We narrowed to 11,210 results for: nar
-
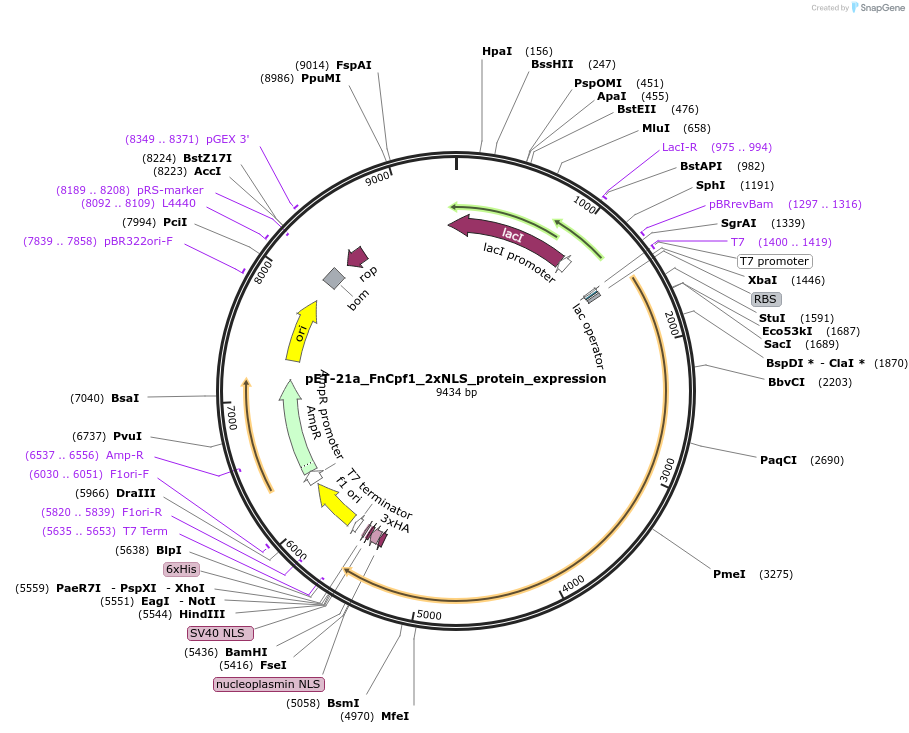
Plasmid#124548PurposeProtein expression plasmid for 2xNLS FnCpf1 in pET-21a backboneDepositorInsertFnCpf1_2xNLS
UseCRISPRExpressionBacterialAvailable SinceApril 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
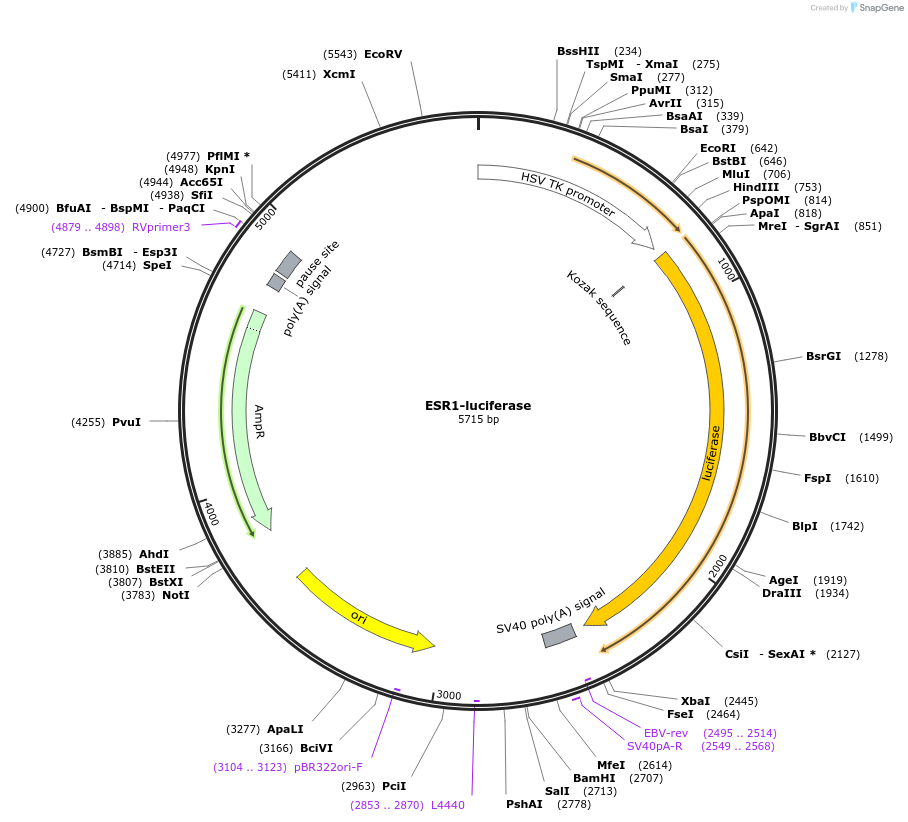
ESR1-luciferase
Plasmid#113351Purposeevaluate FOXA1 DIV motif enhancer activity near 50 kb of ESR1 geneDepositorInsertESR1-enhancer (ESR1 Human)
UseLuciferaseAvailable SinceSept. 12, 2018AvailabilityAcademic Institutions and Nonprofits only -
GFP-DroshaS300/302A
Plasmid#62530PurposeSerines 300 and 302 of Drosha are mutated to alaninesDepositorInserthuman Drosha with serines 300 and 302 changed alanines (DROSHA Human)
TagsGFPExpressionMammalianMutationchanged serines 300 and 302 to alaninesAvailable SinceMarch 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
MSCV-dGFP-JARID1C
Plasmid#74776Purposeretroviral expression of JARID1CDepositorAvailable SinceMay 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
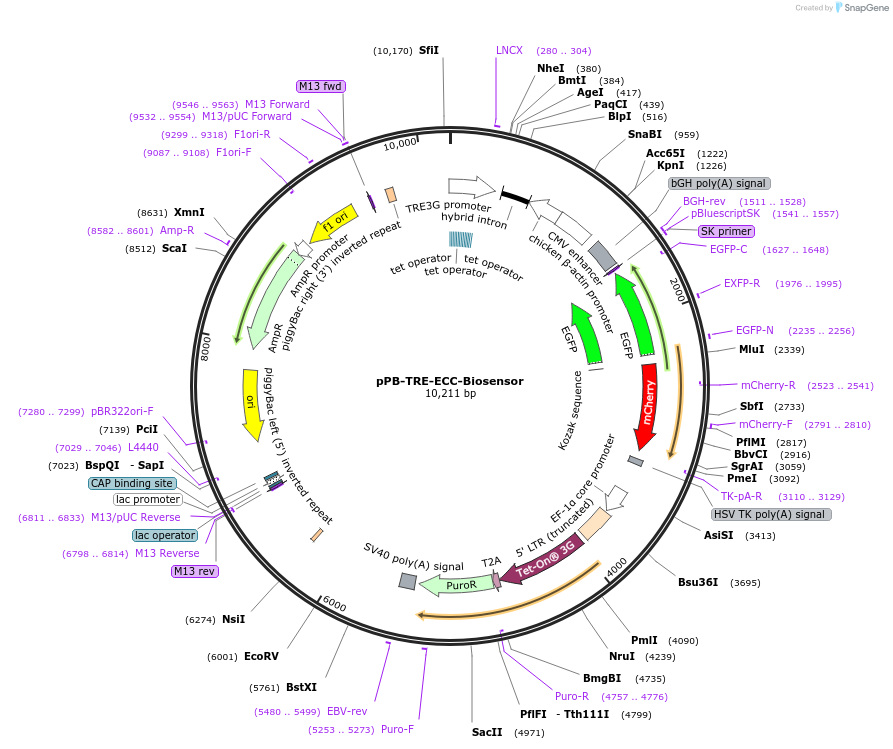
pPB-TRE-ECC-Biosensor
Plasmid#140833PurposeEncoding the TRE promoter based eccDNA biosensorDepositorInsertTRE-ECC Biosensor
UsePiggybac (pb) transposonExpressionMammalianPromoterinverseAvailable SinceJune 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
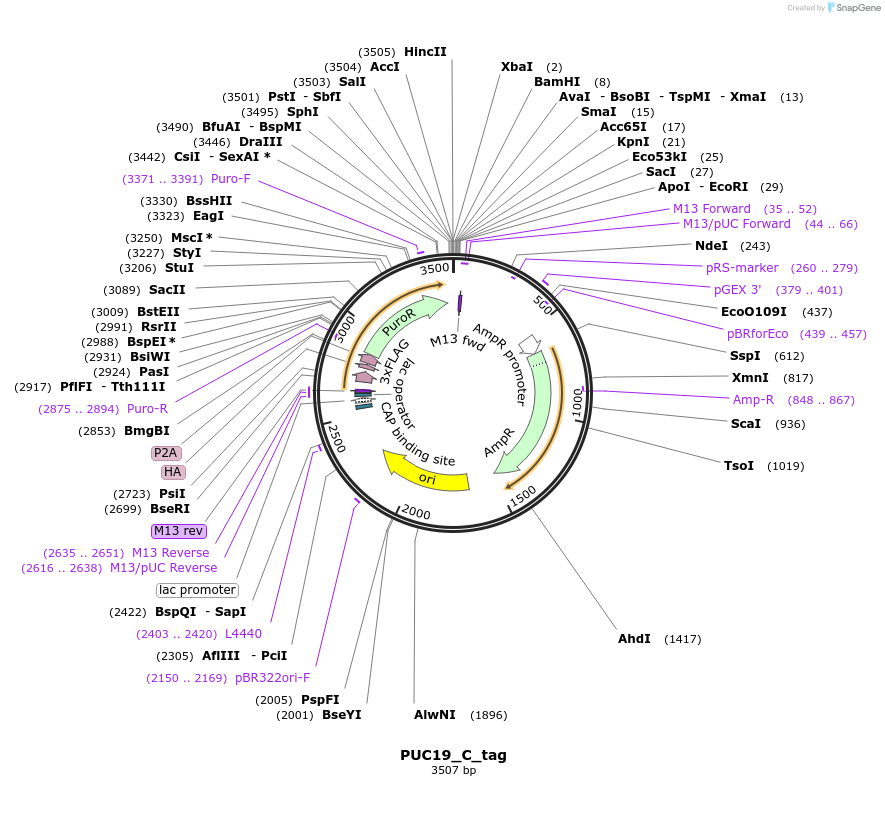
PUC19_C_tag
Plasmid#186659PurposeEmpty donor plasmid for C-terminal tag of 3xFlag-3xHA.DepositorInsertC-terminal 3xFlag-3xHA Tag
Tags3xFlag-3xHAExpressionInsectAvailable SinceJuly 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
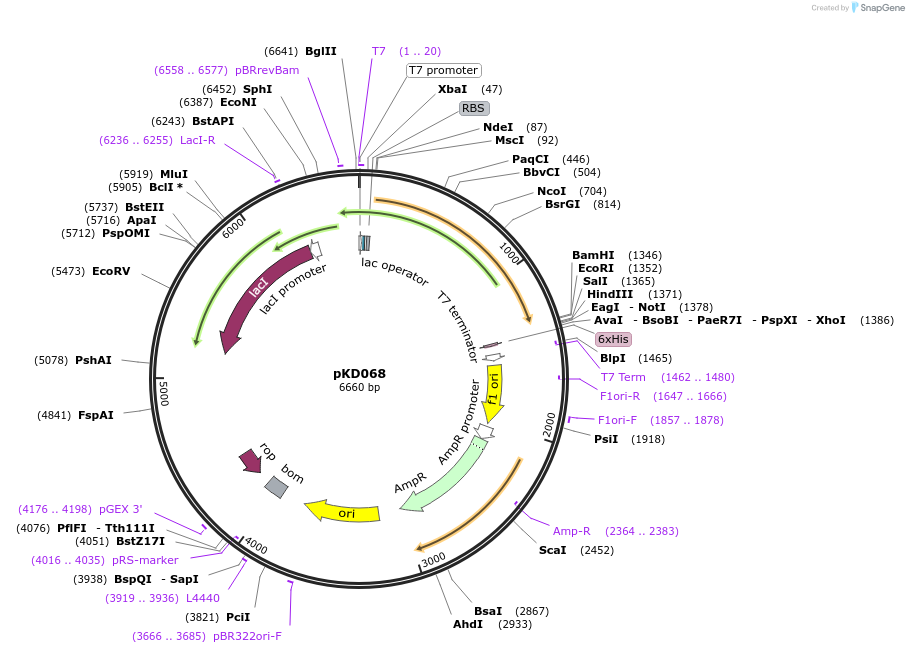
pKD068
Plasmid#136471PurposeSSB-GFP IDL fusion expression plasmidDepositorInsertSSB-GFP (ssb E. coli)
Tagssuperfolder GFPExpressionBacterialMutationsuperfolder GFP inserted between Phe148 and Ser149PromoterT7 promoterAvailable SinceApril 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
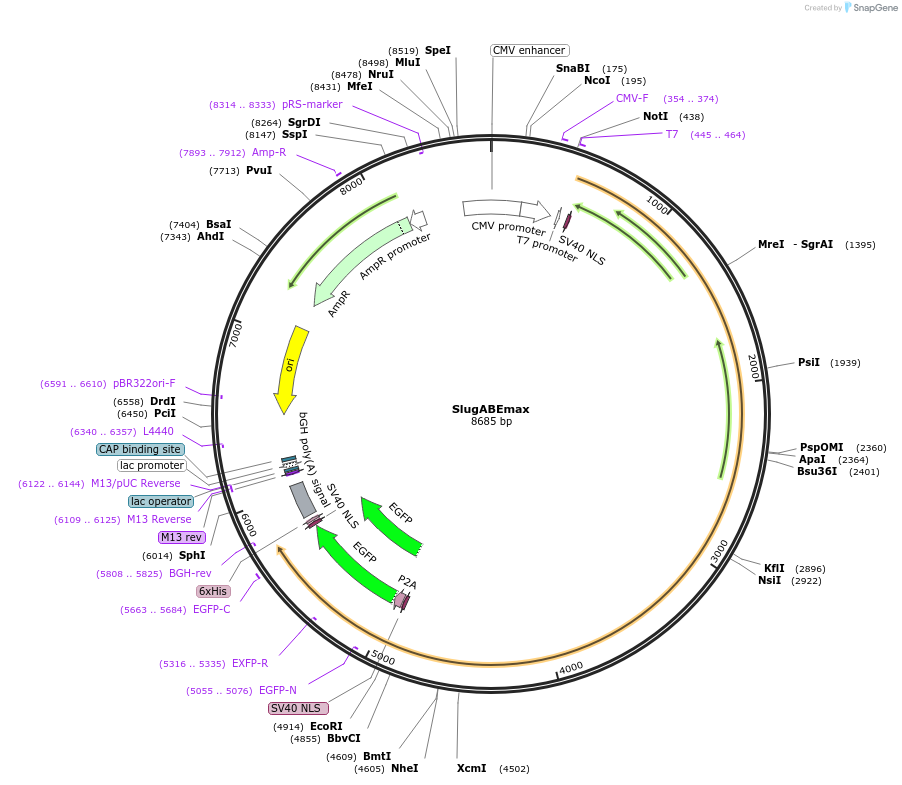
SlugABEmax
Plasmid#163798PurposeExpresses ABEmax and nSlugCas9DepositorTypeEmpty backboneUseCRISPRExpressionMammalianMutationSlugCas9 (D10A)Available SinceApril 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
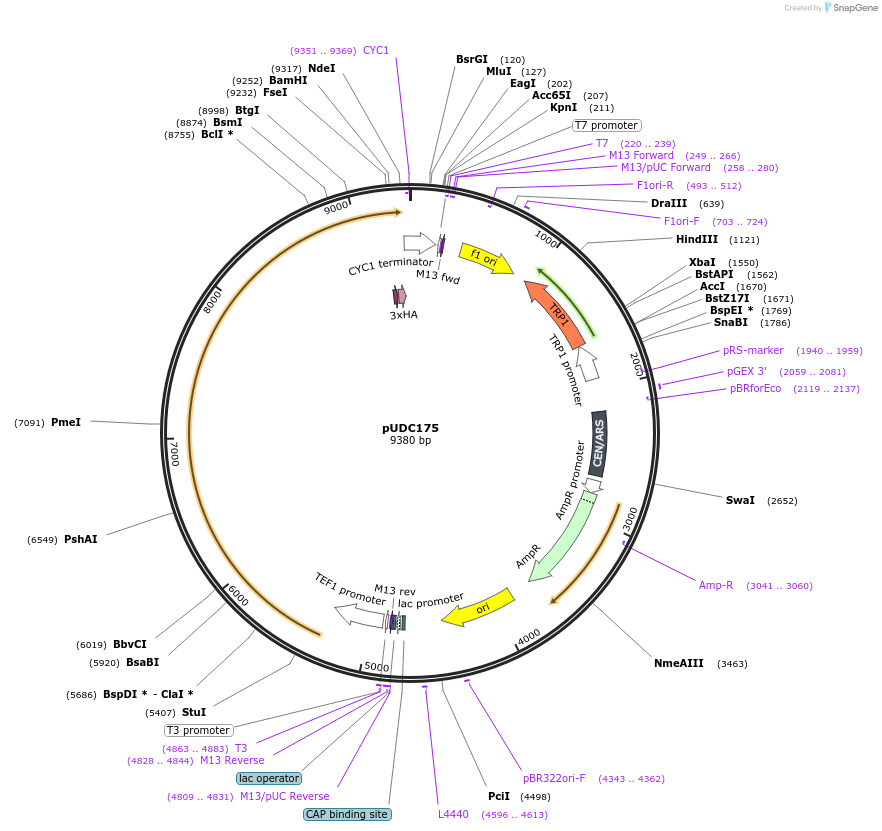
pUDC175
Plasmid#103019PurposeS. cerevisae centromic plasmid harboring Fncpf1 under control of TEF1 promoterDepositorInsertFncpf1
Tags3HA and NLSExpressionYeastMutationhuman codon optimizedPromoterTEF1Available SinceDec. 1, 2017AvailabilityAcademic Institutions and Nonprofits only -
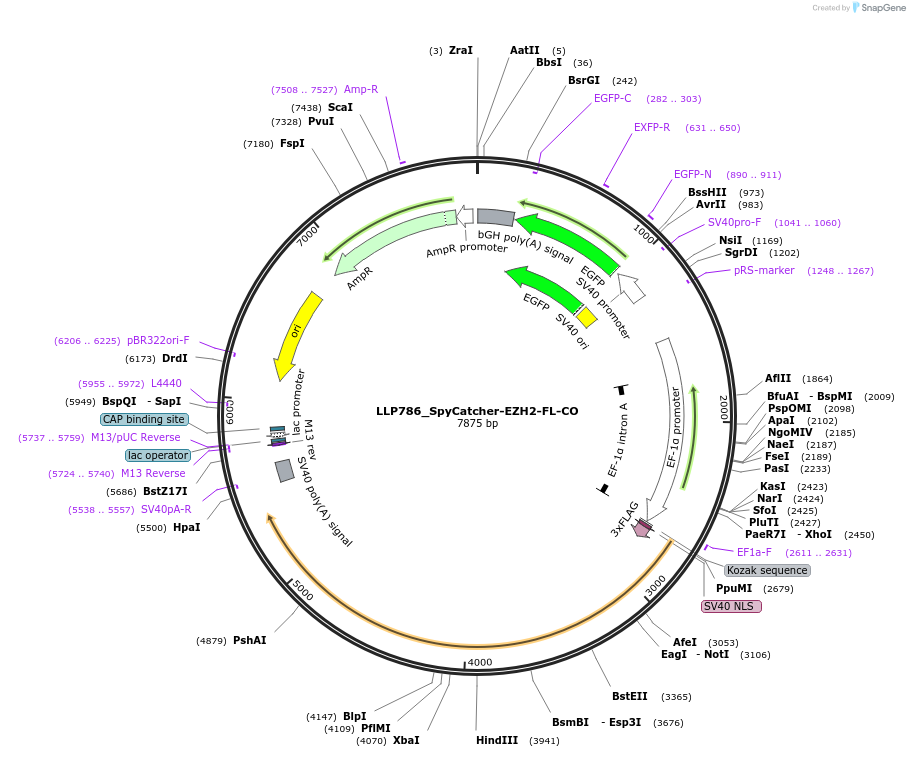
LLP786_SpyCatcher-EZH2-FL-CO
Plasmid#211779PurposeSpyTag counterpart binding domain, SpyCatcher, fused to polycomb repressive complex subunit EZH2, with GFP selectionDepositorInsertaGCN4-KRAB
Tags3xTy1ExpressionMammalianMutationLast 300 bp codon-optimised (CO) to detect KRAB e…PromoterpEF1a and pSV40Available SinceDec. 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
myc-hRAD18
Plasmid#68827PurposeExpresses c-myc-tagged human RAD18DepositorAvailable SinceOct. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
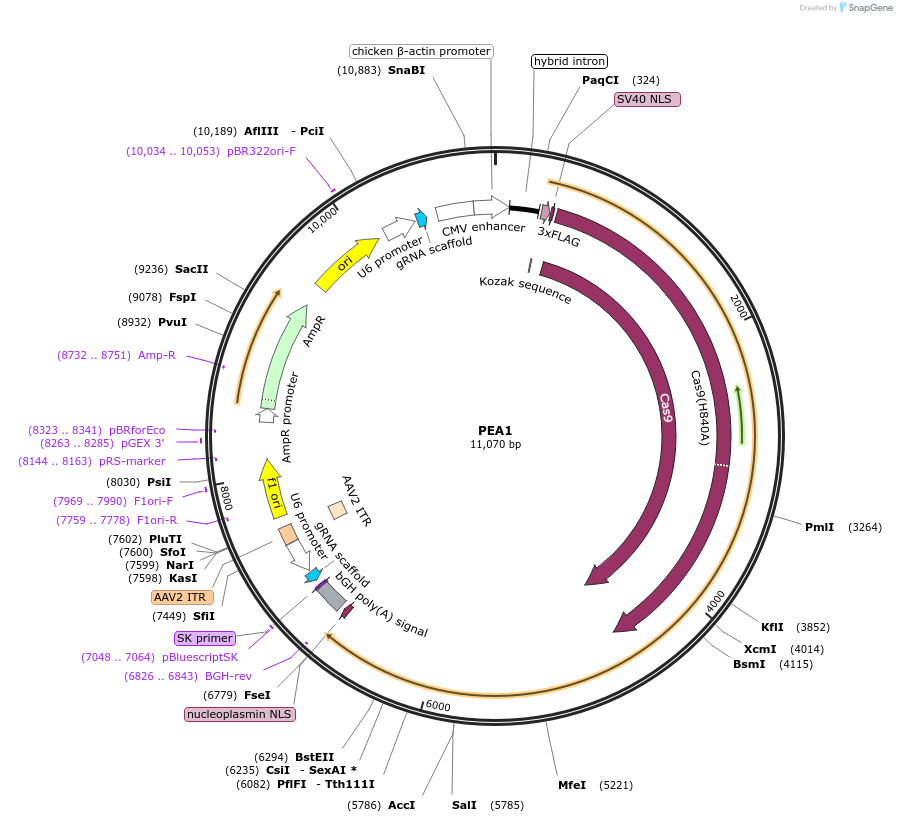
PEA1
Plasmid#176527PurposeDelivers all prime editing nickase components in a single plasmidDepositorInsertCbH-Cas9(H840A)-RT, hU6-pegRNA (Bbs1 gate), hU6-sgRNA (Bbs1 gate)
UseCRISPRExpressionMammalianPromoterCbH for Cas9, hU6 for gRNAsAvailable SinceDec. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
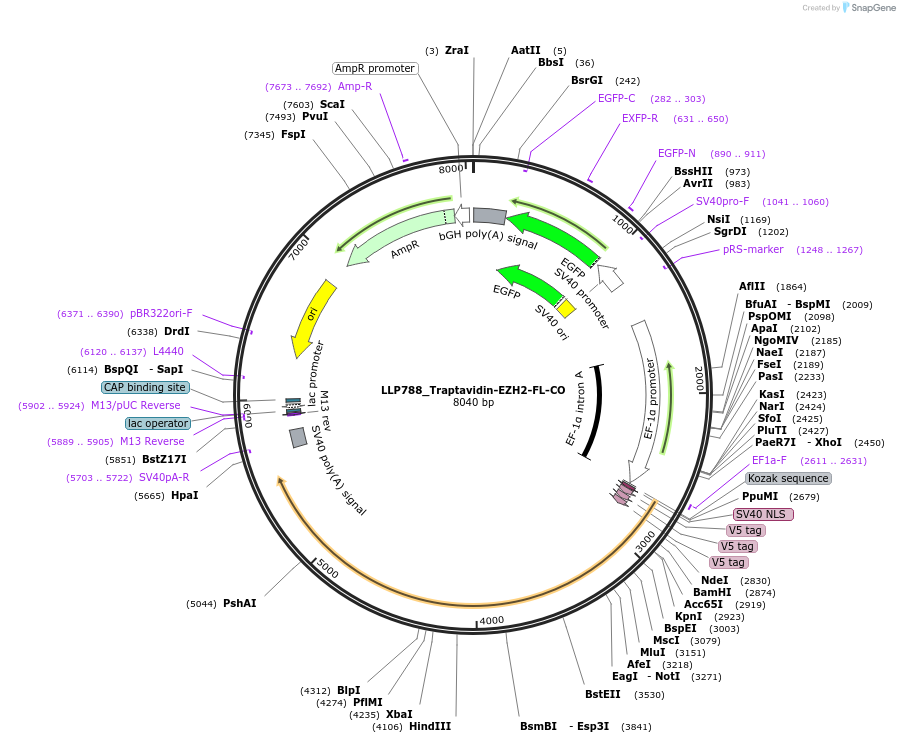
LLP788_Traptavidin-EZH2-FL-CO
Plasmid#211781PurposeAviTag counterpart binding domain, Traptavidin, fused to polycomb repressive complex subunit EZH2, with GFP selectionDepositorInsertaGCN4-EZH2
Tags3xTy1ExpressionMammalianMutationLast 300 bp codon-optimised (CO) to detect EZH2 e…PromoterpEF1a and pSV40Available SinceDec. 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
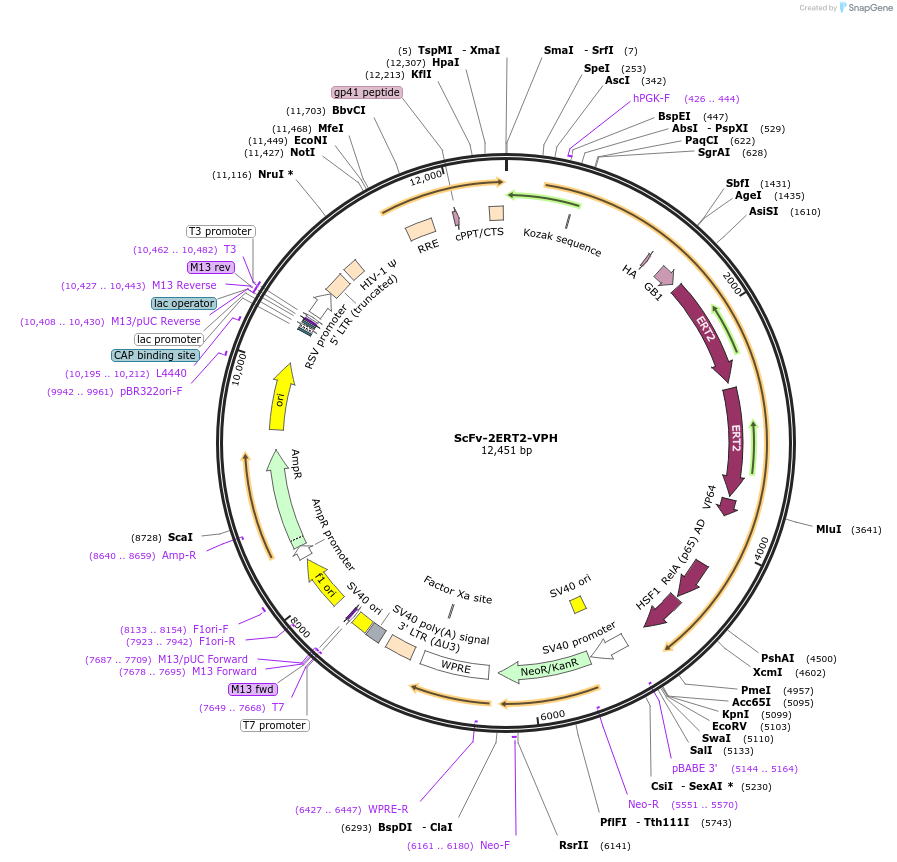
ScFv-2ERT2-VPH
Plasmid#120556PurposeEncode an antibody that binds to the GCN4 peptide from the SunTag system, and is fused to 2 tandem ERT2, VP64, P65 and HSF1DepositorInsertscFvGCN4, GB1, ERT2, VP64, P65 and HSF1
UseLentiviralExpressionMammalianPromoterhPGKAvailable SinceAug. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
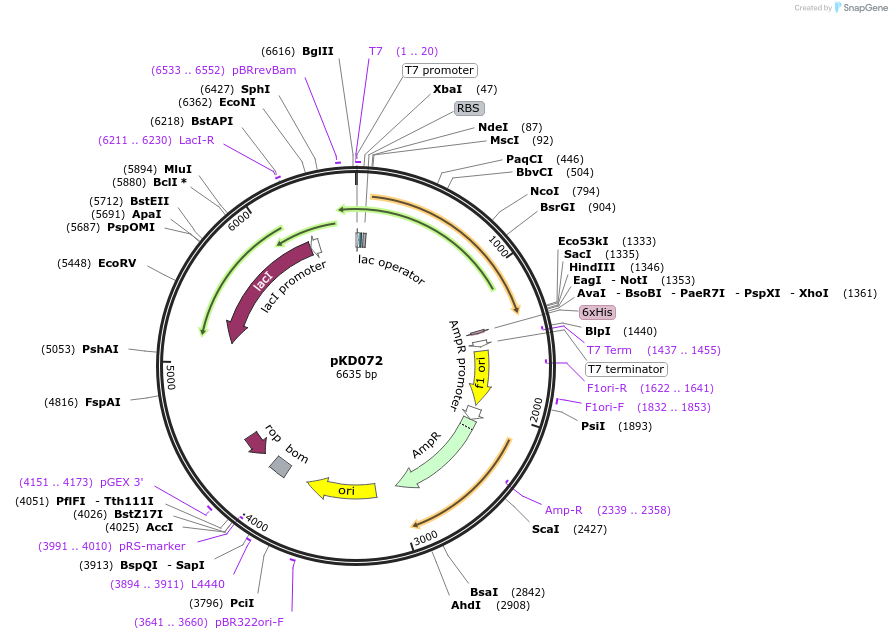
pKD072
Plasmid#136472PurposeSSB-C-term-GFP fusion expression plasmidDepositorInsertSSB-C-term-GFP (ssb E. coli)
Tagssuperfolder GFPExpressionBacterialMutationsuperfolder GFP attached to C-terminus at Phe 178PromoterT7 promoterAvailable SinceApril 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
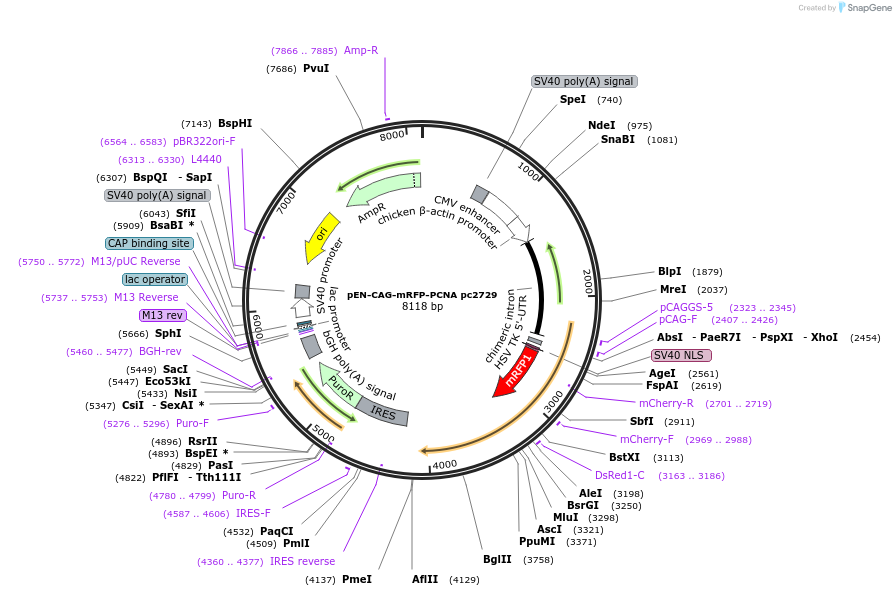
pEN-CAG-mRFP-PCNA pc2729
Plasmid#166040PurposeExpresses mRFP tagged human PCNA in mammalian cells.DepositorAvailable SinceMarch 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
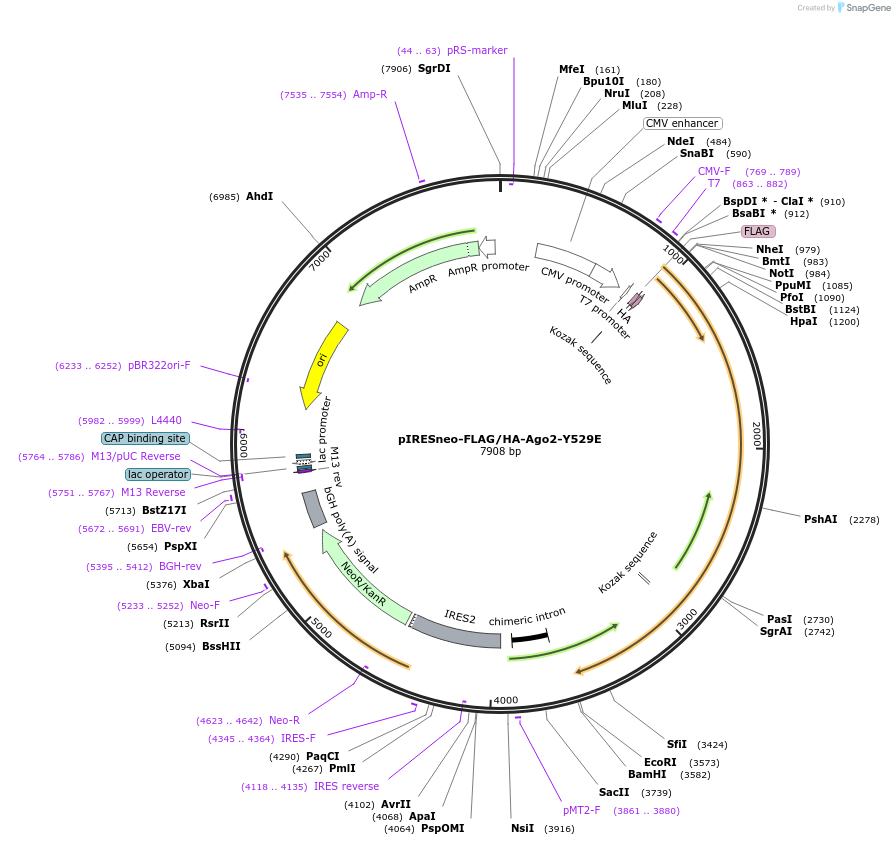
pIRESneo-FLAG/HA-Ago2-Y529E
Plasmid#215842PurposeExpression of AGO2-Y529E mutantDepositorAvailable SinceApril 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
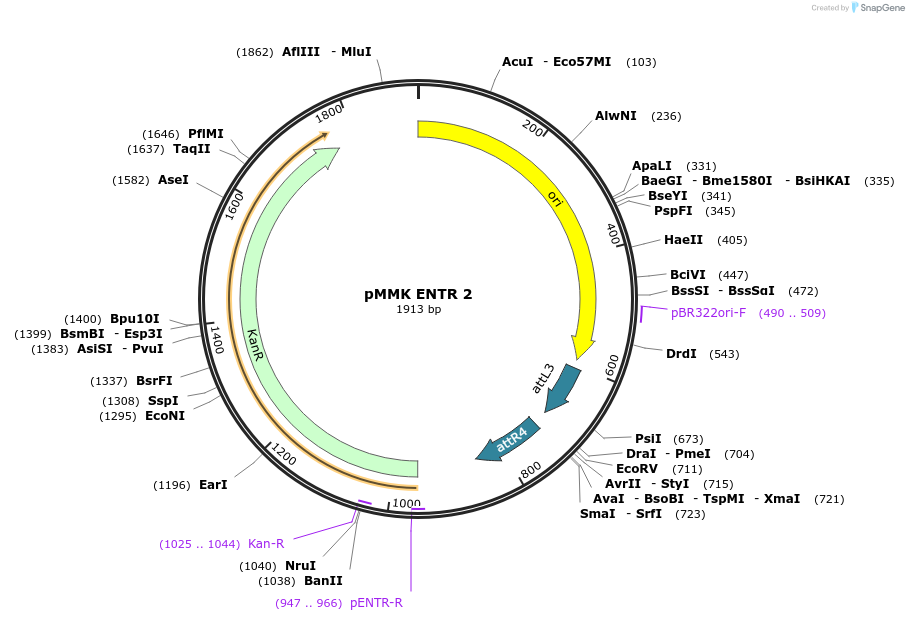
pMMK ENTR 2
Plasmid#206261PurposeENTR Vector 1 for MultiSite Gateway assembly and production of recombinant baculovirus using MultiMate assembly. Empty backbone.DepositorTypeEmpty backboneUseMultimate/gateway entr 2Available SinceSept. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
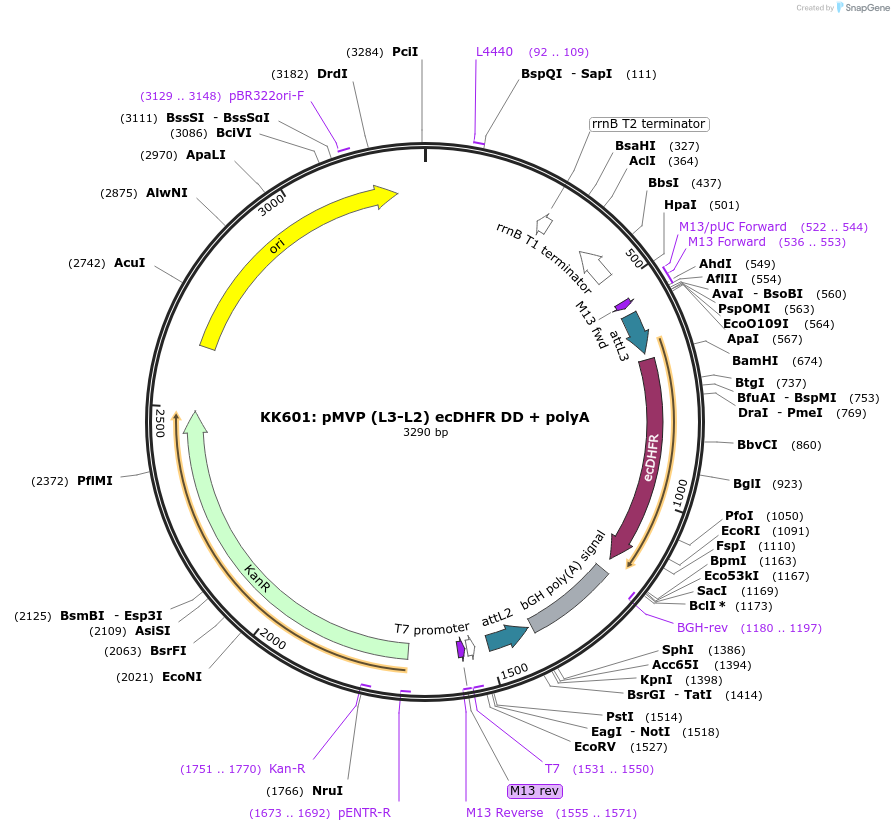
KK601: pMVP (L3-L2) ecDHFR DD + polyA
Plasmid#121790PurposepMVP L3-L2 entry plasmid, contains ecDHFR DD + polyA for 3- or 4-component MultiSite Gateway Pro assembly. Allows fusion of C-term ecDHFR degron domain (stabilized by TMP) to gene of interest.DepositorInsertecDHFR Degron Domain + polyA
UseSynthetic Biology; Pmvp gateway entry plasmidAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
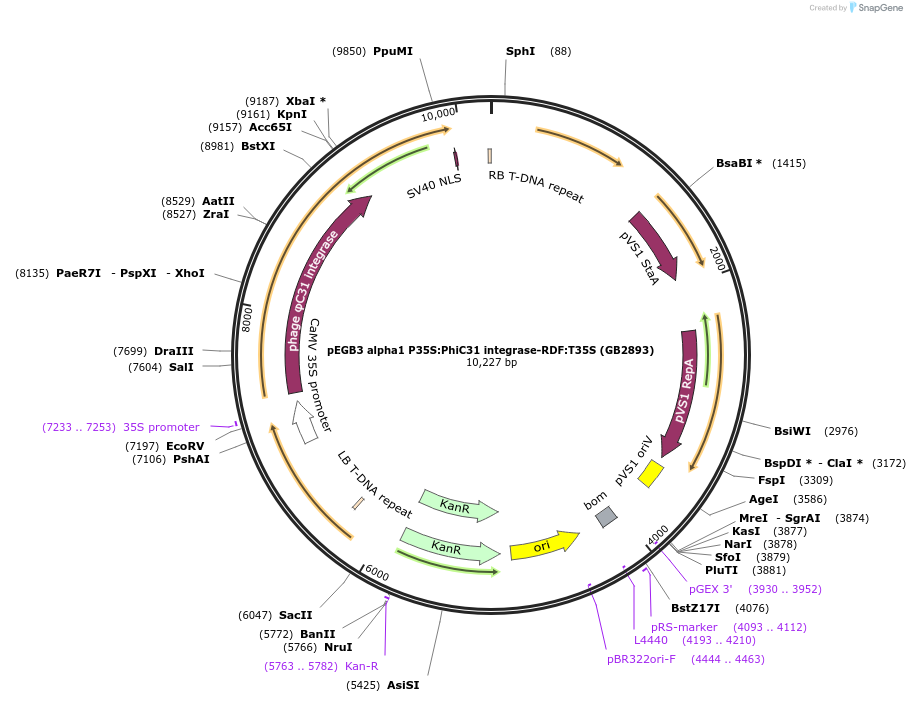
pEGB3 alpha1 P35S:PhiC31 integrase-RDF:T35S (GB2893)
Plasmid#160633PurposeTU for the constitutive expression of the PhiC31 integrase, C-terminally fused with its reversion factor (RDF)DepositorInsertPhiC31-RDF
UseSynthetic BiologyExpressionPlantMutationBsaI and BsmBI sites removedPromoter35SAvailable SinceMarch 29, 2021AvailabilityAcademic Institutions and Nonprofits only