We narrowed to 81,743 results for: MYC;
-
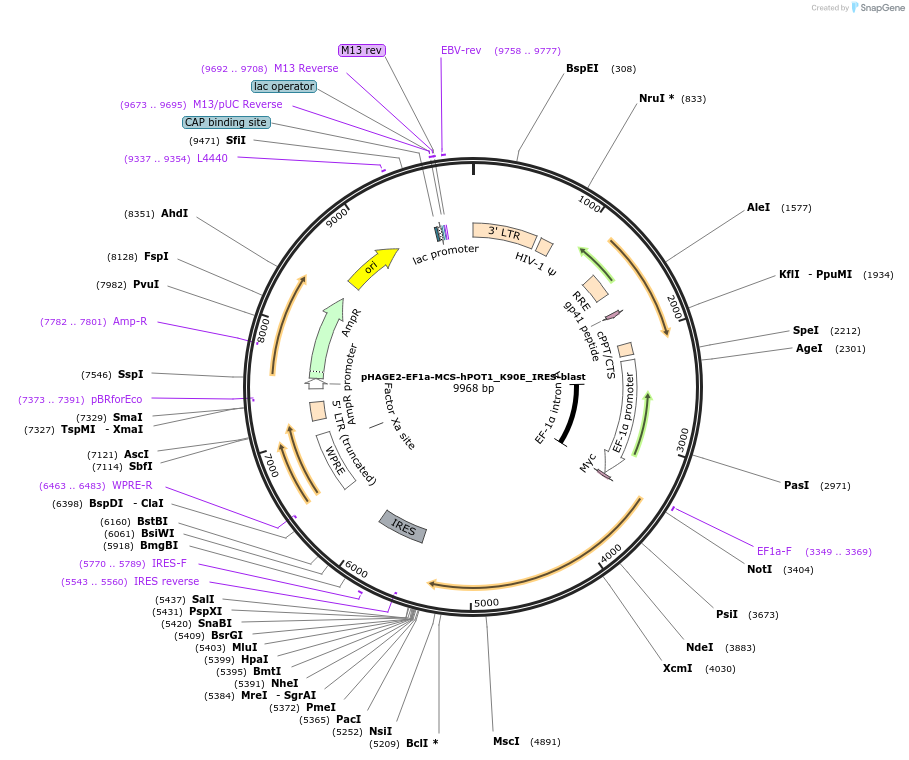
Plasmid#166412Purposeexpress hPOT1 K90E in mammalian cellsDepositorInserthPOT1 K90E (POT1 Human)
UseLentiviralTagsMYCExpressionMammalianMutationK90EPromoterEF-1aAvailable SinceMay 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
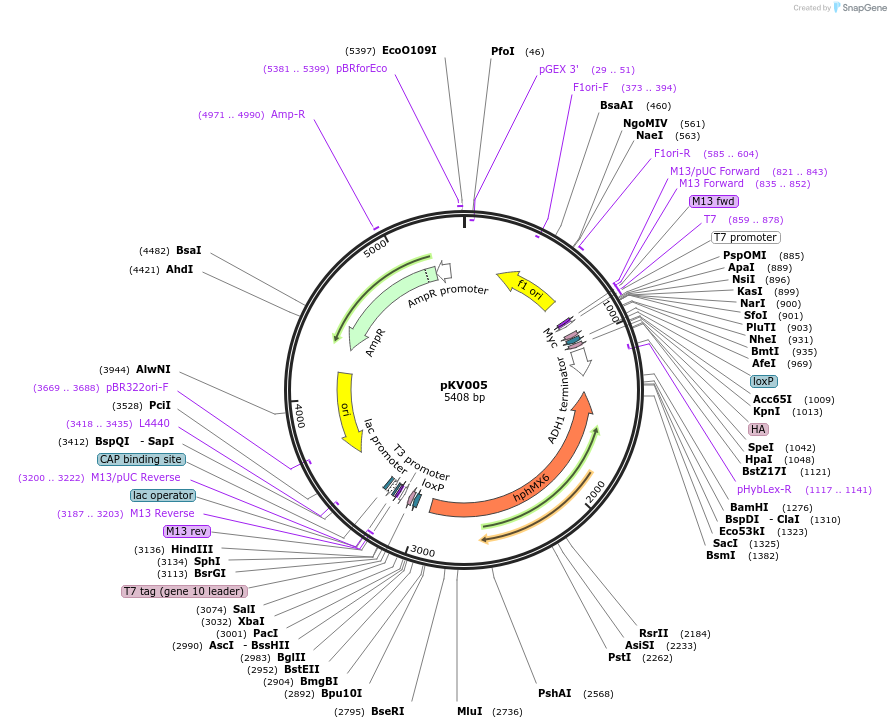
pKV005
Plasmid#64763PurposeRecombination-Induced Tag Exchange (RITE) Cassette, HA to a T7 epitope tag, invariable Myc tag, Hygro selectionDepositorInsertRITE cassette
UseRite cassettePromoternoneAvailable SinceJuly 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
PdarC1
Plasmid#24835DepositorInsertPdarC1
TagsHA and MycExpressionYeastAvailable SinceAug. 23, 2011AvailabilityAcademic Institutions and Nonprofits only -
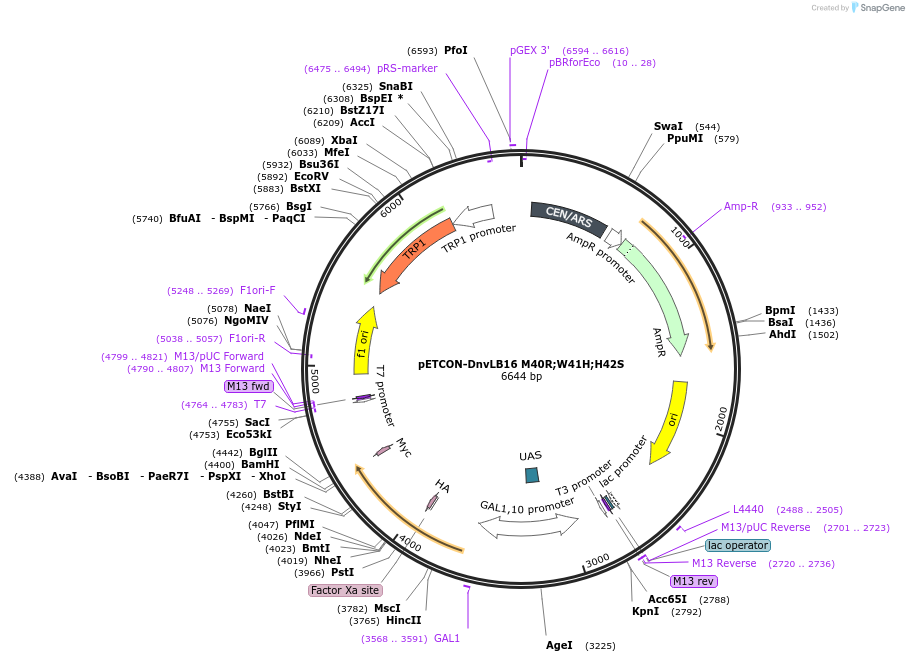
pETCON-DnvLB16 M40R;W41H;H42S
Plasmid#45125DepositorInsertDnvLB16 M40R;W41H;H42S
TagsAga2p for yeast surface display and c-myc epitope…ExpressionYeastMutationM40R;W41H;H42S to DnvLB16PromoterGALAvailable SinceJuly 8, 2013AvailabilityAcademic Institutions and Nonprofits only -
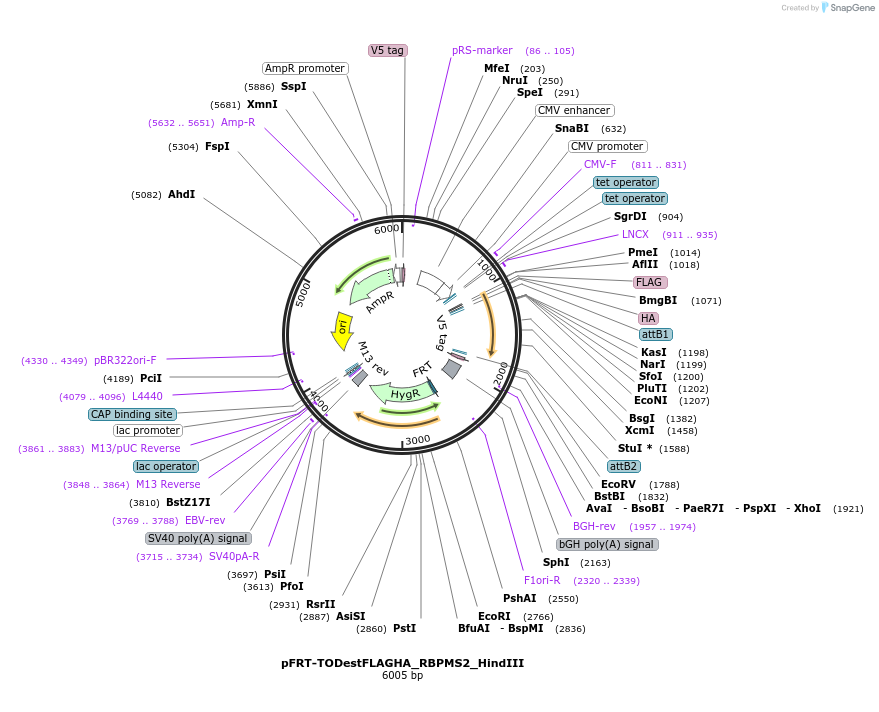
pFRT-TODestFLAGHA_RBPMS2_HindIII
Plasmid#65666PurposeFLAG-HA expression vector with RBPMS2 carrying mutation to introduce HindIII siteDepositorInsertRBPMS2 (RBPMS2 Human)
TagsFLAG HAExpressionMammalianMutationintroduced HindIII site (final goal to exchange F…PromoterCMV, 2x TetAvailable SinceJuly 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
Syn3-27-146+TM
Plasmid#14679DepositorInsertSyntaxin3 (STX3 Human)
Tags2xMyc-HisExpressionMammalianMutationInsert is cloned into a modified vector; aa 27-14…Available SinceApril 20, 2007AvailabilityAcademic Institutions and Nonprofits only -
-
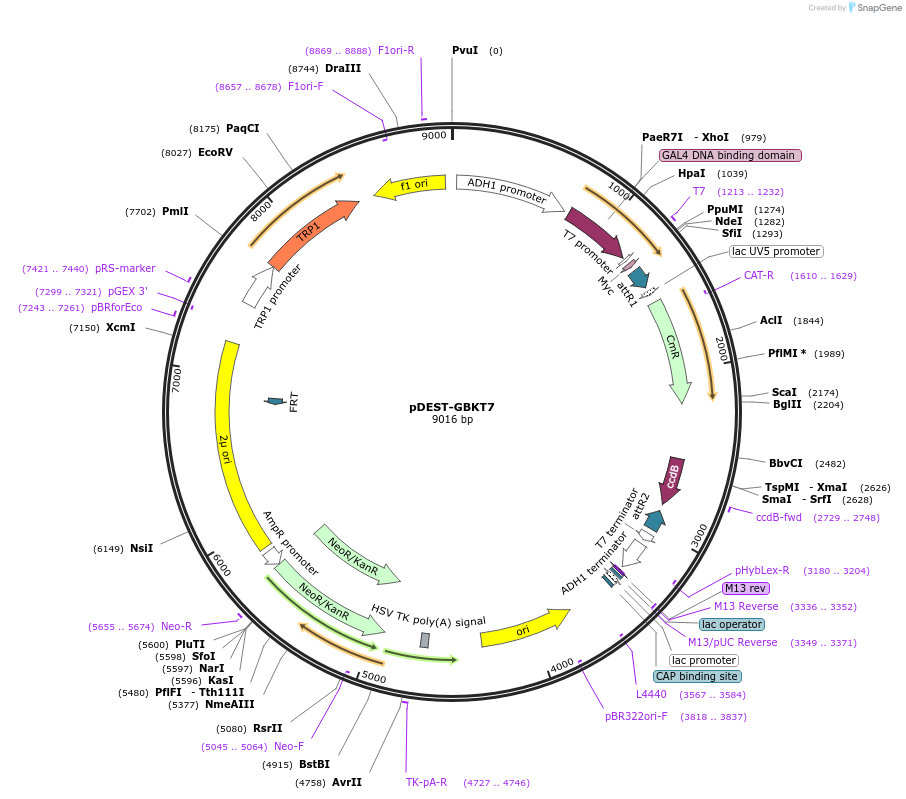
pDEST-GBKT7
Plasmid#248655PurposeEmpty vector for generating yeast 2-hybrid GAL4 DNA binding domain fusion constructs by Gateway cloning. It may also be used as an empty vector experimental control.DepositorTypeEmpty backboneUseYeast 2-hybridTagsGAL4 binding domain, MycExpressionYeastAvailable SinceJan. 7, 2026AvailabilityAcademic Institutions and Nonprofits only -
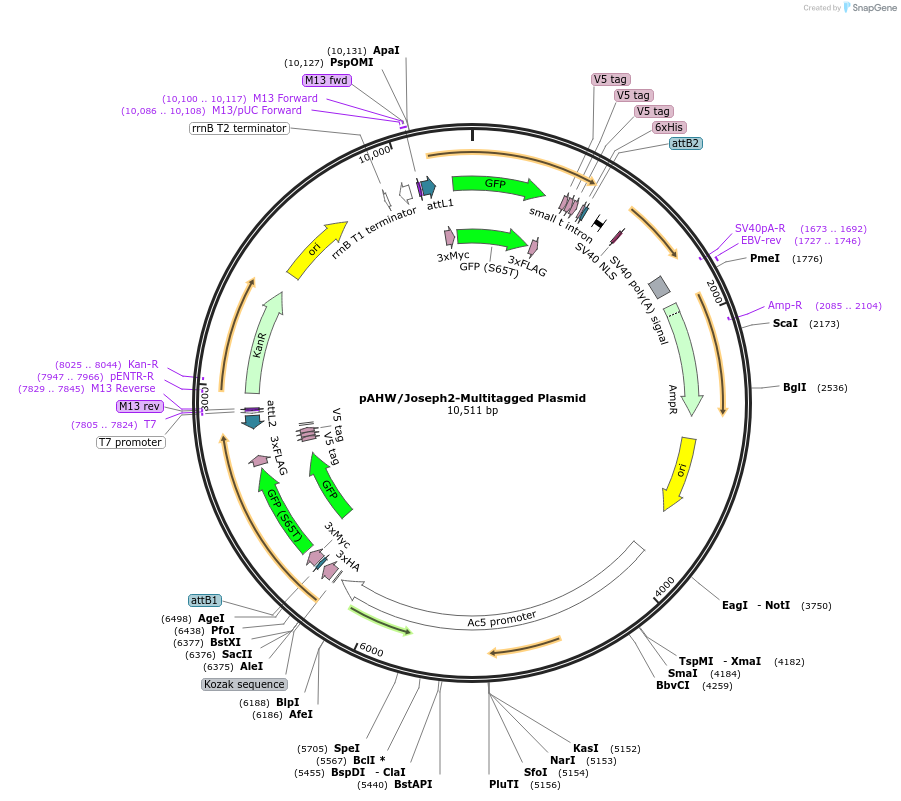
pAHW/Joseph2-Multitagged Plasmid
Plasmid#226725PurposeValidates the expression and detection of epitope-tagged proteins of interest in Drosophila cells.DepositorInsertpAHW/Joseph2
UseDrosophila s2 cell expressionTagsHA, Myc, GFP, Flag, V5, 6xHisExpressionInsectPromoterAct5CAvailable SinceJan. 29, 2025AvailabilityAcademic Institutions and Nonprofits only -
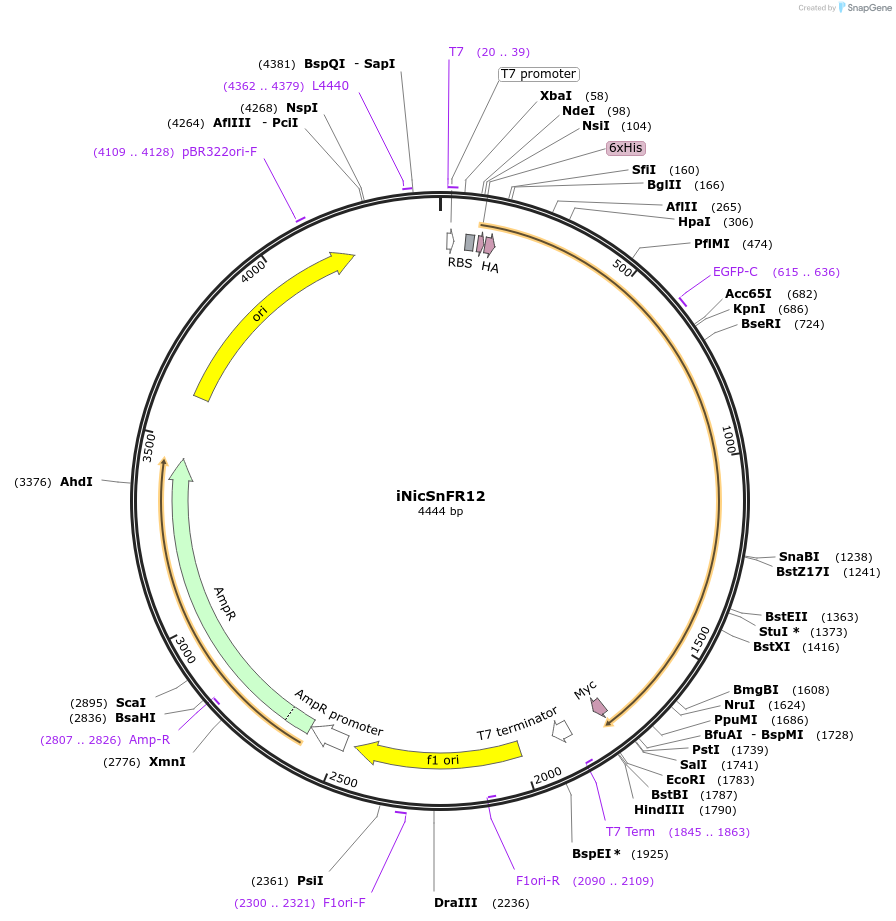
iNicSnFR12
Plasmid#227142PurposeGenetically encoded fluorescent biosensor for nicotine detectionDepositorInsertiNicSnFR12
Tags6xHis-HA and MycExpressionBacterialPromoterT7Available SinceNov. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
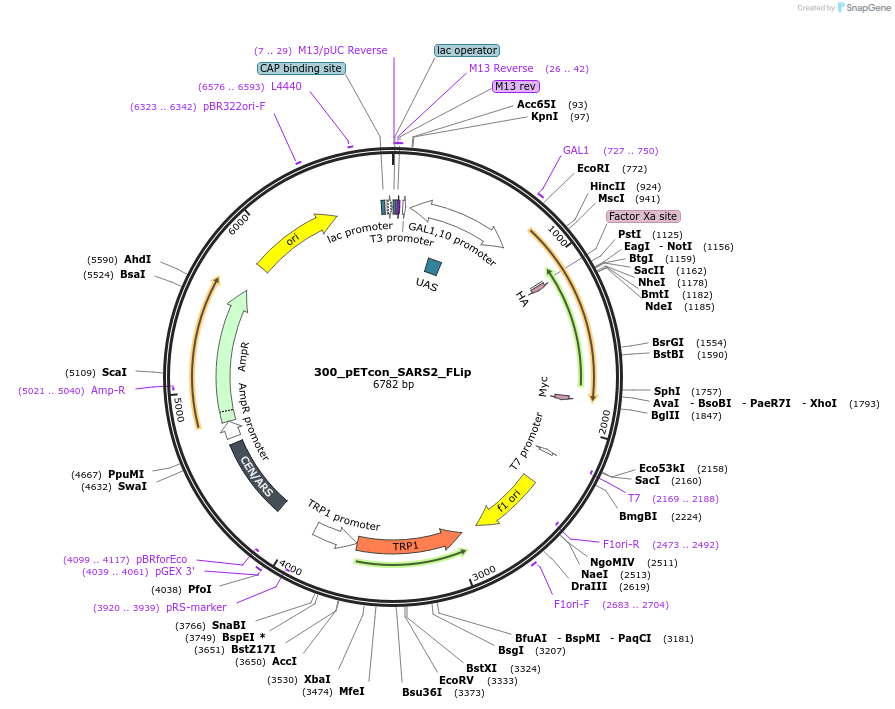
300_pETcon_SARS2_FLip
Plasmid#222230Purposeyeast surface display of the SARS-CoV-2 FLip variant RBDDepositorAvailable SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
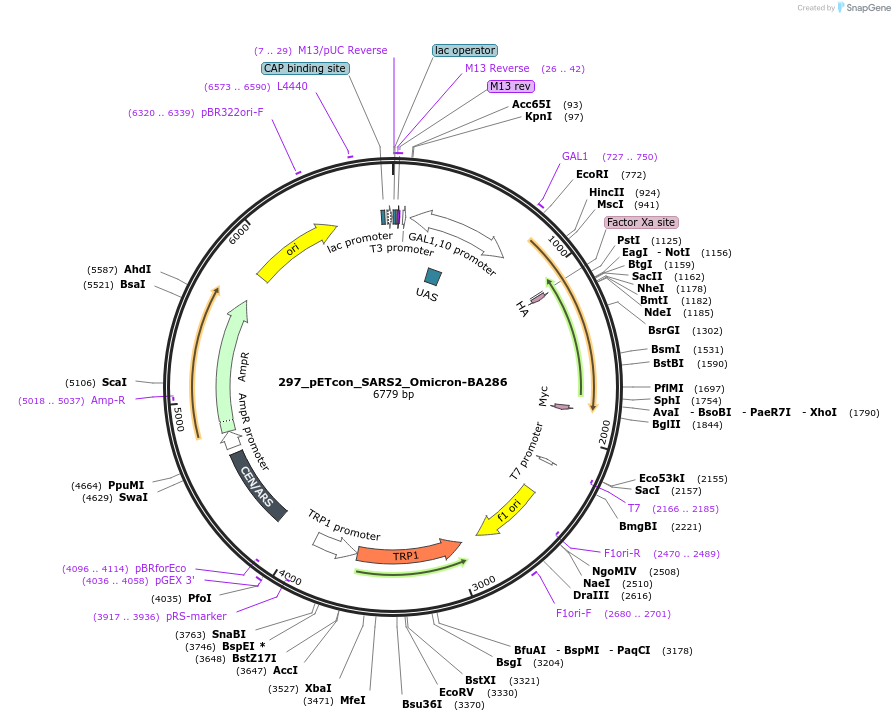
297_pETcon_SARS2_Omicron-BA286
Plasmid#222231Purposeyeast surface display of the SARS-CoV-2 BA.2.86 variant RBDDepositorInsertSARS-CoV-2 BA.2.86 RBD (S Budding Yeast, SARS-CoV-2 virus)
TagsHA and c-MycExpressionYeastAvailable SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
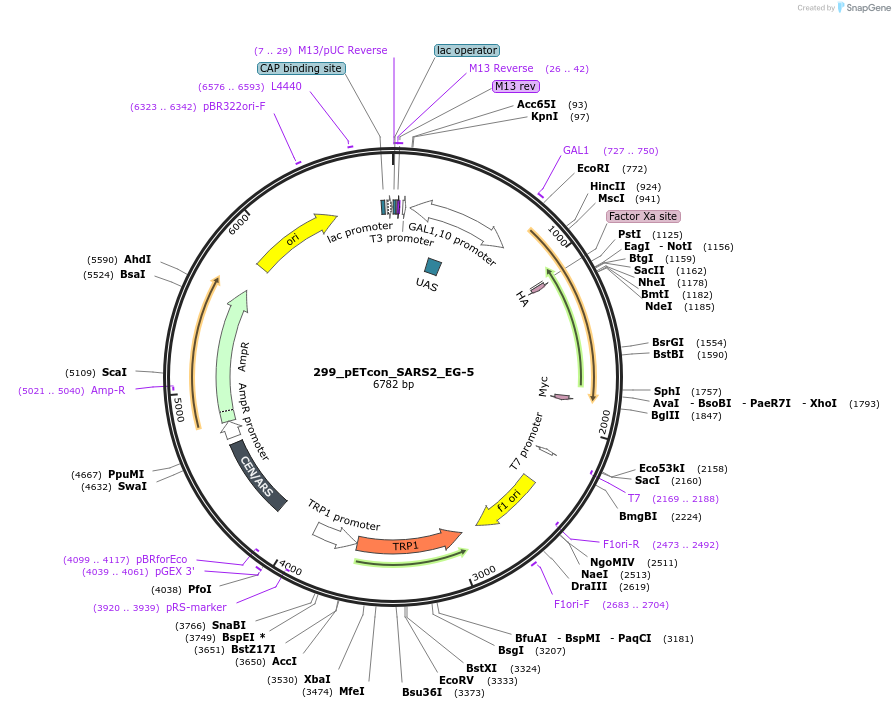
299_pETcon_SARS2_EG-5
Plasmid#222232Purposeyeast surface display of the SARS-CoV-2 EG.5 variant RBDDepositorAvailable SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
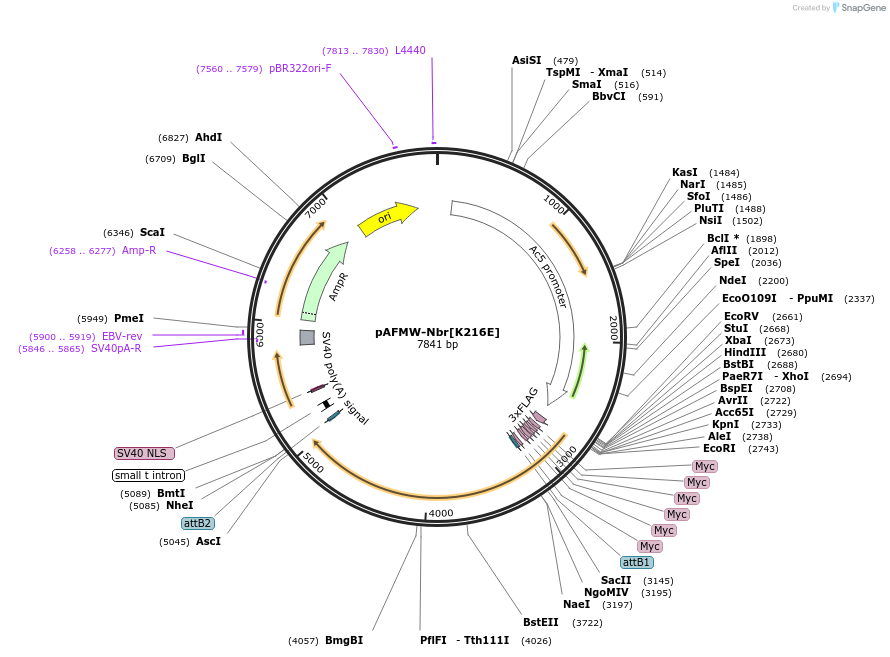
pAFMW-Nbr[K216E]
Plasmid#188600PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
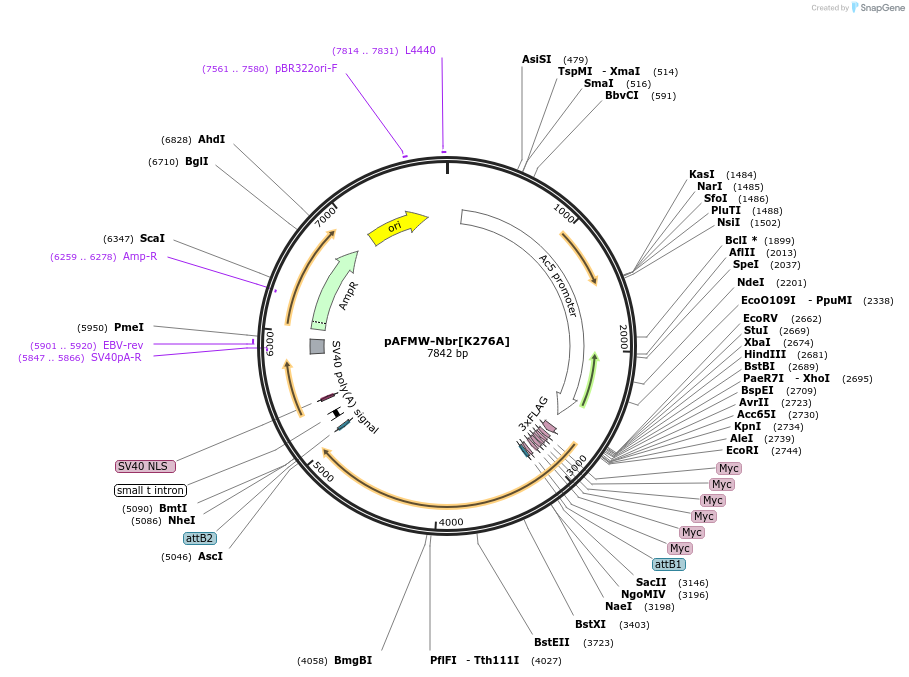
pAFMW-Nbr[K276A]
Plasmid#188597PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
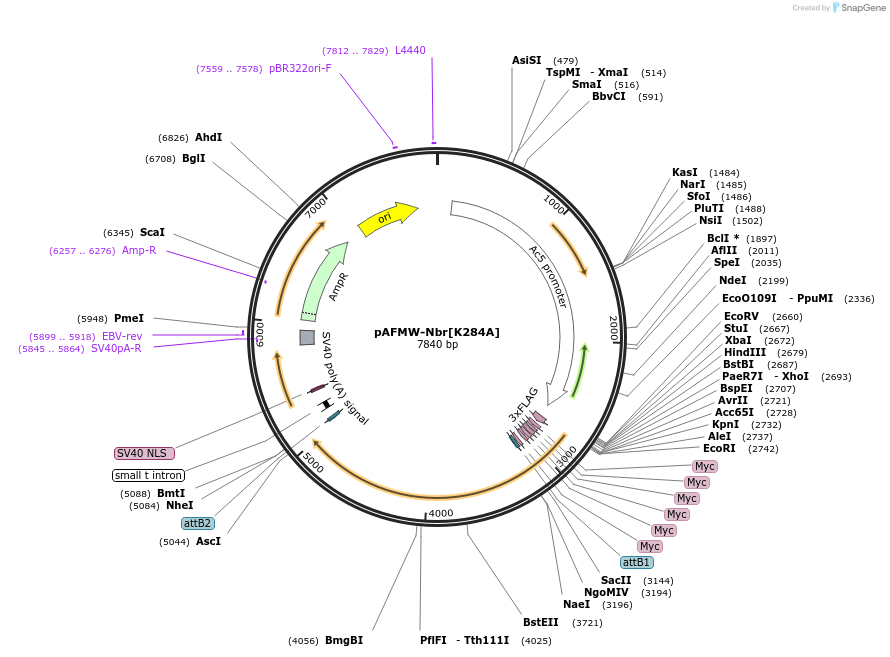
pAFMW-Nbr[K284A]
Plasmid#188595PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
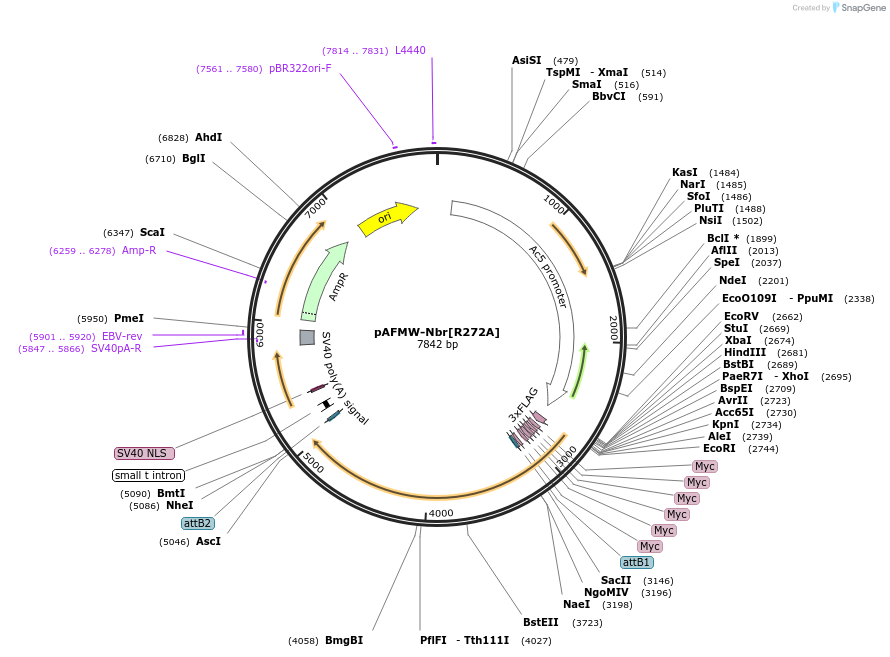
pAFMW-Nbr[R272A]
Plasmid#188594PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
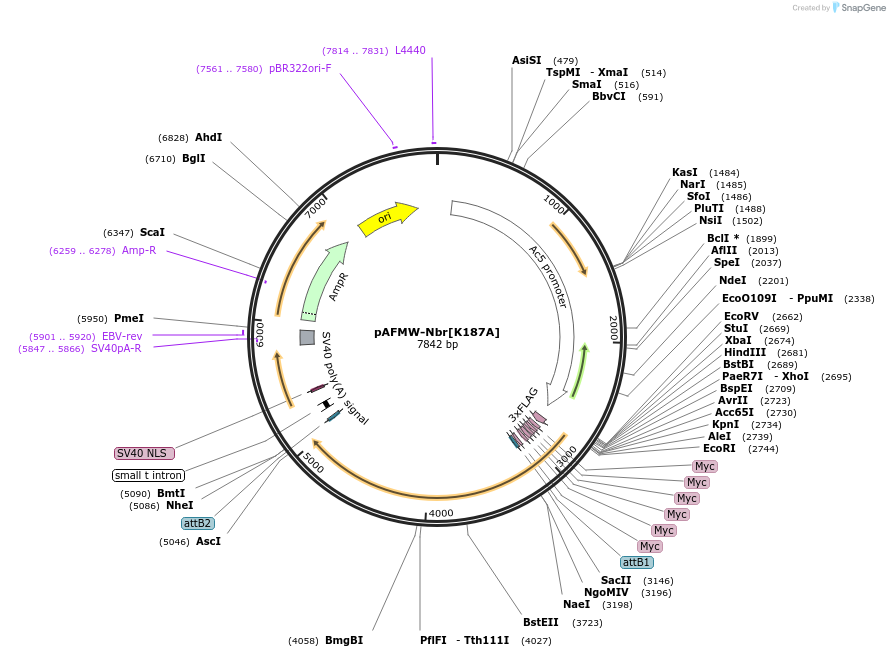
pAFMW-Nbr[K187A]
Plasmid#188593PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
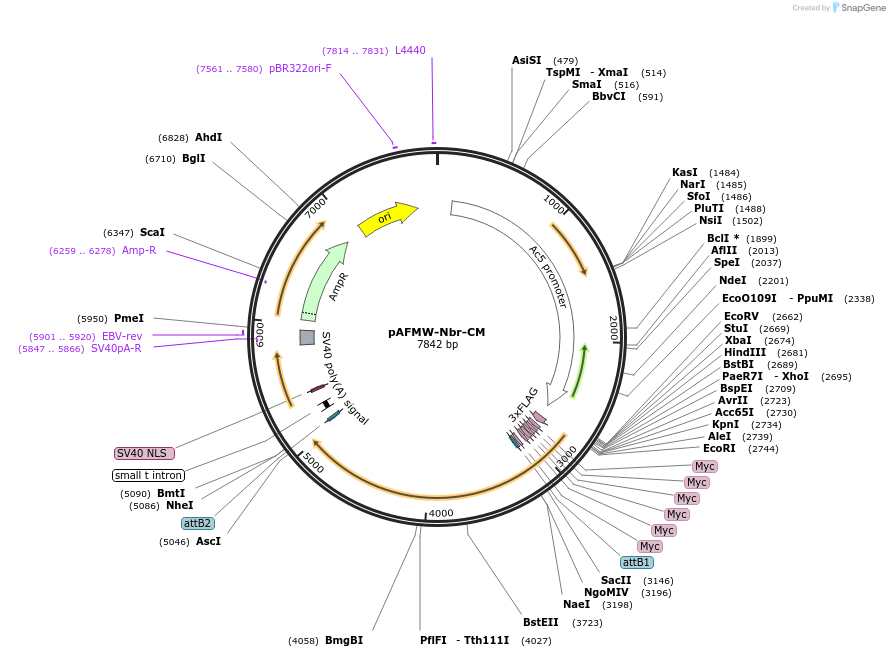
pAFMW-Nbr-CM
Plasmid#188585PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorInsertNibbler Catalytic mutant
TagsFLAG-MycExpressionInsectMutationdeleted first ATG; D435A [GAT=>GCC]; E435A [GA…Available SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
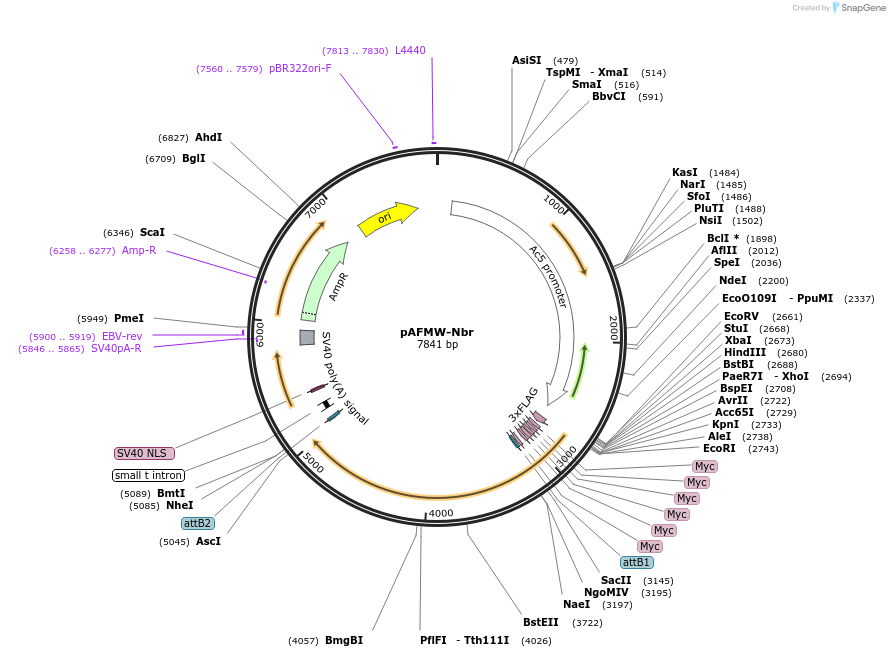
pAFMW-Nbr
Plasmid#188584PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
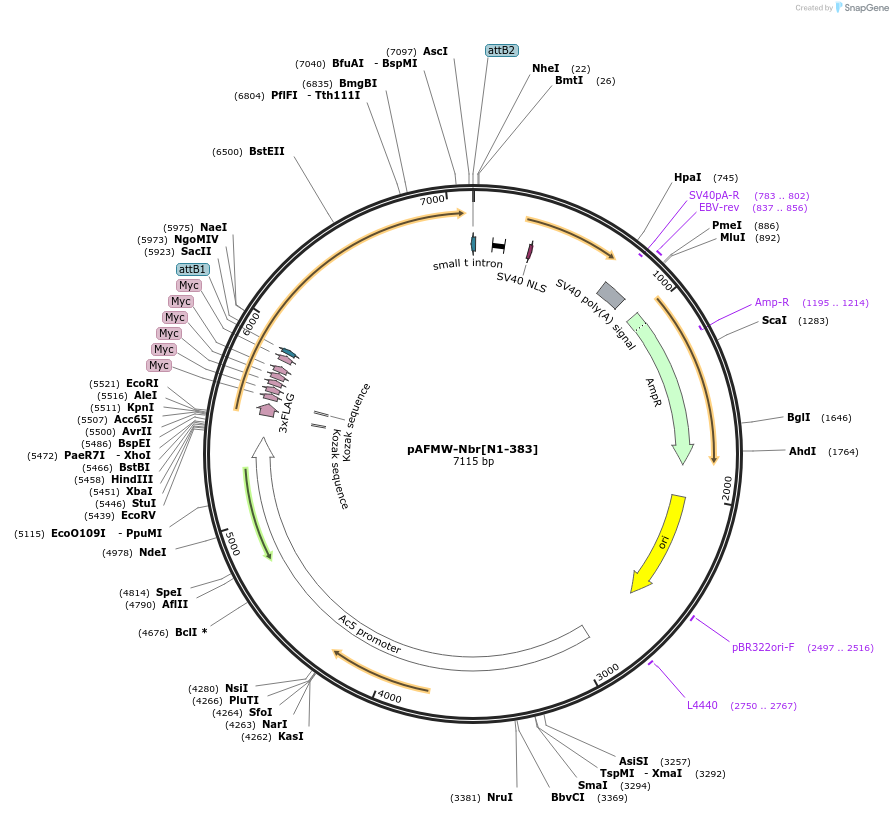
pAFMW-Nbr[N1-383]
Plasmid#188586PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorInsertNibbler N-terminal domain
TagsFLAG-MycExpressionInsectMutationdeleted first ATGAvailable SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
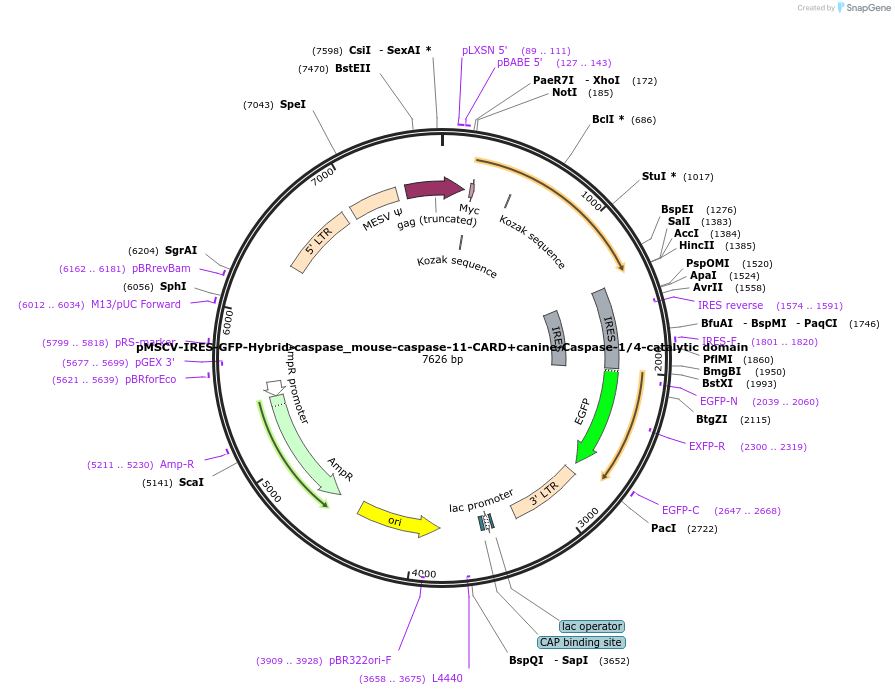
pMSCV-IRES-GFP-Hybrid-caspase_mouse-caspase-11-CARD+canine-Caspase-1/4-catalytic domain
Plasmid#183398PurposeExpression of mammalian inflammatory caspase gene in mammalian cells by retroviral transductionDepositorInsertCARD domain of mouse caspase-11 fused to catalytic domain of canine caspase-1/4
UseRetroviralTagsMycPromoterMESVAvailable SinceMay 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
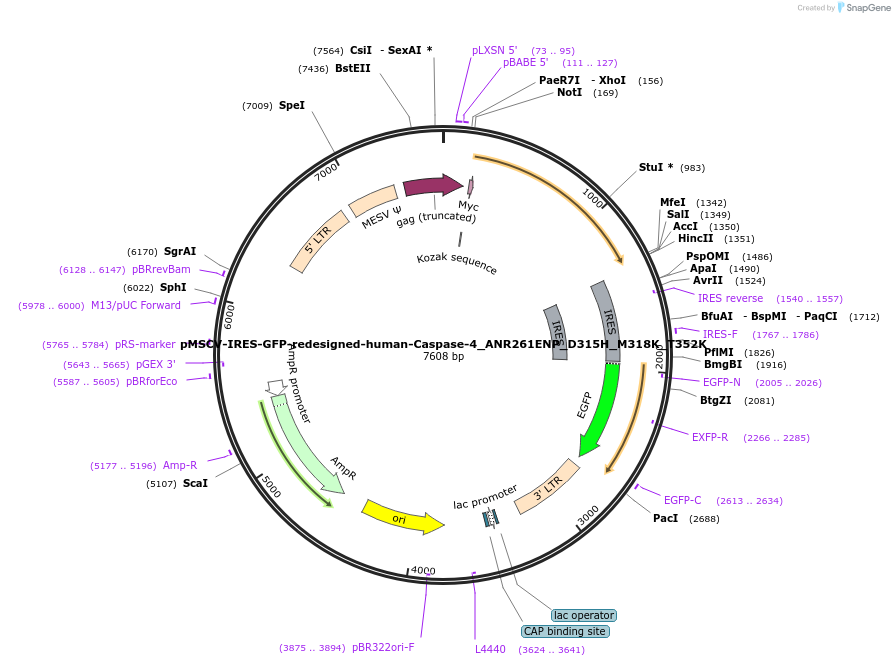
pMSCV-IRES-GFP-redesigned-human-Caspase-4_ANR261ENP_D315H_M318K_T352K
Plasmid#183357PurposeExpression of mammalian inflammatory caspase gene in mammalian cells by retroviral transductionDepositorInsertCaspase-4 with ICE activity (CASP4 Human)
UseRetroviralTagsMycMutationANR261ENP, D315H, M318K, T352KPromoterMESVAvailable SinceMay 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
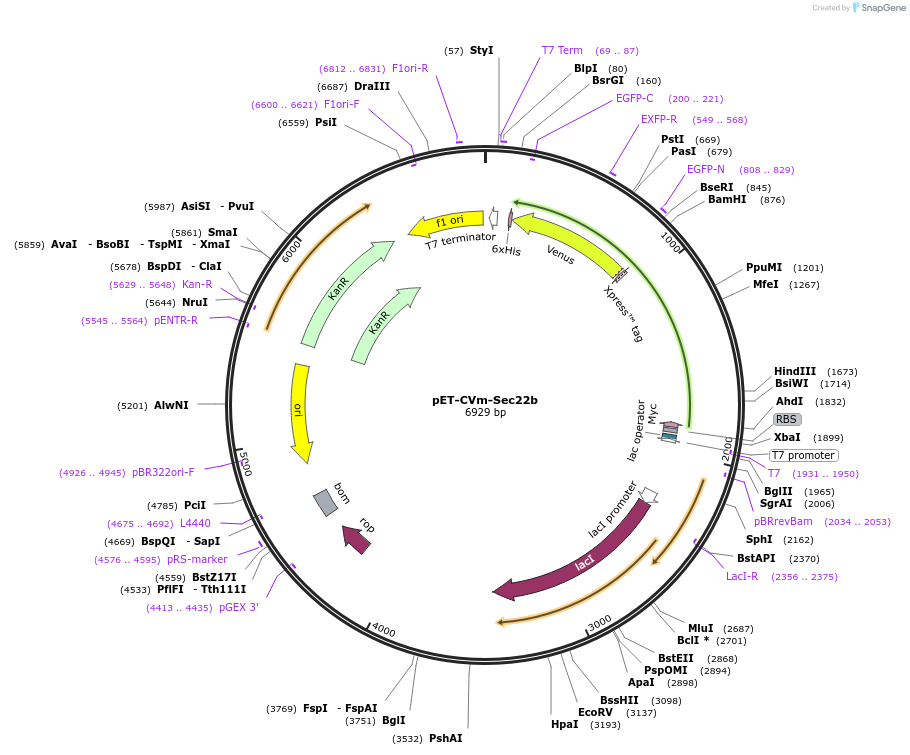
pET-CVm-Sec22b
Plasmid#168029PurposeExpresses eCFP-Sec22b(A131-H190)-mVenus in bacterial cellsDepositorInserteCFP-Sec22b(A131-H190)-mVenus
TagsHisx6 and mycExpressionBacterialAvailable SinceApril 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
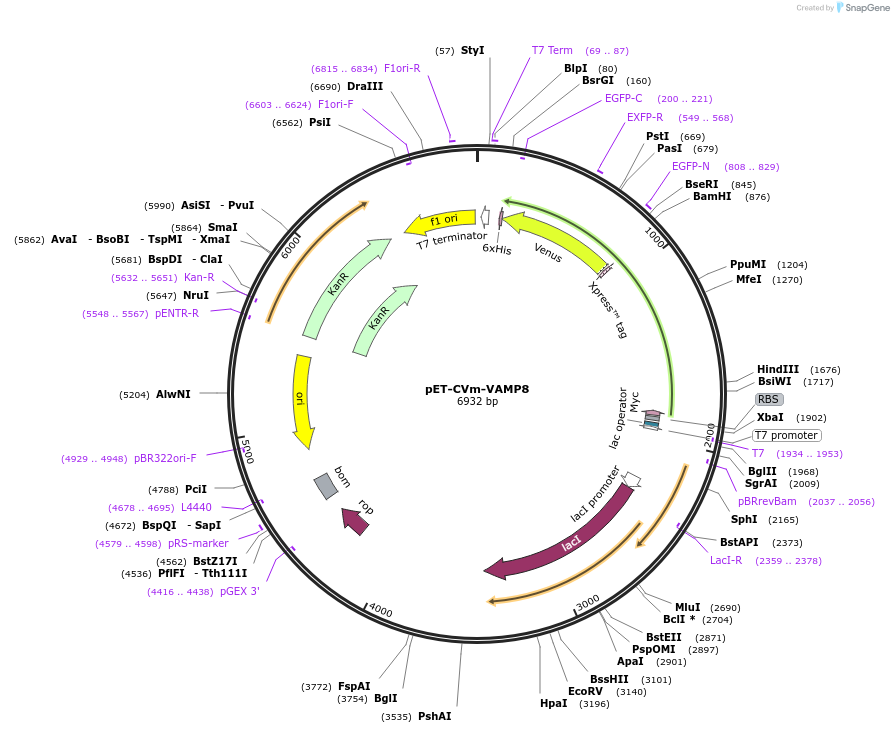
pET-CVm-VAMP8
Plasmid#168028PurposeExpresses eCFP-VAMP8(G7-R67)-mVenus in bacterial cellsDepositorInserteCFP-VAMP8(G7-R67)-mVenus
TagsHisx6 and mycExpressionBacterialAvailable SinceApril 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfE
Bacterial Strain#48935PurposeBase line strain is heat shock induce-able lambda red recombination system for MAGE (Multiplex Recombination Genome Engineering)DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
HE_150
Bacterial Strain#201051PurposeDH10B-derived Escherichia coli isolate obtained from adaptive laboratory evolution to increasing incubation temperatures.DepositorBacterial ResistanceStreptomycinAvailable SinceMay 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfC
Bacterial Strain#48936PurposeBase line strain is heat shock induce-able lambda red recombination system for MAGE (Multiplex Recombination Genome Engineering)DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
HE_100
Bacterial Strain#201050PurposeDH10B-derived Escherichia coli isolate obtained from adaptive laboratory evolution to increasing incubation temperatures.DepositorBacterial ResistanceStreptomycinAvailable SinceMay 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
HE_50
Bacterial Strain#201049PurposeDH10B-derived Escherichia coli isolate obtained from adaptive laboratory evolution to increasing incubation temperatures.DepositorBacterial ResistanceStreptomycinAvailable SinceMay 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC
Bacterial Strain#48937PurposeBase line strain is heat shock induce-able lambda red recombination system for MAGE (Multiplex Recombination Genome Engineering)DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
HB101 endA::frt
Bacterial Strain#45473DepositorBacterial ResistanceStreptomycinAvailable SinceFeb. 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
UQ5871
Bacterial Strain#37170DepositorBacterial ResistanceKanamycinAvailable SinceAug. 23, 2012AvailabilityAcademic Institutions and Nonprofits only -
MM39
Bacterial Strain#42894DepositorBacterial ResistanceStreptomycinAvailable SinceMarch 28, 2013AvailabilityAcademic Institutions and Nonprofits only -
HL 6845
Bacterial Strain#69785PurposeMC4100 + agn43 coding sequence replaced by T7RNAP and camR + Δdam + ΔSgrSDepositorBacterial ResistanceChloramphenicolAvailable SinceApril 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
HL 6846
Bacterial Strain#69786PurposeMC4100 + agn43 coding sequence replaced by T7RNAP and camR + ΔOxyR + ΔSgrSDepositorBacterial ResistanceChloramphenicolAvailable SinceApril 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3E12
Bacterial Strain#48930Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3F12
Bacterial Strain#48931Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3G8
Bacterial Strain#48932Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3H7
Bacterial Strain#48933Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3H12
Bacterial Strain#48934Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M1D4
Bacterial Strain#48921Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M1E2
Bacterial Strain#48922Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M1E6
Bacterial Strain#48923Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M1G7
Bacterial Strain#48924Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3B7
Bacterial Strain#48925Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3B8
Bacterial Strain#48926Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3C3
Bacterial Strain#48927Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3C4
Bacterial Strain#48928Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
EcNGRfEfC-R6M3C8
Bacterial Strain#48929Purposediversity was introduced via homologous recombination to 40 sites to improve fatty acid production. Sequencing of the strains at these sites is ongoing.DepositorBacterial ResistanceSpectinomycinAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only