We narrowed to 17,798 results for: bon
-
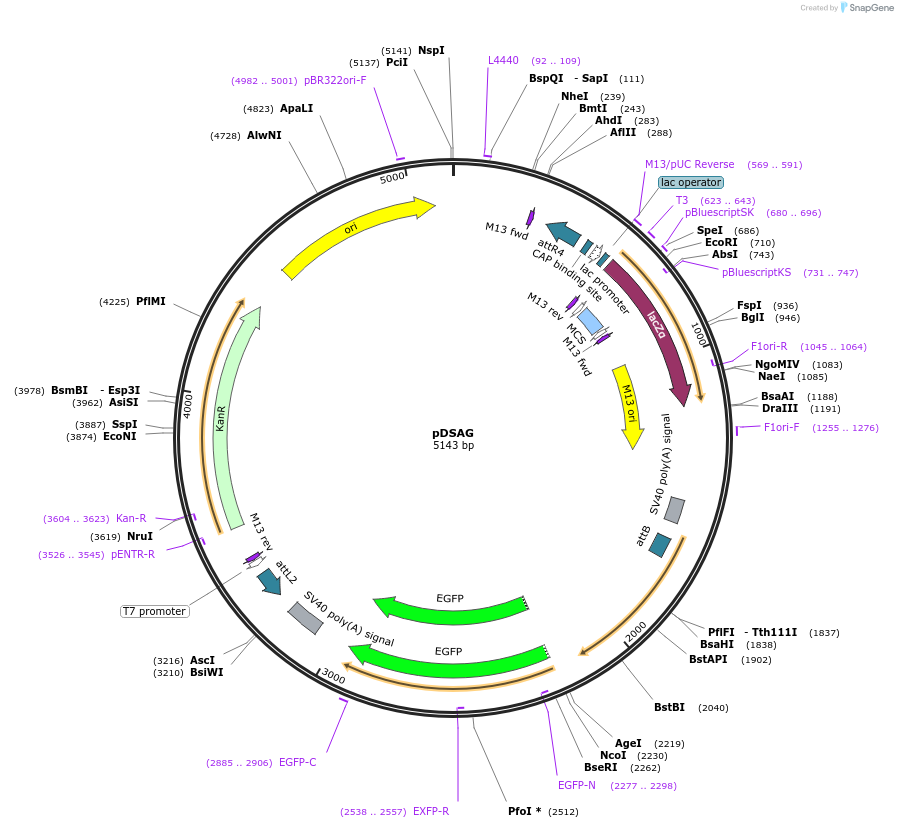
Plasmid#62289Purposedocking site transgenesis plasmid for GoldenGate Cloning and insect transgenesis, GFP markerDepositorTypeEmpty backboneExpressionInsectAvailable SinceMay 18, 2015AvailabilityAcademic Institutions and Nonprofits only
-
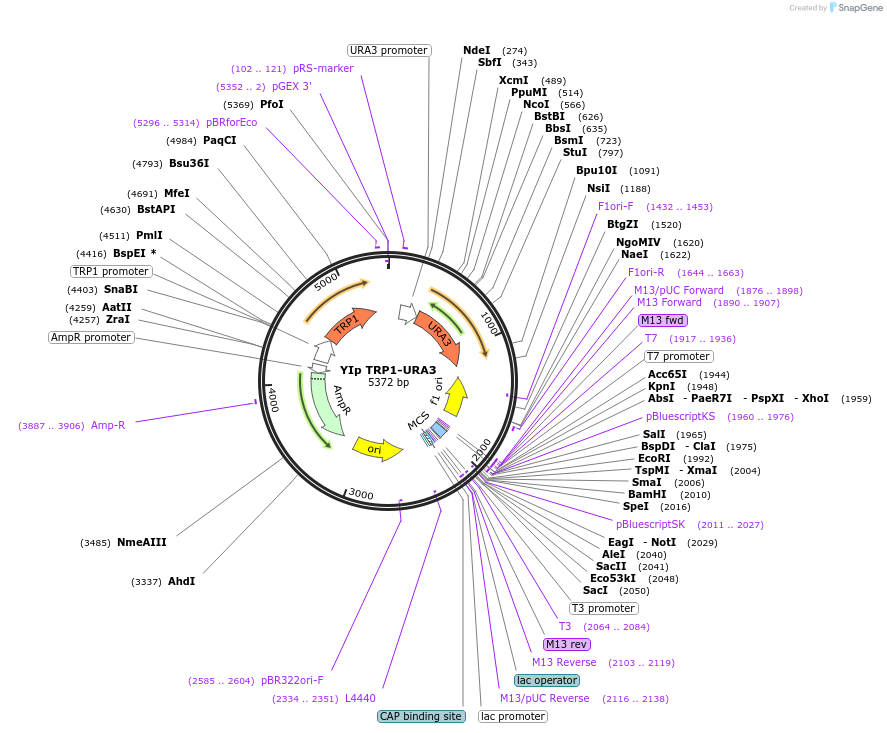
YIp TRP1-URA3
Plasmid#122915PurposeTRP1-URA3 integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailable SinceJuly 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
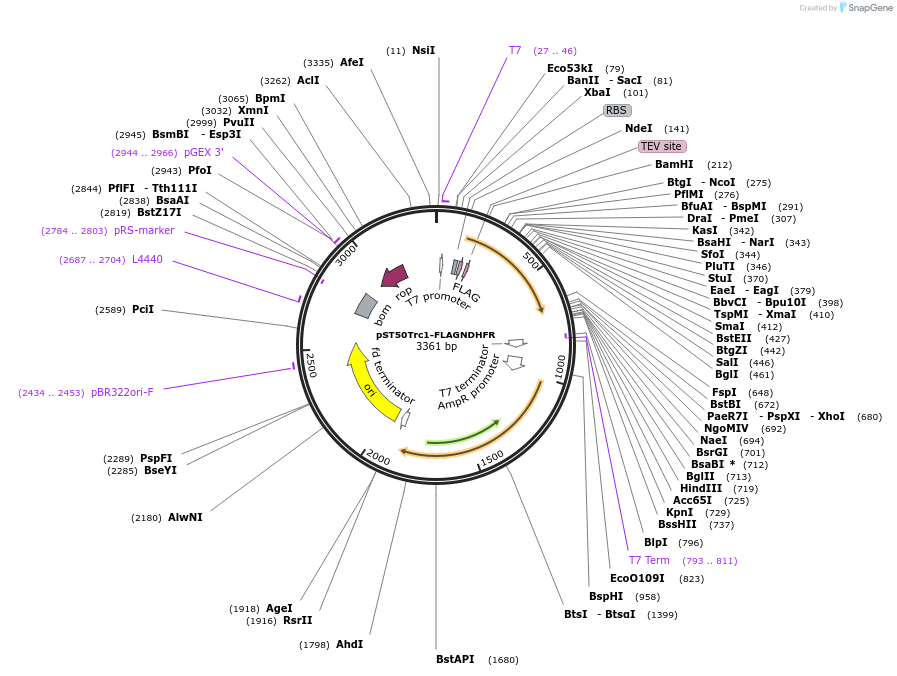
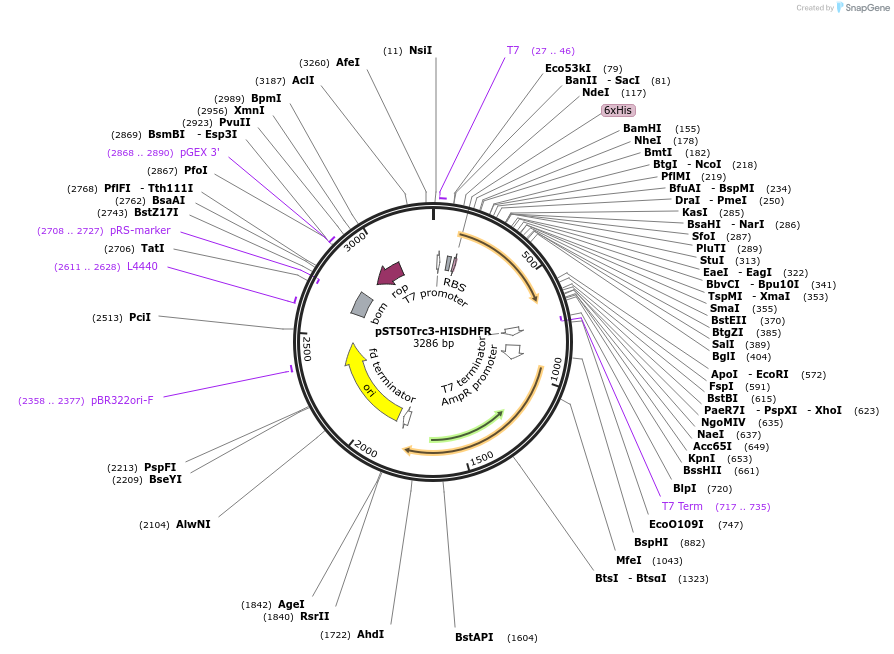
pST50Trc1-FLAGNDHFR
Plasmid#63943PurposePosition 1 transfer plasmid for pST44 polycistronic plasmid suite; N-term cleavable single tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTagsFLAG; TEV cleavableExpressionBacterialPromoterT7Available SinceAug. 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
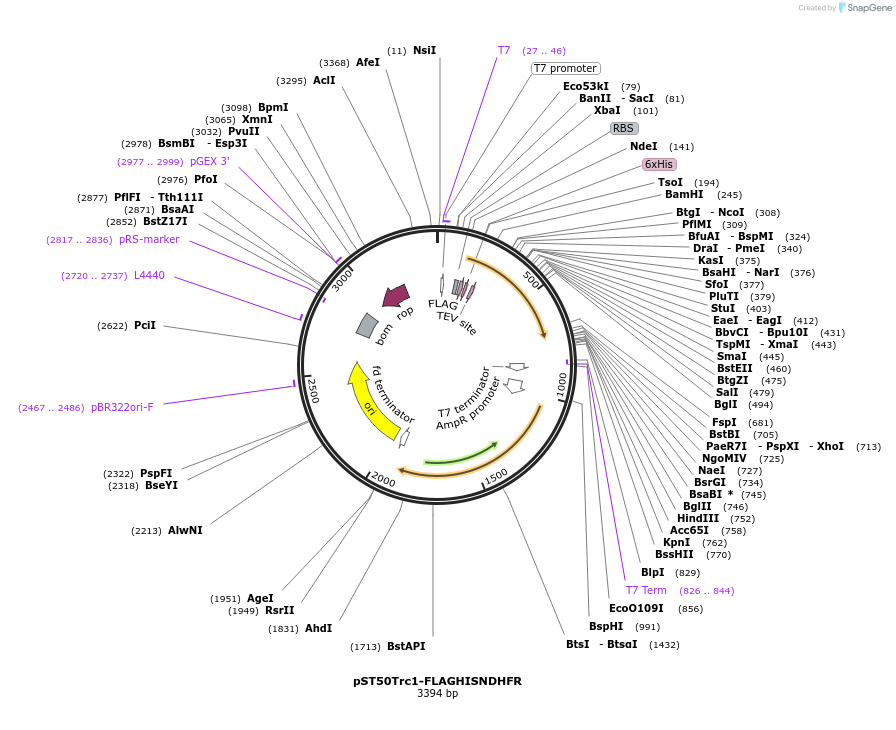
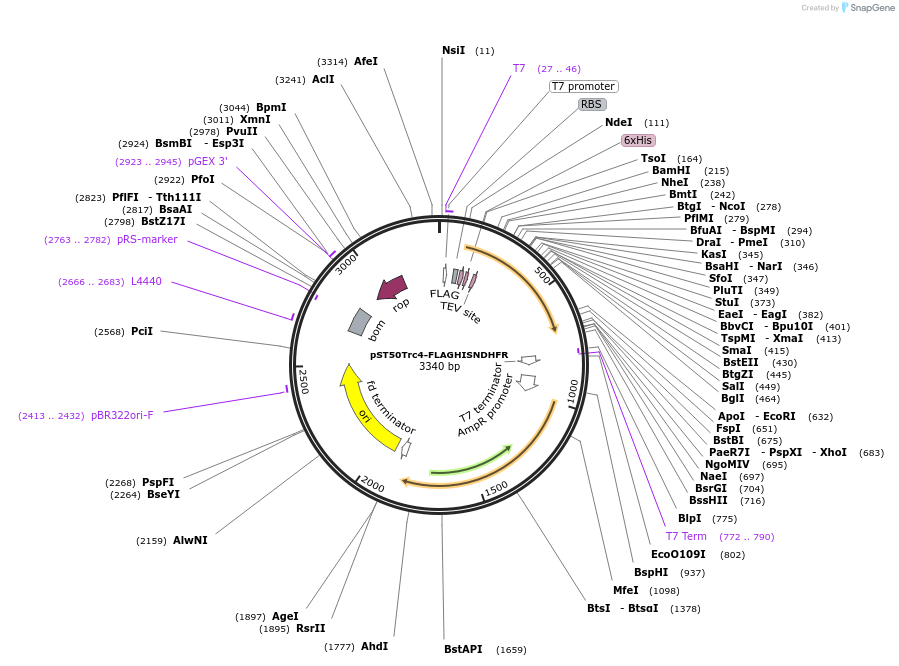
pST50Trc1-FLAGHISNDHFR
Plasmid#63947PurposePosition 1 transfer plasmid for pST44 polycistronic plasmid suite; N-term cleavable double tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTags6xHis & FLAG; TEV cleavableExpressionBacterialPromoterT7Available SinceAug. 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
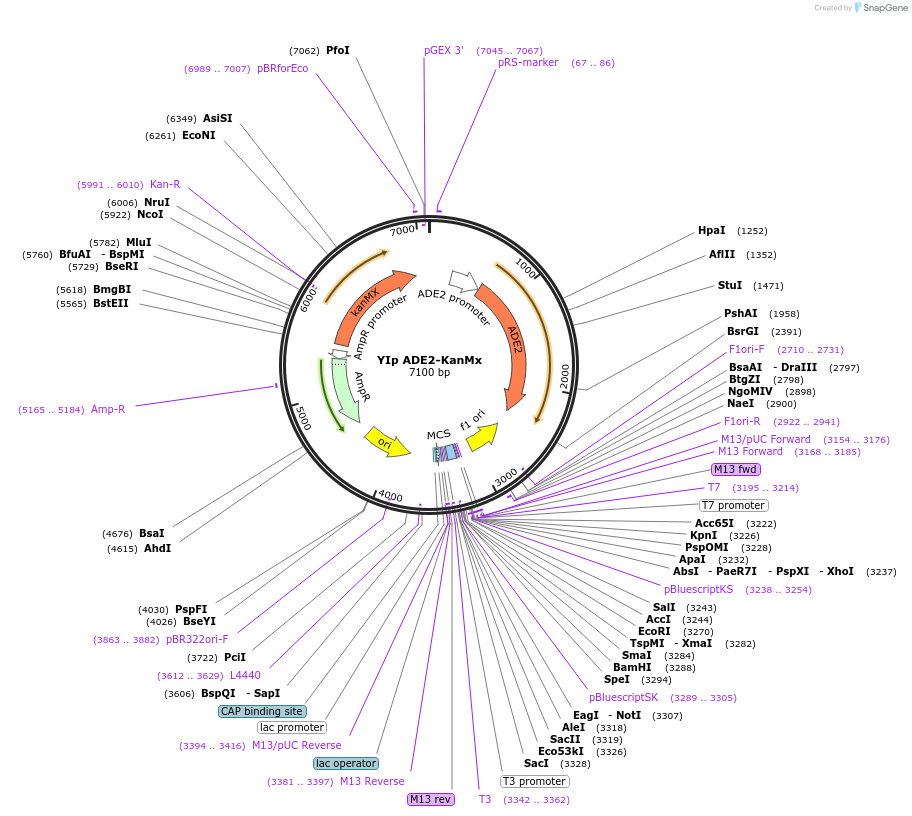
YIp ADE2-KanMx
Plasmid#122917PurposeADE2-KanMx integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailable SinceMay 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
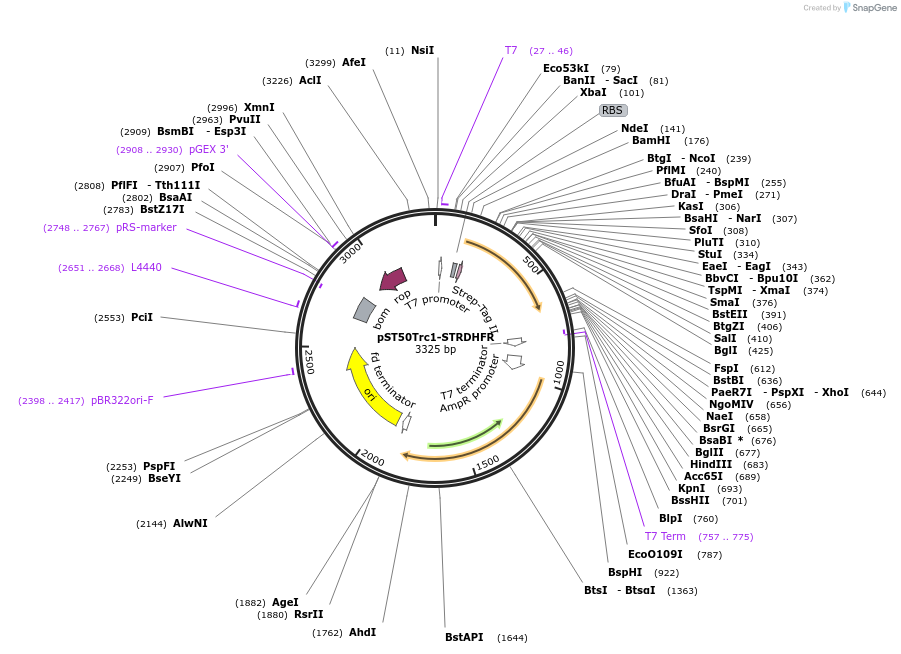
pST50Trc1-STRDHFR
Plasmid#63937PurposePosition 1 transfer plasmid for pST44 polycistronic plasmid suite; N-term non-cleavable single tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTagsStrep II (STR)ExpressionBacterialPromoterT7Available SinceAug. 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
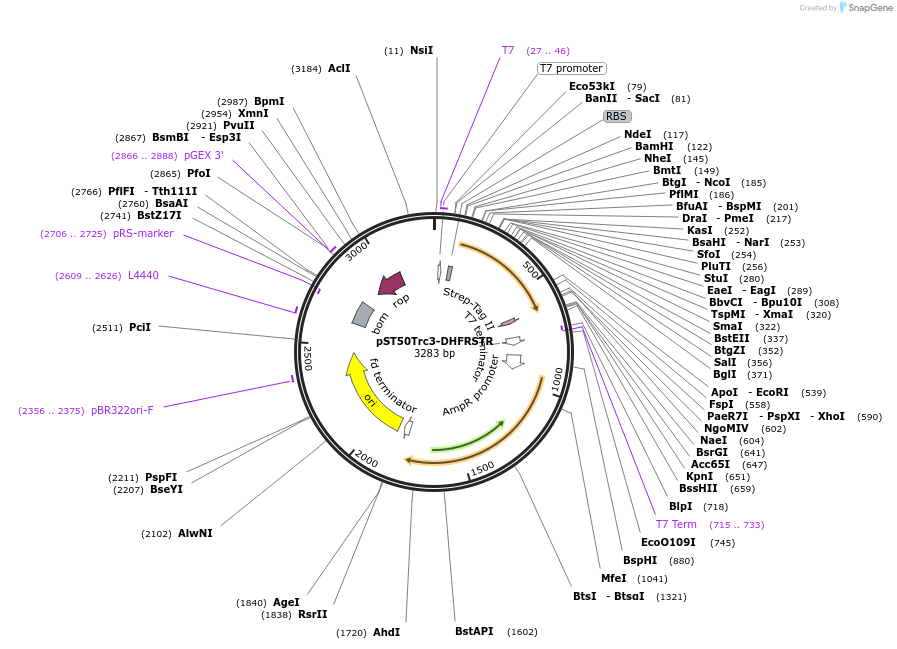
pST50Trc3-DHFRSTR
Plasmid#63986PurposePosition 3 transfer plasmid for pST44 polycistronic plasmid suite; C-term single tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTagsStrep II (STR)ExpressionBacterialPromoterT7Available SinceDec. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAT590
Plasmid#68908PurposeNanoLuc expressed constiutively from PBT1311 and GH049 RBS, pNBU2 backbone, AmpRDepositorInsertNanoLuc
ExpressionBacterialAvailable SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAT780
Plasmid#68917PurposeNanoLuc expressed constiutively from PAM2 and GH022 RBS, pNBU2 backbone, AmpRDepositorInsertNanoLuc
ExpressionBacterialAvailable SinceOct. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAT793
Plasmid#68926PurposeNanoLuc expressed constiutively from PAM4 and GH078 RBS, pNBU2 backbone, AmpRDepositorInsertNanoLuc
ExpressionBacterialAvailable SinceOct. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
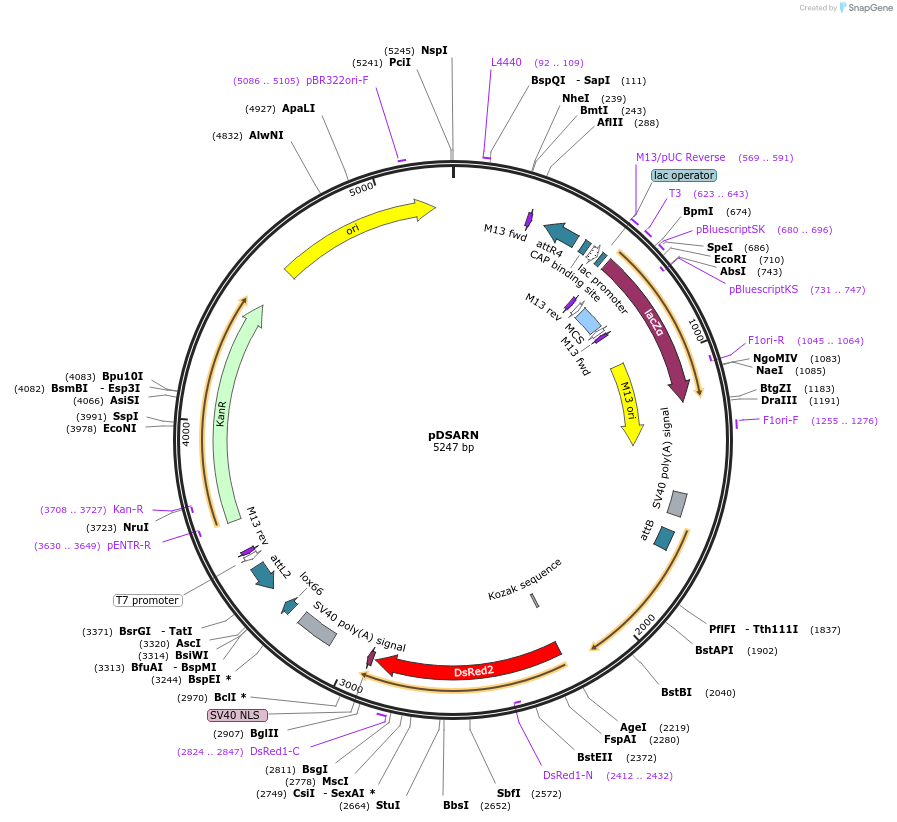
pDSARN
Plasmid#62295Purposedocking site transgenesis plasmid for GoldenGate Cloning and insect transgenesis, DsRed-NLS markerDepositorTypeEmpty backboneExpressionInsectAvailable SinceMarch 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
CA8
Plasmid#38989PurposeBacterial expression for structure determination; may not be full ORFDepositorAvailable SinceMarch 18, 2013AvailabilityAcademic Institutions and Nonprofits only -
pST50Trc3-STRDHFR
Plasmid#63973PurposePosition 3 transfer plasmid for pST44 polycistronic plasmid suite; N-term non-cleavable single tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTagsStrep II (STR)ExpressionBacterialPromoterT7Available SinceDec. 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
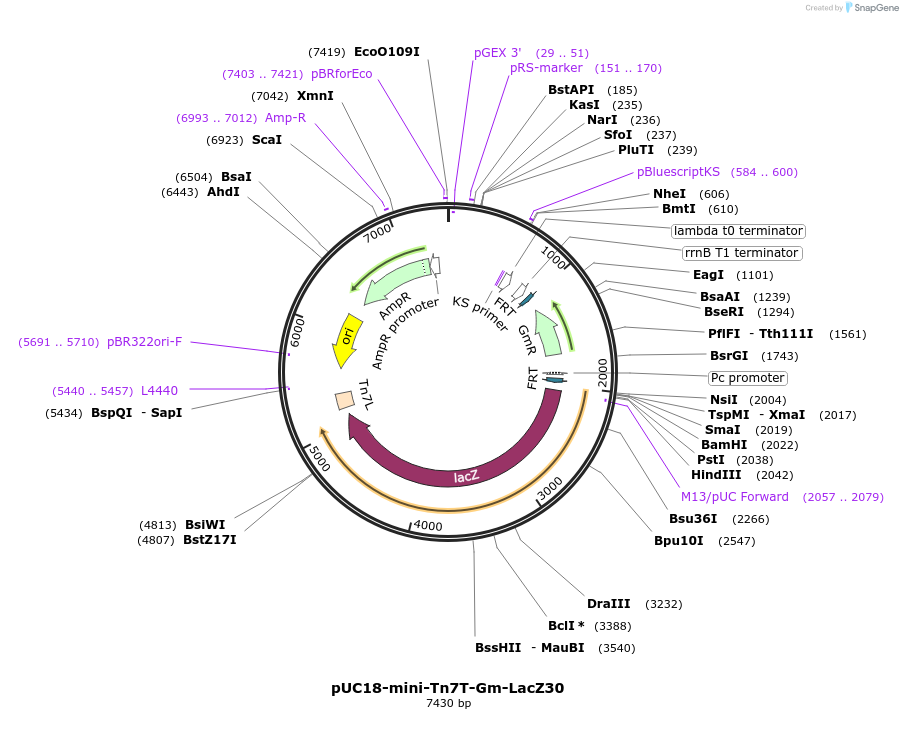
pUC18-mini-Tn7T-Gm-LacZ30
Plasmid#65028PurposeGmR mini-Tn7 vector, for construction of B-galactosidase protein fusions, frame 3. See Depositor Comments below for information regarding the lacZ sequence.DepositorTypeEmpty backboneTagslacZExpressionBacterialAvailable SinceNov. 2, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
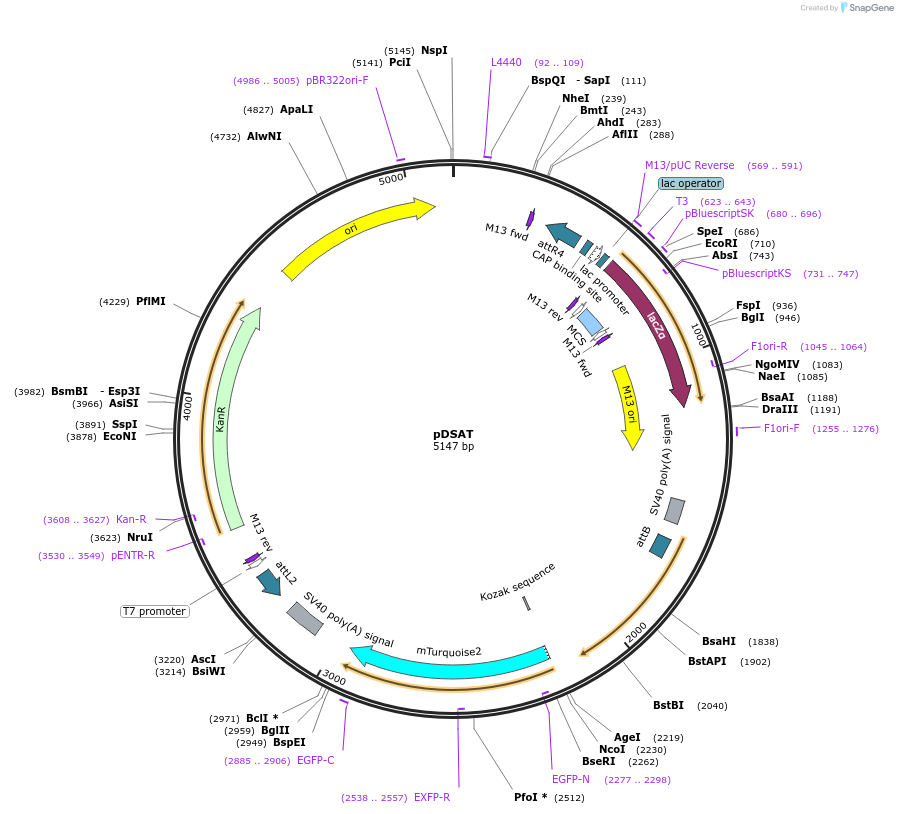
pDSAT
Plasmid#62290Purposedocking site transgenesis plasmid for GoldenGate Cloning and insect transgenesis, mTurquoise CFP markerDepositorTypeEmpty backboneExpressionInsectAvailable SinceMarch 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
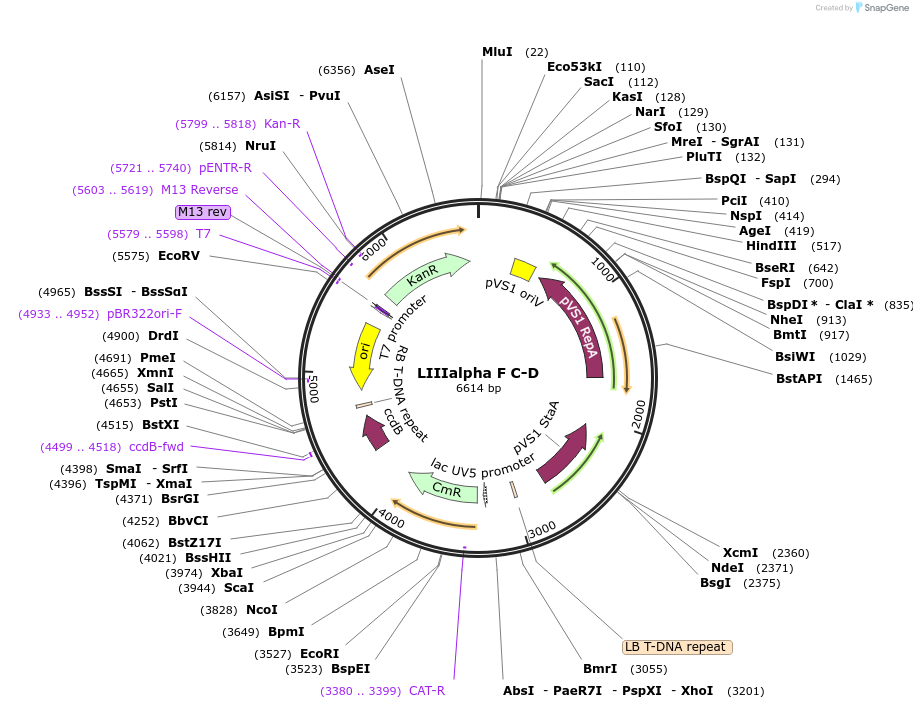
LIIIalpha F C-D
Plasmid#54384DepositorTypeEmpty backboneUsePlant expressionAvailable SinceJuly 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
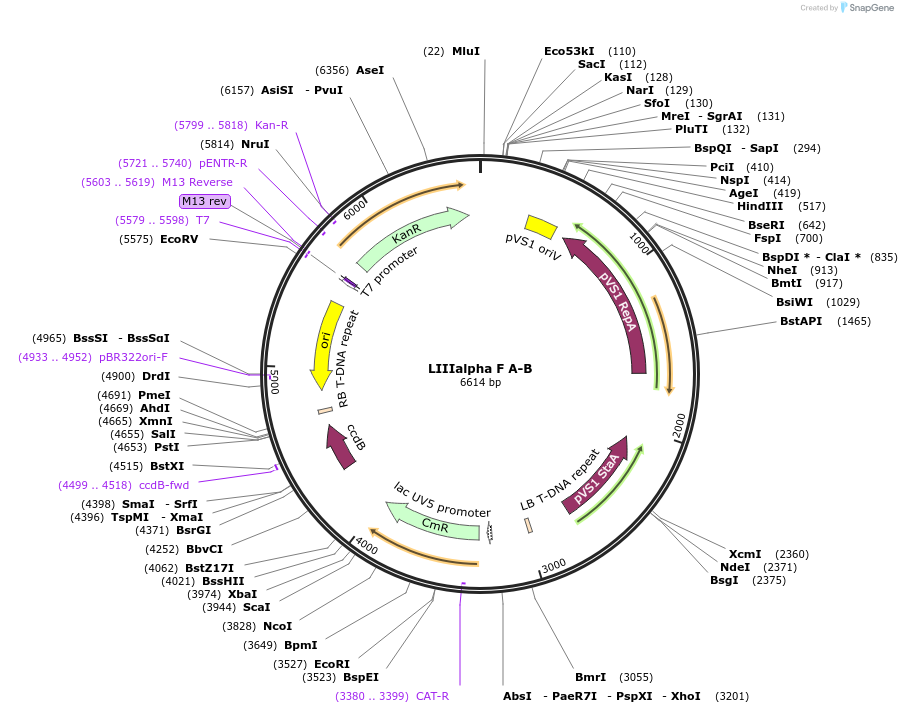
LIIIalpha F A-B
Plasmid#54382DepositorTypeEmpty backboneUsePlant expressionAvailable SinceJune 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
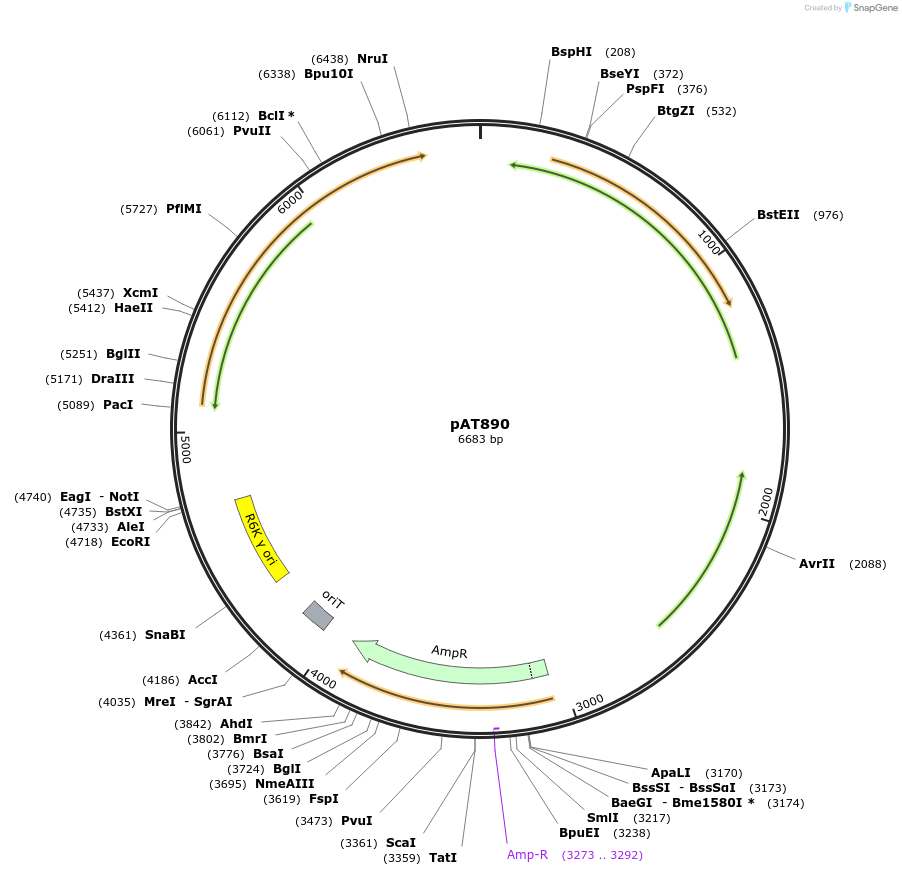
pAT890
Plasmid#68931PurposeInt7 expressed constitutively from PAM4 and the rpiL* RBS, pNBU2 backbone, AmpRDepositorInsertInt7
ExpressionBacterialAvailable SinceOct. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
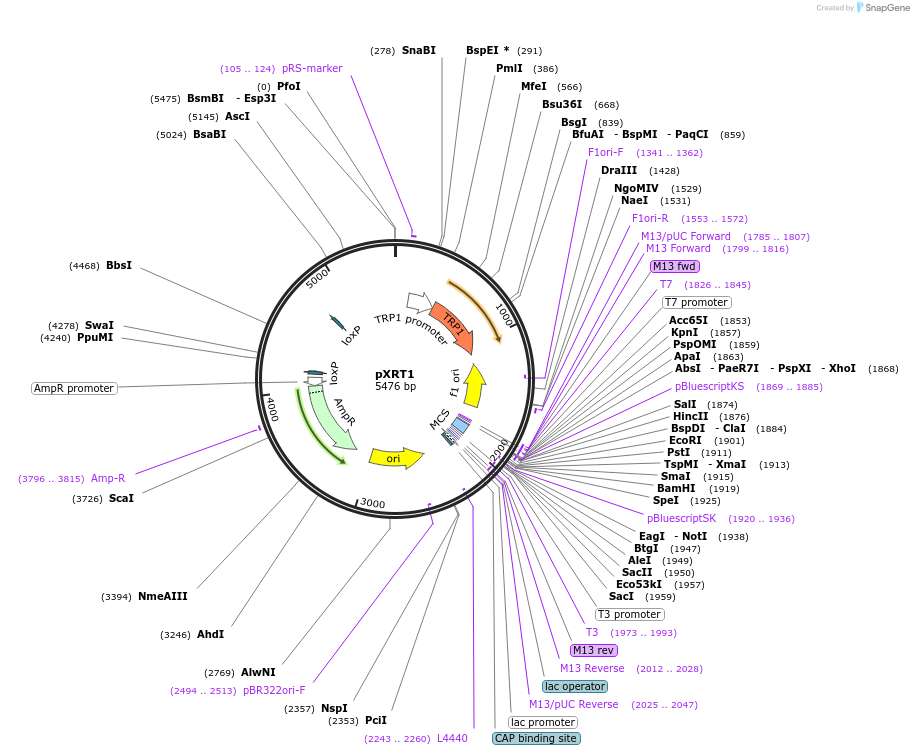
pXRT1
Plasmid#63151PurposeTrp1 marker pXR vector uses for both plasmid assembly in vivo, as well as targeted genomic integration.DepositorTypeEmpty backboneUseCre/LoxExpressionBacterial and YeastAvailable SinceApril 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
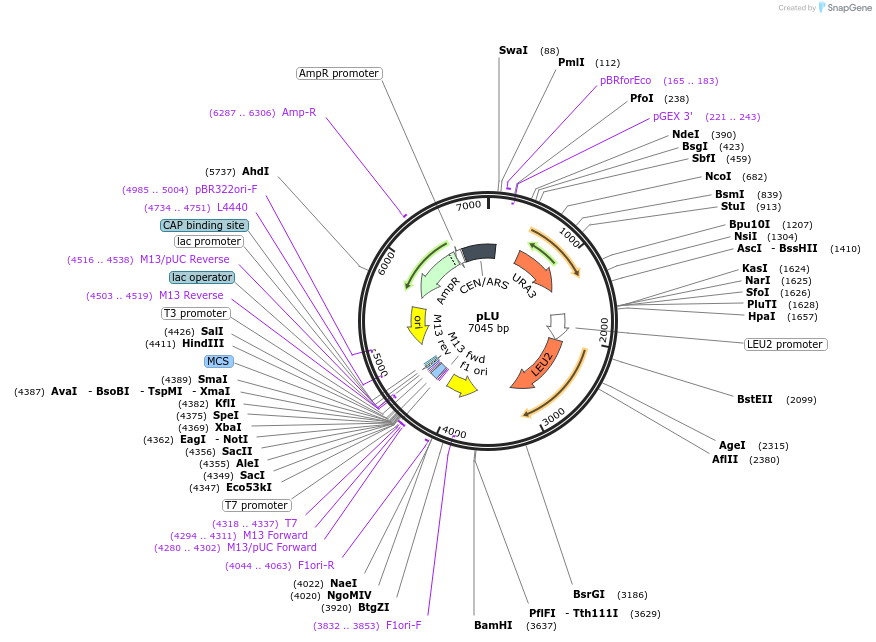
pLU
Plasmid#64174Purposeharbours marker genes LEU2 and URA3DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceAug. 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
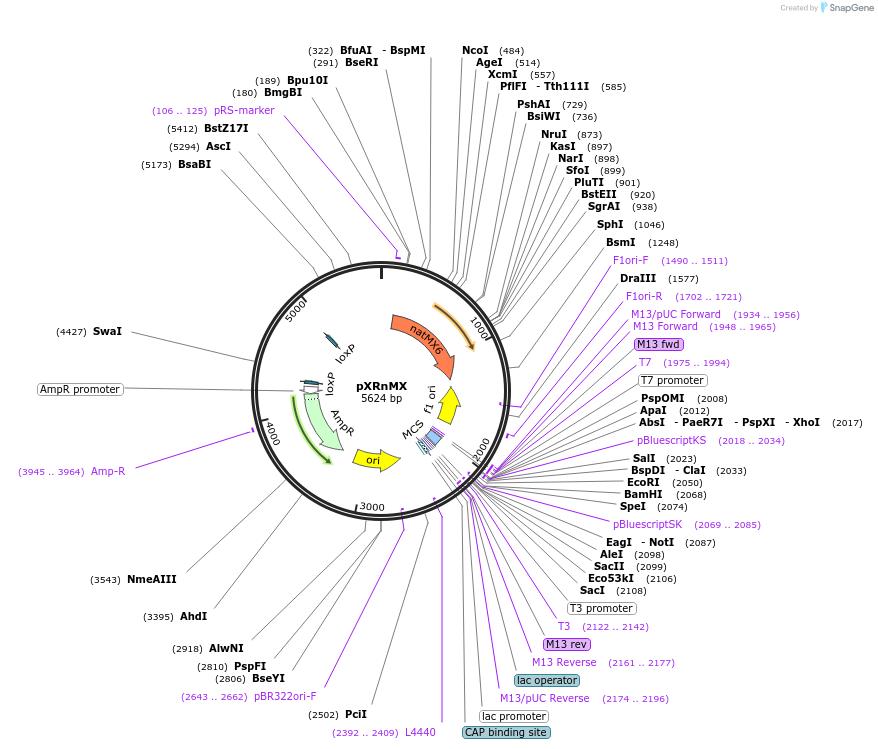
pXRnMX
Plasmid#63153PurposeNat1 marker pXR vector uses for both plasmid assembly in vivo, as well as targeted genomic integration.DepositorTypeEmpty backboneUseCre/LoxExpressionBacterial and YeastAvailable SinceApril 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAT752
Plasmid#68903PurposeNanoLuc expressed constiutively from PAM2, pNBU2 backbone, AmpRDepositorInsertNanoLuc
ExpressionBacterialAvailable SinceOct. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAT754
Plasmid#68905PurposeNanoLuc expressed constiutively from PAM4, pNBU2 backbone, AmpRDepositorInsertNanoLuc
ExpressionBacterialAvailable SinceOct. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
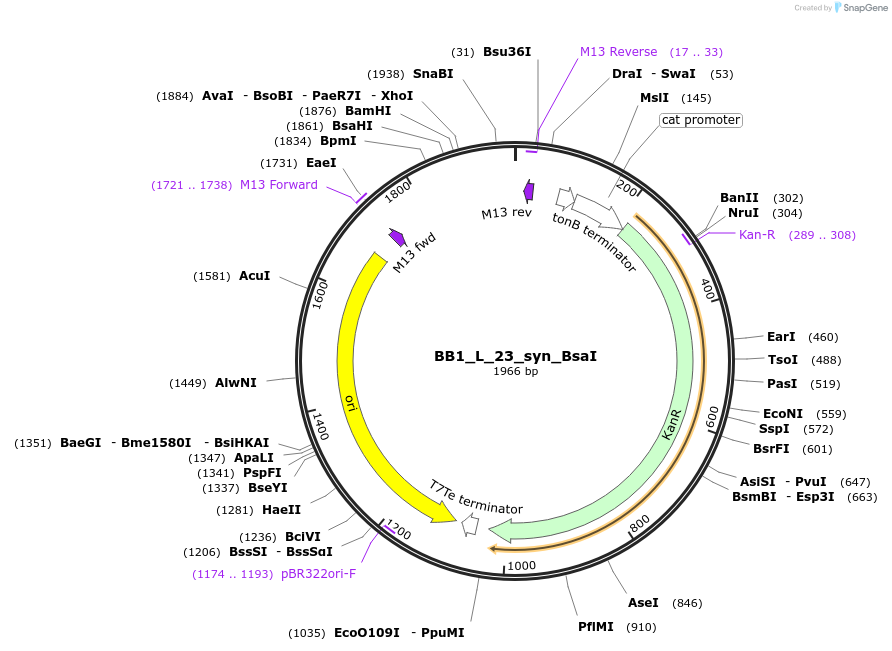
BB1_L_23_syn_BsaI
Plasmid#89915PurposeLinker to construct BB1 from PCR product with FS2 and FS3 (in with BsaI, out with BbsI)DepositorTypeEmpty backboneExpressionBacterialAvailable SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
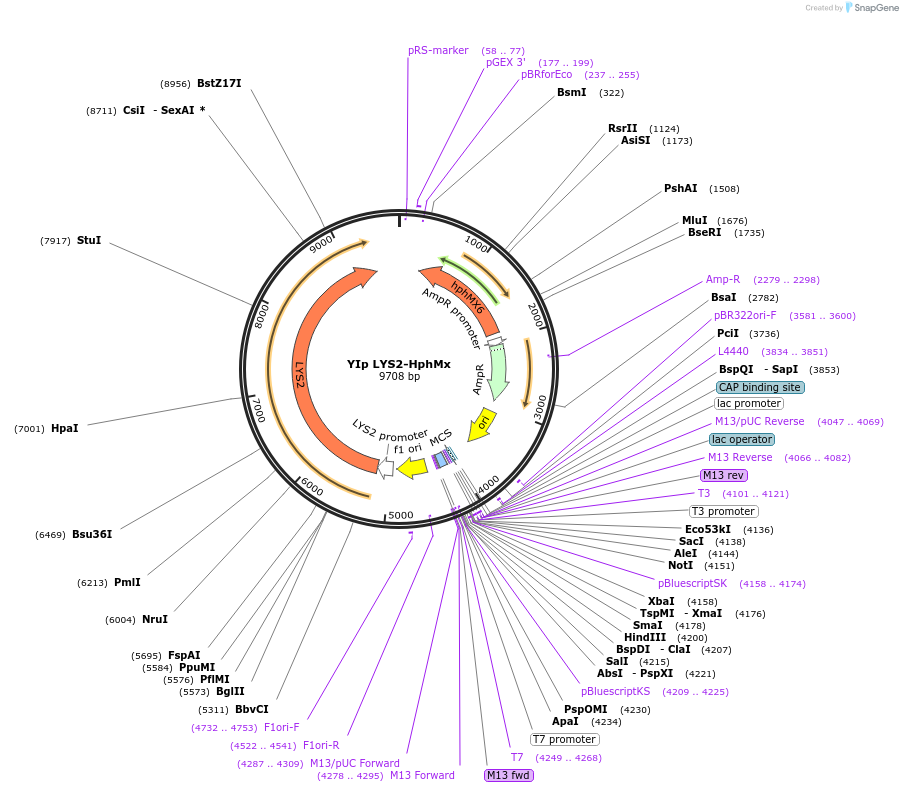
YIp LYS2-HphMx
Plasmid#122919PurposeLYS2-HphMx integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailable SinceOct. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
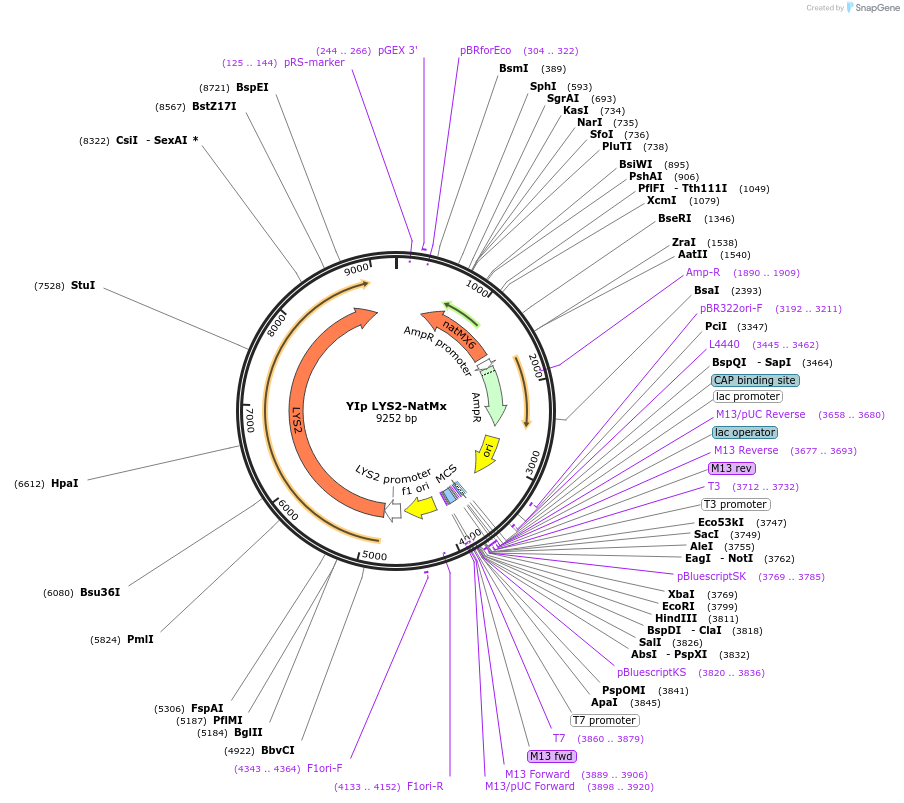
YIp LYS2-NatMx
Plasmid#122921PurposeLYS2-NatMx integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailable SinceOct. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
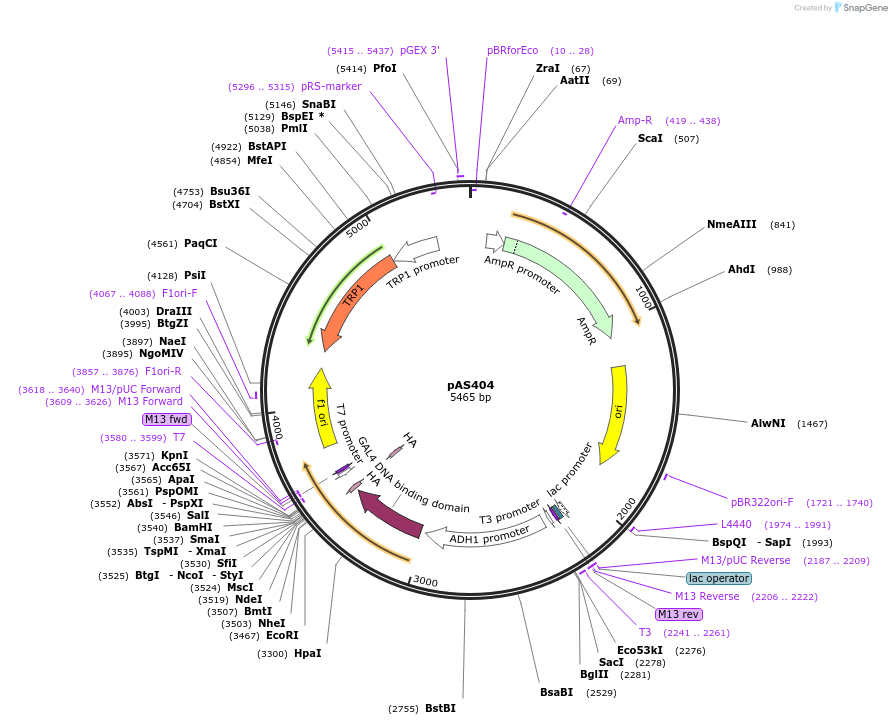
pAS404
Plasmid#15978DepositorTypeEmpty backboneTagsHAExpressionYeastAvailable SinceOct. 31, 2007AvailabilityAcademic Institutions and Nonprofits only -
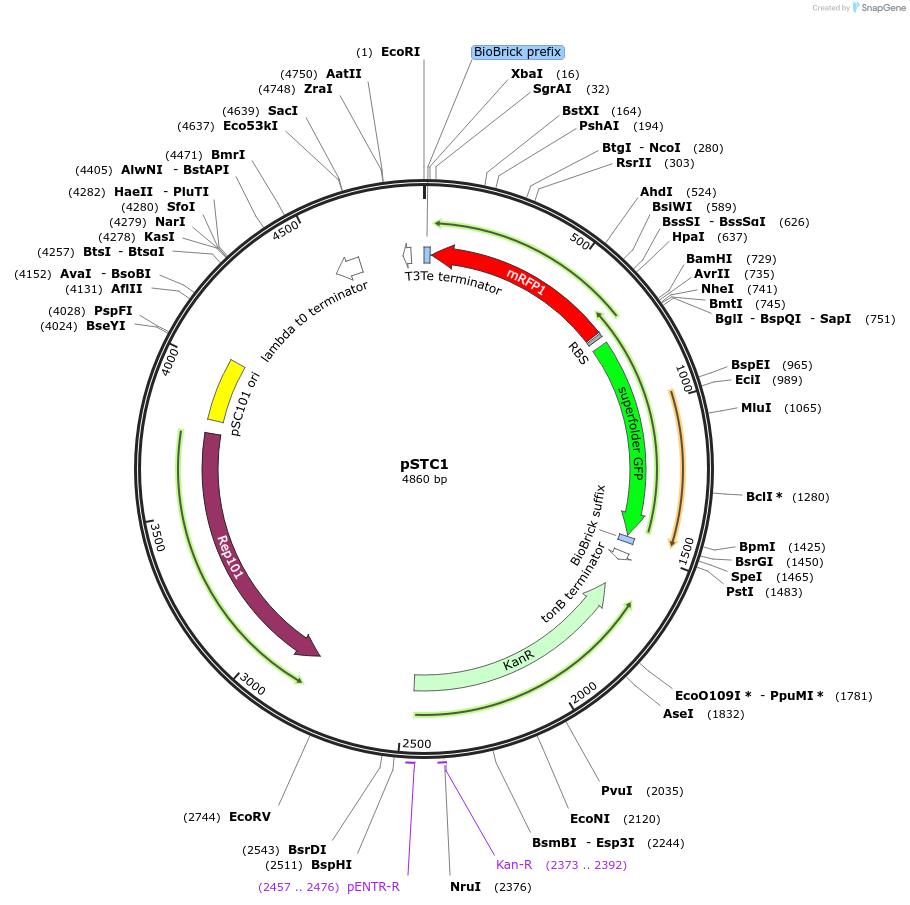
pSTC1
Plasmid#39241DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceSept. 18, 2012AvailabilityAcademic Institutions and Nonprofits only -
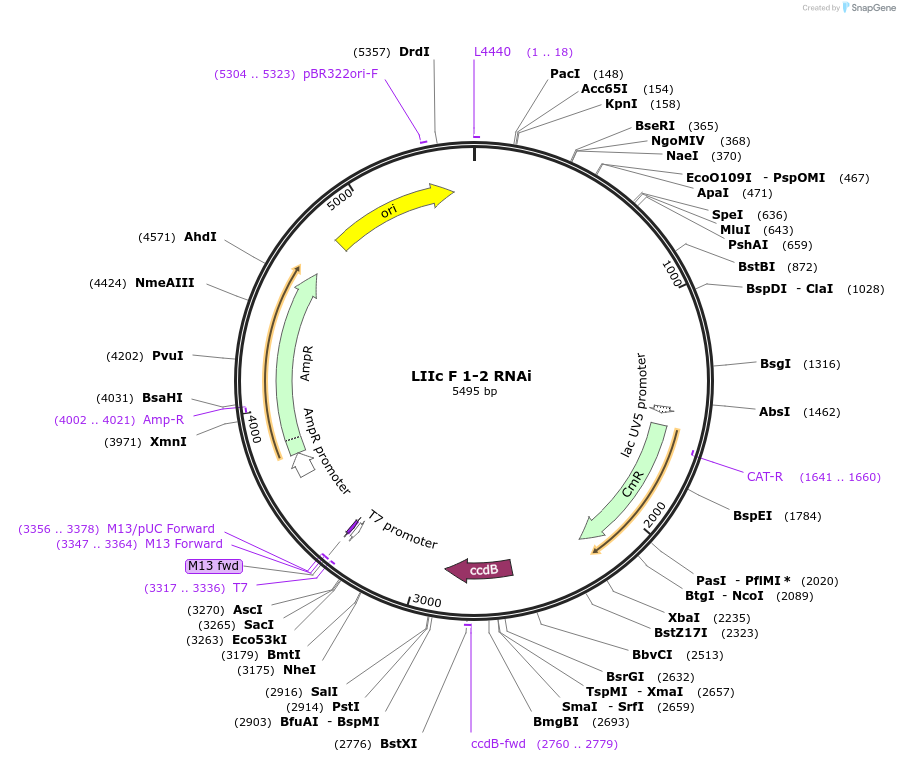
LIIc F 1-2 RNAi
Plasmid#54402DepositorTypeEmpty backboneUseCloning vectorAvailable SinceJuly 1, 2014AvailabilityAcademic Institutions and Nonprofits only -
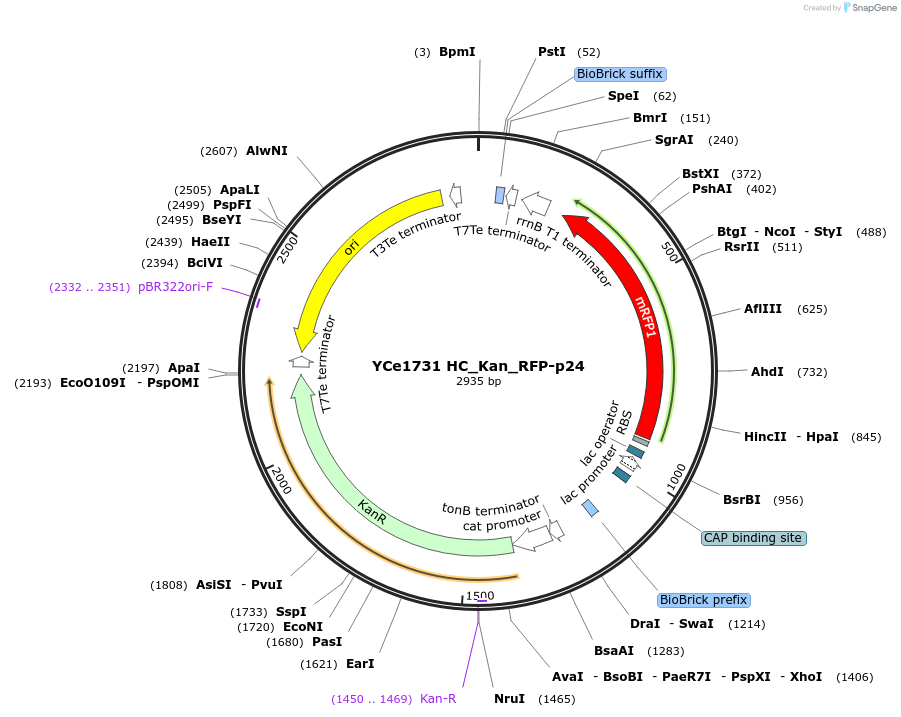
YCe1731 HC_Kan_RFP-p24
Plasmid#100634PurposeEMMA toolkit part-entry vector position 24DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
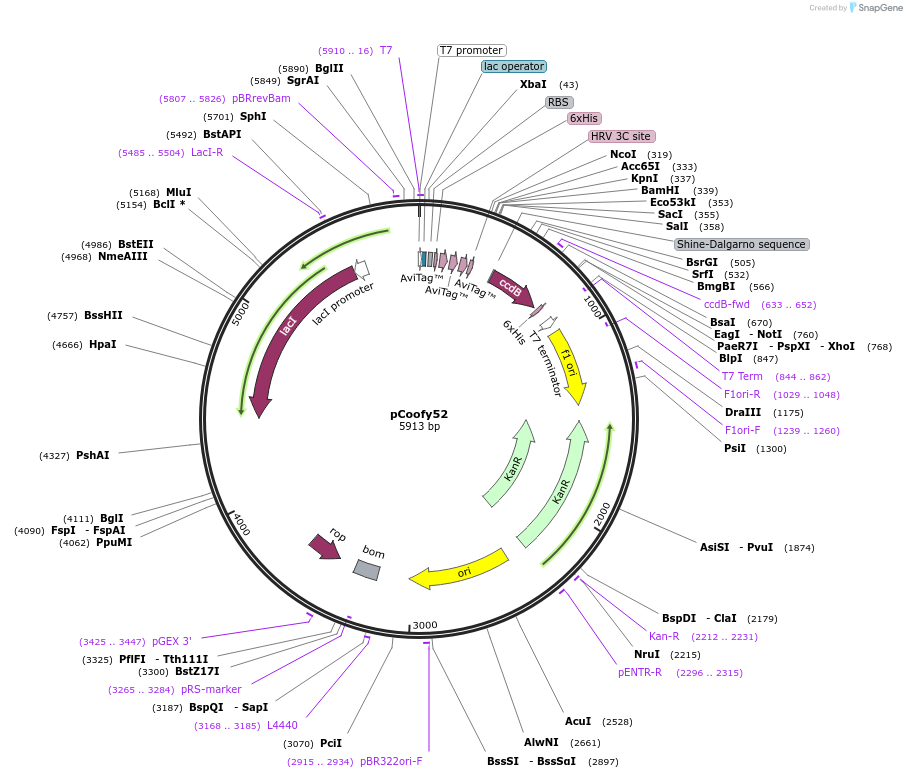
pCoofy52
Plasmid#122006PurposeE.coli vector for parallel SLIC cloning containing N-terminal His6-3xAvi with PreScission Cleavage siteDepositorTypeEmpty backboneTagsHis6-3xAviExpressionBacterialAvailable SinceJuly 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
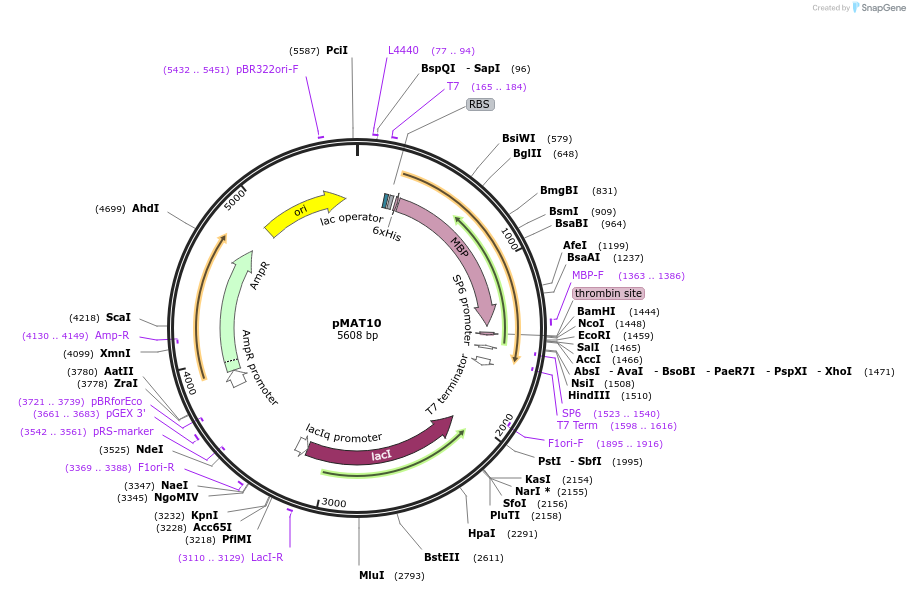
pMAT10
Plasmid#112591PurposeUsed for expression of protein in E. coli as Thrombin cleavable N-termina 6His-tag-MBP fusions. Like pMAT9, but without the second BamHI site in the polylinkerDepositorTypeEmpty backboneTags6His-MBPExpressionBacterialAvailable SinceAug. 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
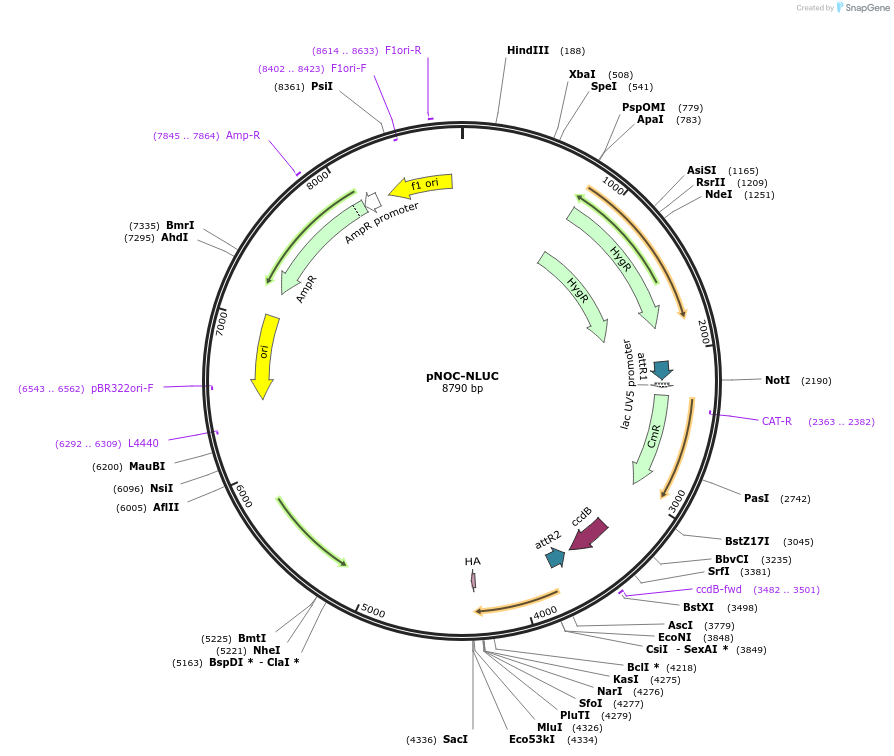
pNOC-NLUC
Plasmid#98141PurposeGateway compatible construct for N' terminal fusion or promoter driven NanoLuciferase reporter, hygromycin resistance cassetteDepositorTypeEmpty backboneUseAlgae, nannochloropsisTagsHAAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
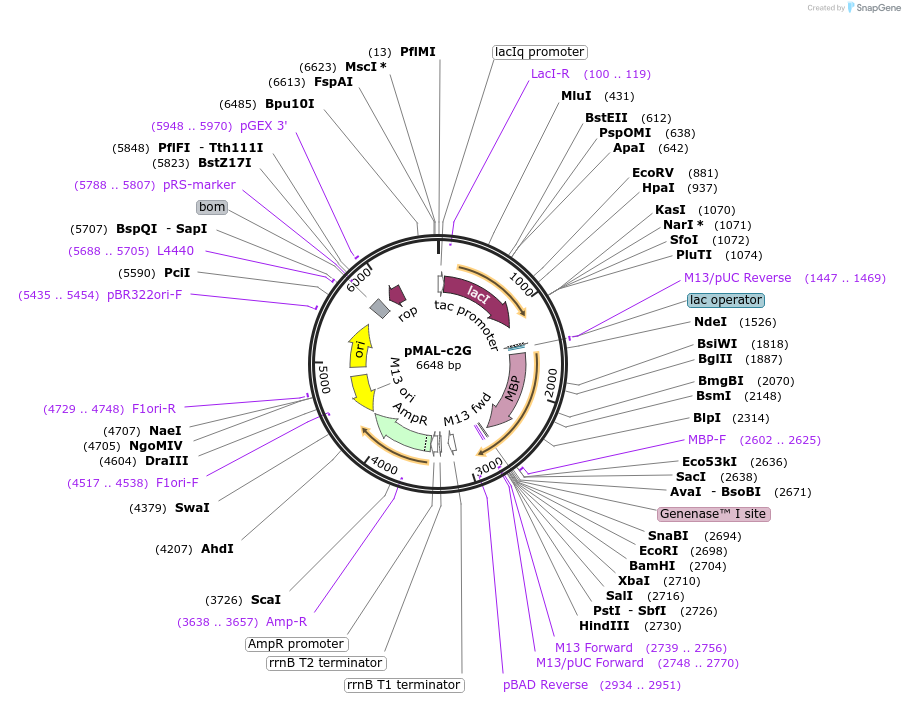
pMAL-c2G
Plasmid#75290PurposeExpresses proteins in the cytoplasm as fusions to maltose-binding proteinDepositorTypeEmpty backboneTagsmaltose-binding proteinExpressionBacterialPromotertacAvailable SinceJuly 20, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
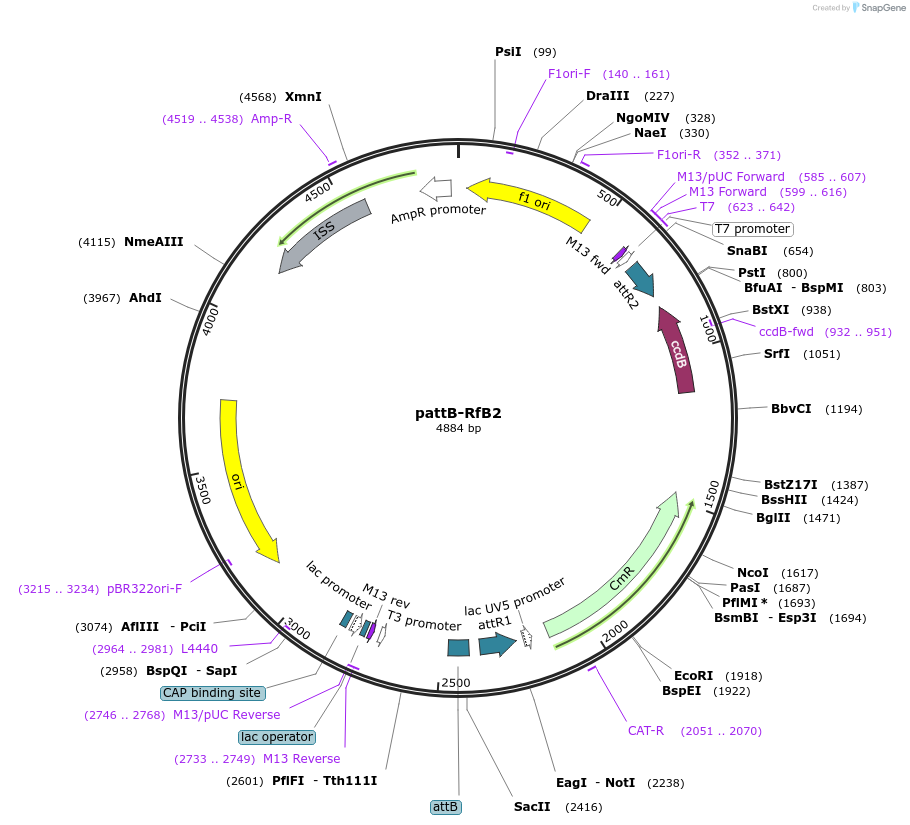
pattB-RfB2
Plasmid#62296Purposedocking site transgenesis plasmid for Gateway Cloning and insect transgenesisDepositorTypeEmpty backboneUseUnspecifiedAvailable SinceMay 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
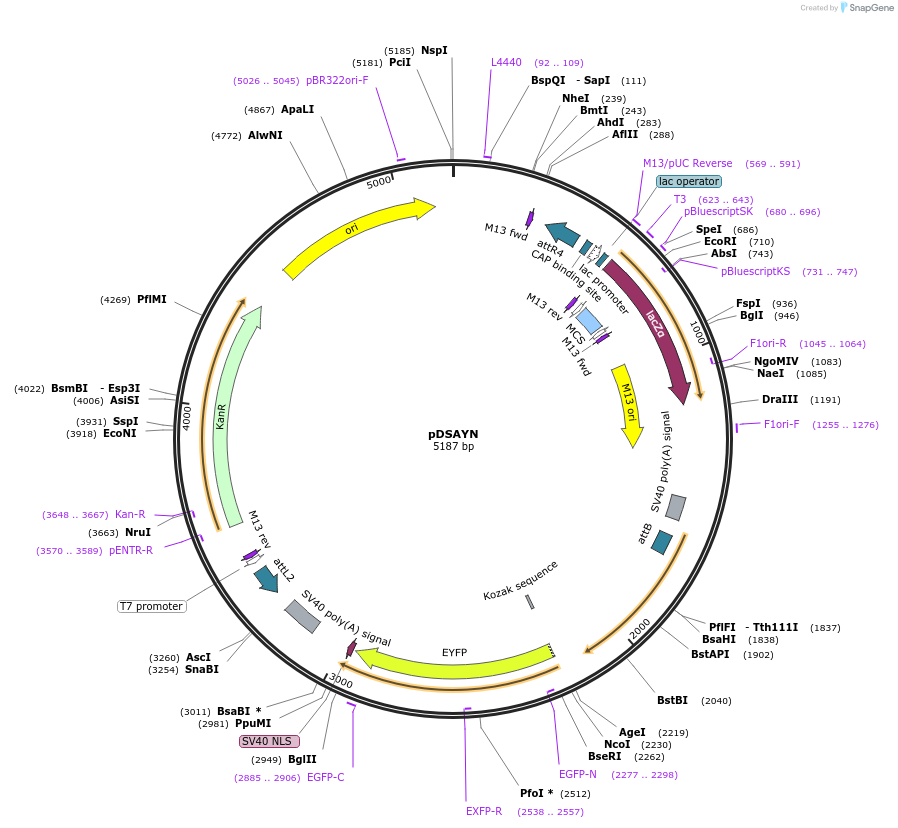
pDSAYN
Plasmid#62294Purposedocking site transgenesis plasmid for GoldenGate Cloning and insect transgenesis, YFP-NLS markerDepositorTypeEmpty backboneExpressionInsectAvailable SinceMarch 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
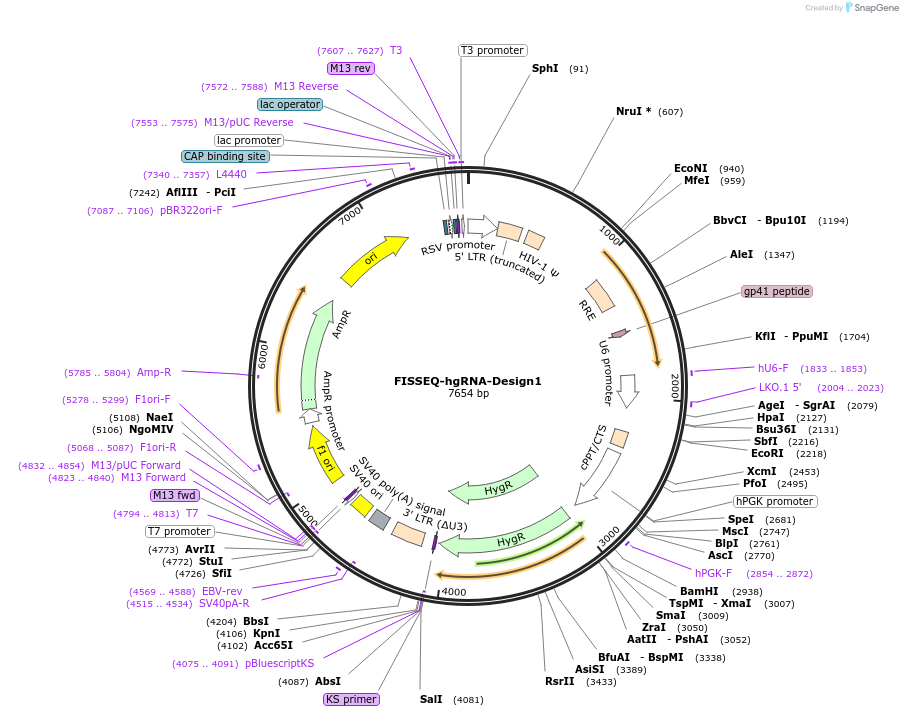
FISSEQ-hgRNA-Design1
Plasmid#100571PurposeFISSEQ-hgRNA-Design1 in a lentiviral backboneDepositorInsertFISSEQ-hgRNA-Design1
UseCRISPR, Lentiviral, and Synthetic BiologyExpressionMammalianAvailable SinceAug. 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
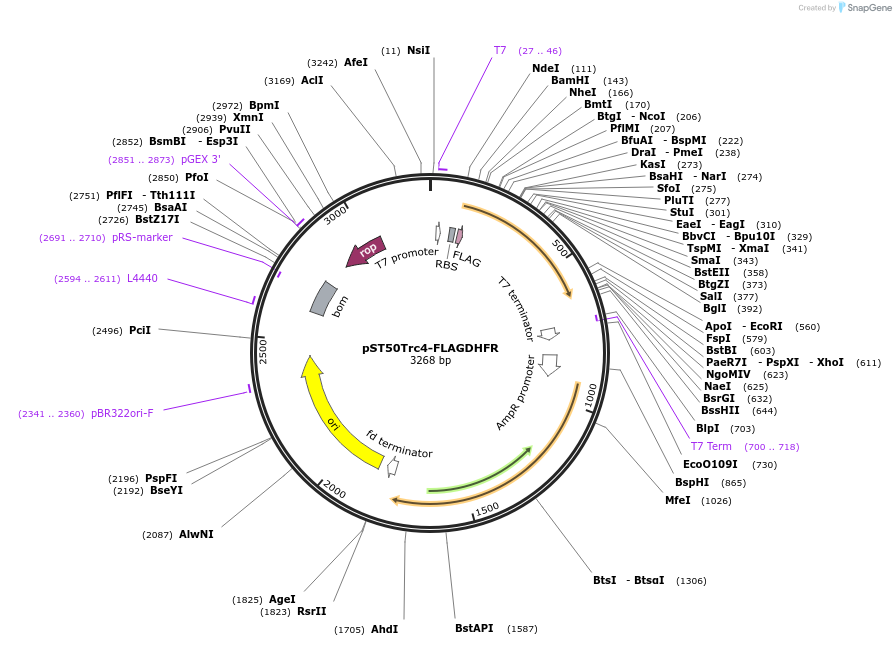
pST50Trc4-FLAGHISNDHFR
Plasmid#64001PurposePosition 4 transfer plasmid for pST44 polycistronic plasmid suite; N-term cleavable double tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTags6xHis & FLAG; TEV cleavableExpressionBacterialPromoterT7Available SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pST50Trc4-FLAGDHFR
Plasmid#63992PurposePosition 4 transfer plasmid for pST44 polycistronic plasmid suite; N-term non-cleavable single tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTagsFLAGExpressionBacterialPromoterT7Available SinceDec. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
pST50Trc3-HISDHFR
Plasmid#63971PurposePosition 3 transfer plasmid for pST44 polycistronic plasmid suite; N-term non-cleavable single tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTags6xHisExpressionBacterialPromoterT7Available SinceDec. 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
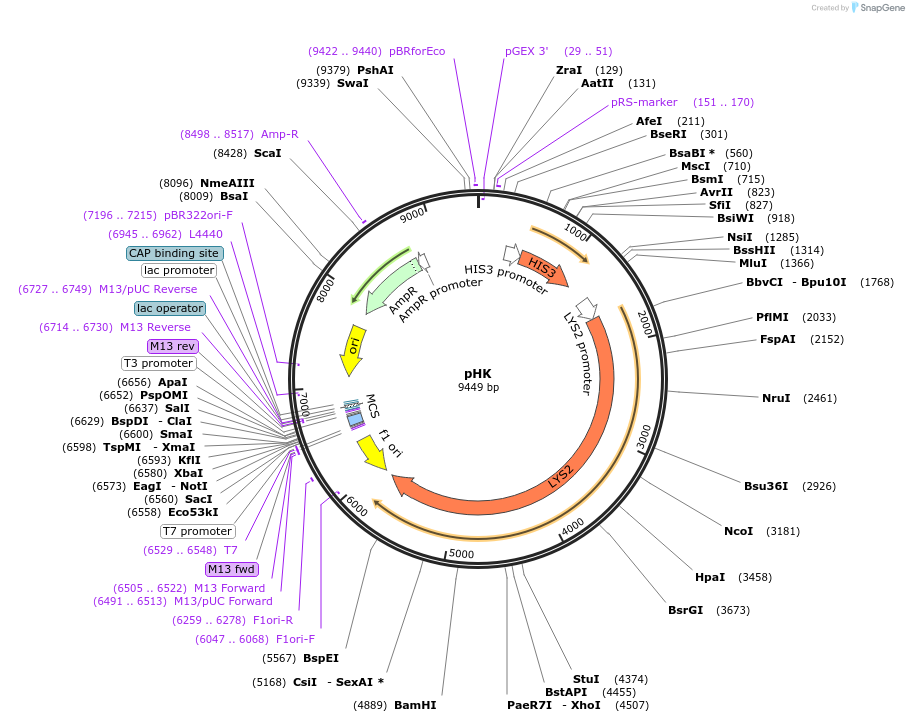
pHK
Plasmid#64187Purposeharbours marker genes HIS3 and LYS2DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceSept. 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
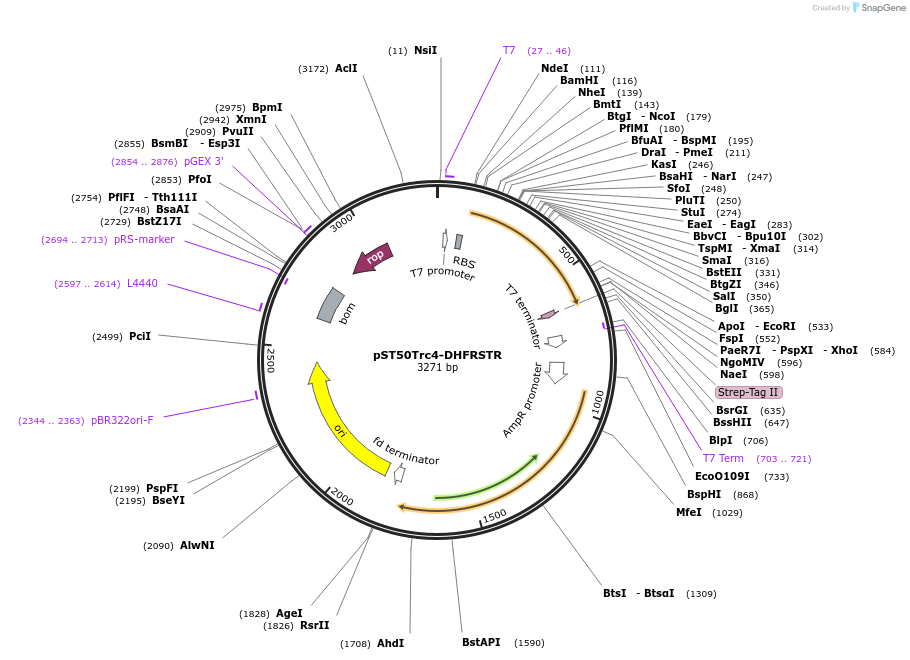
pST50Trc4-DHFRSTR
Plasmid#64004PurposePosition 4 transfer plasmid for pST44 polycistronic plasmid suite; C-term single tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTagsStrep II (STR)ExpressionBacterialPromoterT7Available SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
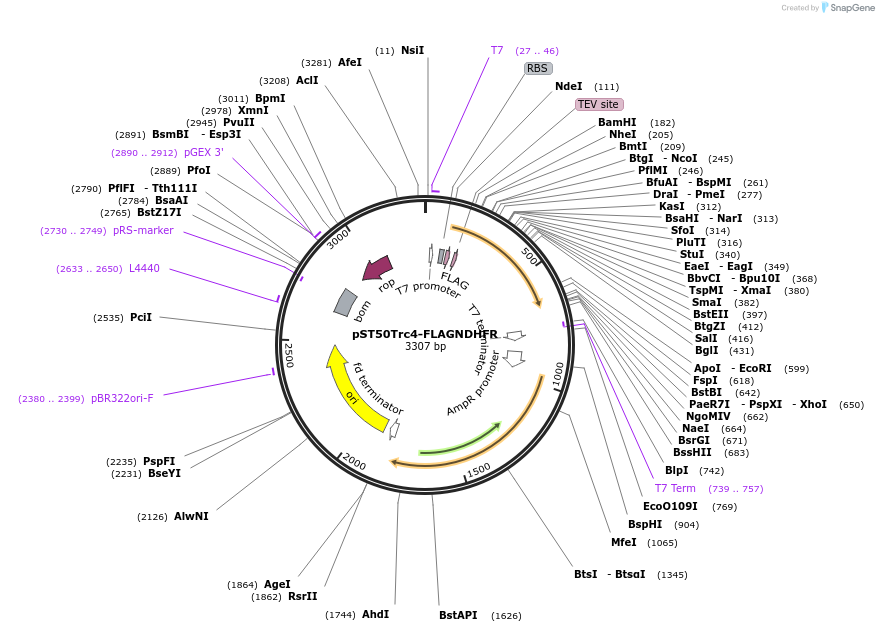
pST50Trc4-FLAGNDHFR
Plasmid#63997PurposePosition 4 transfer plasmid for pST44 polycistronic plasmid suite; N-term cleavable single tag; contains DHFR gene in BamHI-NgoMIV cassette as positive controlDepositorTypeEmpty backboneTagsFLAG; TEV cleavableExpressionBacterialPromoterT7Available SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
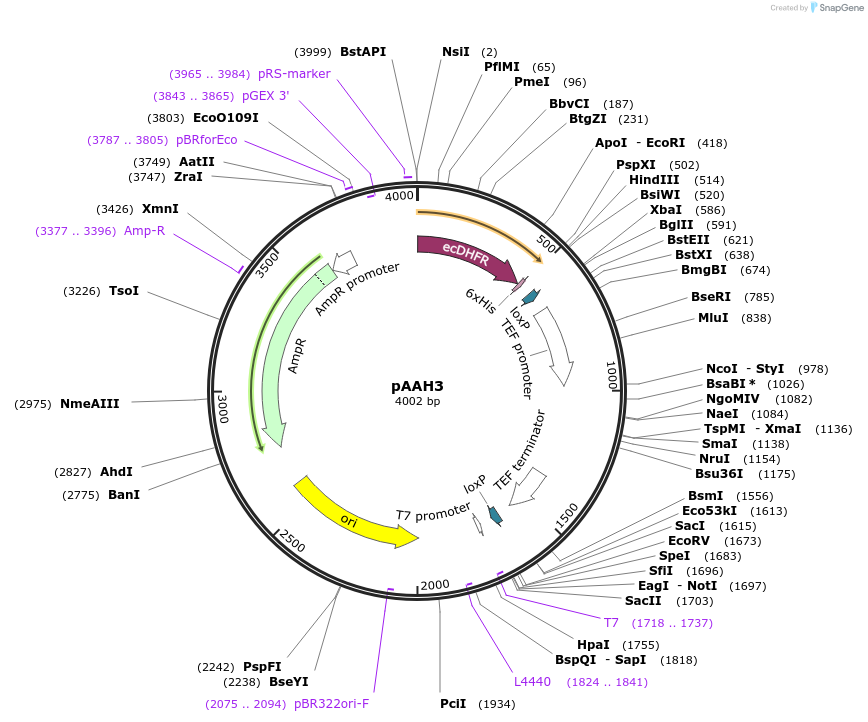
pAAH3
Plasmid#41827PurposeUsed as a PCR template for tagging yeast proteins with the DHFR tag (c-terminal histag) by homologous recombination and selection for phleomycin resistance.DepositorTypeEmpty backboneUsePcr templateExpressionYeastPromoterTEFAvailable SinceOct. 29, 2013AvailabilityAcademic Institutions and Nonprofits only -
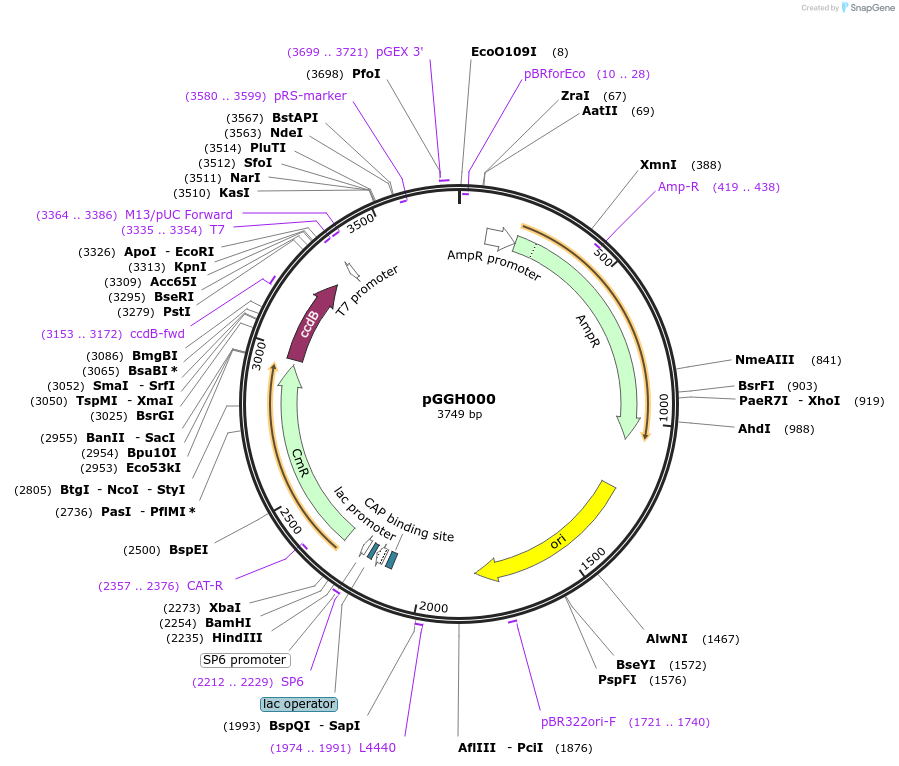
pGGH000
Plasmid#48862PurposeEmpty entry vector for creating GreenGate N-terminal tag + coding sequence + C-terminal tag + terminator modules.DepositorTypeEmpty backboneUseGolden gate compatible cloning vectorAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
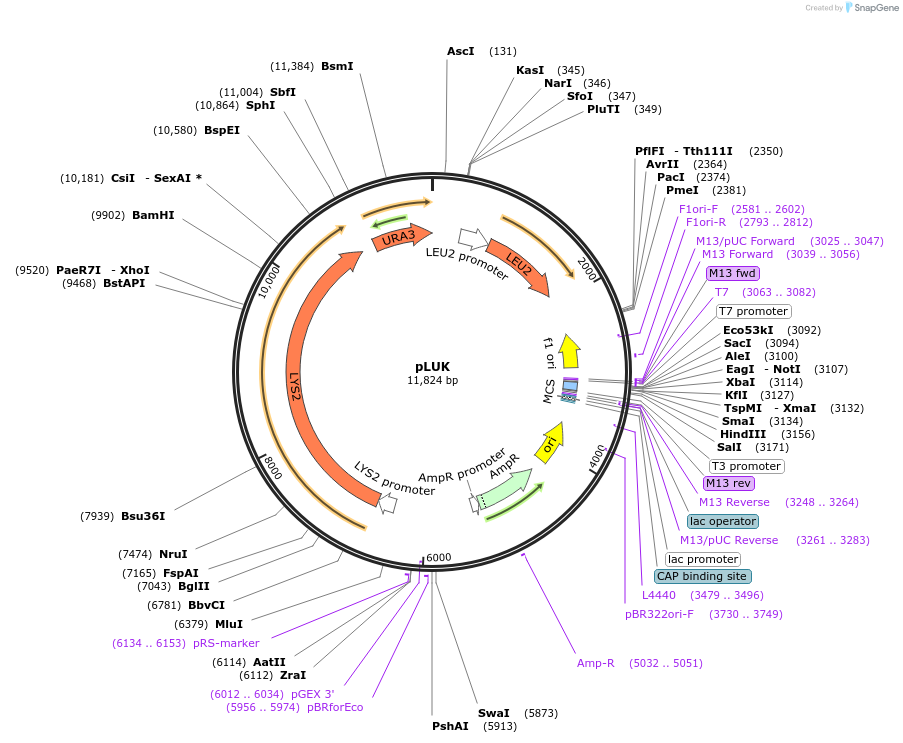
pLUK
Plasmid#64184Purposeharbours marker genes LEU2, URA3 and LYS2DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceAug. 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
TAL-3TA4
Plasmid#41472DepositorInsert3TA4
UsePcr cloning vectorAvailable SinceJan. 2, 2013AvailabilityAcademic Institutions and Nonprofits only -
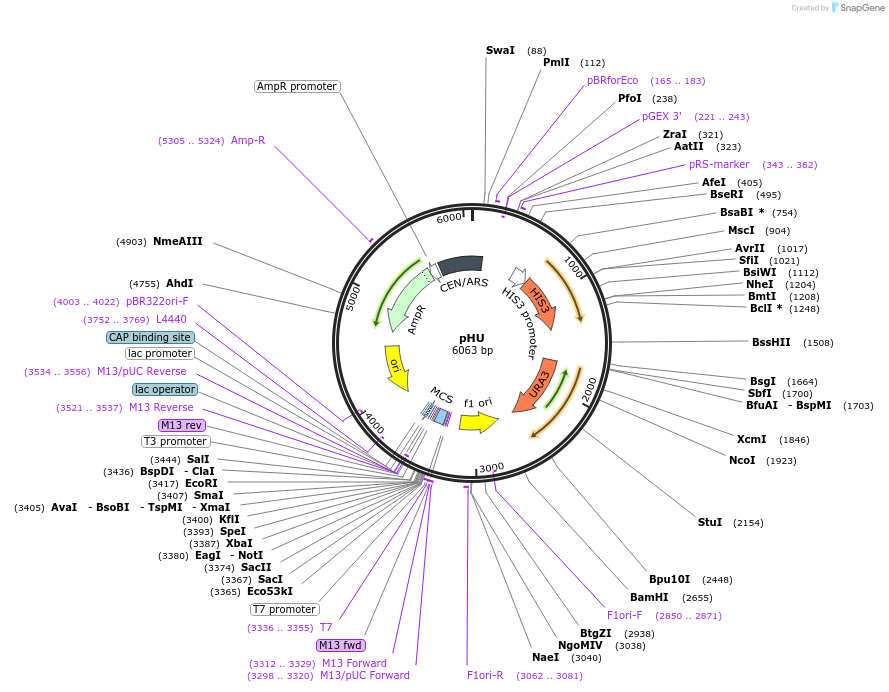
pHU
Plasmid#64172Purposeharbours marker genes HIS3 and URA3DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceSept. 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
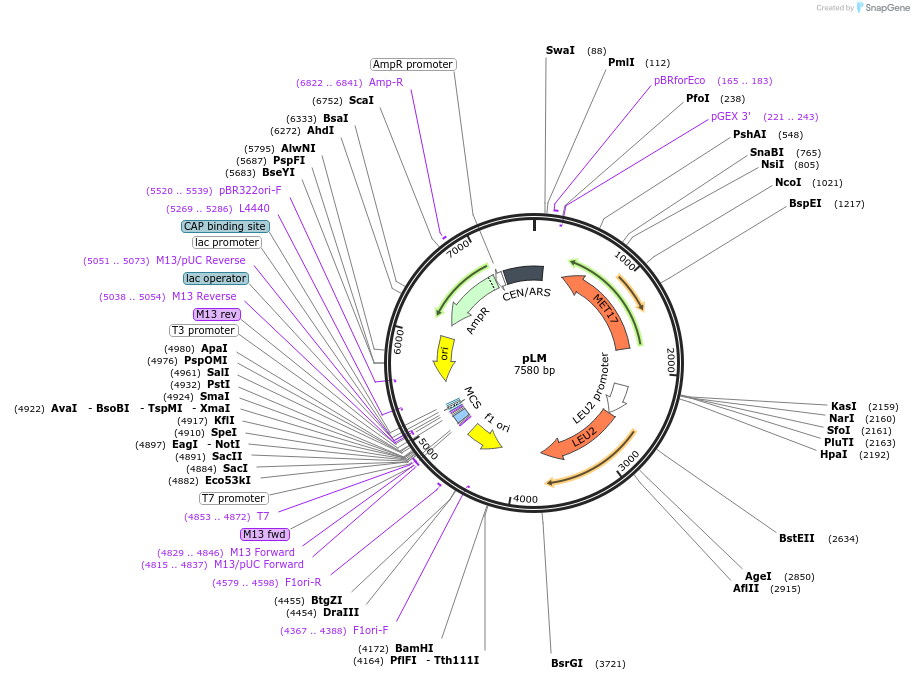
pLM
Plasmid#64175Purposeharbours marker genes LEU2 and MET17DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceAug. 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
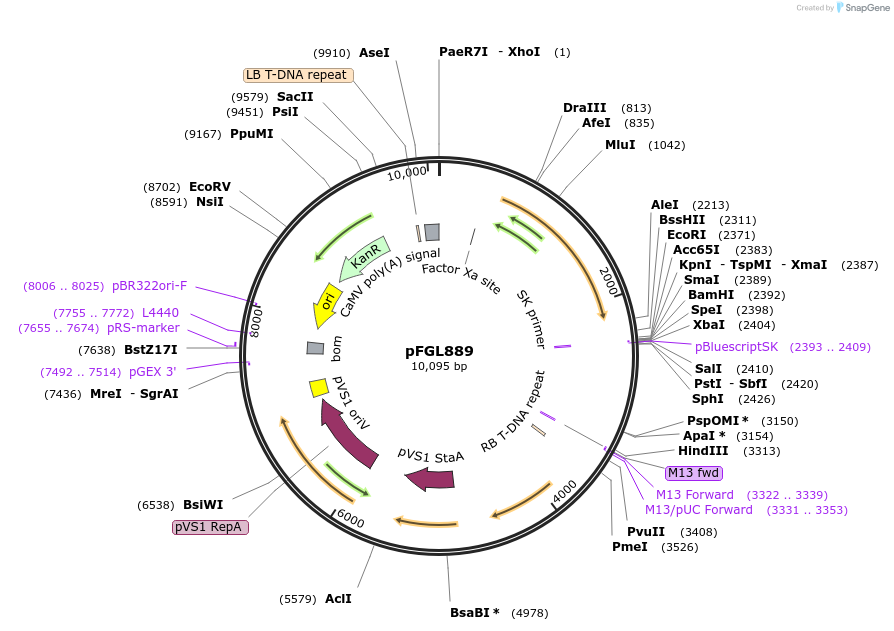
pFGL889
Plasmid#52321PurposeIntroducing ectopic integration at the defined fungal ILV2-locus, based on SRR (Sulfonylurea Resistance Reconstitution).DepositorTypeEmpty backboneUseAgrobacteriumExpressionBacterialAvailable SinceJuly 2, 2014AvailabilityAcademic Institutions and Nonprofits only