We narrowed to 694 results for: MOM
-
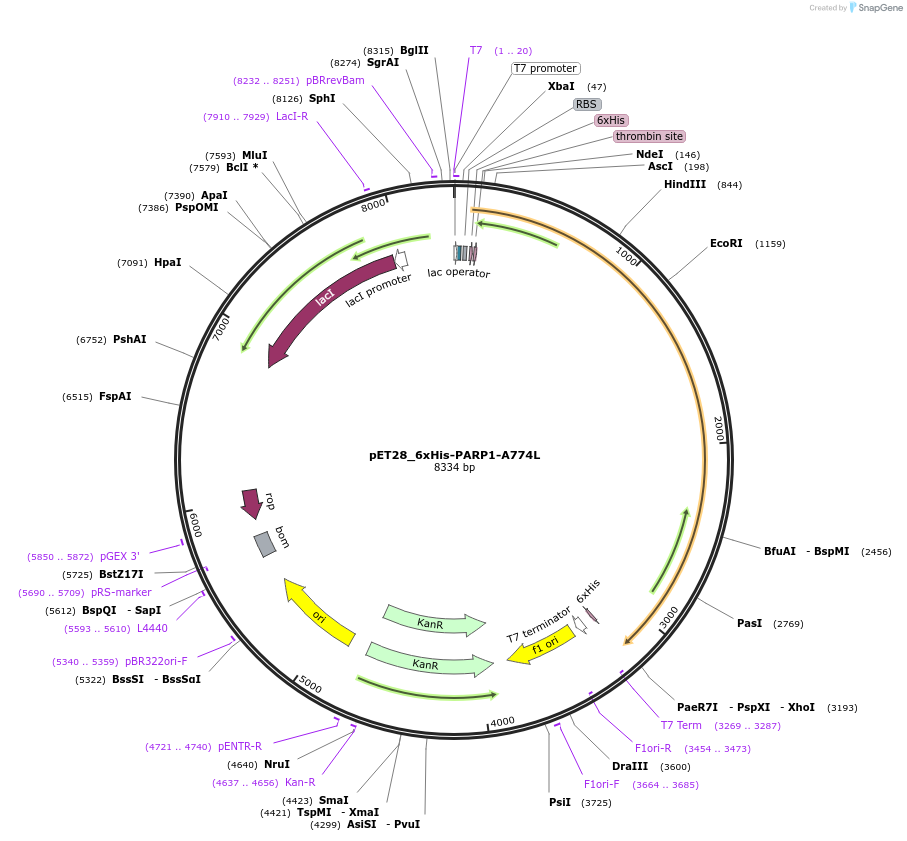
Plasmid#174796PurposeBacterial expression of a hyperactive PARP1 mutant (A774L destabilizes the autoinhibitory HD subdomain)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only
-
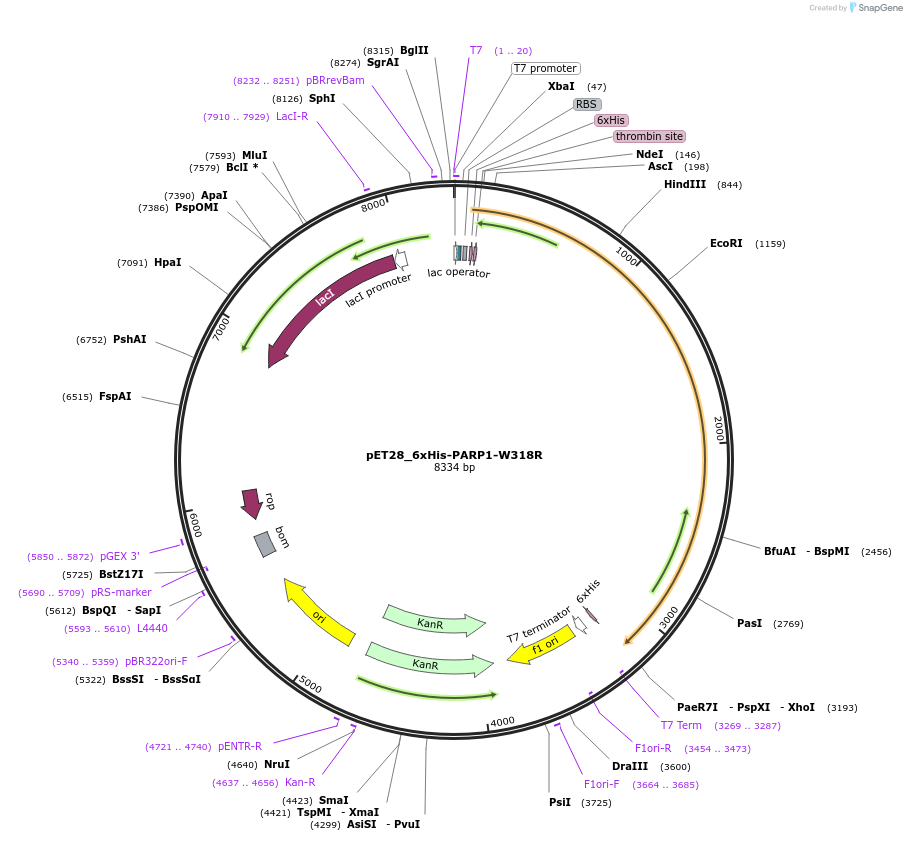
pET28_6xHis-PARP1-W318R
Plasmid#174793PurposeBacterial expression of an inactive PARP1 (W318R disrupts interdomain communication and HD subdomain unfolding)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
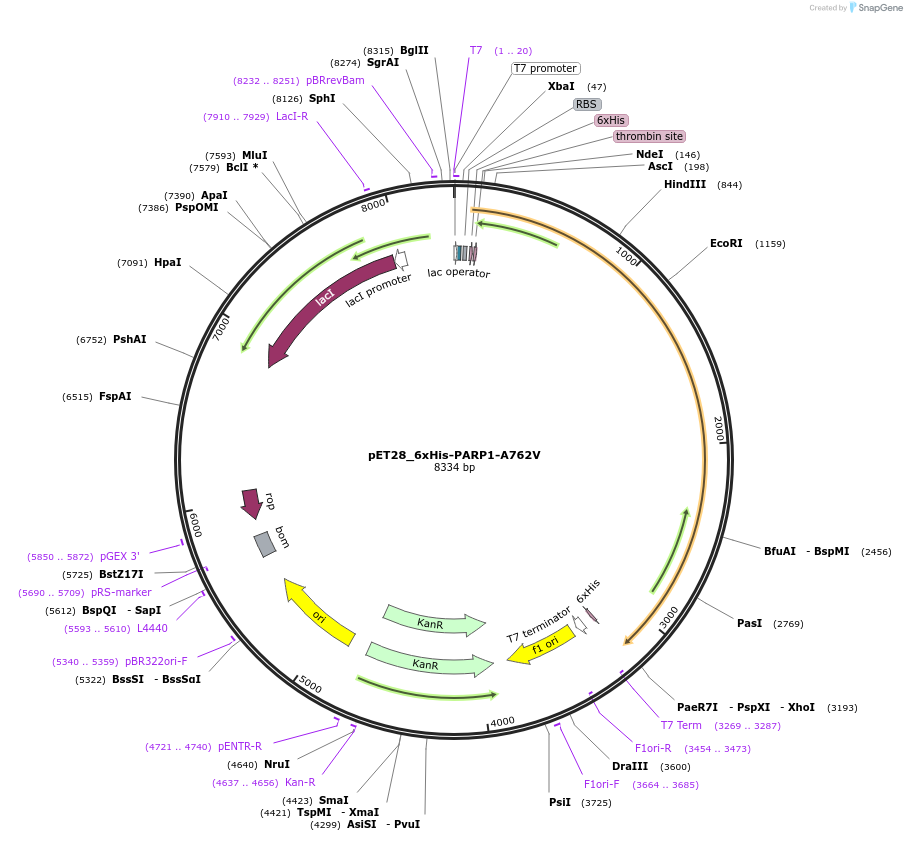
pET28_6xHis-PARP1-A762V
Plasmid#174795PurposeBacterial expression of wild type PARP1 mutant (A762V is reversion of the common V762A polymorphism)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
pcDNA5-F-Med26-K74A K75A
Plasmid#49253PurposeMammalian expression of flag-tagged human Med26-K74A K75ADepositorAvailable SinceDec. 5, 2013AvailabilityAcademic Institutions and Nonprofits only -
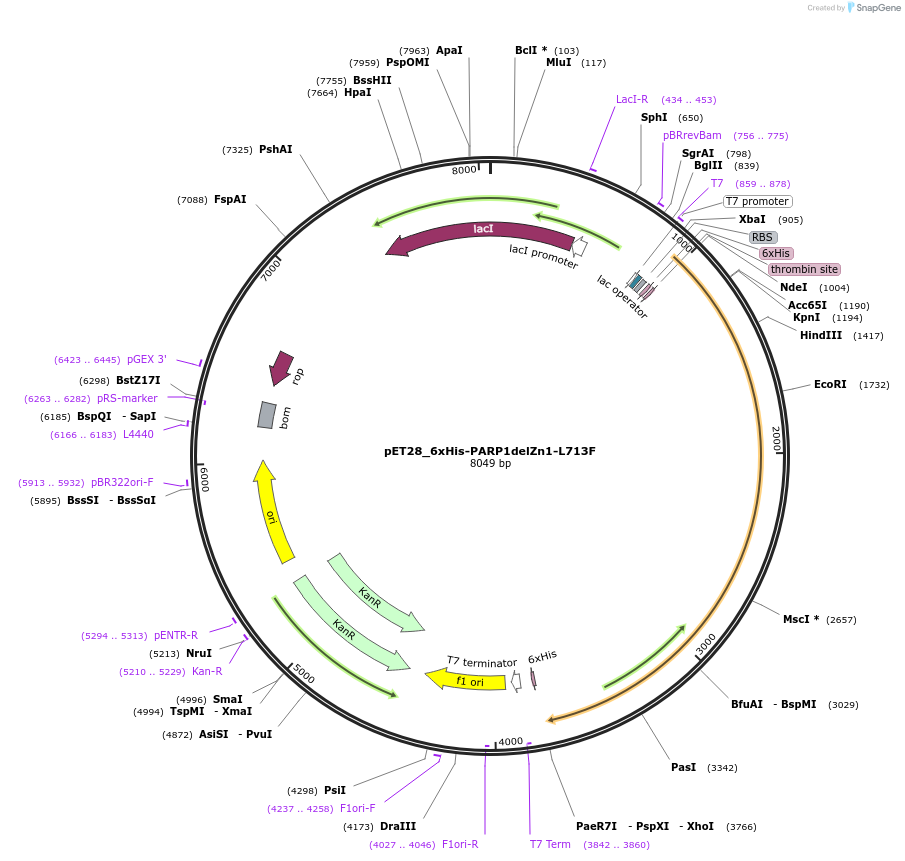
pET28_6xHis-PARP1delZn1-L713F
Plasmid#173937PurposeBacterial expression of PARP1 lacking Zn1 domain but containing L713F gain-of-function mutationDepositorInsertPARP1delZn1-L713F (PARP1 Human)
Tags6xHisExpressionBacterialMutationDeletion of residues 1-96; L713F, V762AAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
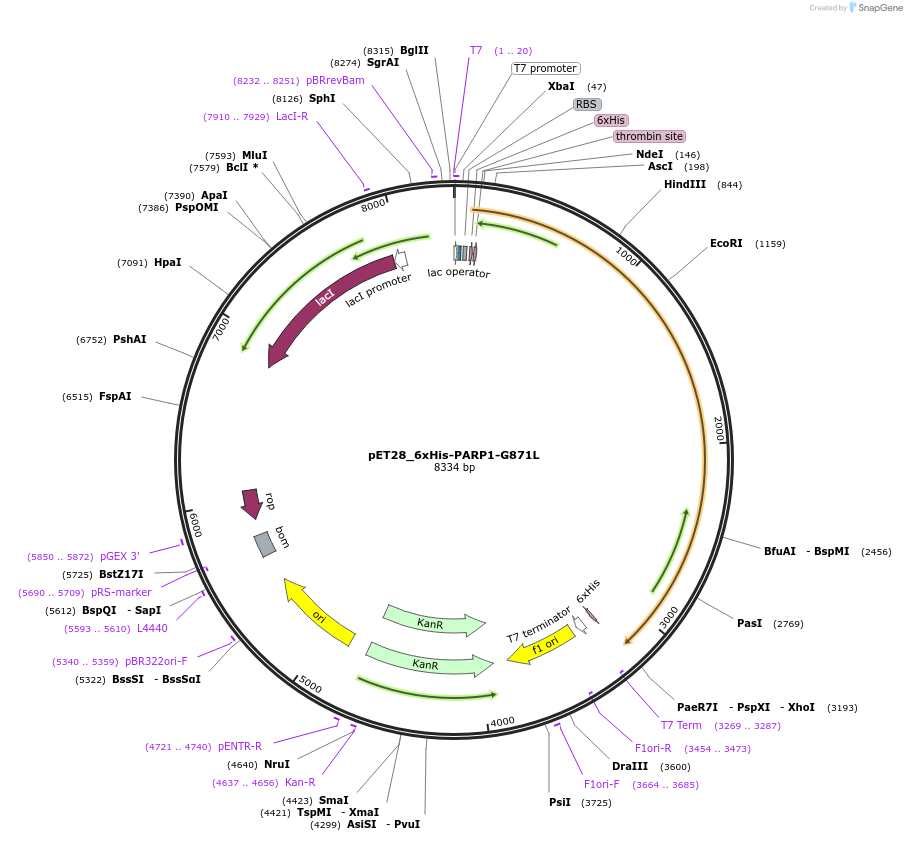
pET28_6xHis-PARP1-G871L
Plasmid#174798PurposeBacterial expression of a hyperactive PARP1 mutant (G871L destabilizes the autoinhibitory HD subdomain)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
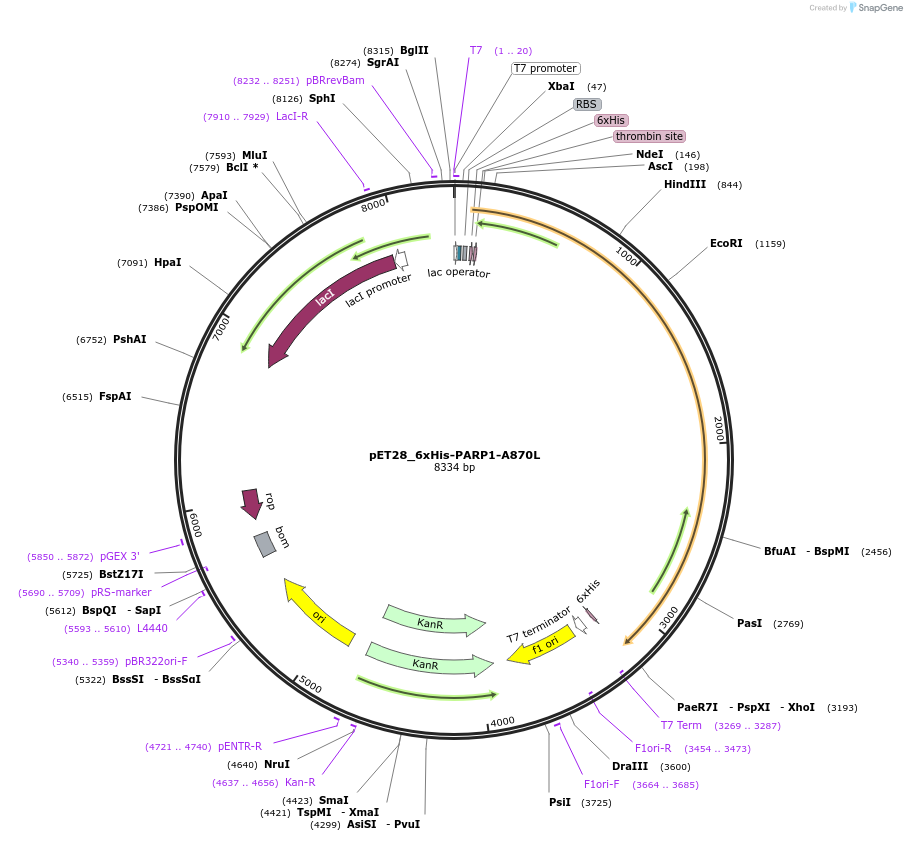
pET28_6xHis-PARP1-A870L
Plasmid#174797PurposeBacterial expression of a hyperactive PARP1 mutant (A870L destabilizes the autoinhibitory HD subdomain)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
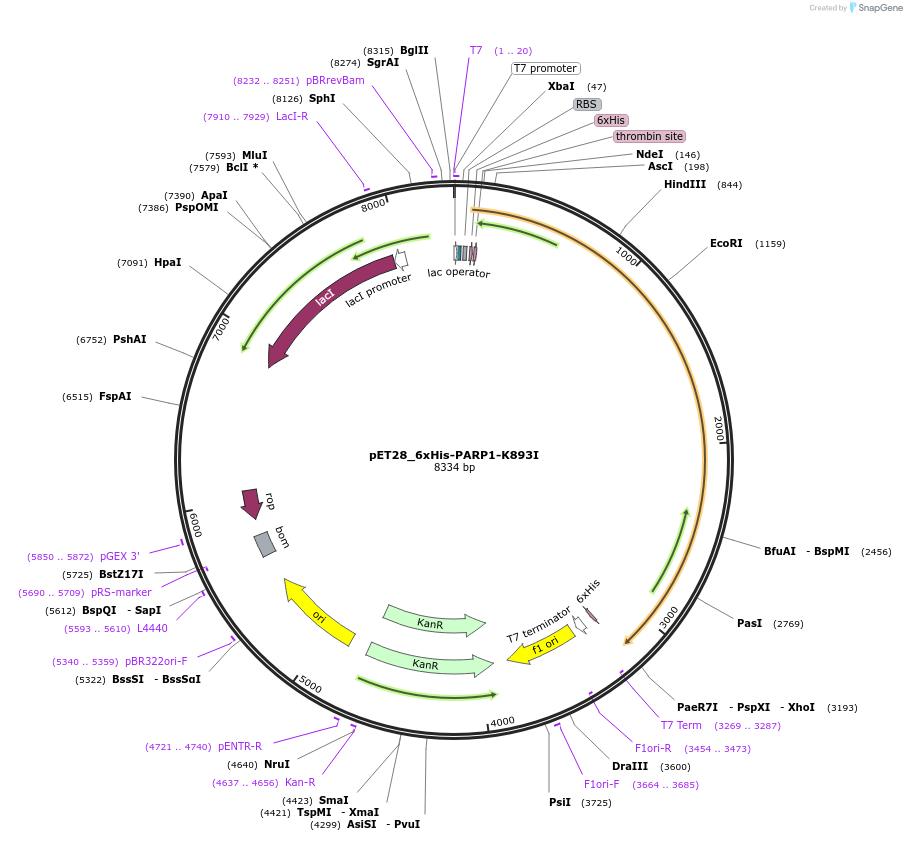
pET28_6xHis-PARP1-K893I
Plasmid#174799PurposeBacterial expression of an inactive mutant (K893I may disrupt NAD+ binding)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
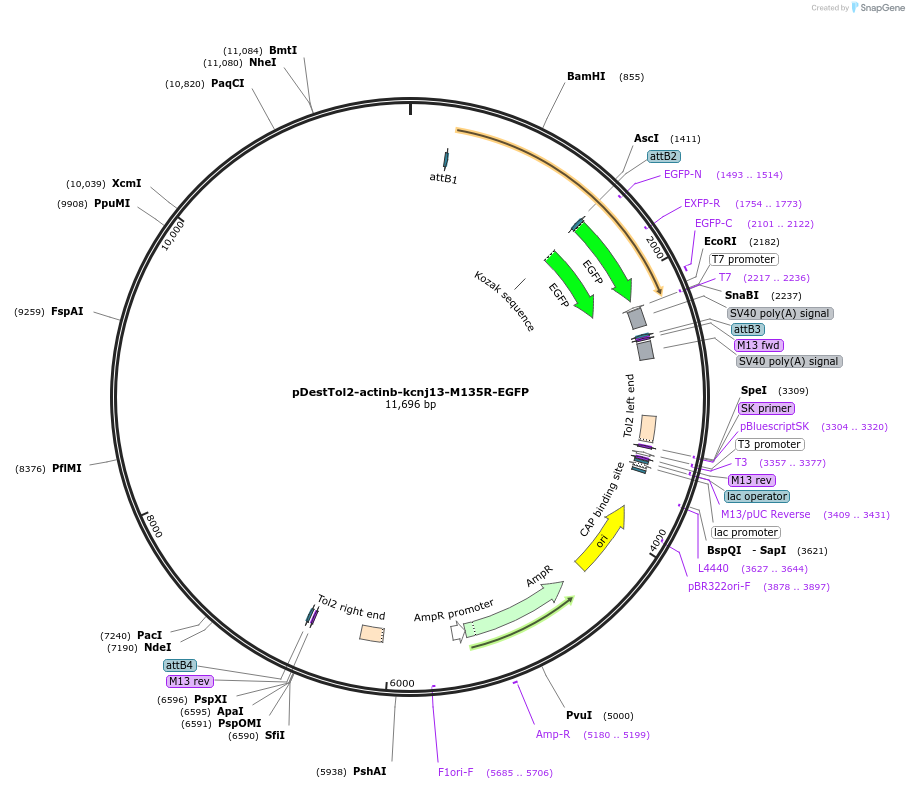
pDestTol2-actinb-kcnj13-M135R-EGFP
Plasmid#164944PurposeTol2 construct: actinb (actb2) promoter driven zebrafish gain-of-function kcnj13 mutant, M135RDepositorInsertkcnj13 (kcnj13 Zebrafish)
UseTol2 transposon destination vectorTagsEGFPMutationStop codon deleted; M135RPromoteractinb (actb2) promoterAvailable SinceJune 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
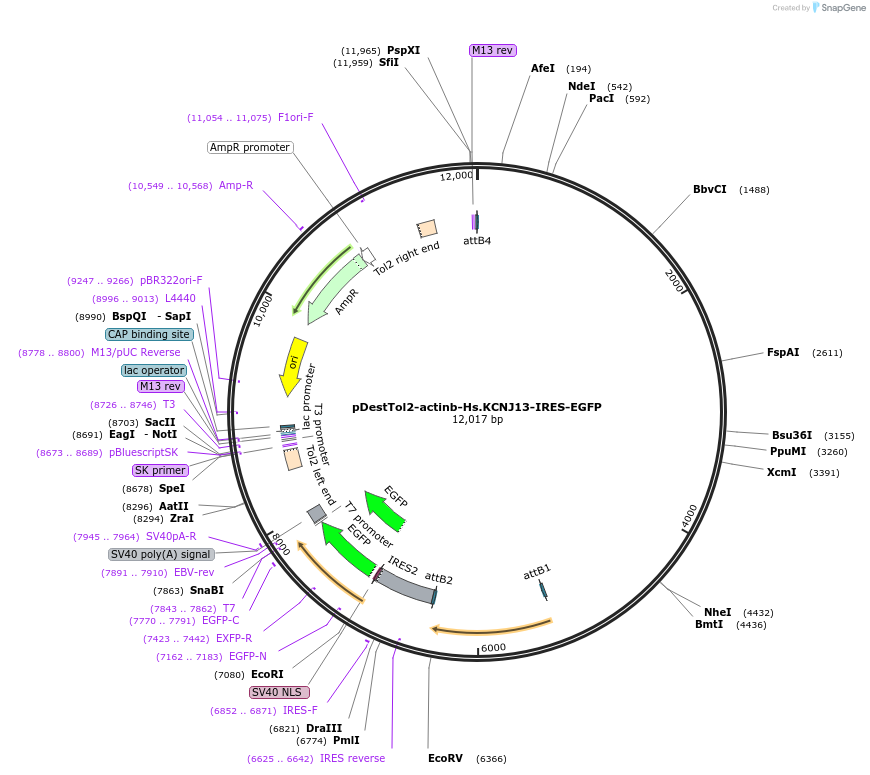
pDestTol2-actinb-Hs.KCNJ13-IRES-EGFP
Plasmid#164945PurposeTol2 construct: actinb (actb2) promoter driven human KCNJ13 and EGFP”. This will reflect the linkage of KCNJ13 and EGFPDepositorInsertKCNJ13 (KCNJ13 Human)
UseTol2 transposon destination vectorTagsIRES-EGFPMutationStop codon deletedPromoteractinb (actb2) promoterAvailable SinceMarch 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
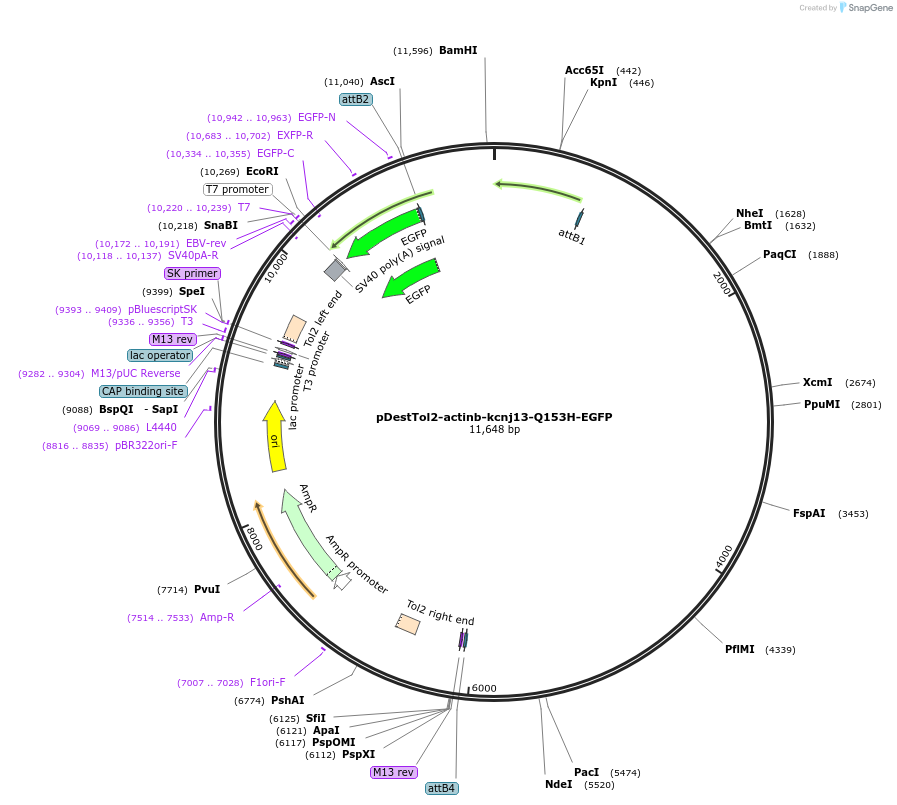
pDestTol2-actinb-kcnj13-Q153H-EGFP
Plasmid#164943PurposeTol2 construct: actinb (actb2) promoter driven zebrafish loss-of-functionkcnj13 mutant, Q153H and M154IDepositorInsertkcnj13 (kcnj13 Zebrafish)
UseTol2 transposon destination vectorTagsEGFPMutationStop codon deleted; Q153H; M154IPromoteractinb (actb2) promoterAvailable SinceMarch 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
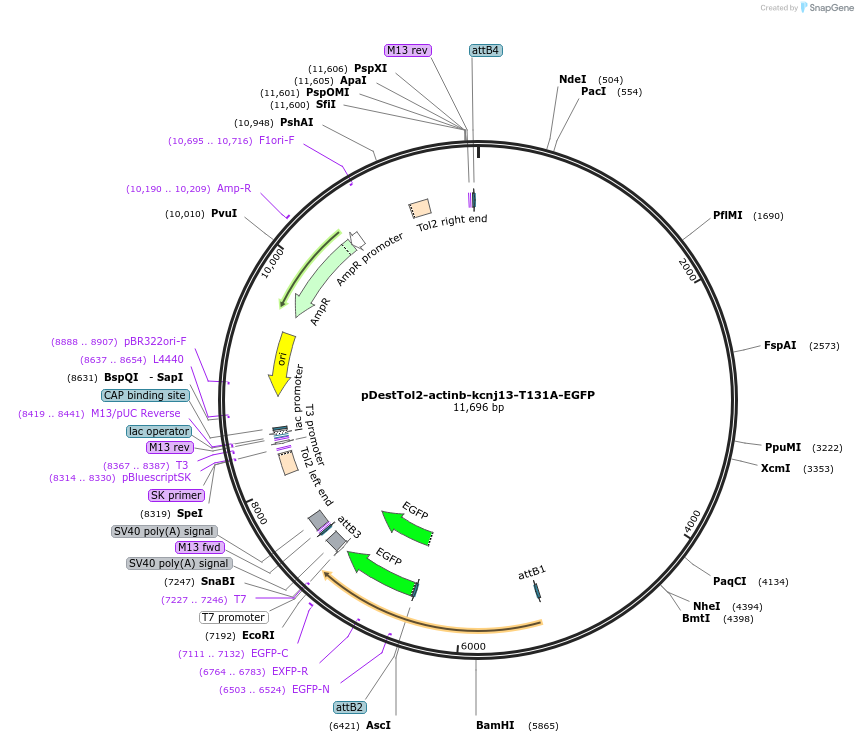
pDestTol2-actinb-kcnj13-T131A-EGFP
Plasmid#164942PurposeTol2 construct: actinb (actb2) promoter driven zebrafish loss-of-function kcnj13 mutant, T131A and G133RDepositorInsertkcnj13 (kcnj13 Zebrafish)
UseTol2 transposon destination vectorTagsEGFPMutationStop codon deleted; T131A; G133RPromoteractinb (actb2) promoterAvailable SinceMarch 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
pcDNA5-F-Med26-NTD R61A K62A
Plasmid#49256PurposeMammalian expression of flag-tagged human Med26-NTD R61A K62ADepositorAvailable SinceFeb. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
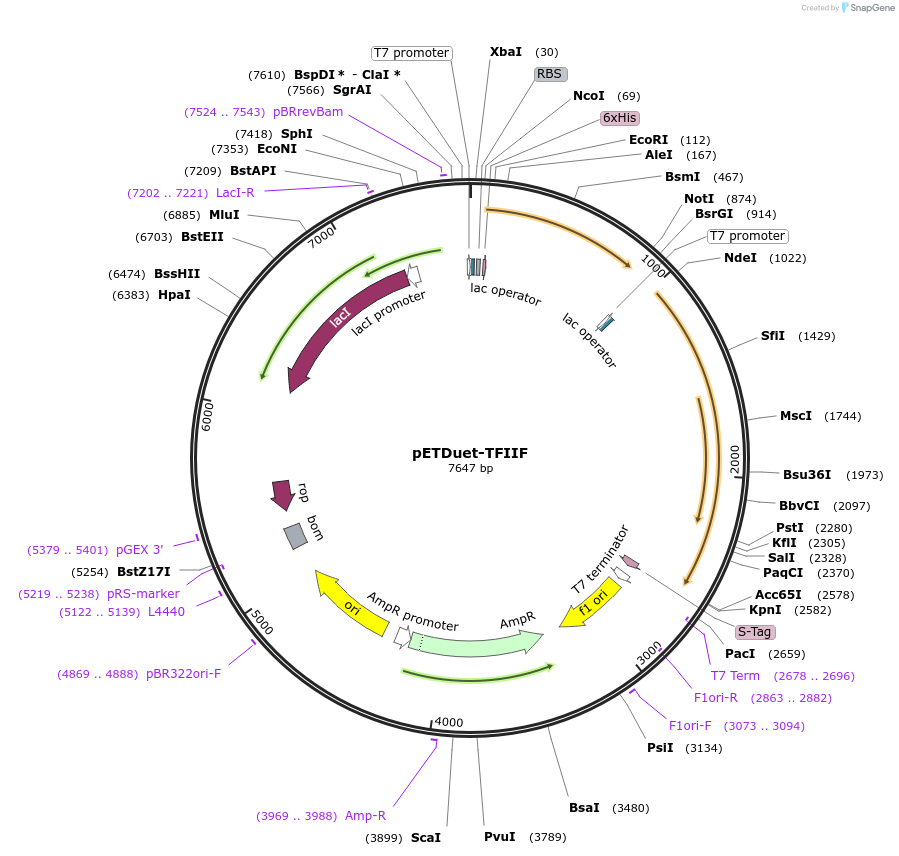
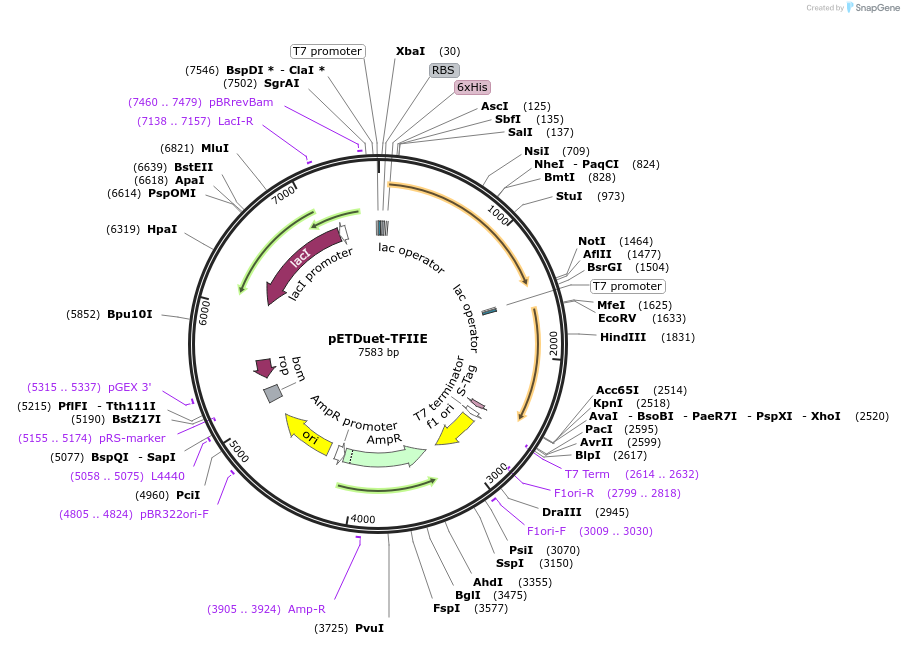
pETDuet-TFIIF
Plasmid#171082PurposeCo-expresses human TFIIF. The resulting plasmid was used to generate a expression construct encoding 2 subunits, with His-tag on RAP30. Originally from MN Gonzalez, was submitted with permissionDepositorTagsHisExpressionBacterialPromoterT7 promoterAvailable SinceJuly 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET28_6xHis-PARP1delWGR-L713F
Plasmid#173941PurposeBacterial expression of PARP1 lacking WGR domain but containing L713F gain-of-function mutationDepositorInsertPARP1delWGR-L713F (PARP1 Human)
Tags6xHisExpressionBacterialMutationDeletion of residues 521-660; L713F, V762AAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
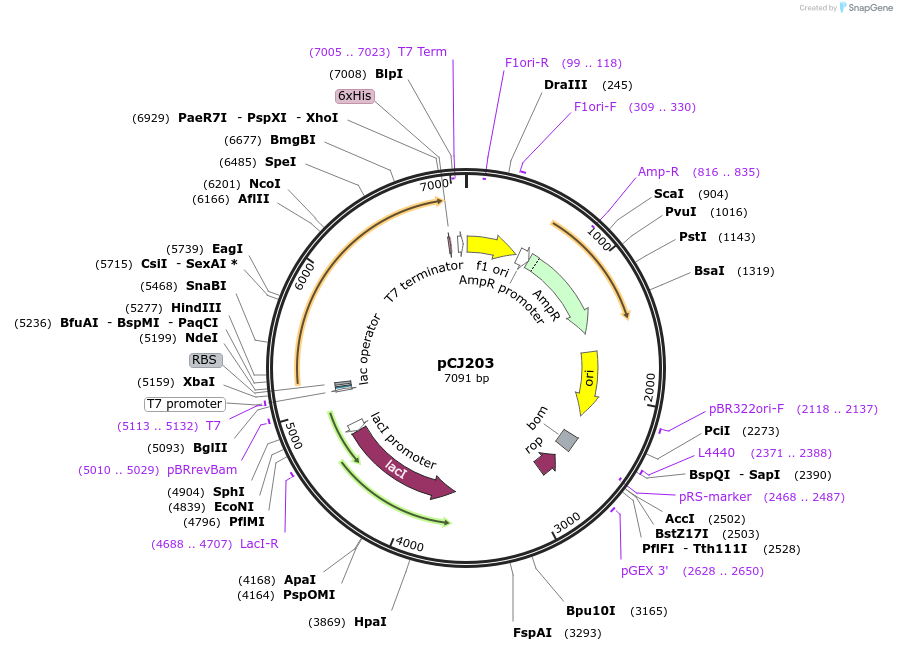
pCJ203
Plasmid#162678PurposepET-21b(+) based plasmid for expression of the putative MHET hydrolase from Comomonas thiooxydans (Genbank WP_080747404.1) with deletion of the predicted signal peptide (Phe2:Ala75)DepositorInsertPutative MHETase from Comomonas thiooxydans (Genbank WP_080747404.1) without 75 residue signal peptide
TagsHisExpressionBacterialMutationdeltaF2-A75; codon optimized for expression in E.…PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
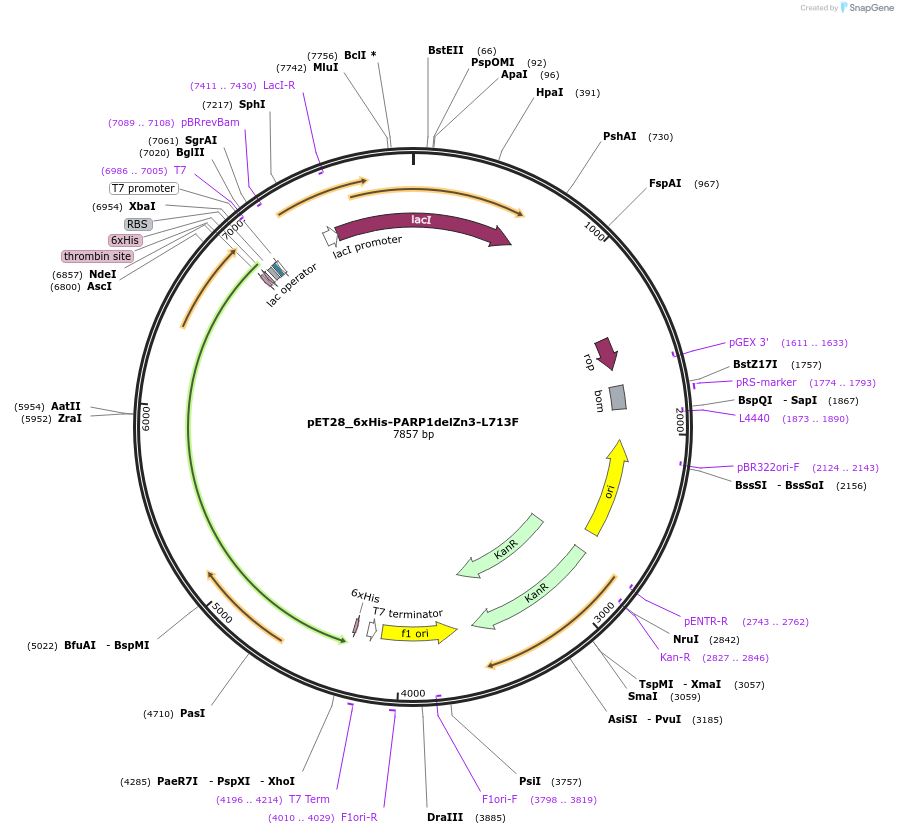
pET28_6xHis-PARP1delZn3-L713F
Plasmid#173934PurposeBacterial expression of PARP1 lacking Zn3 domain but containing L713F gain-of-function mutationDepositorInsertPARP1delZn3-L713F (PARP1 Human)
Tags6xHisExpressionBacterialMutationDeletion of residues 214-373; L713F, V762AAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
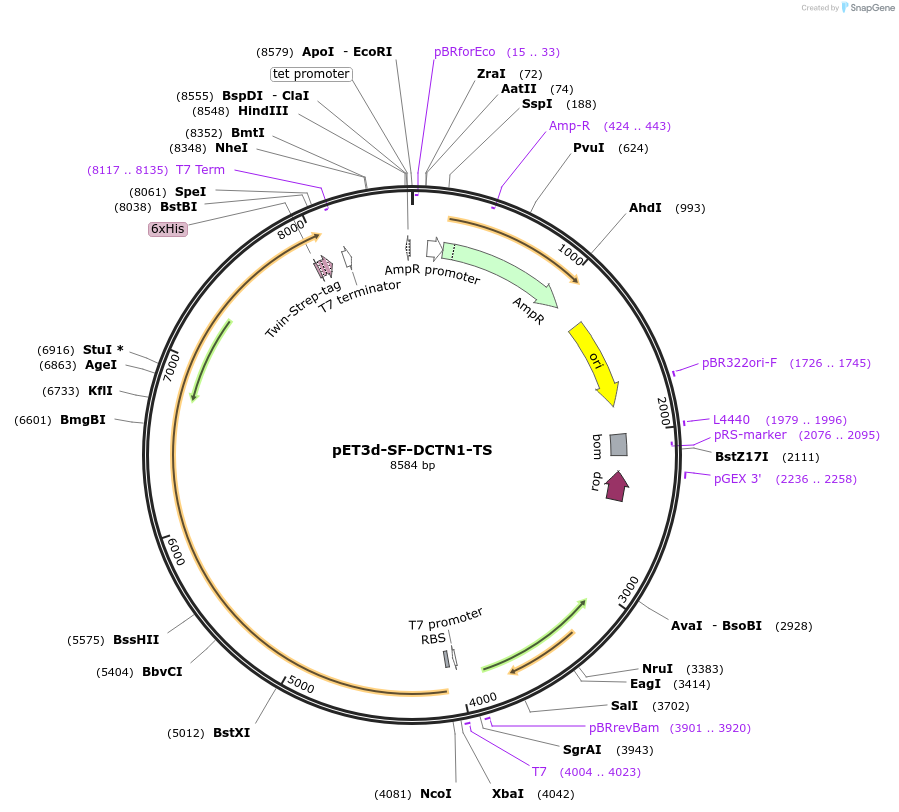
pET3d-SF-DCTN1-TS
Plasmid#169375PurposeE. coli expression plasmid for human DCTN1 (longest transcript variant p150glued/isoform 1), with an N-terminal splitFlAsH tag and a C-terminal TwinStrep tag.DepositorInsertdynactin subunit 1 isoform 1 (DCTN1 Human)
Tags6xHis Tag, SplitFlAsH Tag, and TwinStrep TagExpressionBacterialMutationPoint mutations: A139T, I304T, Q322R, T516A, E577…Available SinceMay 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
pcDNA5-F-Med26-NTD- K74A K75A
Plasmid#49252PurposeMammalian expression of flag-tagged human Med26-NTD- K74A K75ADepositorAvailable SinceDec. 30, 2013AvailabilityAcademic Institutions and Nonprofits only -
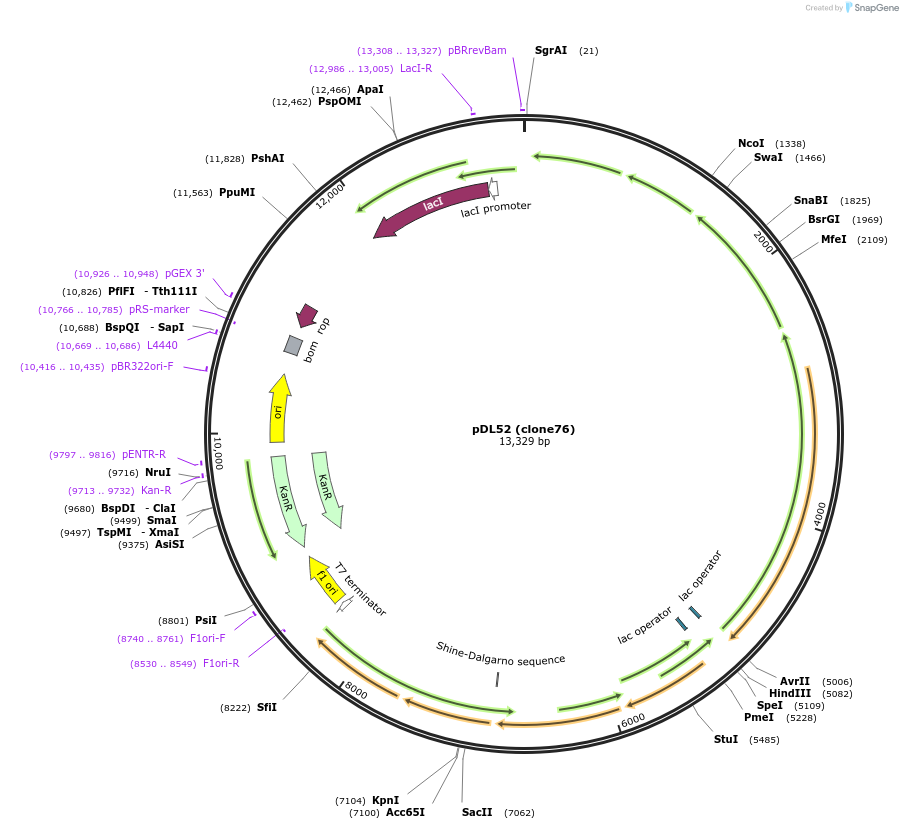
pDL52 (clone76)
Plasmid#188579PurposeHigh-yield production of coenzyme F420 in E. coliDepositorInsertsribA
ribD
yigB
fbiC
cofC
cofD
cofI
cofE
ExpressionBacterialMutationCodon optimization, synthetic ribosome binding si…PromoterT7 variantsAvailable SinceSept. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
pETDuet-TFIIE
Plasmid#171083PurposeCo-expresses human TFIIE. The resulting plasmid was used to generate a single expression construct encoding 2 subunits, with His-tag on TF2E1.Originally from MN Gonzalez, was submitted with permissionDepositorTagsHisExpressionBacterialPromoterT7 promoterAvailable SinceJuly 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
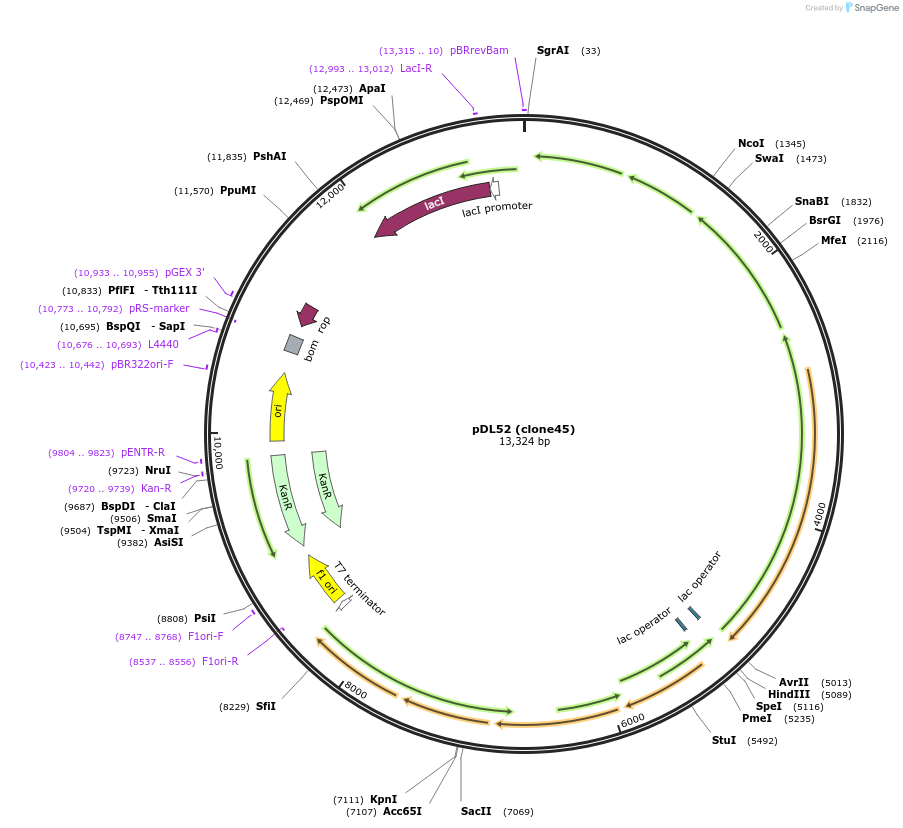
pDL52 (clone45)
Plasmid#188578PurposeHigh-yield production of coenzyme F420 in E. coliDepositorInsertsribA
ribD
yigB
fbiC
cofC
cofD
cofI
cofE
ExpressionBacterialMutationCodon optimization, synthetic ribosome binding si…PromoterT7 variantsAvailable SinceSept. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
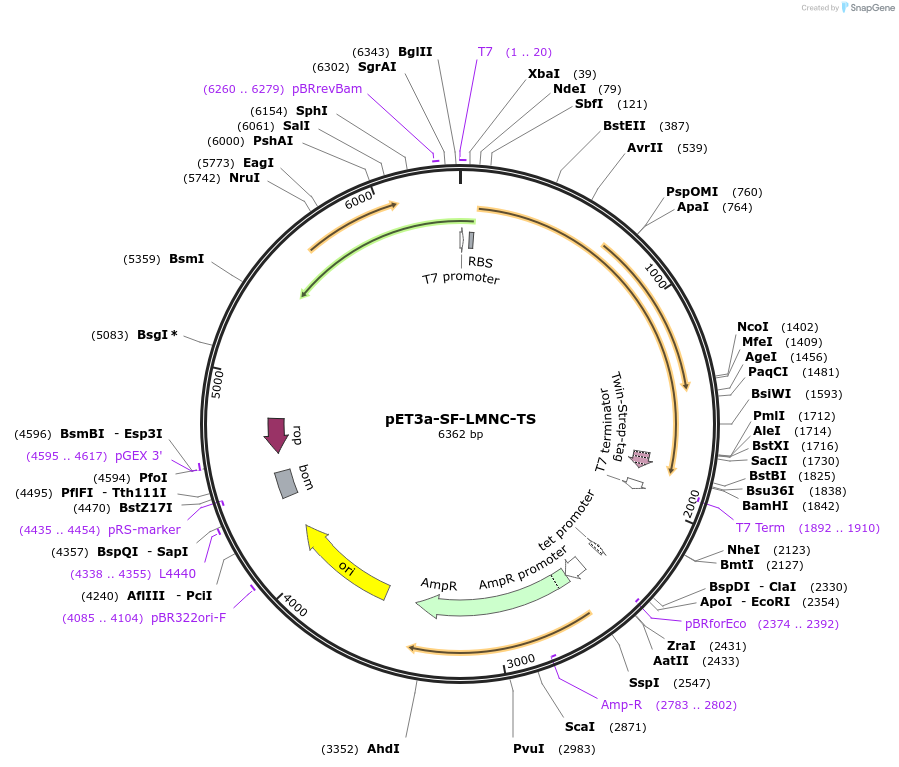
pET3a-SF-LMNC-TS
Plasmid#169374PurposeE. coli expression plasmid for human LMNA residues 31-542, corresponding to the shorter lamin C isoform lacking the unstructured head, with an N-terminal splitFlAsH tag and a C-terminal TwinStrep tag.DepositorInsertLMNA residues 31-542 (LMNA Human)
TagsSplit-FlAsH Tag and TwinStrep TagExpressionBacterialMutationdeleted amino acid 1-30 (unstructured head domain…PromoterNative pET3a promotorAvailable SinceMay 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLENTI-PGK-HYGRO-GATA6mut-HA
Plasmid#65476PurposeLentiviral expression of GATA6 Znc-finger mutantDepositorInsertGATA6mut (GATA6 Human)
UseLentiviralTagsHAExpressionMammalianMutationlacking the eight most highly conserved bases of …PromoterPGKAvailable SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
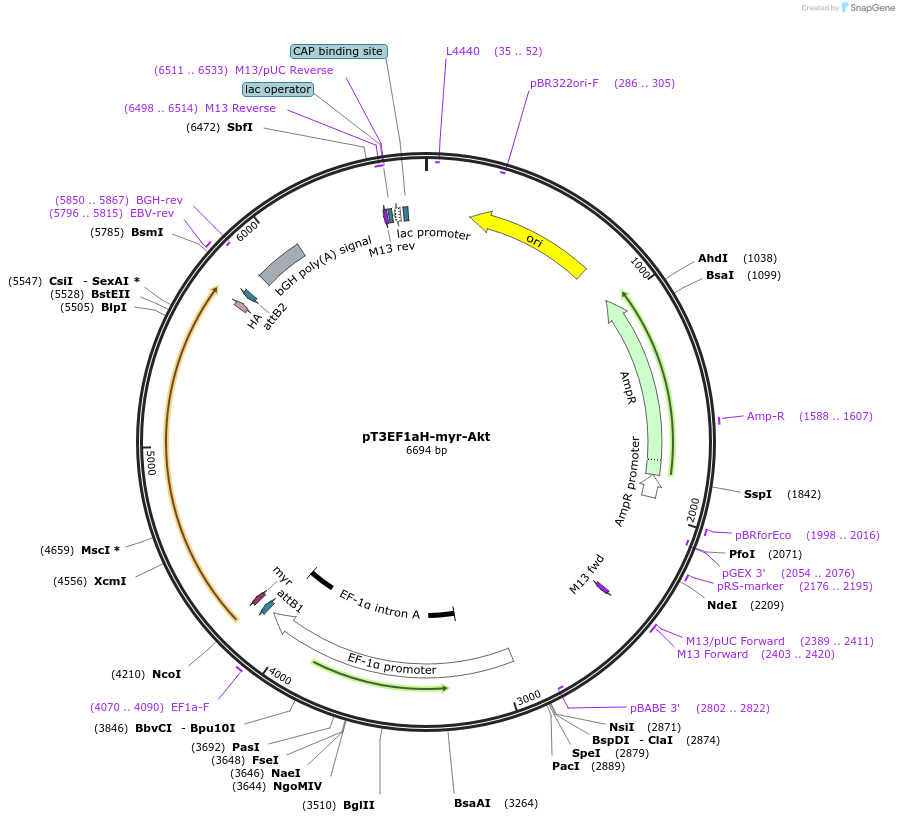
pT3EF1aH-myr-Akt
Plasmid#179909PurposeThis plasmid is in the pT3-EF1a vector without loxP sites flanking the inverted repeats of SB (sleeping beauty) sequence. Therefore this plasmid can be used with Cre for in vivo studies.DepositorAvailable SinceApril 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
pQ-Elo-HB-C-A(1-772)
Plasmid#171085PurposeCo-expresses of WT-ELOA containing Elongin complex in bacteria. The resulting plasmid was used to generate a single expression construct encoding all 3Elongin subunits, with a 6-His tag on ELOBDepositorTagsHisExpressionBacterialMutationDeleted N-term 26 amino acids for expression purp…PromoterT5 promoterAvailable SinceJuly 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
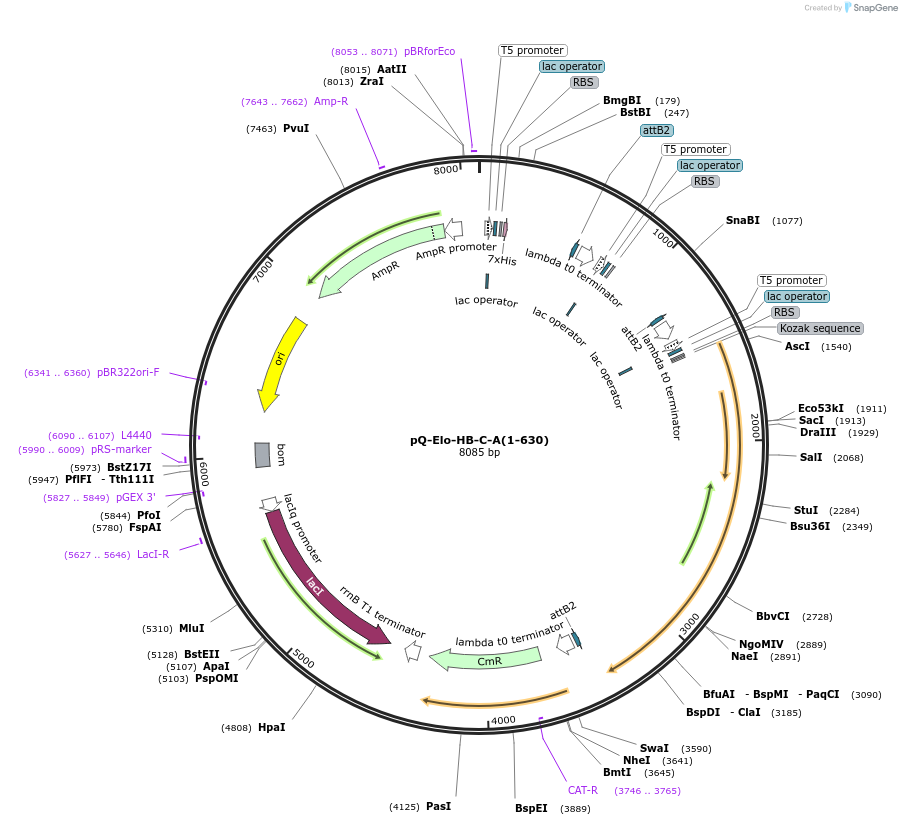
pQ-Elo-HB-C-A(1-630)
Plasmid#171086PurposeCo-expresses of ΔC-ELOA containing Elongin complex in bacteria. The resulting plasmid was used to generate a single expression construct encoding all 3Elongin subunits, with a 6-His tag on ELOBDepositorTagsHisExpressionBacterialMutationDeleted N-term 26aa and C-term 142aa for mutant e…PromoterT5 promoterAvailable SinceJuly 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
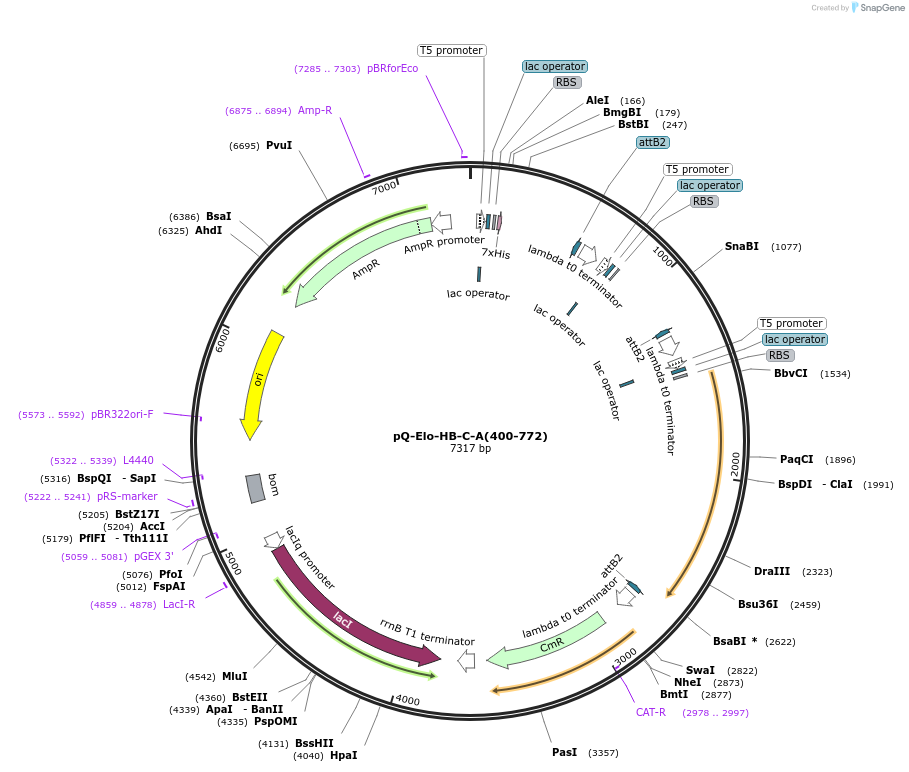
pQ-Elo-HB-C-A(400-772)
Plasmid#171087PurposeCo-expresses of ΔN-ELOA containing Elongin complex in bacteria. The resulting plasmid was used to generate a single expression construct encoding all 3Elongin subunits, with a 6-His tag on ELOBDepositorTagsHisExpressionBacterialMutationDeleted N-term 26+399 amino acids for mutant expr…PromoterT5 promoter and T7 promoterAvailable SinceJuly 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
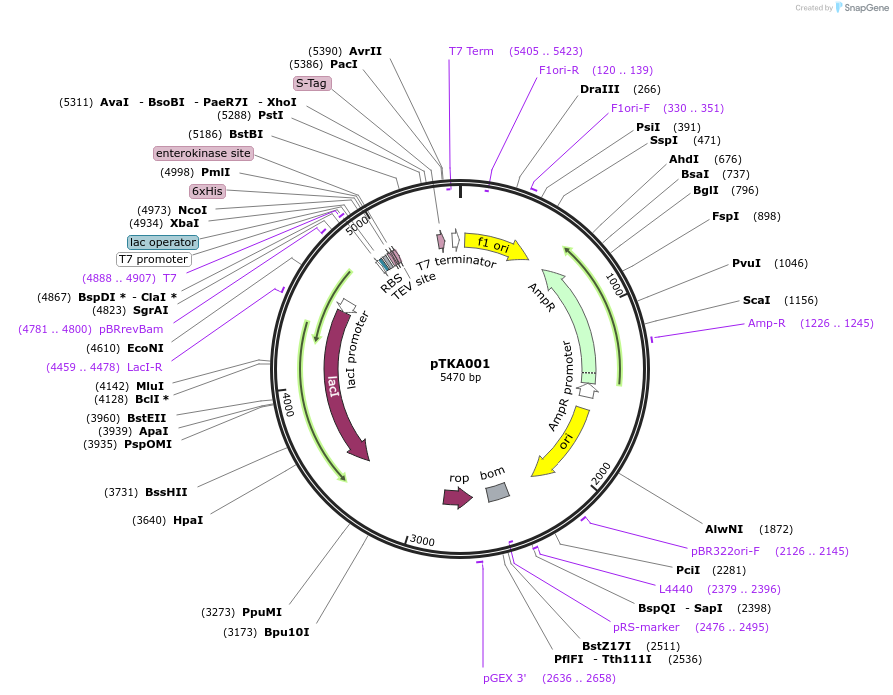
pTKA001
Plasmid#164577PurposeExpresses SARS-CoV-2 nsp7 from a pET46 backbone for IPTG-inducible bacterial expressionDepositorAvailable SinceJan. 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
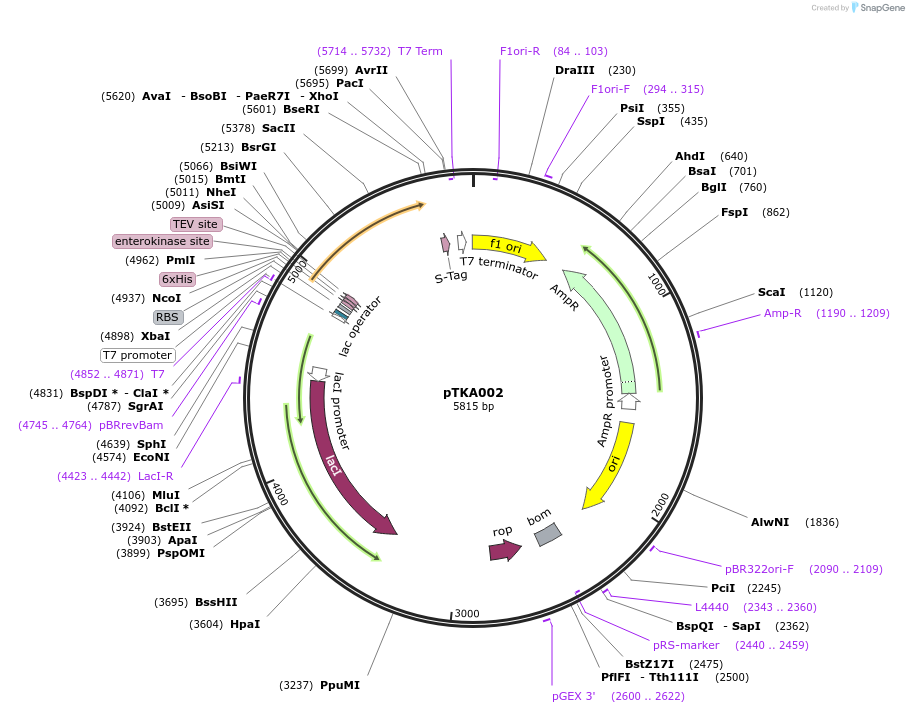
pTKA002
Plasmid#164578PurposeExpresses SARS-CoV-2 nsp8 from a pET46 backbone for IPTG-inducible bacterial expressionDepositorInsertnsp8
Tags6xHis, Ek, TEVExpressionBacterialPromoterT7 PromoterAvailable SinceFeb. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
-
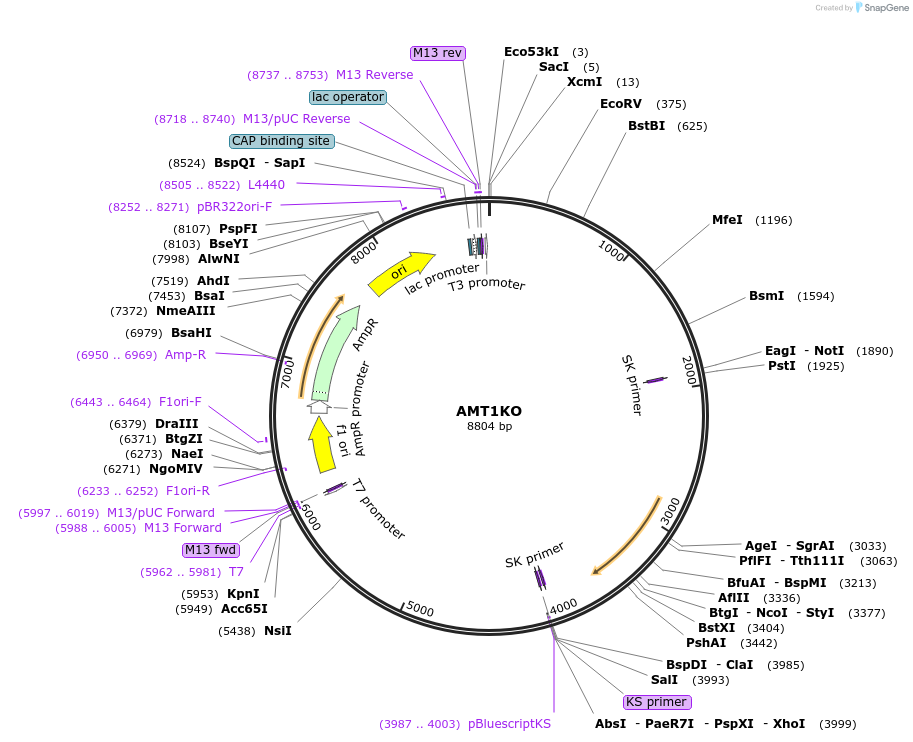
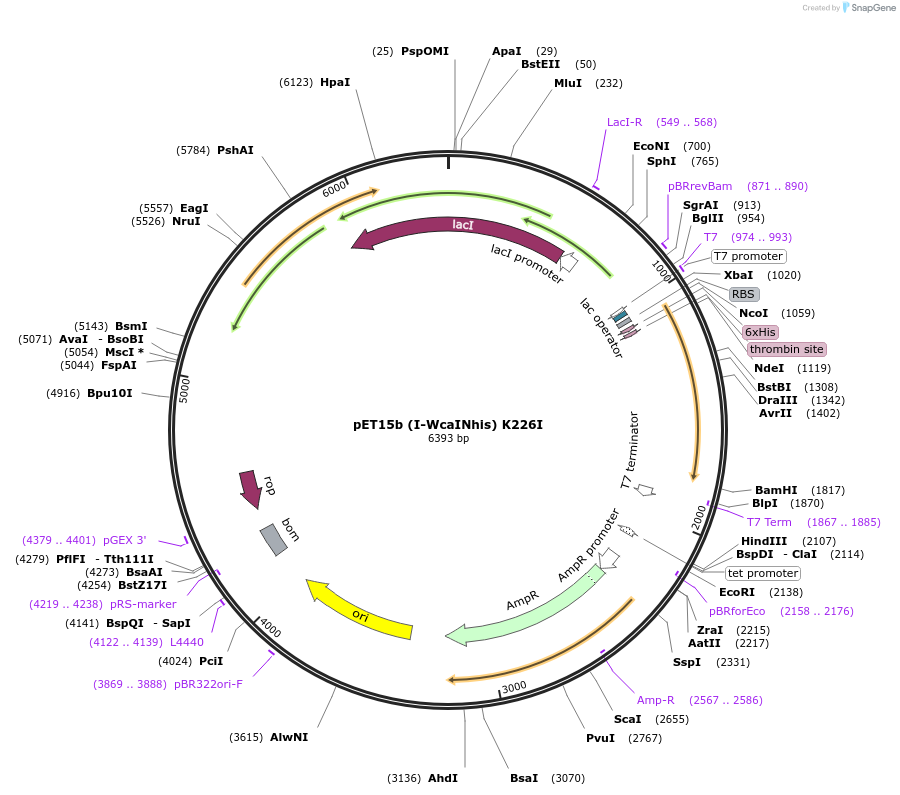
pET15b (I-WcaINhis) K226I
Plasmid#195620PurposeExpresses I-WcaI homing endonuclease from Wickerhamomyces canadensis containing the K226I mutationDepositorInsertI-WcaI
ExpressionBacterialMutationA1930T, A1931TPromoterT7Available SinceFeb. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
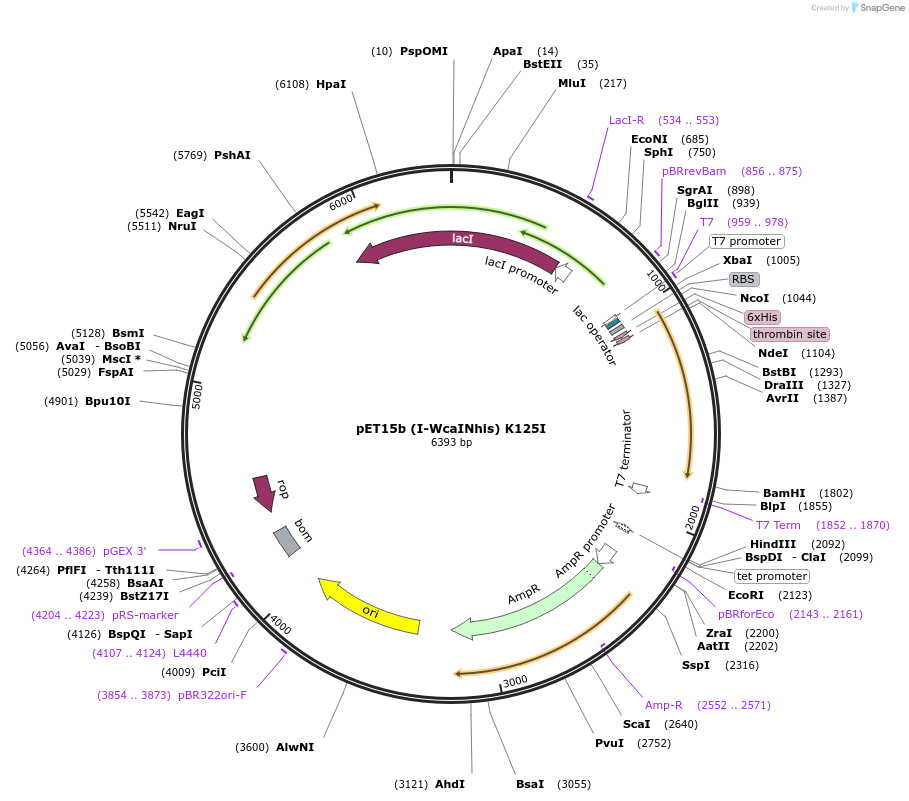
pET15b (I-WcaINhis) K125I
Plasmid#195619PurposeExpresses I-WcaI homing endonuclease from Wickerhamomyces canadensis containing the K125I mutationDepositorInsertI-WcaI
ExpressionBacterialMutationA2030T, A2031TPromoterT7Available SinceFeb. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
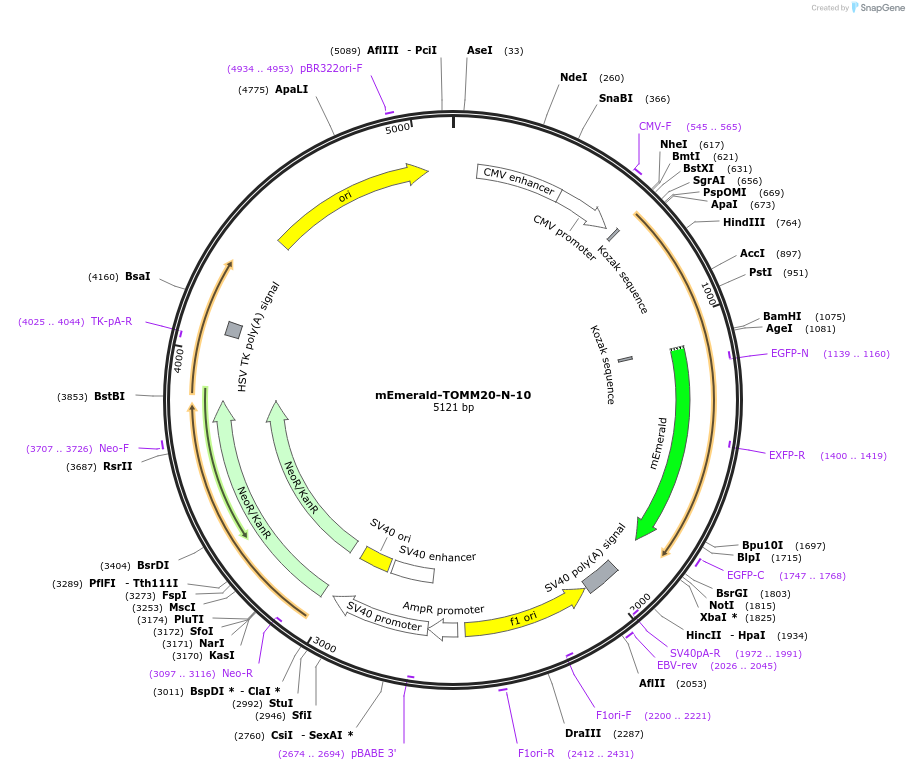
mEmerald-TOMM20-N-10
Plasmid#54282PurposeLocalization: Mitochondrial Membrane, Excitation: 487, Emission: 509DepositorAvailable SinceJuly 10, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
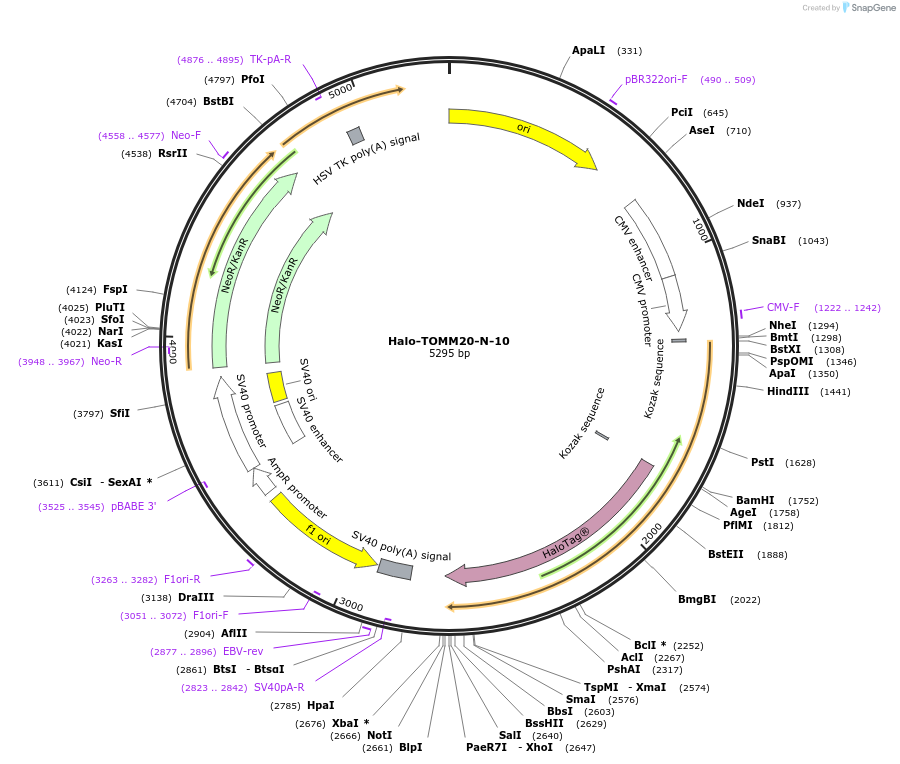
Halo-TOMM20-N-10
Plasmid#123284PurposeLocalization: Outer Mitochondrial MembraneDepositorAvailable SinceApril 24, 2019AvailabilityAcademic Institutions and Nonprofits only -
mEmerald-TOMM20-C-10
Plasmid#54281PurposeLocalization: Nucleus, Excitation: 487, Emission: 509DepositorAvailable SinceJuly 10, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
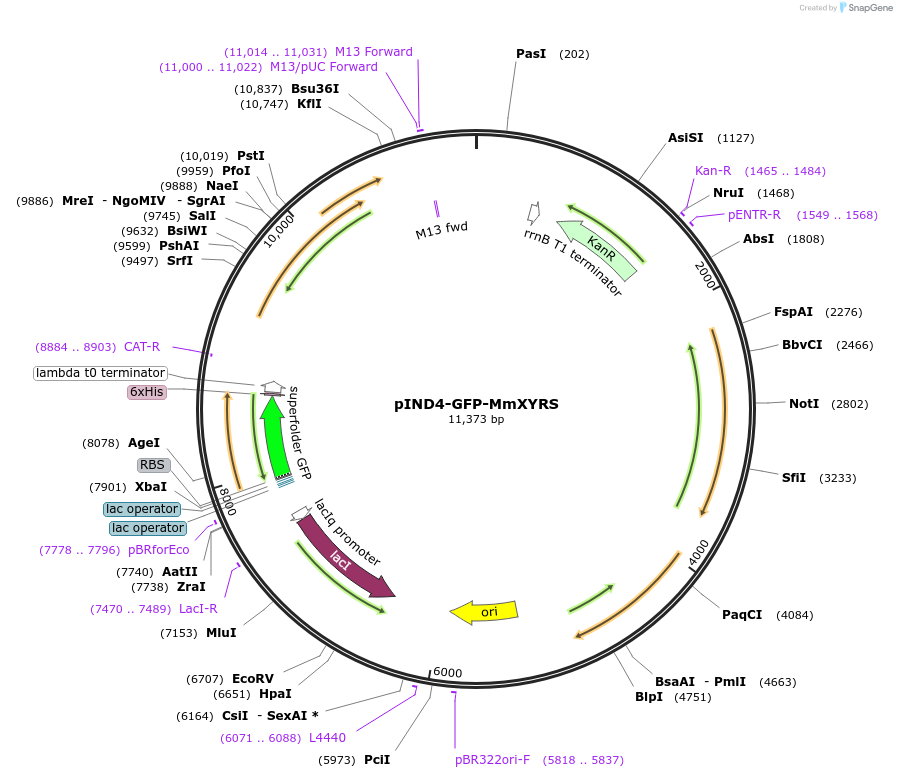
pIND4-GFP-MmXYRS
Plasmid#222745PurposeExpresses sfGFP and aaRS-tRNA pair for incorporating haloalkane crosslinkerDepositorInsertssfGFP
MmXYRS
Mm Pyl tRNA
Tags6xHisMutationContains the following substitutions: V346A, W348…PromoterrrnBAvailable SinceSept. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
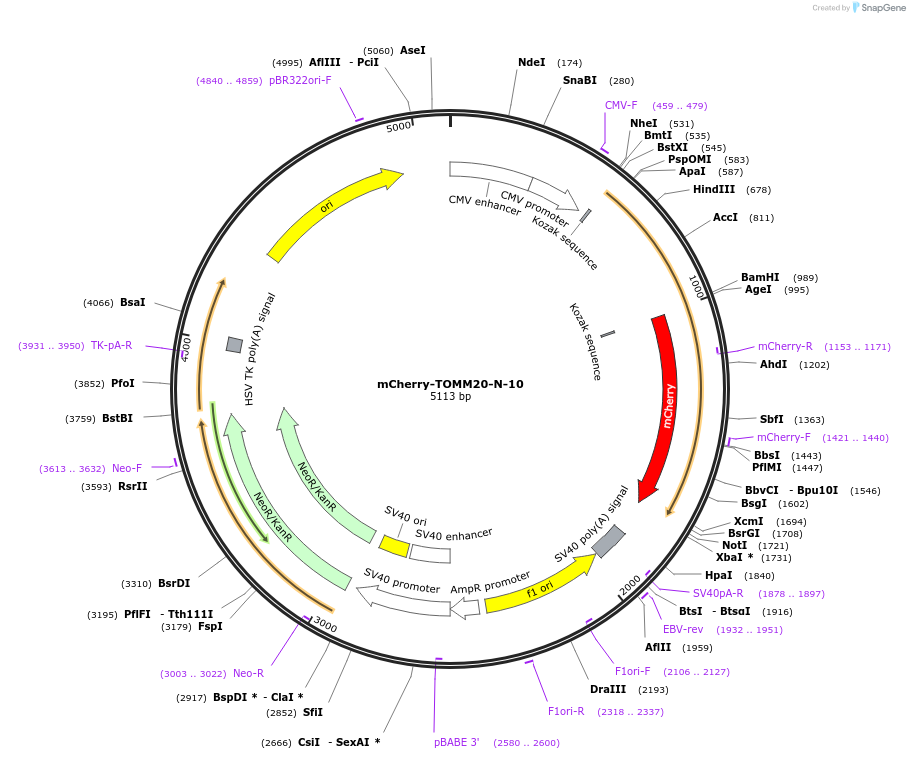
mCherry-TOMM20-N-10
Plasmid#55146PurposeLocalization: Outer Mitochondrial Membrane, Excitation: 587, Emission: 610DepositorAvailable SinceSept. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
tdTomato-TOMM20-N-10
Plasmid#58137PurposeLocalization: Outer Mitochondrial Membrane, Excitation: 554, Emission: 581DepositorAvailable SinceDec. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
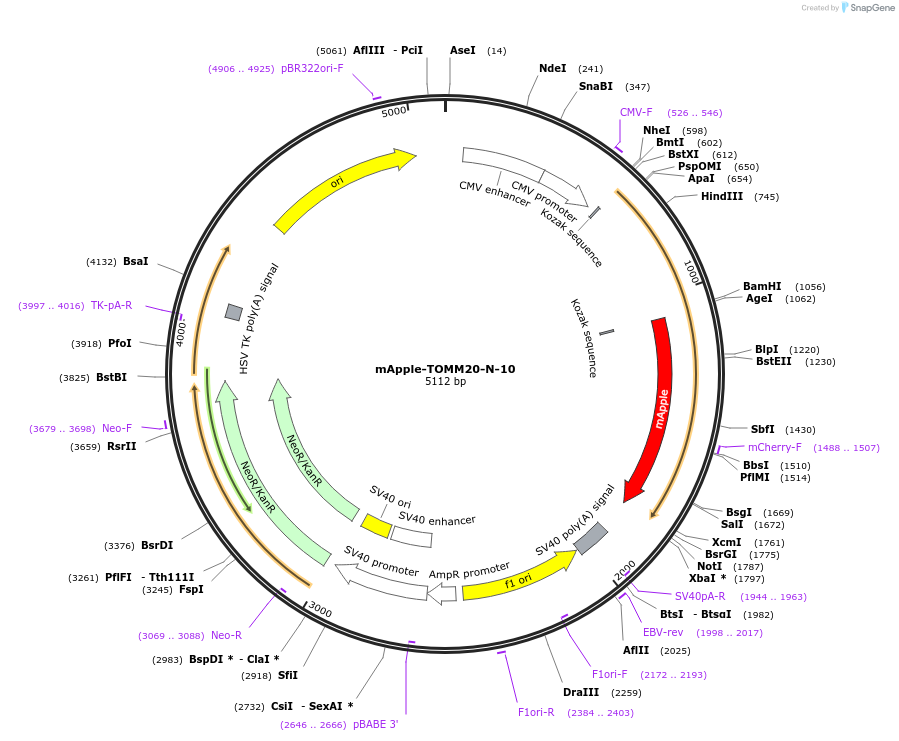
mApple-TOMM20-N-10
Plasmid#54955PurposeLocalization: Outer Mitochondrial Membrane, Excitation: 568, Emission: 592DepositorAvailable SinceAug. 7, 2014AvailabilityAcademic Institutions and Nonprofits only