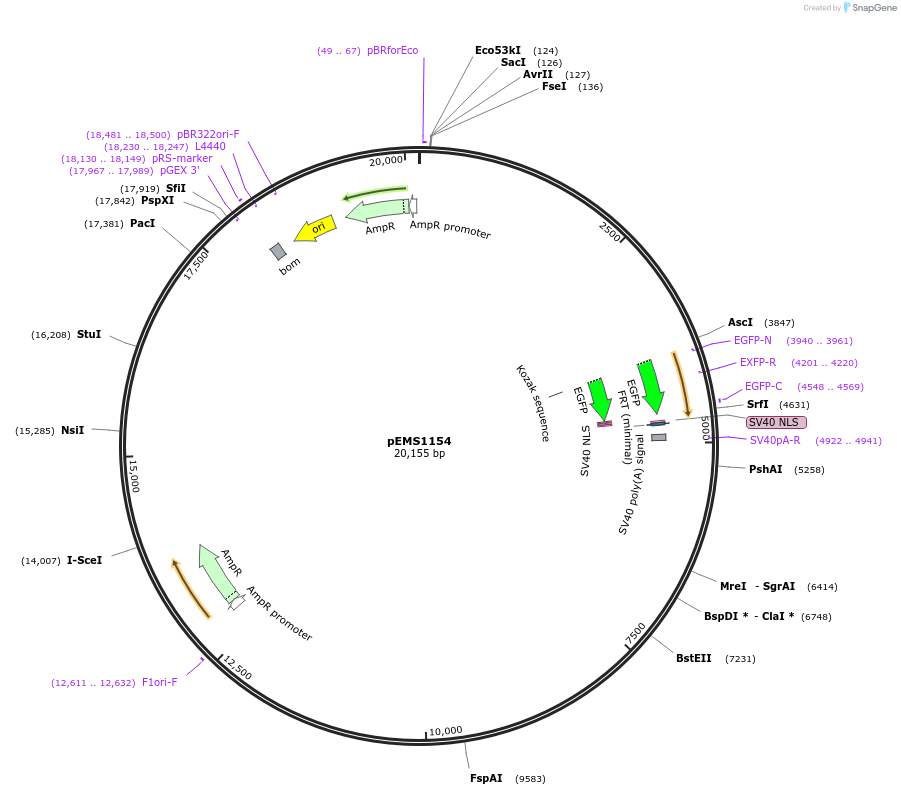
pEMS1154
Plasmid#29069PurposeExpression of the reporter gene under the control of the MiniPromoter in this plasmid has not been testedDepositorAvailable SinceJan. 9, 2012AvailabilityAcademic Institutions and Nonprofits only -
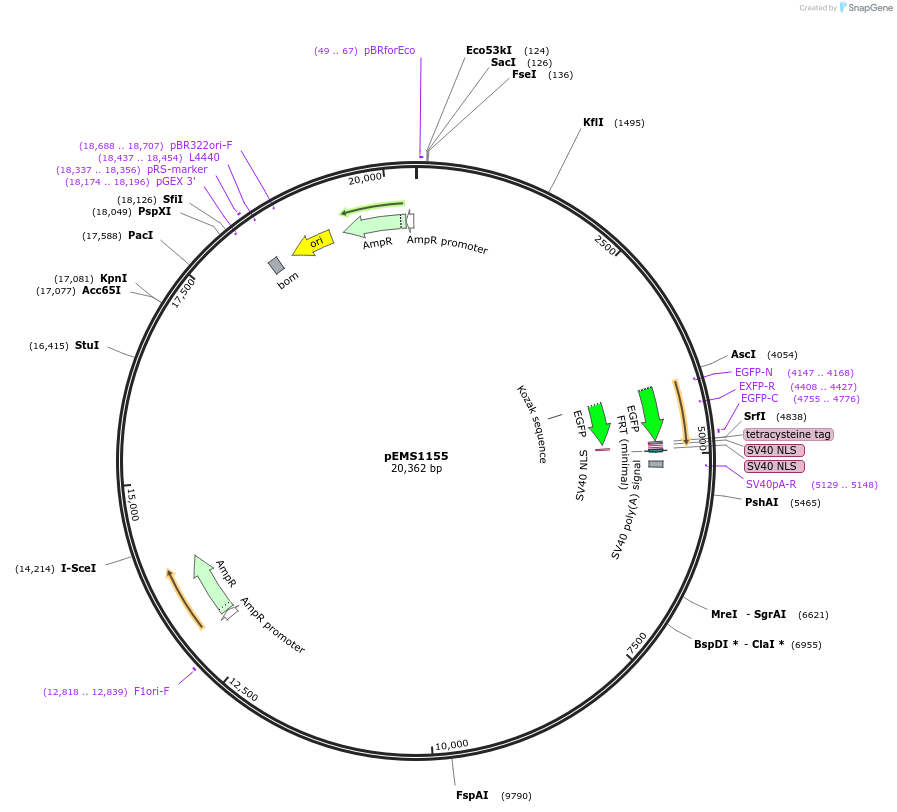
pEMS1155
Plasmid#29070PurposeExpression of the reporter gene under the control of the MiniPromoter in this plasmid has not been testedDepositorAvailable SinceJan. 9, 2012AvailabilityAcademic Institutions and Nonprofits only -
XanthoMoClo Xanthobacter Genetic Toolkit
Plasmid Kit#1000000259PurposePlasmids and parts for modular assembly of single and multi-gene plasmids for any Xanthobacter or Roseixanthobacter strain.DepositorAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
E. coli MEV20
Bacterial Strain#197113PurposeE. coli BL21(DE3) derivative for diterpenoid production with genome-integrated yeast MEV pathway, redox enzyme array, and Marionette clusterDepositorBacterial ResistanceNoneAvailable SinceJune 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
E. coli MEV15
Bacterial Strain#197112PurposeE. coli BL21(DE3) derivative for sesquiterpenoid production with genome-integrated yeast MEV pathway, redox enzyme array, and Marionette clusterDepositorBacterial ResistanceNoneAvailable SinceJune 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
rEcoli XpS
Bacterial Strain#192872PurposeMG1655-C321 -- All 321 UAG codons changed to UAADepositorBacterial ResistanceBleocin (Zeocin)SpeciesEscherichia coliAvailable SinceDec. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
E. coli ER1821ΔlacI
Bacterial Strain#141407PurposeLacI-deficient E. coli host bacterium that produces β-galactosidase constitutivelyDepositorBacterial ResistanceNoneAvailable SinceAug. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655 RARE
Bacterial Strain#61440PurposeThis E. coli chassis displays reduced endogenous reduction of aromatic and aliphatic aldehydesDepositorBacterial ResistanceNoneAvailable SinceFeb. 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
Escherichia coli DH10B-MOB
Bacterial Strain#229395PurposeEscherichia coli DH10B DAP-auxotrophic strain containing an integrated RK2/RP4 mobilization vector to conjugate plasmids with an IncP origin of transfer (modified pTA-Mob 2.0, AddGene Plasmid #149662)DepositorBacterial Resistance60 ug/mL of DAPSpeciesEscherichia coliAvailable SinceMarch 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
E. coli BL21 (DE3) ΔEntD
Bacterial Strain#192874PurposeDeletion of native E. coli EntD, a 4'-PPTase required for native enterobactin synthesis. Allows expression of NRPS carrier proteins that do not harbor a phosphopantetheinyl group.DepositorBacterial ResistanceNoneSpeciesEscherichia coliAvailable SinceSept. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
E. coli BW25113 ΔtnaA ΔtrpR
Bacterial Strain#205015PurposeUsed for the characterization of the TrpR1-PtrpO1 biosensor system with the deletion of tnaA gene and trpR gene (no antibiotics marker)DepositorBacterial ResistanceNoneSpeciesEscherichia coliAvailable SinceAug. 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
E. coli K-12 MG1655_rLP5(nth-ydgR)
Bacterial Strain#110245PurposeE. coli K-12 MG1655 with Landing Pad engineered into the nth-ydgR intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_fLP4(essQ-cspB)
Bacterial Strain#110244PurposeE. coli K-12 MG1655 with Landing Pad engineered into the essQ-cspB intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_rLP6(ygcE-ygcF)
Bacterial Strain#110246PurposeE. coli K-12 MG1655 with Landing Pad engineered into the ygcE-ygcF intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_rLP1(atpI-gidB)
Bacterial Strain#110241PurposeE. coli K-12 MG1655 with Landing Pad engineered into the atpl-gidB intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only