We narrowed to 9,887 results for: Coli
-
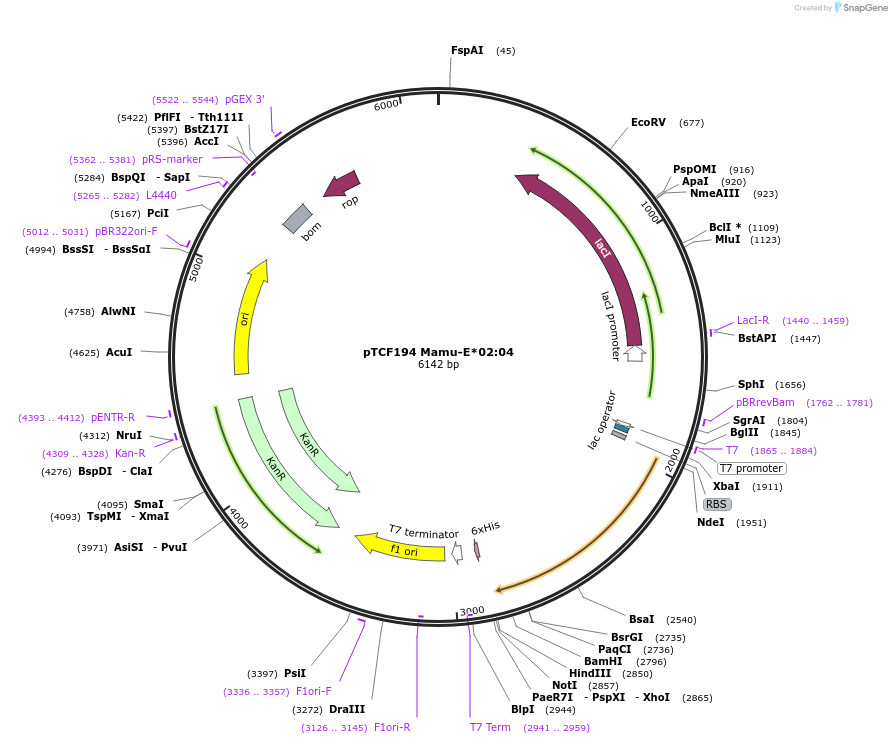
Plasmid#180461PurposeE. coli expression of soluble MHC proteinDepositorInsertMamu-E*02:04
TagsBSP41ExpressionBacterialMutationReplacement of transmembrane domain with enzymati…Available SinceFeb. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
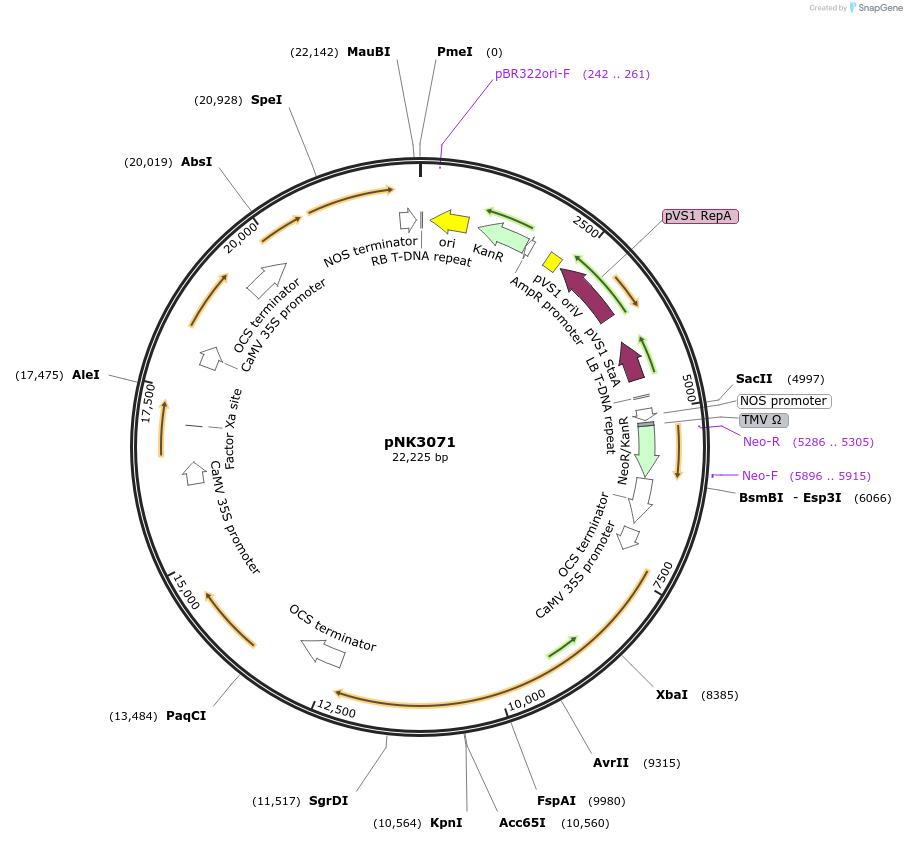
pNK3071
Plasmid#219755PurposeMoClo-compatible Level P vector for improved autonomous bioluminescence in plants encoding kanamycin resistance cassette, mcitHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH.DepositorInsertmcitHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH
ExpressionPlantAvailable SinceOct. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
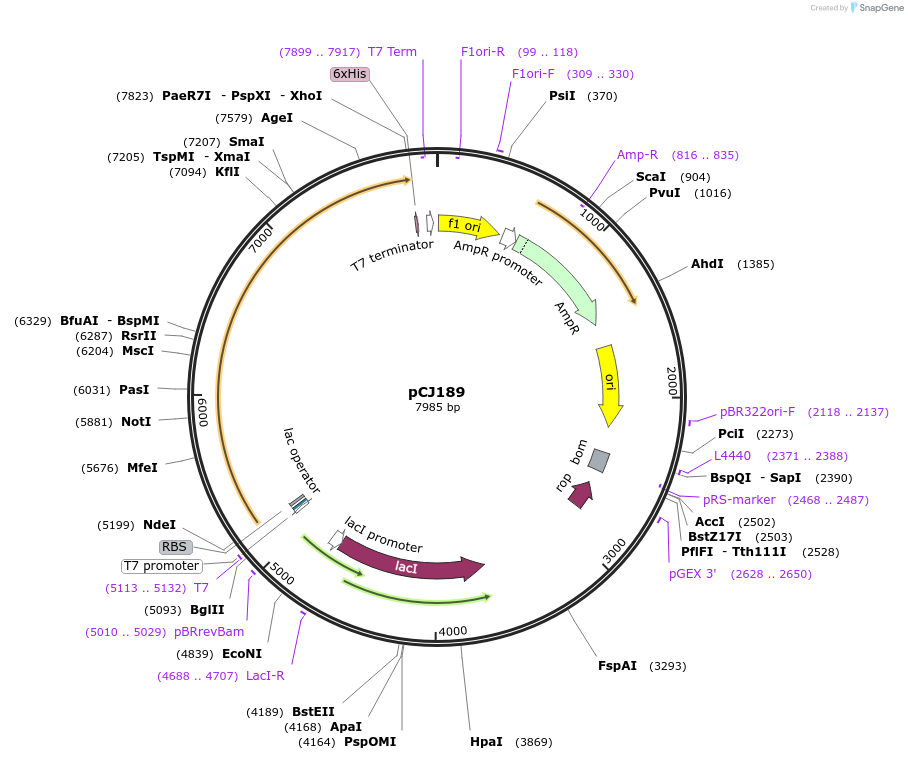
pCJ189
Plasmid#162666PurposepET-21b(+) based plasmid for expresion of MHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by an 8 residue Gly/Ser linker; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by an 8 residue Gly/Ser linker
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
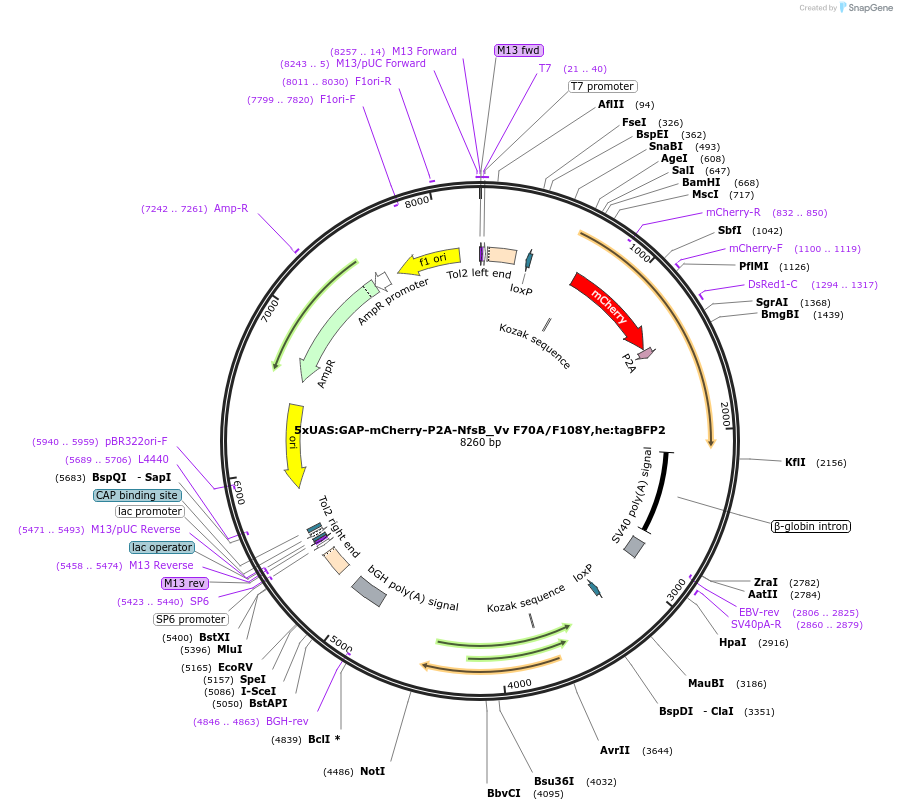
5xUAS:GAP-mCherry-P2A-NfsB_Vv F70A/F108Y,he:tagBFP2
Plasmid#158653Purposemaking transgenics expressing a UAS driven mCherry and NTR 2.0DepositorInsert5xUAS:GAP-mCherry-P2A-NfsB_Vv F70A/F108Y;he:BFP
UseTol2-based transgenic expression in zebrafish/oth…MutationF70A/F108YAvailable SinceSept. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
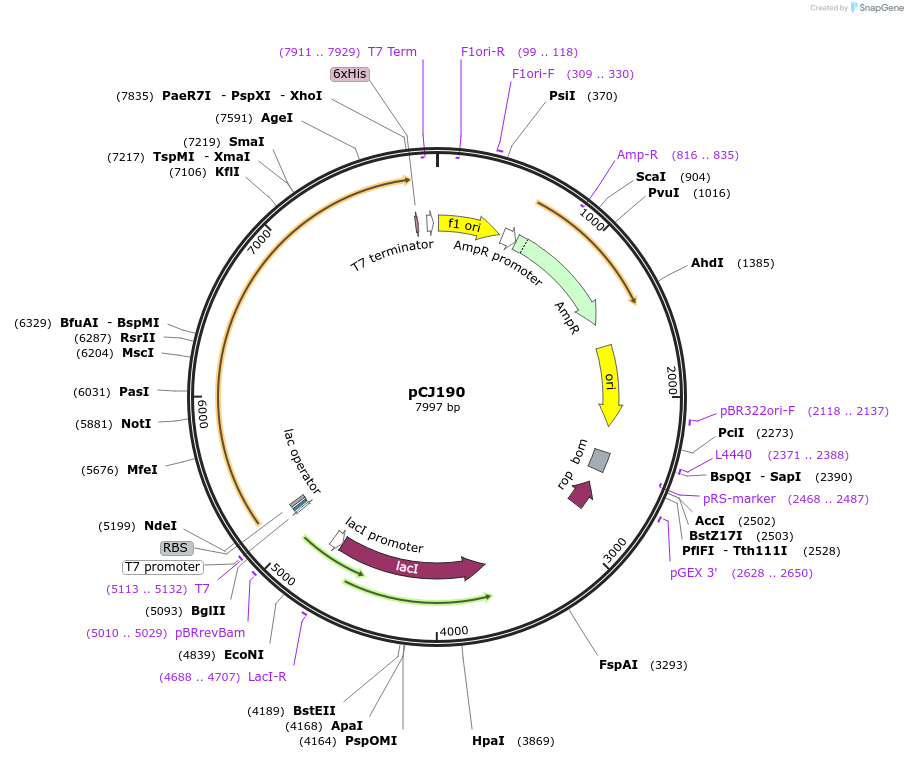
pCJ190
Plasmid#162667PurposepET-21b(+) based plasmid for expresion of MHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 12 residue Gly/Ser linker; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 12 residue Gly/Ser linker
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
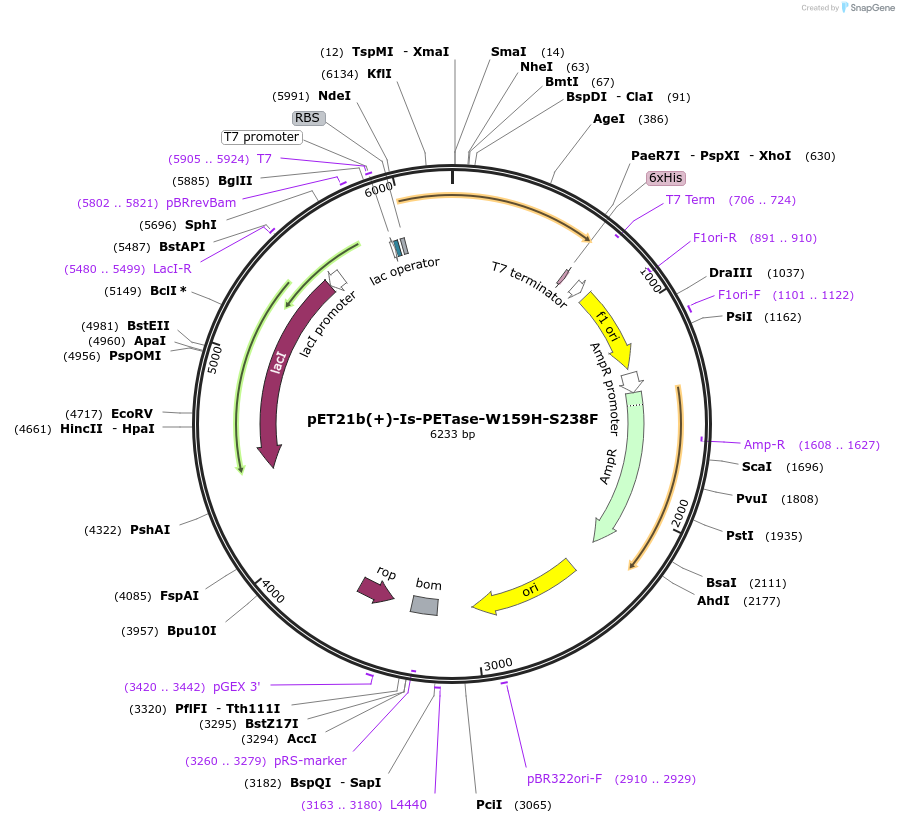
pET21b(+)-Is-PETase-W159H-S238F
Plasmid#112203PurposepET-21b(+) based plasmid for expression of PETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1) with W159H and S238F mutations, codon optimized for expression in E. coli K12DepositorInsertPETase gene from Ideonella sakaiensis 201-F6 with W159H and S238F mutations, codon optimized for expression in E. coli K12
Tags6XHISExpressionBacterialMutationW159H, S238FPromoterN/AAvailable SinceJuly 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
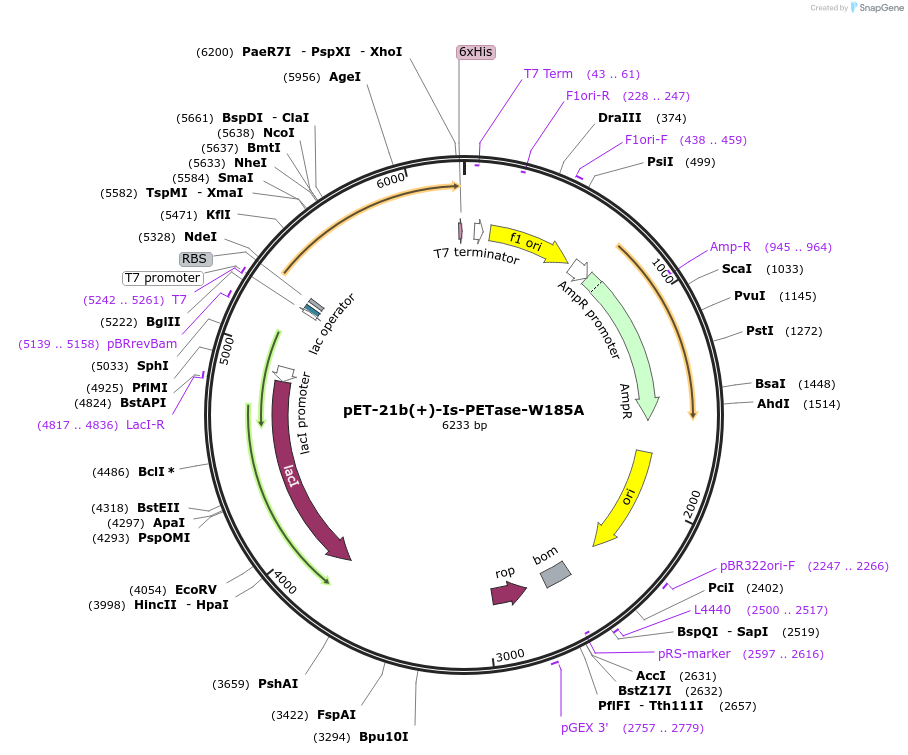
pET-21b(+)-Is-PETase-W185A
Plasmid#112204PurposepET-21b(+) based plasmid for expression of PETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1) with W185A mutations, codon optimized for expression in E. coli K12DepositorInsertPETase gene from Ideonella sakaiensis 201-F6 with W185A mutation, codon optimized for expression in E. coli K12
Tags6XHISExpressionBacterialMutationW185APromoterN/AAvailable SinceJuly 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
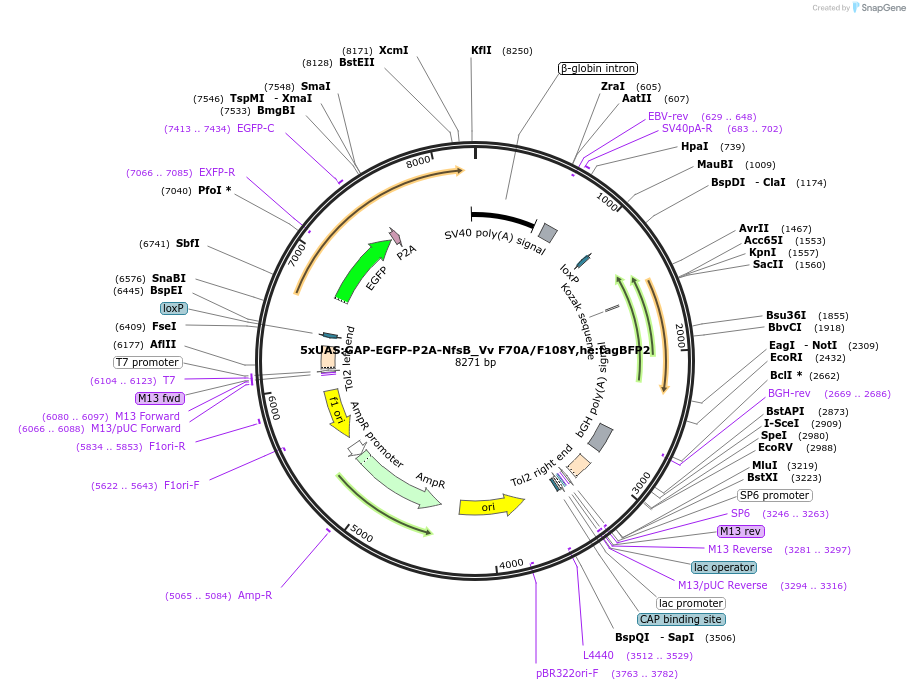
5xUAS:GAP-EGFP-P2A-NfsB_Vv F70A/F108Y,he:tagBFP2
Plasmid#158652Purposemaking transgenics expressing a UAS driven eGFP and NTR 2.0DepositorInsert5xUAS:GAP-eGFP-P2A-NfsB_Vv F70A/F108Y;he:BFP
UseTol2-based transgenic expression in zebrafish/oth…MutationF70A/F108YAvailable SinceSept. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
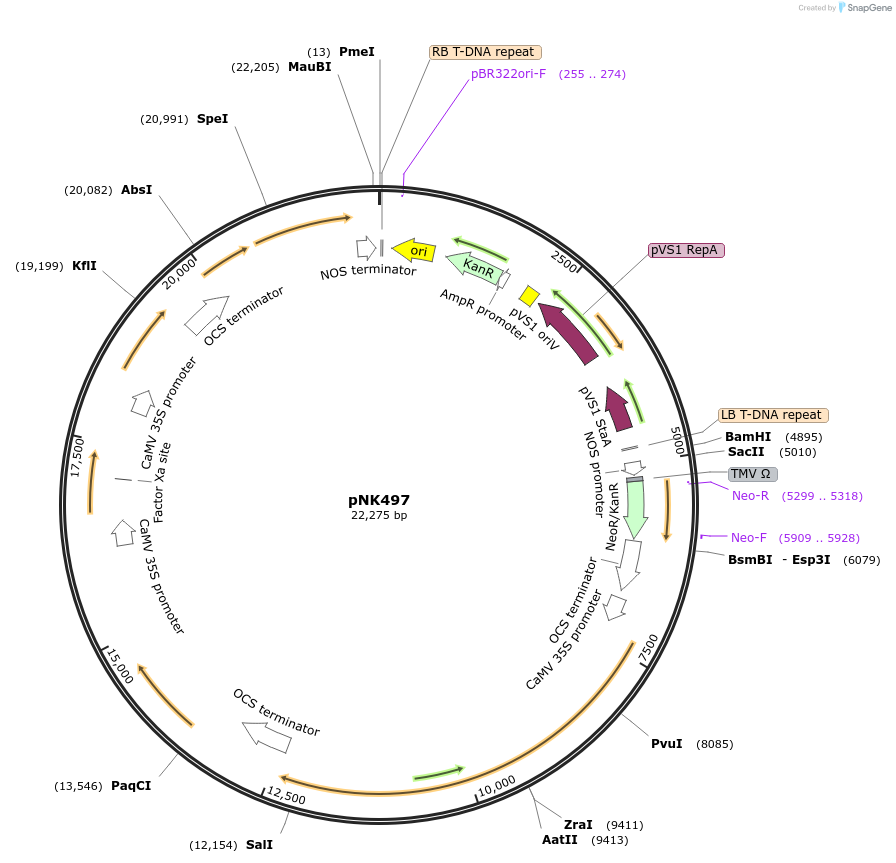
pNK497
Plasmid#219754PurposeMoClo-compatible Level P vector for improved autonomous bioluminescence in plants encoding kanamycin resistance cassette, nnHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH.DepositorInsertnnHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH
ExpressionPlantAvailable SinceJan. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
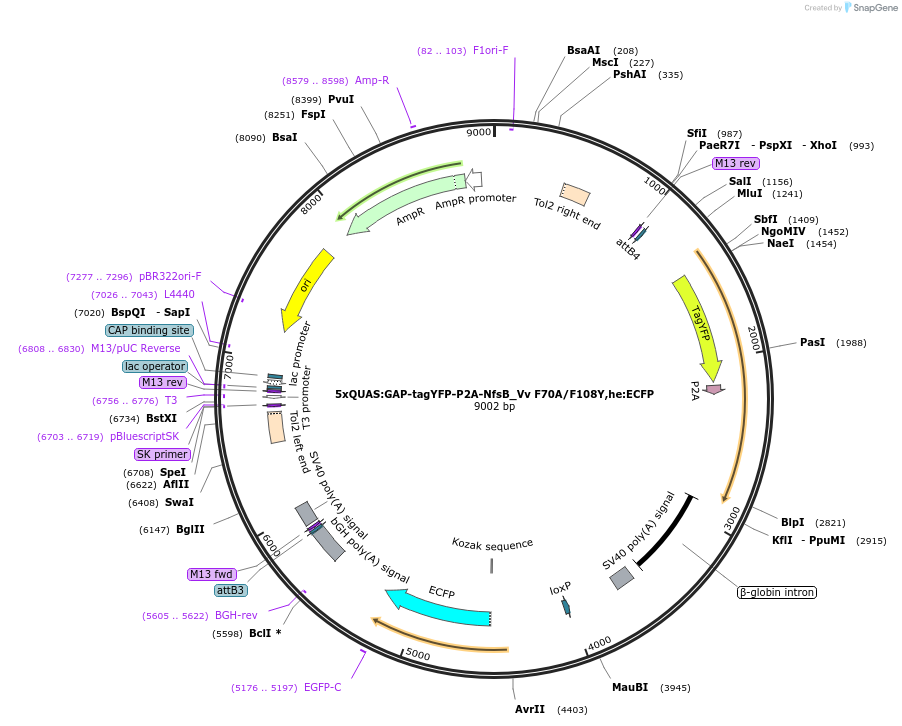
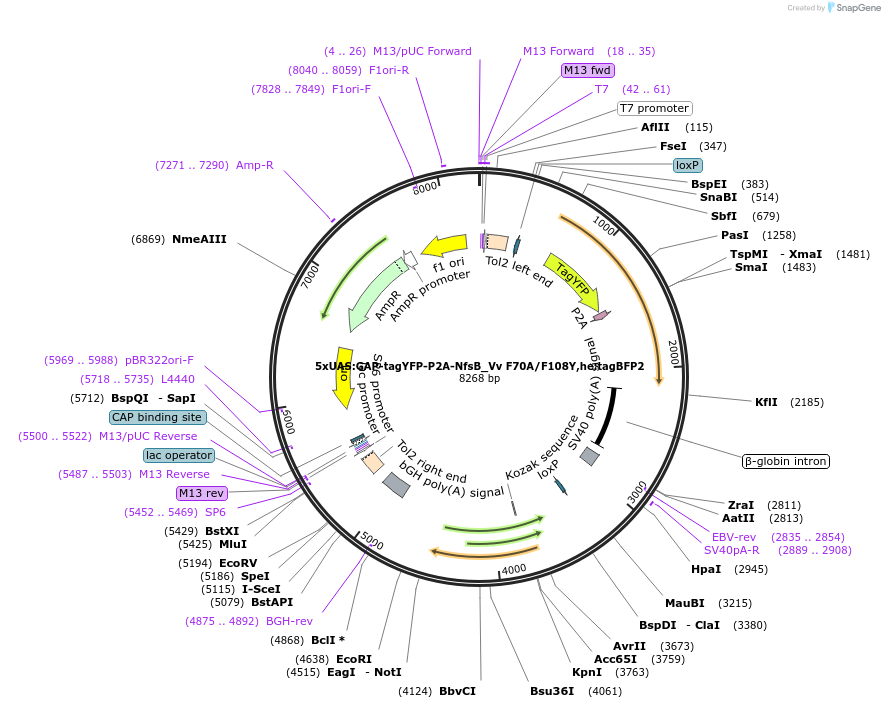
5xQUAS:GAP-tagYFP-P2A-NfsB_Vv F70A/F108Y,he:ECFP
Plasmid#158654Purposemaking transgenics expressing a QUAS driven YFP and NTR 2.0DepositorInsert5xQUAS:GAP-tagYFP-P2A-NfsB_Vv F70A/F108Y;he:eCFP
UseTol2-based transgenic expression in zebrafish/oth…MutationF70A/F108YAvailable SinceSept. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
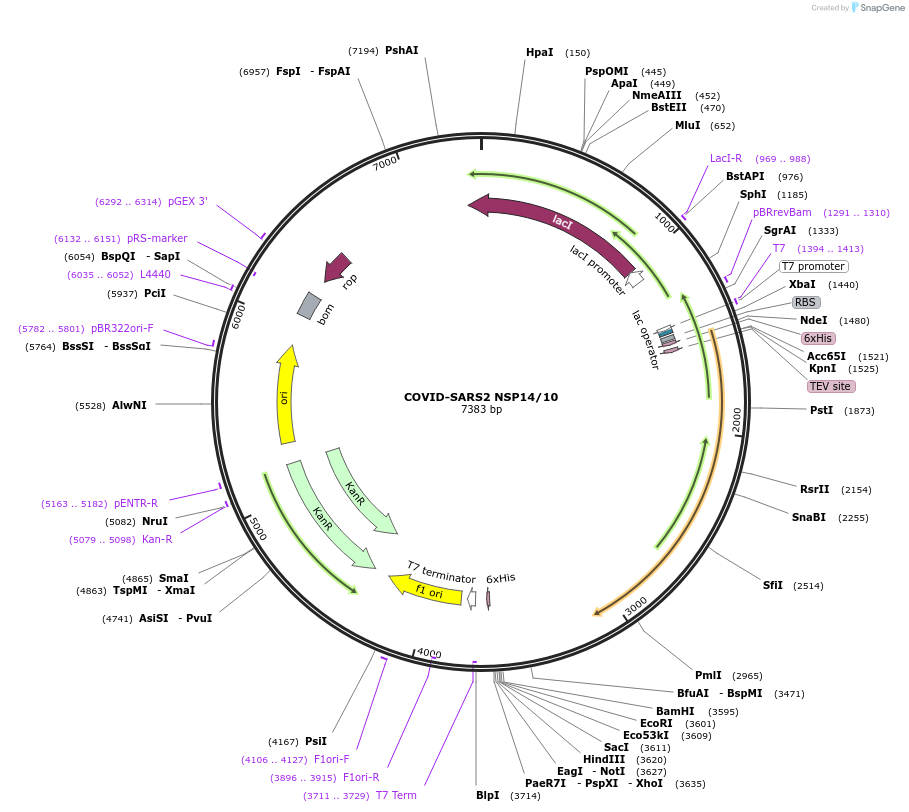
COVID-SARS2 NSP14/10
Plasmid#159613PurposeBacterial co-expression vector fo Covid-SARS2 NSP14 and NSP10DepositorInsertsTagsHis6, TEV cleavage site and noneExpressionBacterialMutationCodon-optimized for E. coli expressionPromoter(Bicistronic) and T7 - LacOAvailable SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
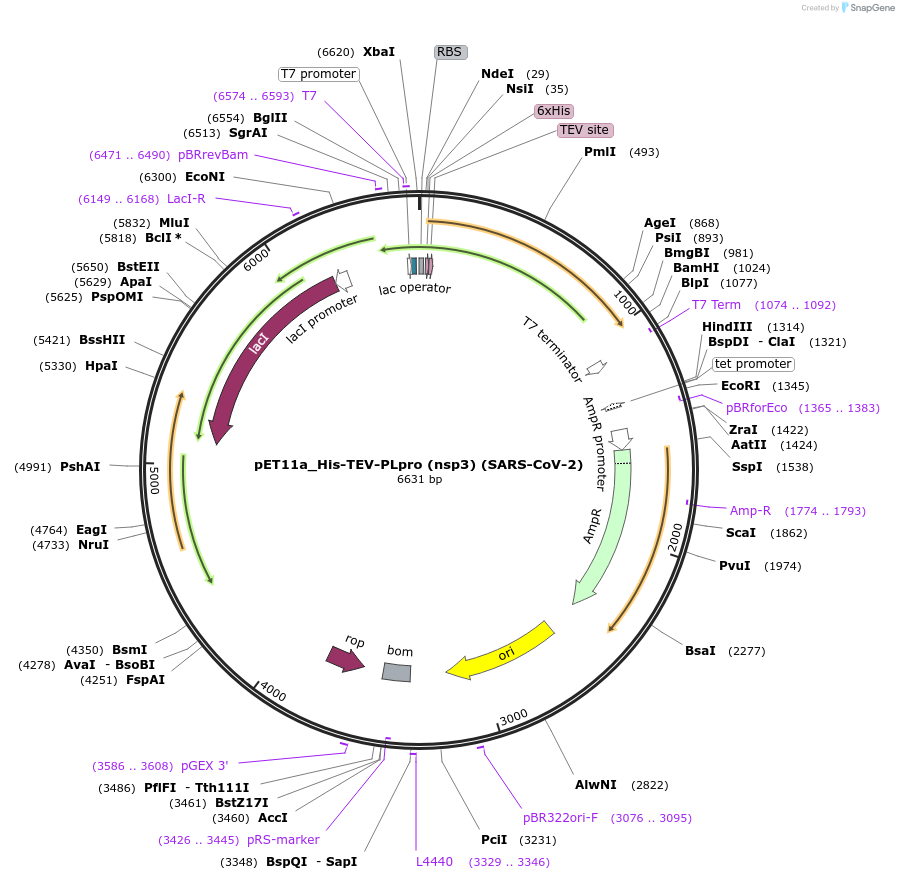
pET11a_His-TEV-PLpro (nsp3) (SARS-CoV-2)
Plasmid#169192PurposeTo express SARS-CoV-2 Papain-like protease in E. coliDepositorInsert6His-TEV-nsp3_E746-K1060 (pp1ab_E1564-K1878) (ORF1ab Synthetic, SARS-CoV-2)
UseUnspecifiedTags6His-TEVMutationCodon optimised for E. coliPromoterT7Available SinceJuly 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
5xUAS:GAP-tagYFP-P2A-NfsB_Vv F70A/F108Y,he:tagBFP2
Plasmid#158651Purposemaking transgenics expressing a UAS driven YFP and NTR 2.0DepositorInsert5xUAS:GAP-tagYFP-P2A-NfsB_Vv F70A/F108Y;he:BFP
UseTol2-based transgenic expression in zebrafish/oth…MutationF70A/F108YAvailable SinceSept. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
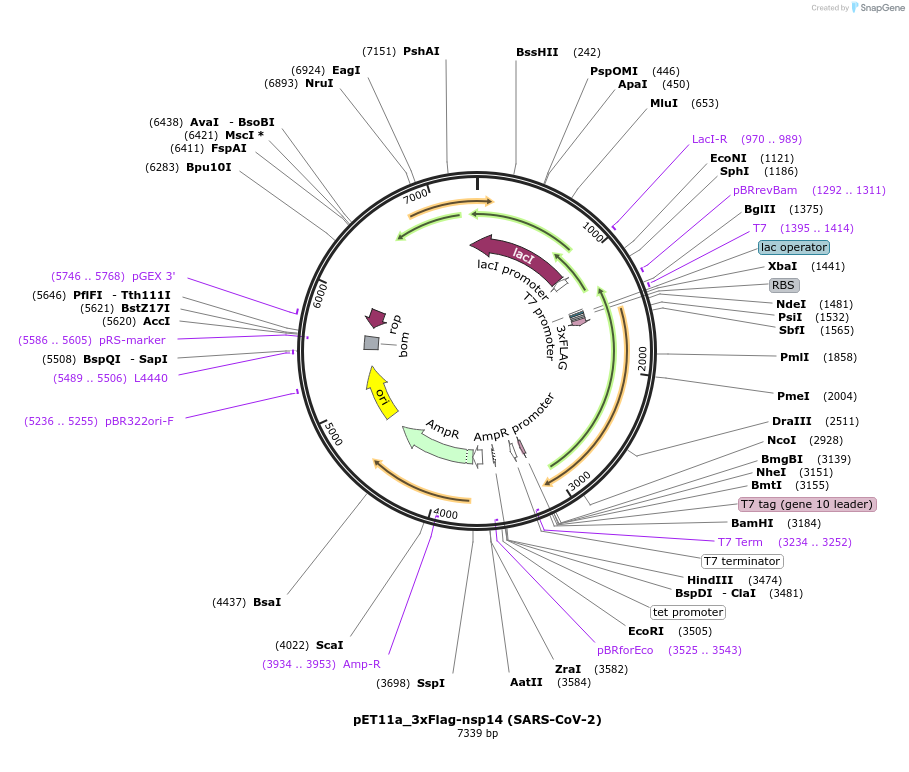
pET11a_3xFlag-nsp14 (SARS-CoV-2)
Plasmid#169159PurposeTo express SARS-CoV-2 nsp14 in E. coliDepositorInsert3xFlag-nsp5CS[VATLQ]-nsp14 (ORF1ab SARS-CoV-2)
Tags3xFlag and nsp5CS[VATLQ]: predicted nsp5 protease…ExpressionBacterialMutationCodon optimised for E. coliPromoterT7Available SinceJuly 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
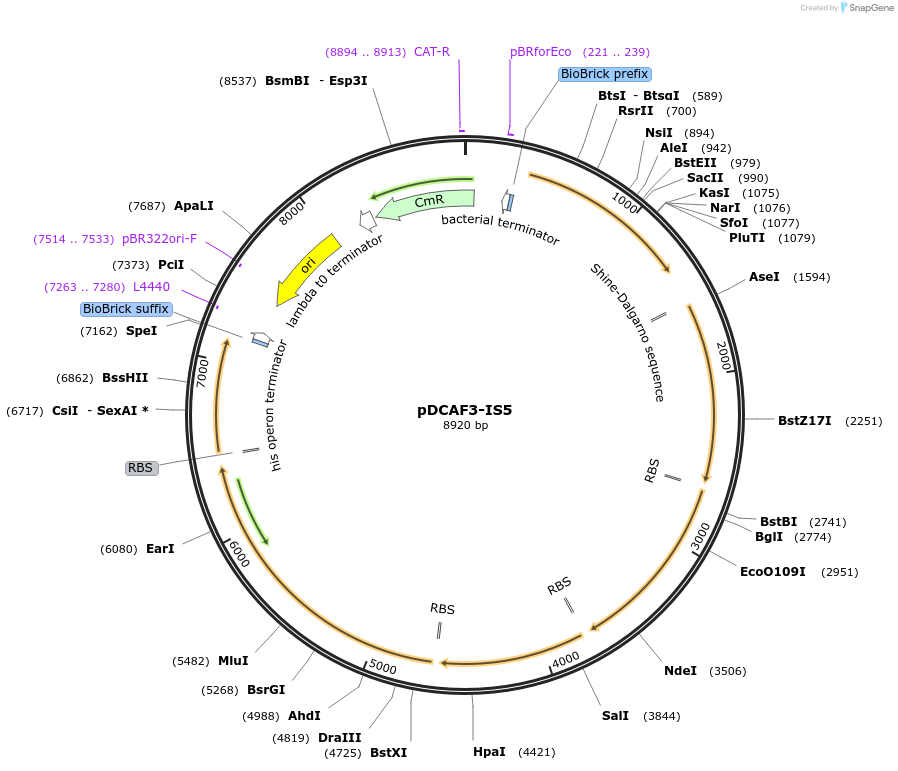
pDCAF3-IS5
Plasmid#65220PurposeExpresses N-demethylases for the degredation of caffeine and related methylxanthines. Can be used to measure caffeine content as described in the publication listed below.DepositorInsertsndmA
ndmB
ndmC
ndmD
gst9
chlR
UseSynthetic BiologyExpressionBacterialMutationChanged start codon from gtg to atg and Changed s…PromoterBBa_J23100Available SinceJan. 8, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
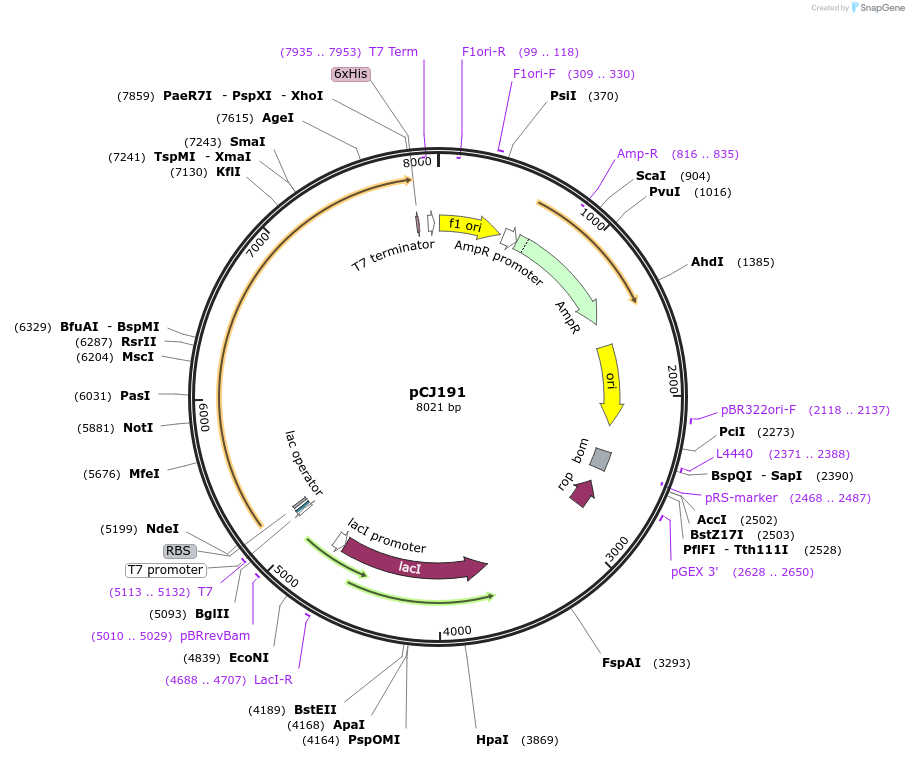
pCJ191
Plasmid#162668PurposepET-21b(+) based plasmid for expresion of MHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 20 residue Gly/Ser linker; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 20 residue Gly/Ser linker
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
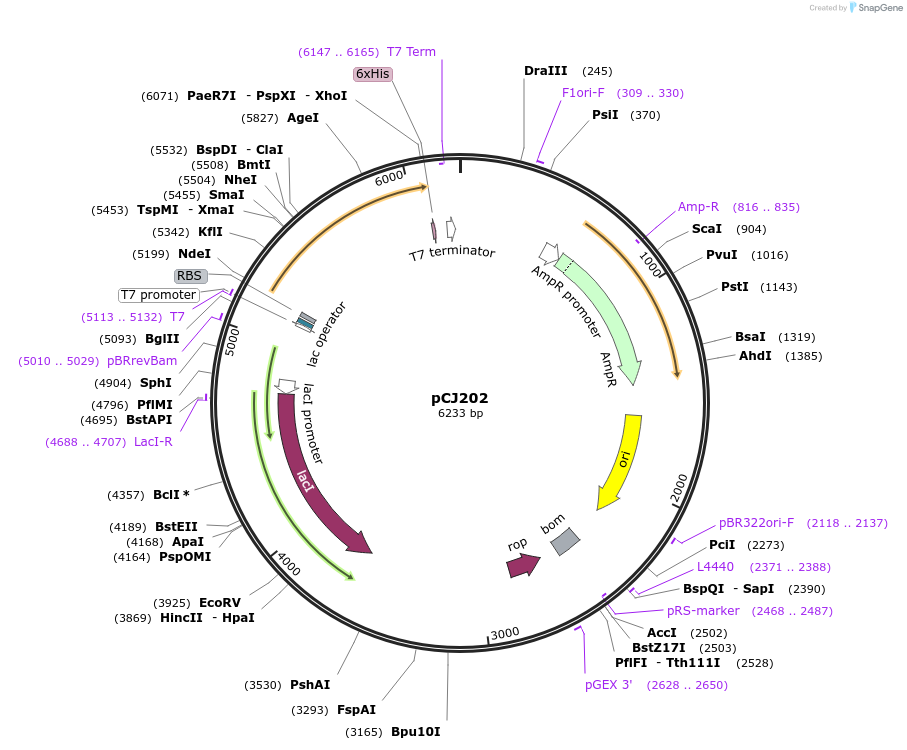
pCJ202
Plasmid#162675PurposepET-21b(+) based plasmid for expression of PETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating W159C and S238C mutations.DepositorInsertPETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1)
TagsHisExpressionBacterialMutationW159C, S238C; codon optimized for expression in E…PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
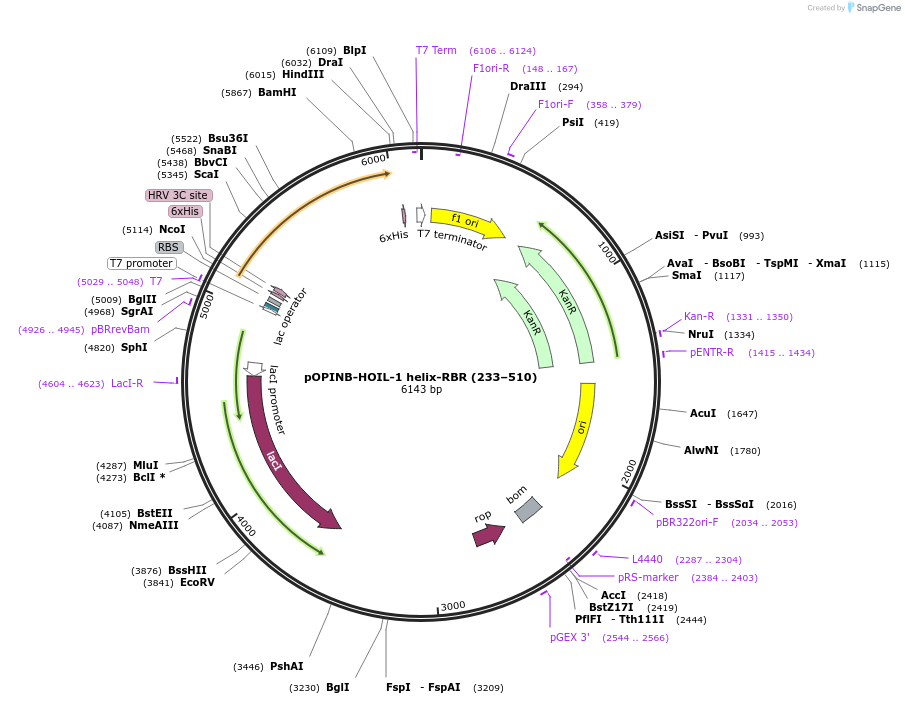
pOPINB-HOIL-1 helix-RBR (233–510)
Plasmid#193859PurposeExpression of human His-tagged HOIL-1 (RBCK1) helix-RBR (aa 233–510) codon optimized for E. coliDepositorInsertHOIL-1 (RBCK1 Human)
Tags3C protease cleavage site and 6x-His tagExpressionBacterialMutationResidues 233–510, codon optimised for expression …PromoterT7Available SinceJan. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
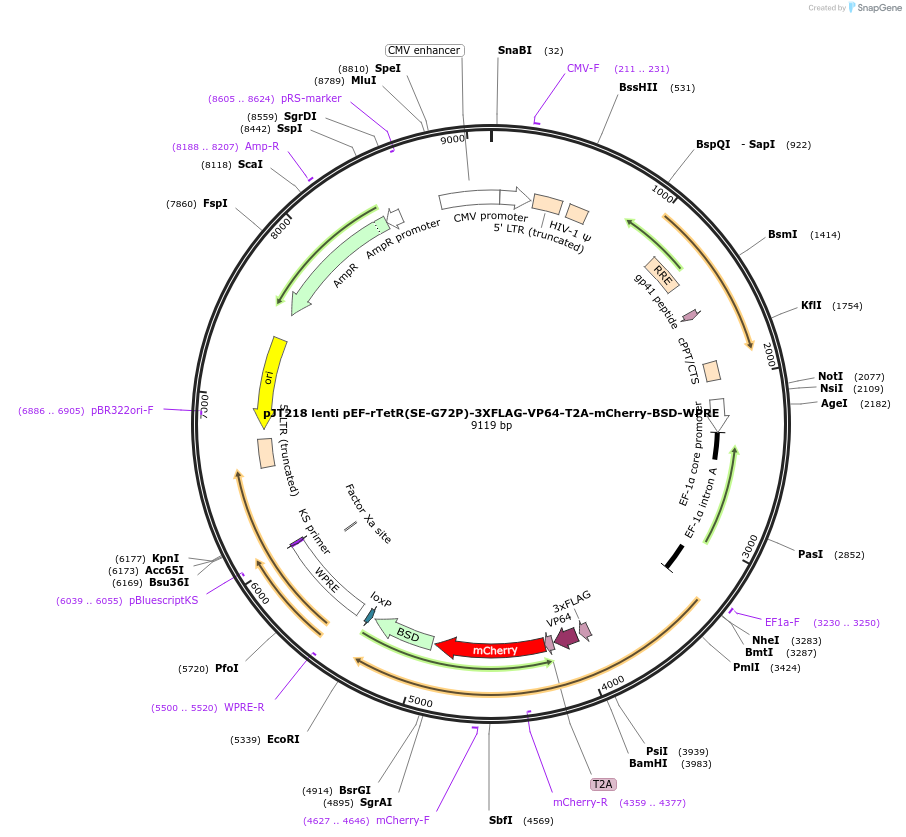
pJT218 lenti pEF-rTetR(SE-G72P)-3XFLAG-VP64-T2A-mCherry-BSD-WPRE
Plasmid#188758PurposeLentiviral backbone containing VP64 as a fusion with rTetR(SE-G72P)-3XFLAGDepositorInsertVP64
UseLentiviralTagsrTetR(SE-G72P)-3xFLAGExpressionMammalianPromoterpEF1aAvailable SinceMay 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
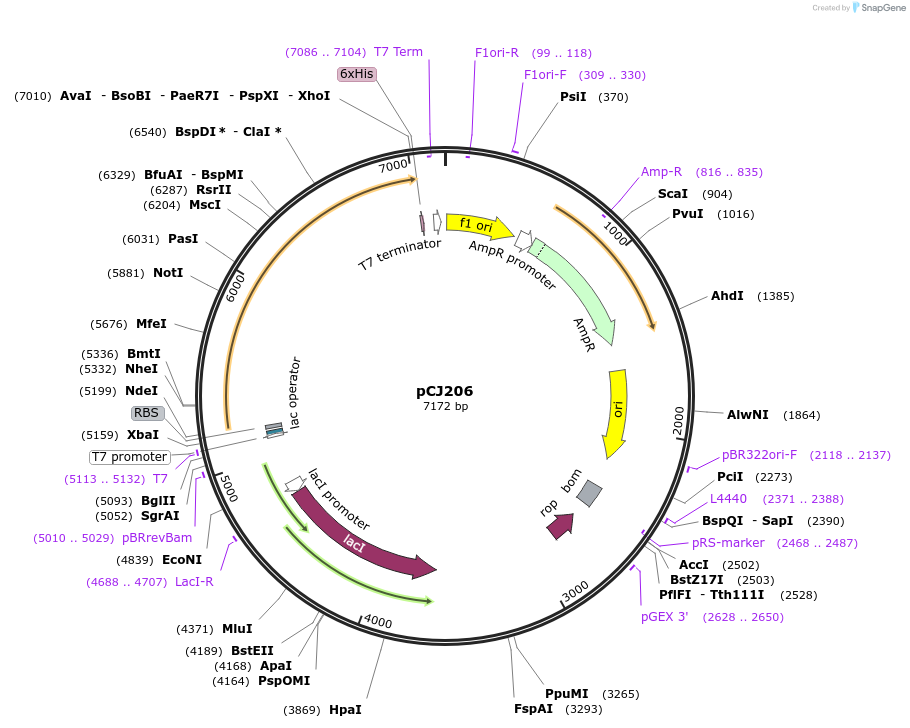
pCJ206
Plasmid#162679PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating the E226T mutation to the putative lipase box.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), incorporating the E226T mutation to the putative lipase box
TagsHisExpressionBacterialMutationE226T; codon optimized for expression in E. coli …PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only