We narrowed to 20,614 results for: ACE
-
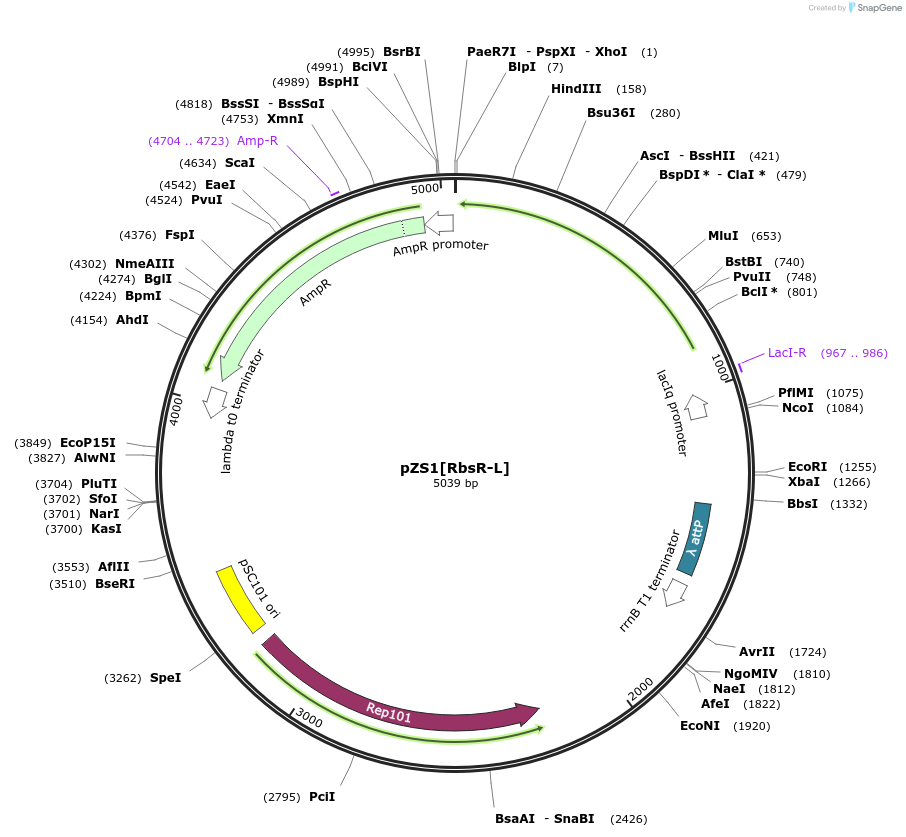
Plasmid#60741PurposeContains PIq driving expression RbsR-L, the Ribose inducible chimera with the wt Lac DBD.DepositorInsertRbsR-L
UseSynthetic BiologyExpressionBacterialMutationLacI/GalR repressor chimera. LacI DBD with RbsR L…Available SinceApril 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
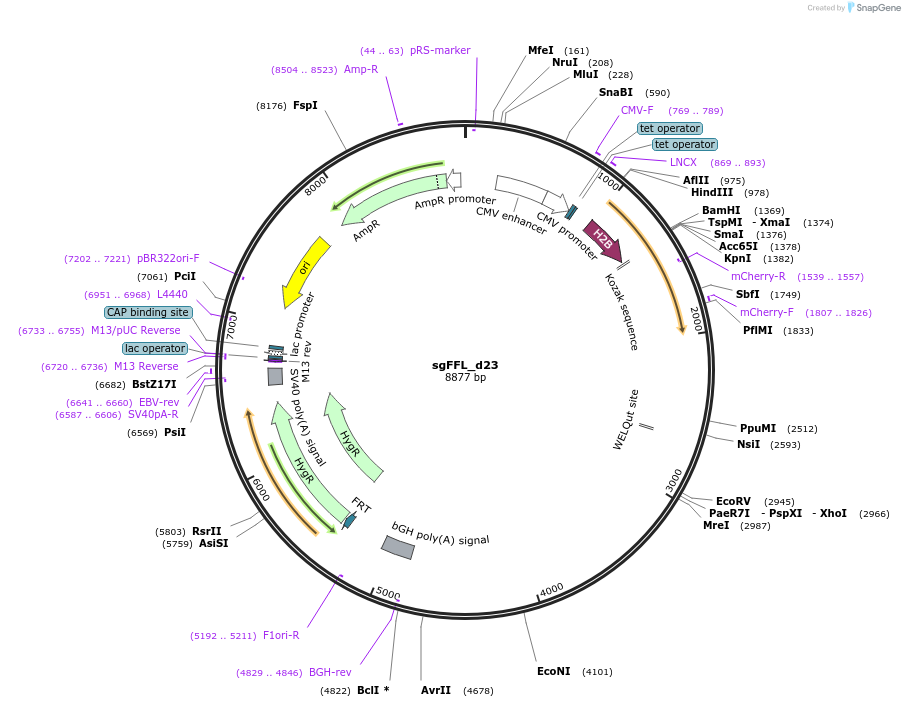
sgFFL_d23
Plasmid#59892Purposesynthetic autoregulatory gene circuit made by inserting an intron containing the mouse mir-124–3 gene into mCherry. Contains the miR-124-regulated 3′UTR of the Vamp3 gene with two seed sites mutatedDepositorInsertmCherry with intron containing the mouse mir-124–3
TagsPEST and VAMP 3 UTRExpressionMammalianMutationVAMP 3 UTR--3rd and fourth seed site removedPromoterdoxycycline (Dox)-inducible promoter (CMV/TO)Available SinceApril 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
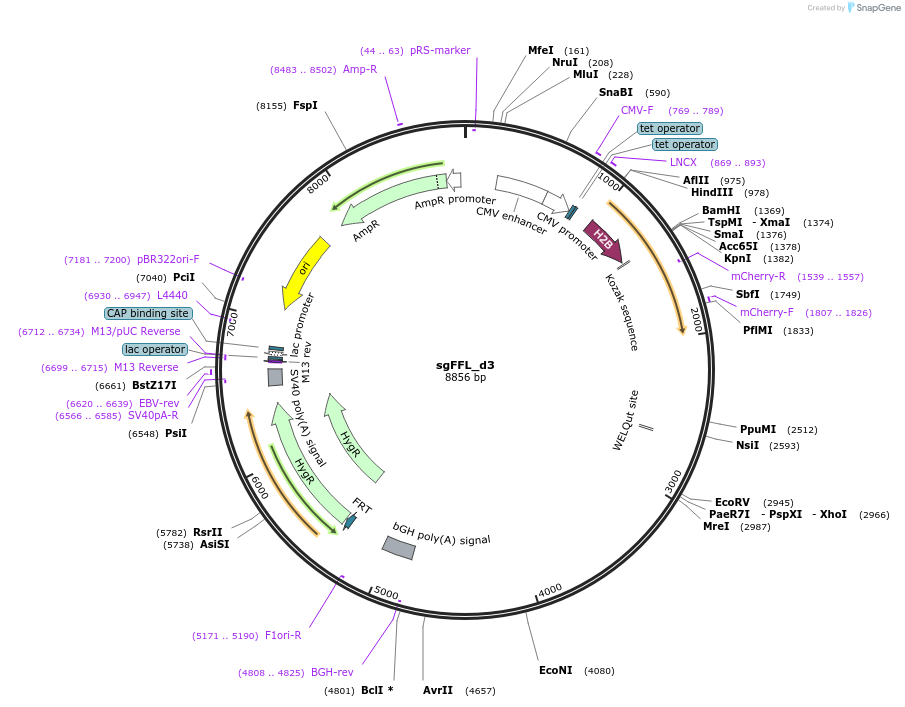
sgFFL_d3
Plasmid#59891Purposesynthetic autoregulatory gene circuit made by inserting an intron containing the mouse mir-124–3 gene into mCherry. Contains the miR-124-regulated 3′UTR of the Vamp3 gene with one seed site mutated.DepositorInsertmCherry with intron containing the mouse mir-124–3
TagsPEST and VAMP 3 UTRExpressionMammalianMutationVAMP 3 UTR--3rd seed site removedPromoterdoxycycline (Dox)-inducible promoter (CMV/TO)Available SinceApril 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
5mARF17 pBluescript
Plasmid#12069DepositorInsert5mARF17 (ARF17 Mustard Weed)
ExpressionBacterialMutationmiR160-resistant version of ARF17 has five silent…Available SinceJune 2, 2006AvailabilityAcademic Institutions and Nonprofits only -
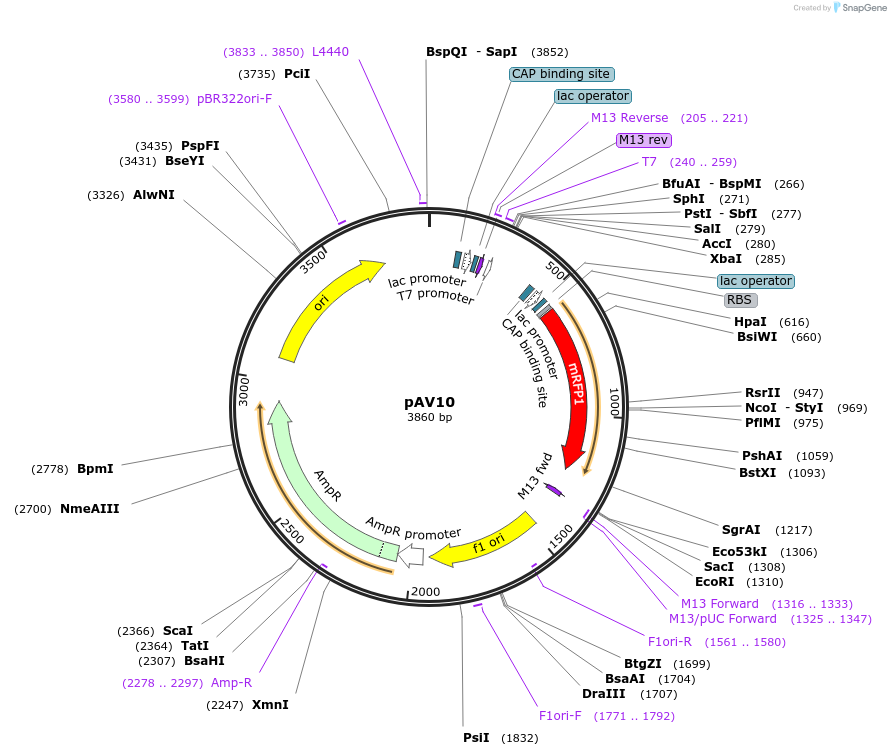
pAV10
Plasmid#63213PurposepUC19 backbone (Amp), contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. NotI or FseI sites for TU digest.DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailabilityAcademic Institutions and Nonprofits only -
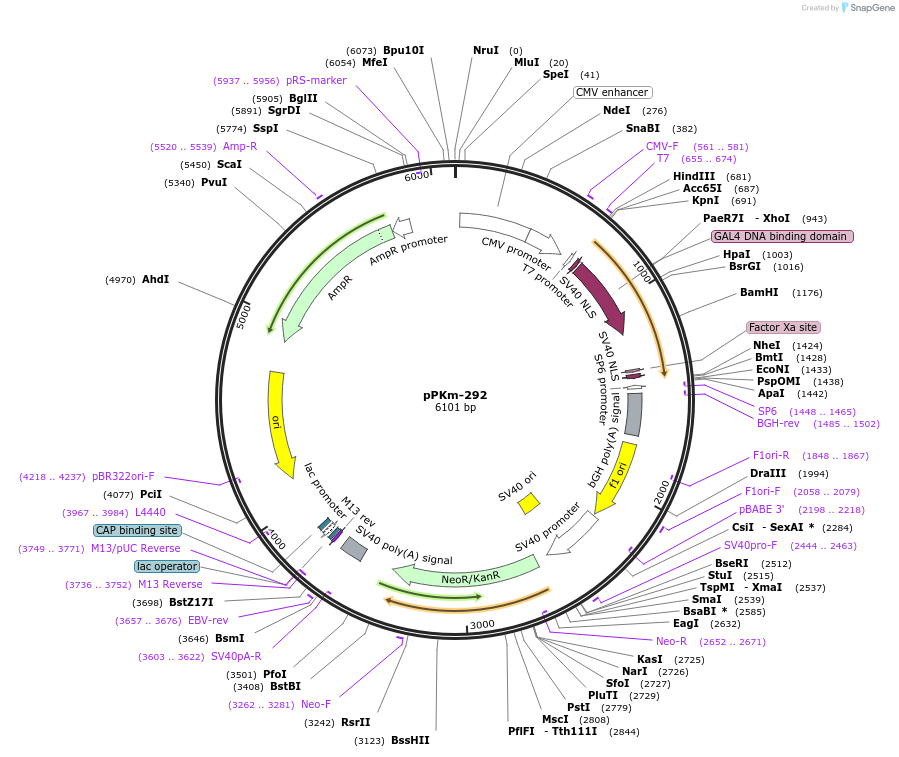
pPKm-292
Plasmid#105816PurposepcDNA - GAL4 DBD - MTAD. Expresses Gal4 DNA binding domain fused to MTADDepositorInsertGAL4 DBD and MTAD
ExpressionMammalianPromoterCMVAvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 1 (positions 333-436)
Pooled Library#174293PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
C-5545 ∆cosδε
Bacterial Strain#209018PurposeDeficient in the P2 bacteriophage packaging cos site and encoding rhamnose-inducible P4 delta (δ) and epsilon (ε) genesDepositorBacterial ResistanceHygromycinSpeciesEscherichia coliAvailable SinceJan. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
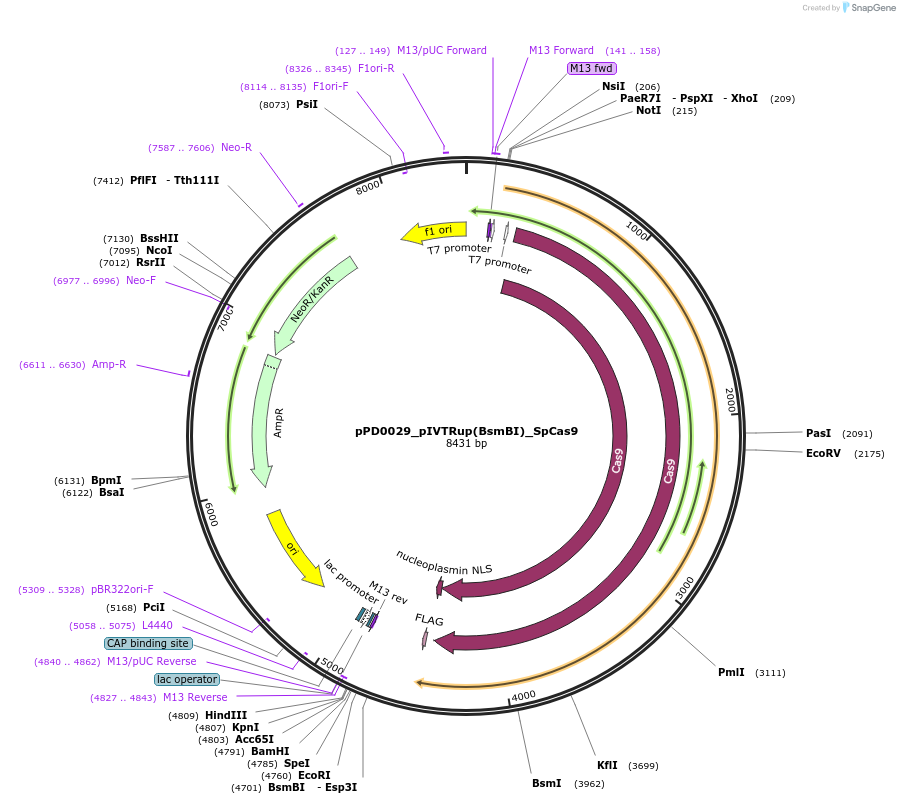
pPD0029_pIVTRup(BsmBI)_SpCas9
Plasmid#242535PurposeIVTDepositorInsertSpCas9
UseCRISPRAvailable SinceNov. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
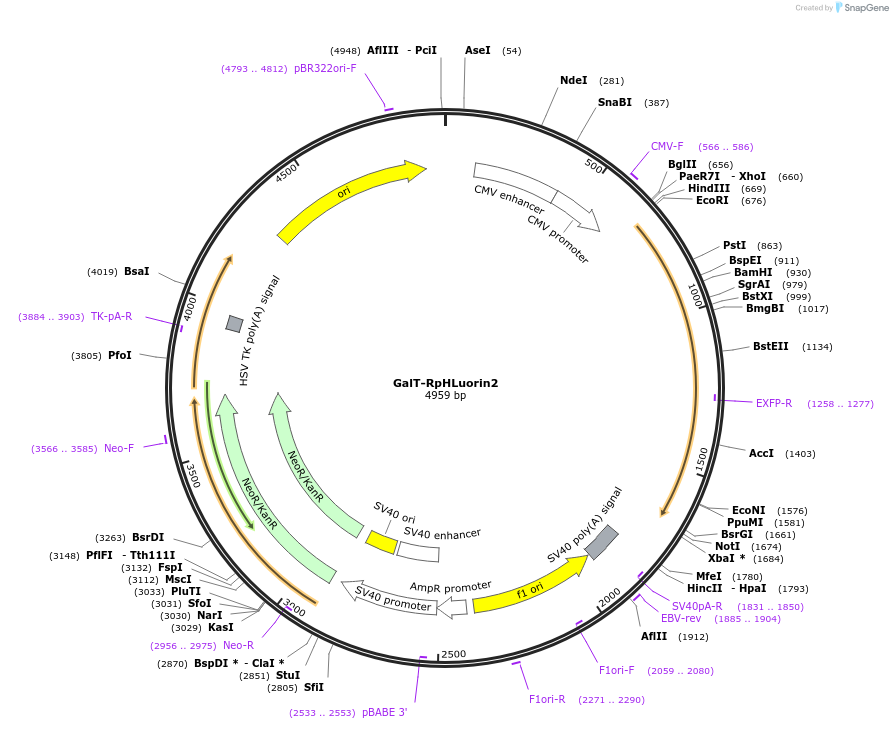
GalT-RpHLuorin2
Plasmid#171719PurposeTrans-Golgi expression of ratiometric pHLuorin2DepositorInsertRatiometric pHLuorin2 fused to B4GALT1 (B4GALT1 Human)
Tagsratiometric pHluorin2ExpressionMammalianPromoterCMVAvailable SinceMarch 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
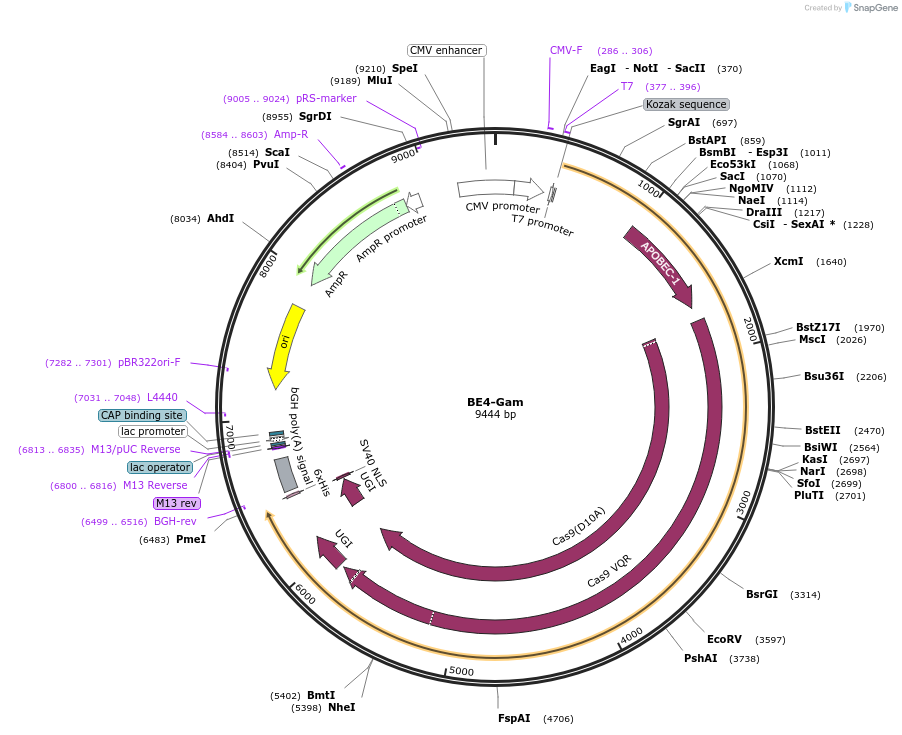
BE4-Gam
Plasmid#100806PurposeExpresses BE4-Gam in mammalian cellsDepositorInsertBE4-Gam
ExpressionMammalianPromoterCMVAvailable SinceAug. 31, 2017AvailabilityAcademic Institutions and Nonprofits only -
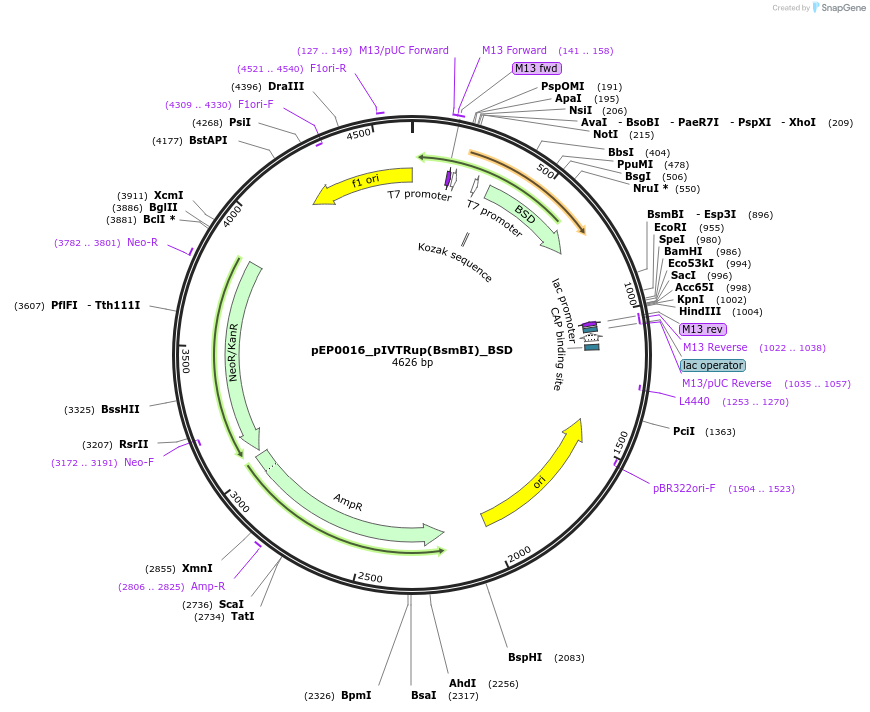
pEP0016_pIVTRup(BsmBI)_BSD
Plasmid#242546PurposeIVTDepositorInsertBSD (Blasticidin S deaminase)
UseOtherAvailable SinceNov. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
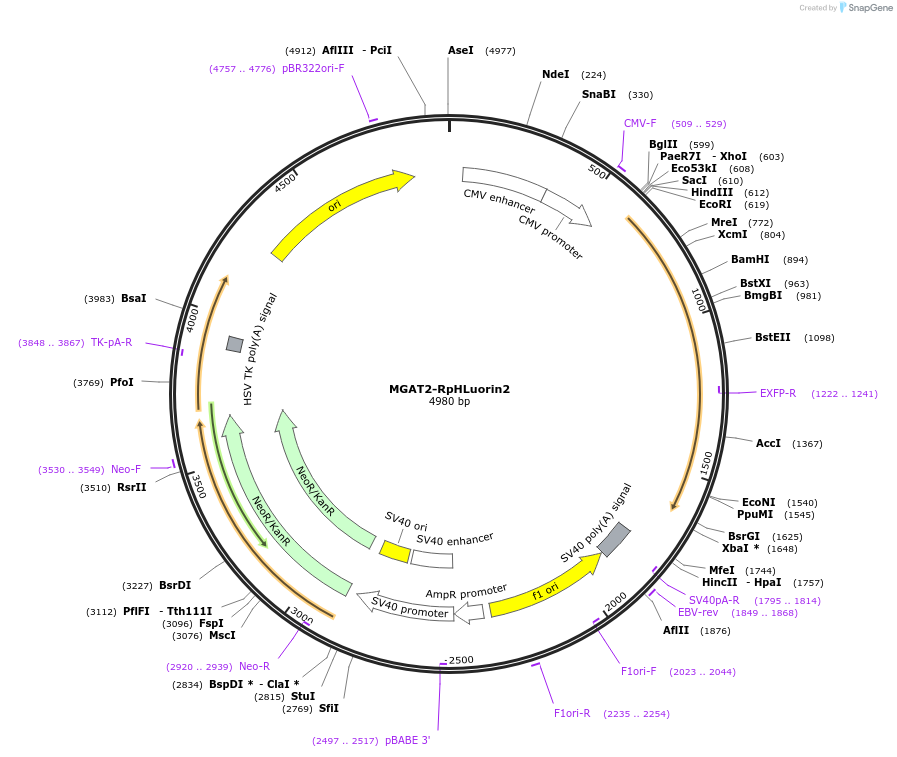
MGAT2-RpHLuorin2
Plasmid#171718PurposeCis-/medial-Golgi expression of ratiometric pHLuorin2DepositorInsertRatiometric pHLuorin2 fused to MGAT2 (MGAT2 Human)
Tagsratiometric pHluorin2ExpressionMammalianPromoterCMVAvailable SinceMarch 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
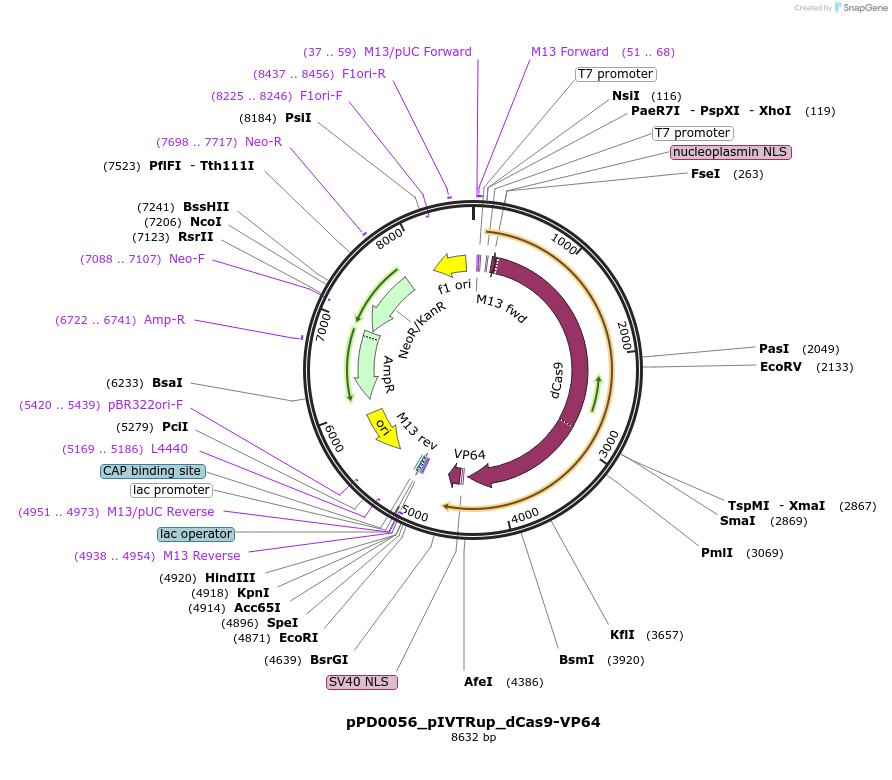
pPD0056_pIVTRup_dCas9-VP64
Plasmid#242542PurposeIVTDepositorInsertdCas9-VP64
UseCRISPRMutationD10AAvailable SinceNov. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
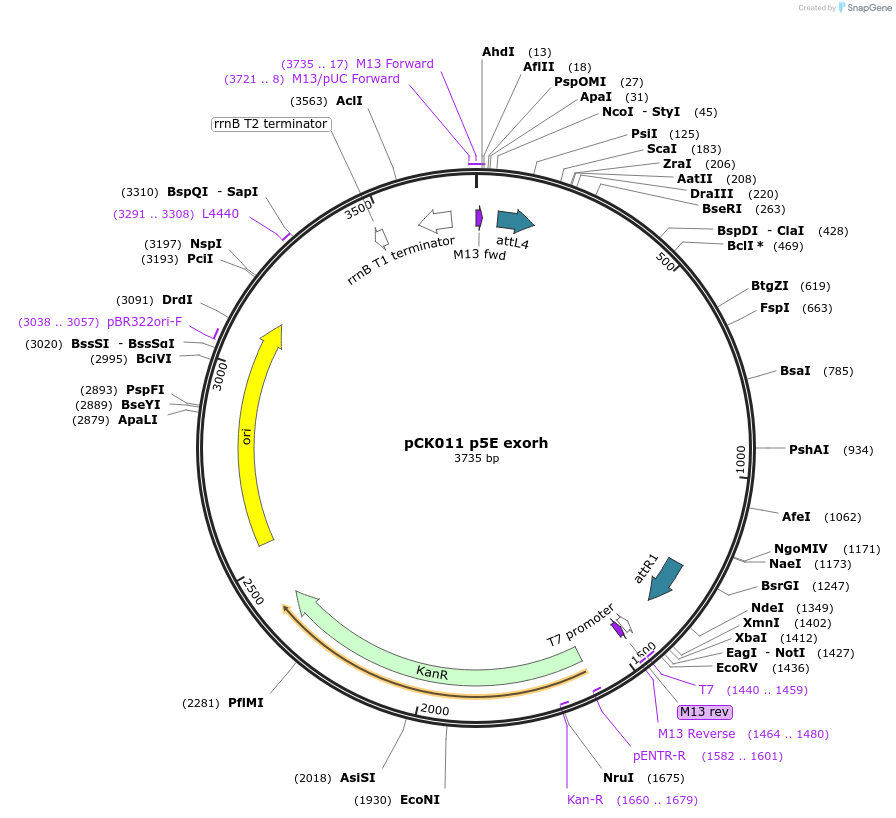
pCK011 p5E exorh
Plasmid#195949PurposeGateway p5E vectorDepositorInsertexorh regulatory elements
UseOtherAvailable SinceFeb. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
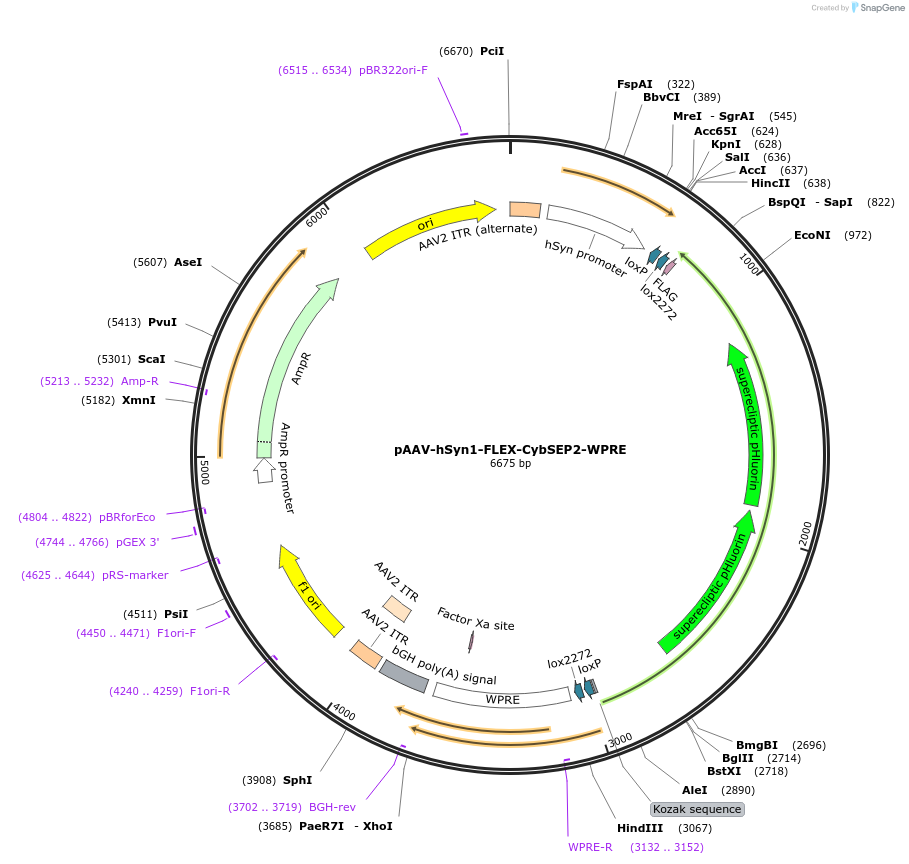
pAAV-hSyn1-FLEX-CybSEP2-WPRE
Plasmid#207661PurposeMonitoring neuropeptide releaseDepositorInsertCytochrome b-561 (Cyb561 Mouse)
UseAAVMutationHistidine 86 and 159 to alanines, inserted 2 X SE…PromoterhSynapsinAvailable SinceDec. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
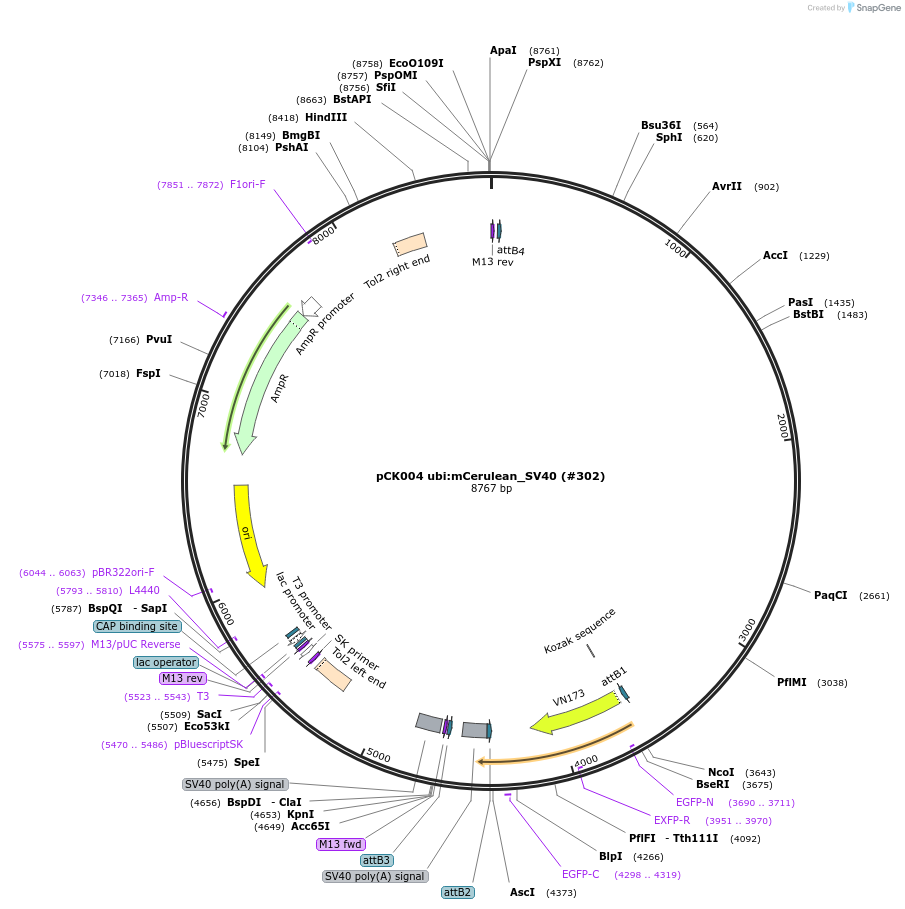
pCK004 ubi:mCerulean_SV40 (#302)
Plasmid#195978PurposeExpression constructDepositorInsertubi:mCerulean
UseOtherAvailable SinceFeb. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
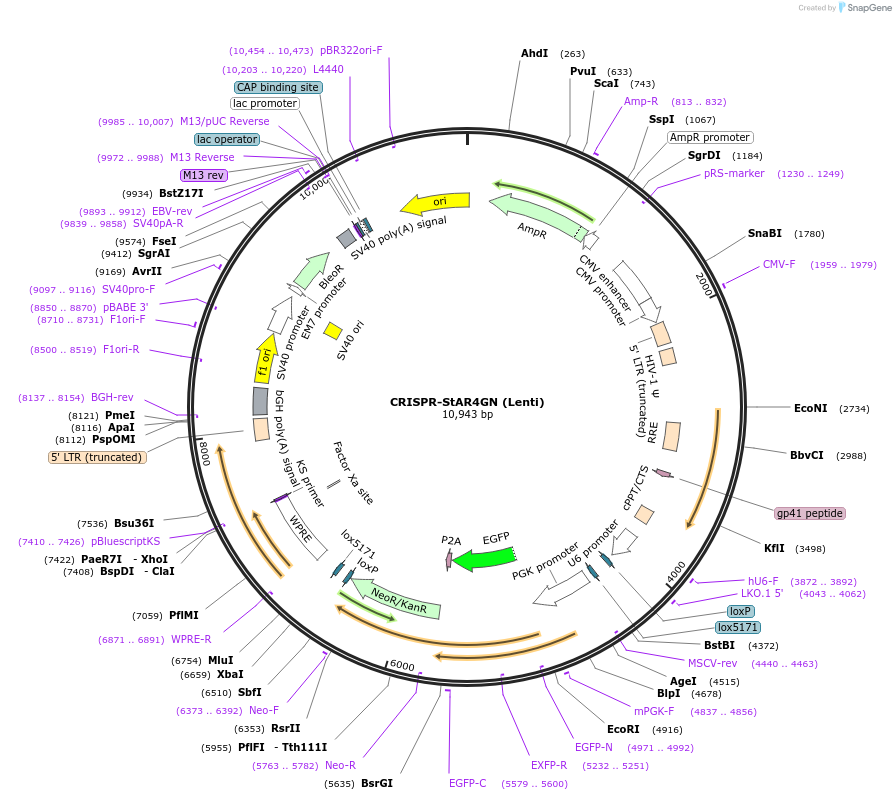
CRISPR-StAR4GN (Lenti)
Plasmid#222693PurposeCRISPR-screeningDepositorTypeEmpty backboneUseCRISPR and LentiviralTagsGFPExpressionMammalianMutationPGK without BsmBI cutsiteAvailable SinceDec. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
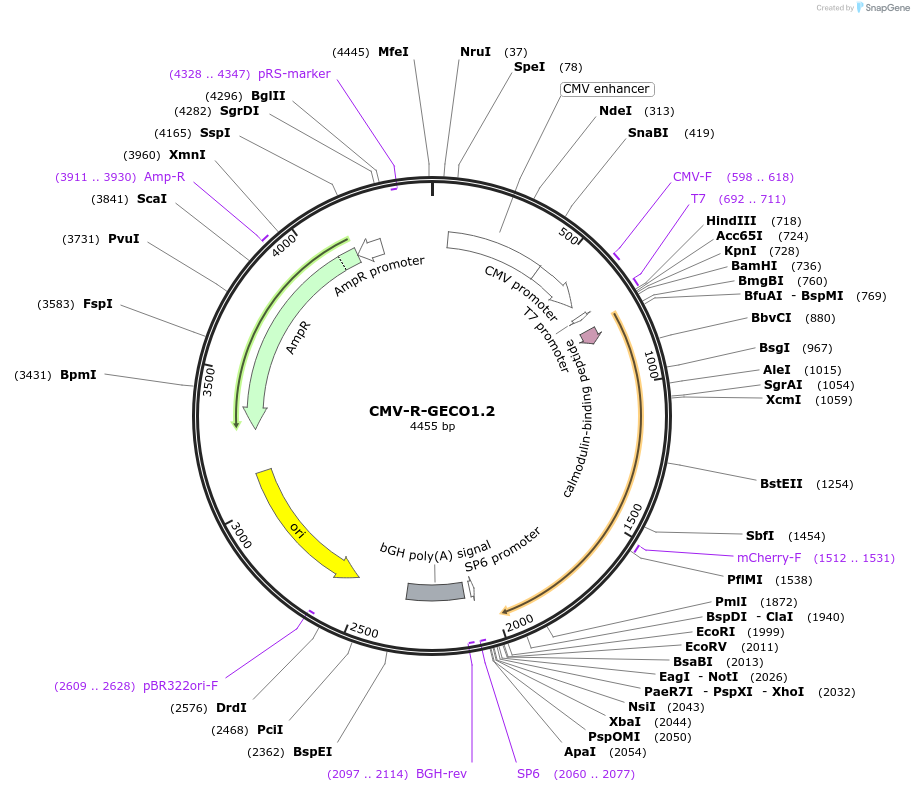
CMV-R-GECO1.2
Plasmid#45494PurposeRed intensiometric genetically encoded Ca2+-indicators for optical imaging 1.2DepositorInsertR-GECO1.2
ExpressionMammalianMutationR-GECO1 M164R/I166V/V174L/F222L/N267S/S270T/I330M…PromoterCMVAvailable SinceJuly 23, 2013AvailabilityAcademic Institutions and Nonprofits only -
1477 pcDNA3 flag HA Akt1
Plasmid#9021DepositorAvailable SinceNov. 2, 2005AvailabilityAcademic Institutions and Nonprofits only -
pX330-PITCh-ARL13B
Plasmid#227276PurposeExpresses SpCas9, the PITCh gRNA, and a sgRNA targeting the C-terminus of ARL13B for knock-in.DepositorInsertsgRNA Targeting C-terminus of ARL13B (ARL13B Human)
UseCRISPRExpressionMammalianPromoterU6Available SinceNov. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
pX330-PITCh-TOMM20
Plasmid#207789PurposeExpresses SpCas9, the PITCh gRNA, and a sgRNA targeting the C-terminus of TOMM20 for knock-in.DepositorInsertsgRNA Targeting C-terminus of TOMM20 (TOMM20 Human)
UseCRISPRExpressionMammalianPromoterU6Available SinceDec. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
pSC-cUnaG-P2A-nUnaG-ST-P2A-mCh
Plasmid#207634PurposeA plasmid encoding photocaged SpyCatcher (pSC)-cUnaG, nUnaG-SpyTag (ST), and mCherry separated by P2A cleaving sequences for mammalian expression.DepositorInsertphotocaged SpyCatcher-cUnaG-P2A-nUnaG-SpyTag-P2A-mCh
ExpressionMammalianMutationAmber stop codon at SC's critical lysinePromoterCMVAvailable SinceNov. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
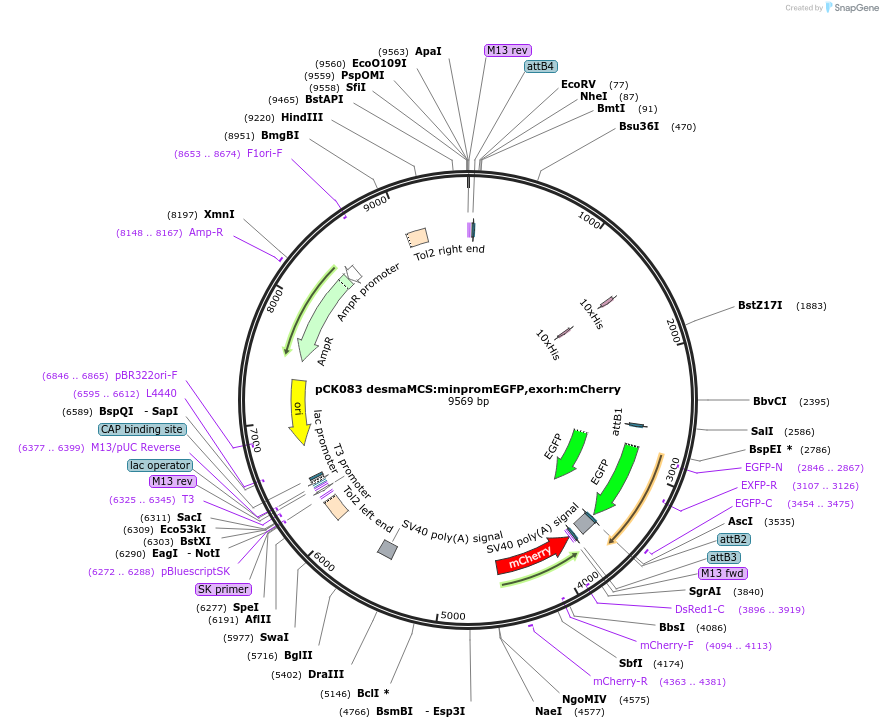
pCK083 desmaMCS:minpromEGFP,exorh:mCherry
Plasmid#200016PurposeExpression construct with desmin a regulatory elements, mouse beta-globin minimal promoter EGFP, exorh:mCherryDepositorTypeEmpty backboneUseZebrafish expressionAvailable SinceDec. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
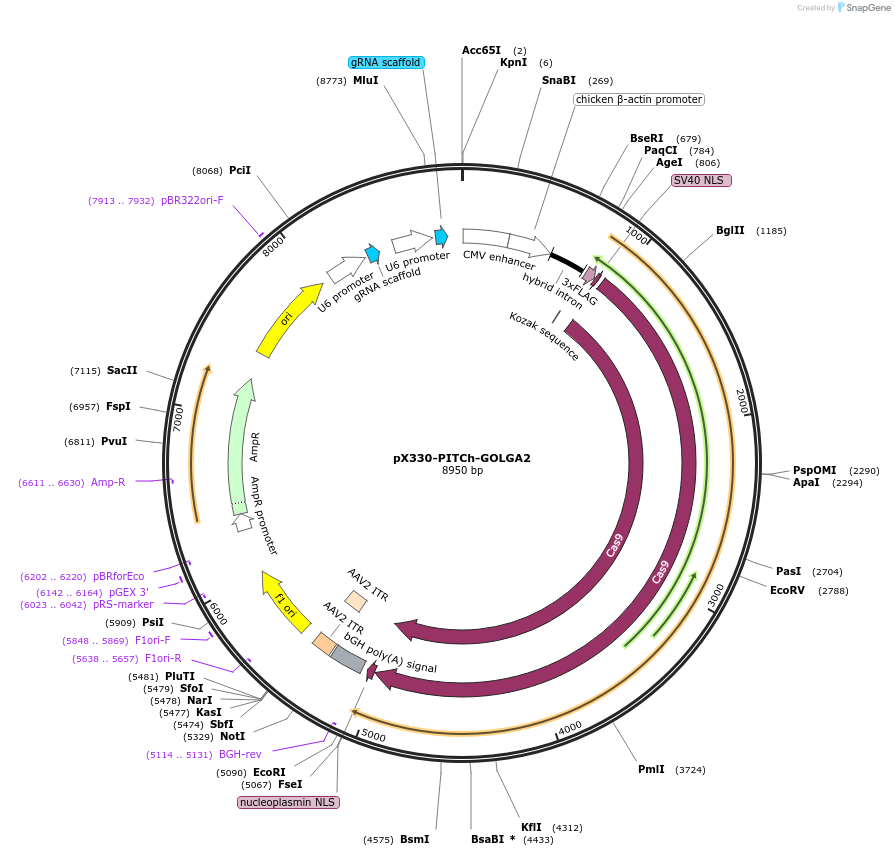
pX330-PITCh-GOLGA2
Plasmid#207791PurposeExpresses SpCas9, the PITCh gRNA, and a sgRNA targeting the N-terminus of GOLGA2 for knock-in.DepositorInsertsgRNA Targeting N-terminus of GOLGA2 (GOLGA2 Human)
UseCRISPRExpressionMammalianPromoterU6Available SinceDec. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
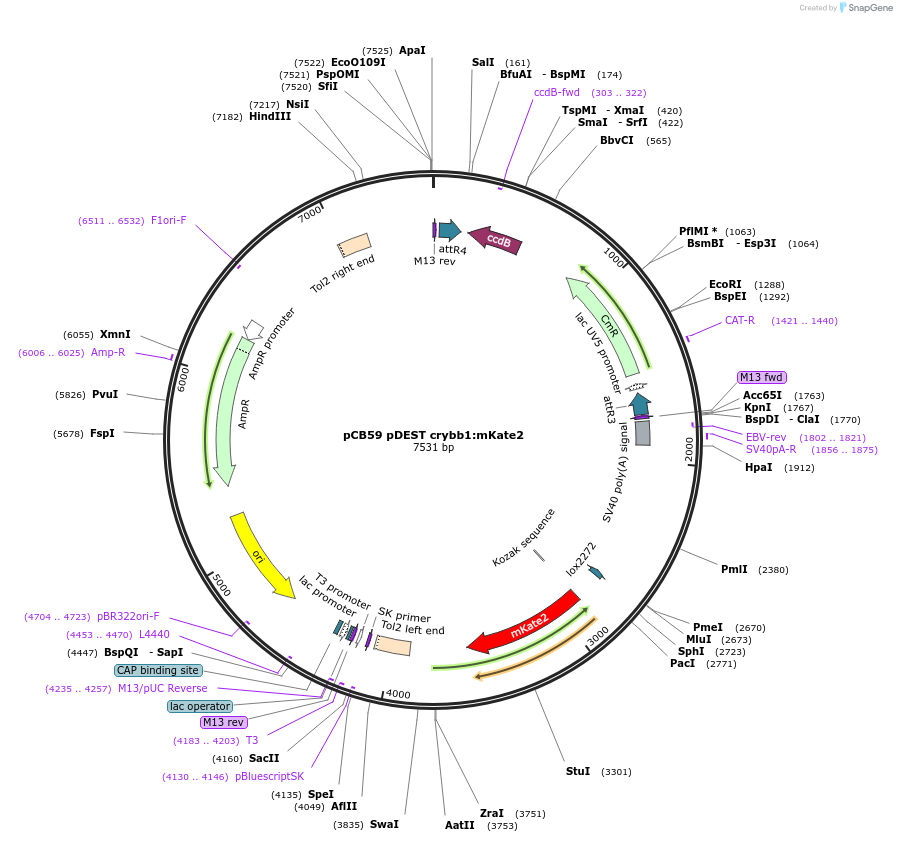
pCB59 pDEST crybb1:mKate2
Plasmid#195986PurposeTransgenesis backboneDepositorTypeEmpty backboneUseOtherAvailable SinceFeb. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
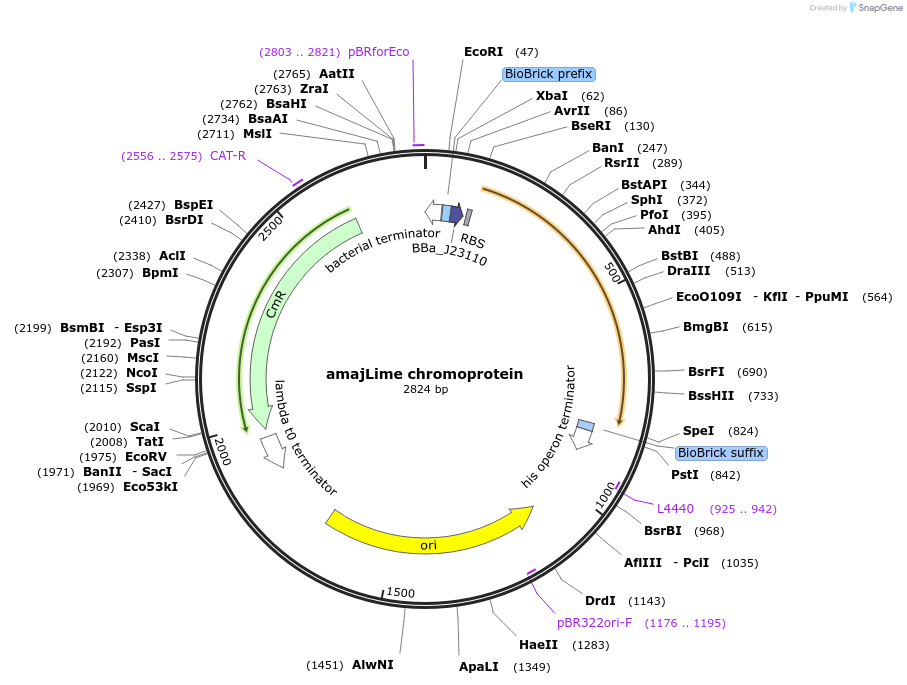
amajLime chromoprotein
Plasmid#117843PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amajLime chromoprotein in E. coliDepositorInsertpromoter, RBS, amajLime
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
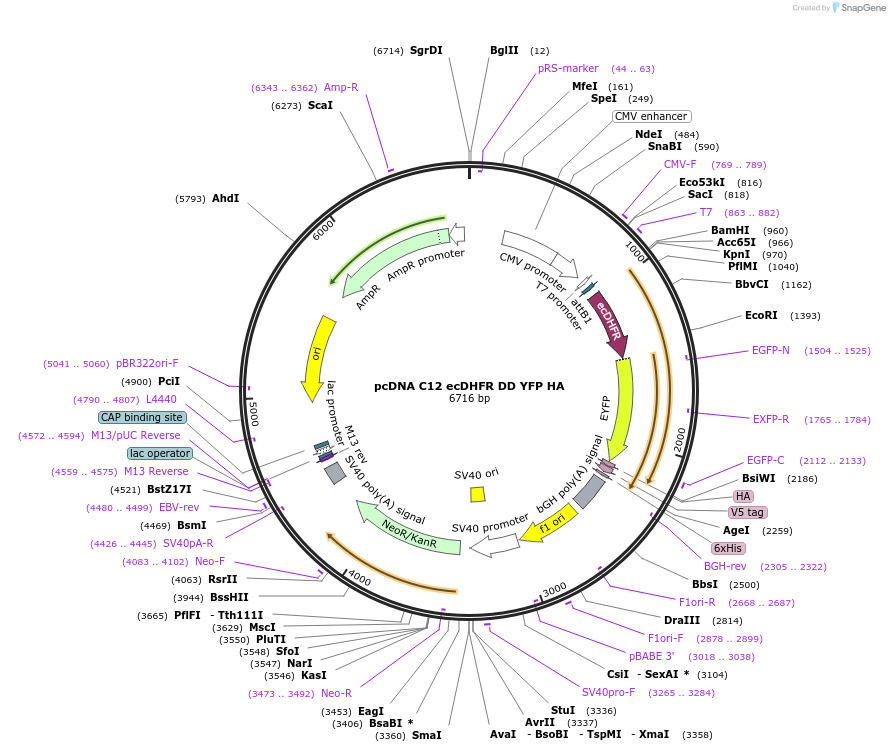
pcDNA C12 ecDHFR DD YFP HA
Plasmid#192815PurposeExpresses an N-terminal enhanced destabilizing domain (DD) version of E. coli DHFR with higher basal turnover in mammalian systems. Contains missense mutations W74R/T113S/E120D/Q146L.DepositorInsertE. coli dihydrofolate reductase
Tagsenhanced yellow fluorescent protein and hemagglut…ExpressionMammalianMutationW74R/T113S/E120D/Q146LPromoterCMVAvailable SinceNov. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
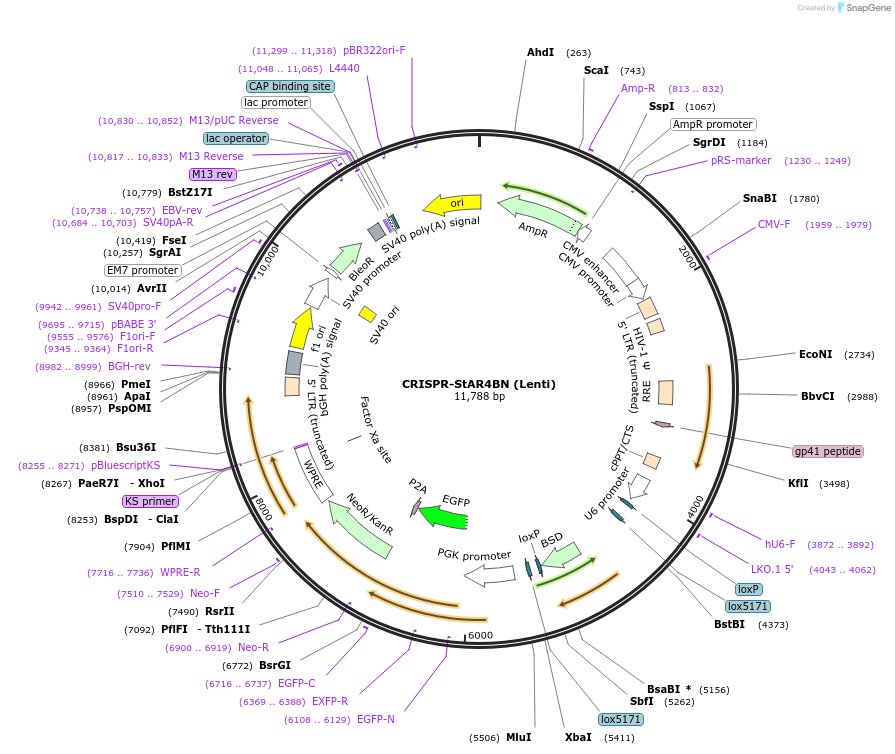
CRISPR-StAR4BN (Lenti)
Plasmid#222692PurposeCRISPR-screeningDepositorTypeEmpty backboneUseCRISPR and LentiviralTagsGFPExpressionMammalianMutationPGK without BsmBI cutsiteAvailable SinceDec. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
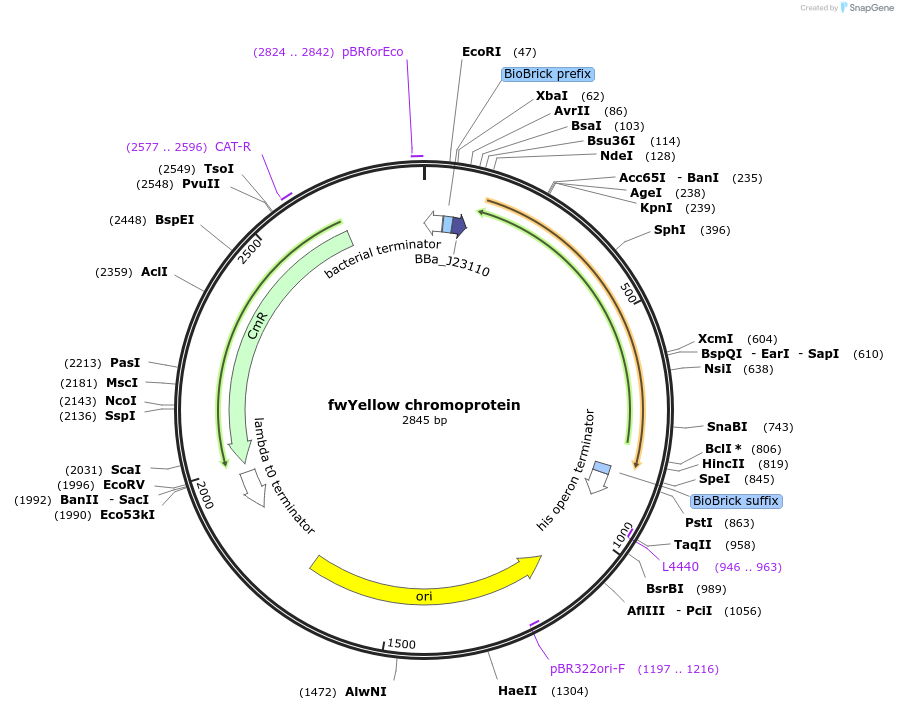
fwYellow chromoprotein
Plasmid#117841PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses fwYellow chromoprotein in E. coliDepositorInsertpromoter, RBS, fwYellow
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
scOrange chromoprotein
Plasmid#117840PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses scOrange chromoprotein in E. coliDepositorInsertpromoter, RBS, scOrange
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
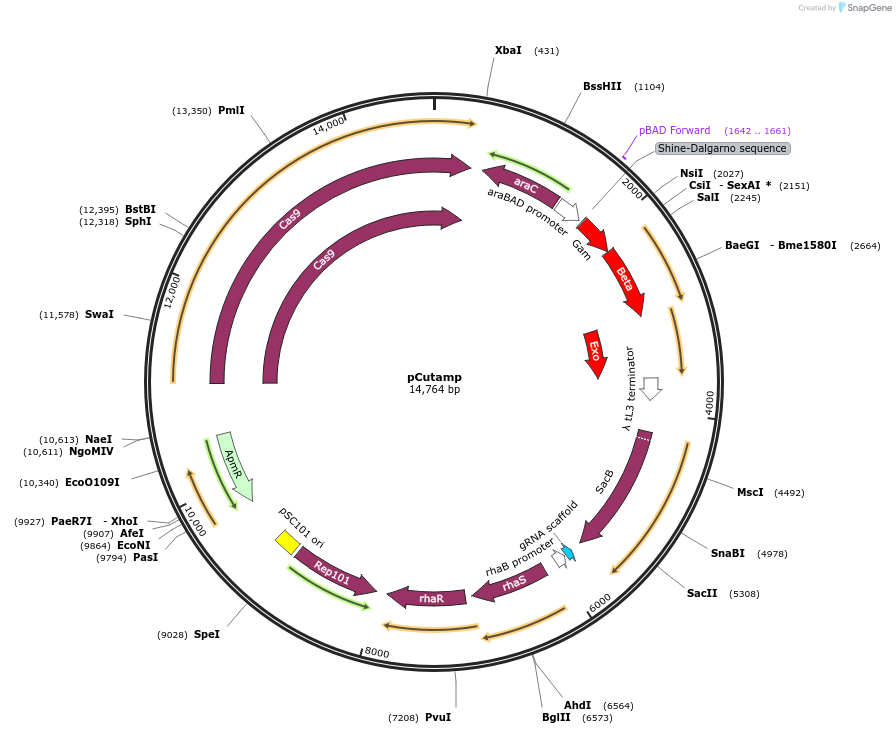
pCutamp
Plasmid#140632PurposePlasmid-curing in Escherichia coli by targeting the AmpR promoterDepositorInsertSpCas9_lambda-RED system, SacB, Rha induction system, sgRNA targeting AmpR promoter
UseCRISPRExpressionBacterialAvailable SinceAug. 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
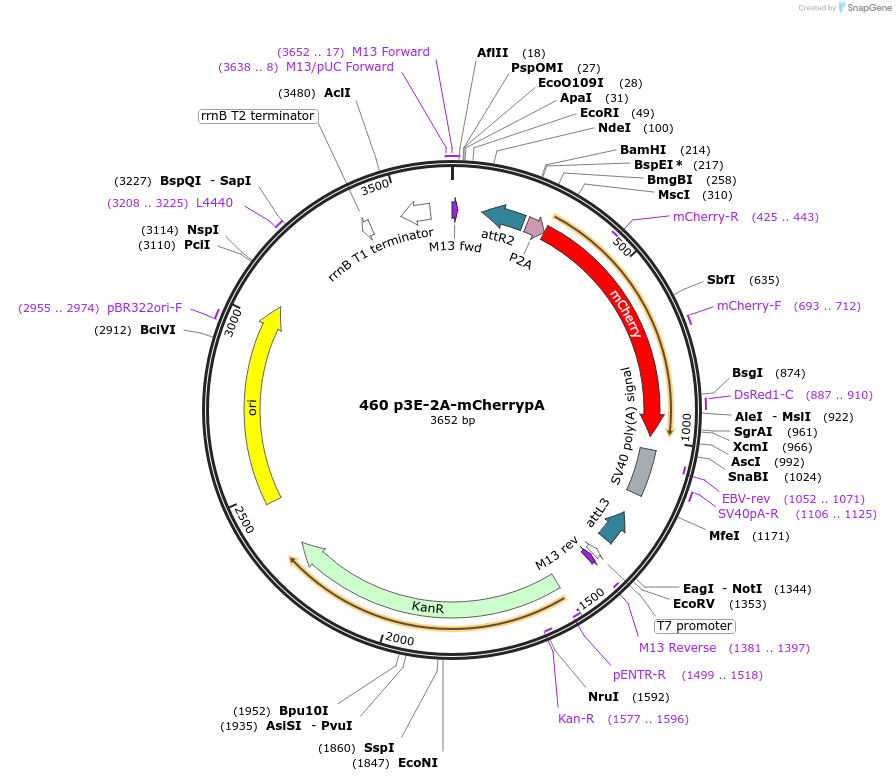
460 p3E-2A-mCherrypA
Plasmid#195970PurposeGateway p3E 2A fluorophoreDepositorInsert2A-mCherry
UseOtherAvailable SinceFeb. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
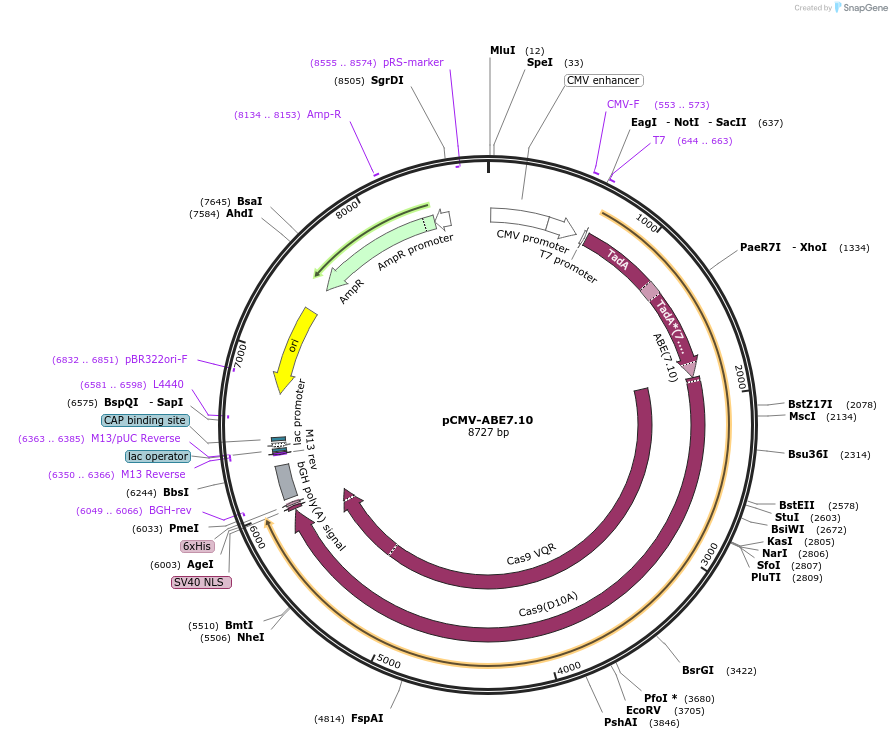
pCMV-ABE7.10
Plasmid#102919Purposeexpresses ABE7.10 in mammalian cellsDepositorInsertABE7.10
TagsNLSExpressionMammalianMutationsee manuscriptPromoterCMVAvailable SinceNov. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
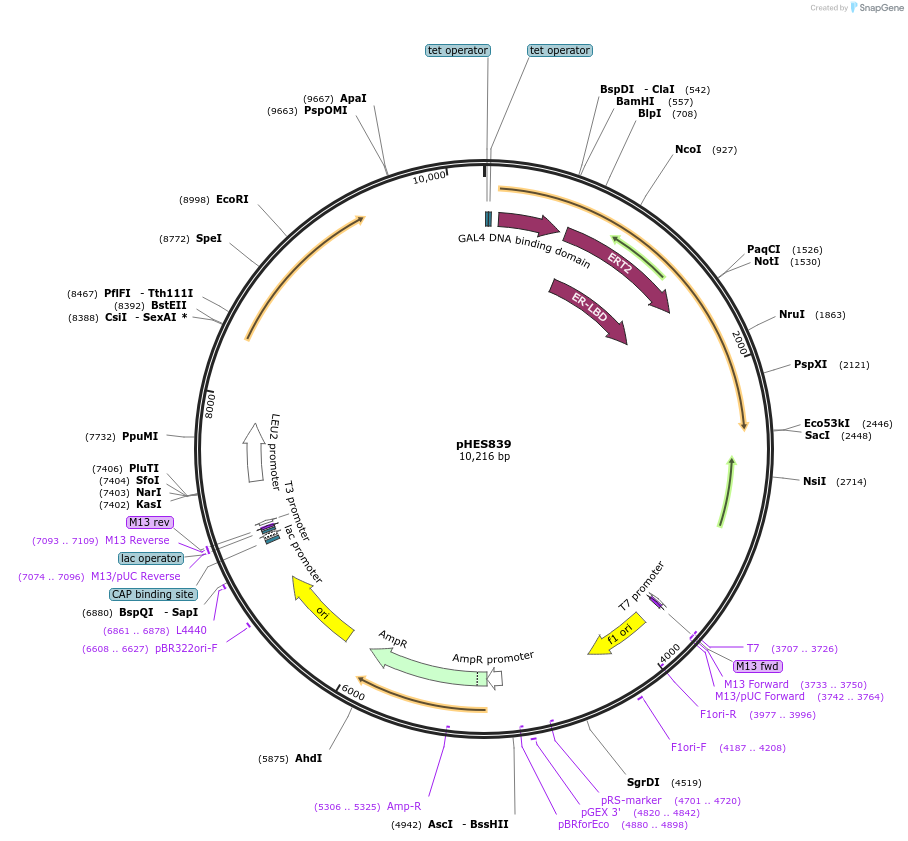
pHES839
Plasmid#87941PurposeConstitutively expresses estradiol-inducible synthetic transcriptional regulator in yeast. Transcriptional regulator activates transcription of promoters containing Gal4 binding sites.DepositorInsertGAL4 DBD - hER LBD - MSN2 AD (ESR1 Synthetic, Budding Yeast, Human)
UseSynthetic BiologyExpressionYeastPromoterADH1 (crippled)Available SinceDec. 4, 2017AvailabilityAcademic Institutions and Nonprofits only -
amilGFP chromoprotein
Plasmid#117842PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amilGFP chromoprotein in E. coliDepositorInsertpromoter, RBS, amilGFP
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
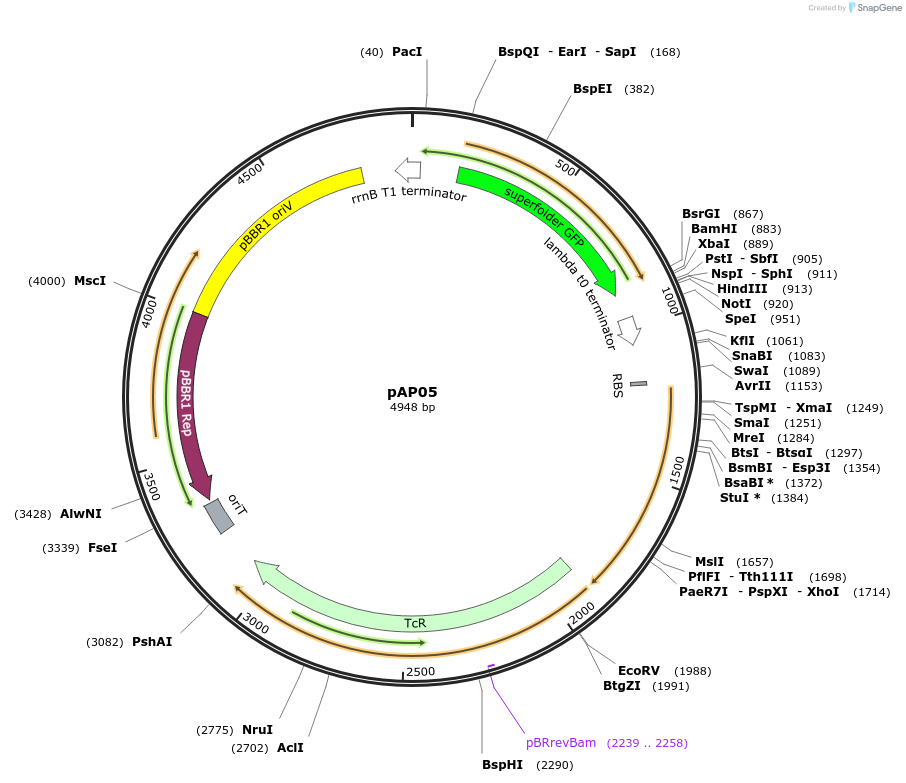
pAP05
Plasmid#186261PurposesfGFP under light light responsive PEL222 promoter and EL222 under constitutively active BBa_J2305 promoterDepositorInsertsExpressionBacterialPromoterBBa_23105 (GGCTAGCTCAGTCCTAGGTACTATGCTAGC) and PE…Available SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only