We narrowed to 11,082 results for: CHL
-
Plasmid#85719PurposeGateway destination vectorDepositorTypeEmpty backboneUseMammalian/avian/xenopus/zebrafishTags3xHAAvailable SinceJan. 9, 2017AvailabilityAcademic Institutions and Nonprofits only
-
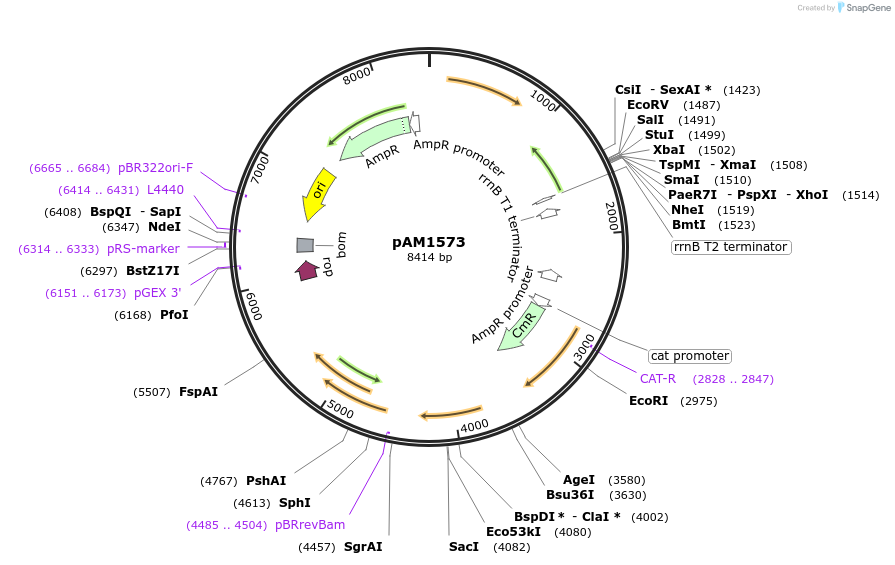
pAM1573
Plasmid#40239DepositorInsertCm Resistance, neutral site II integration platform for Synechococcus
UseCyanobacteria cloning vectorPromotern/aAvailable SinceMarch 25, 2014AvailabilityAcademic Institutions and Nonprofits only -
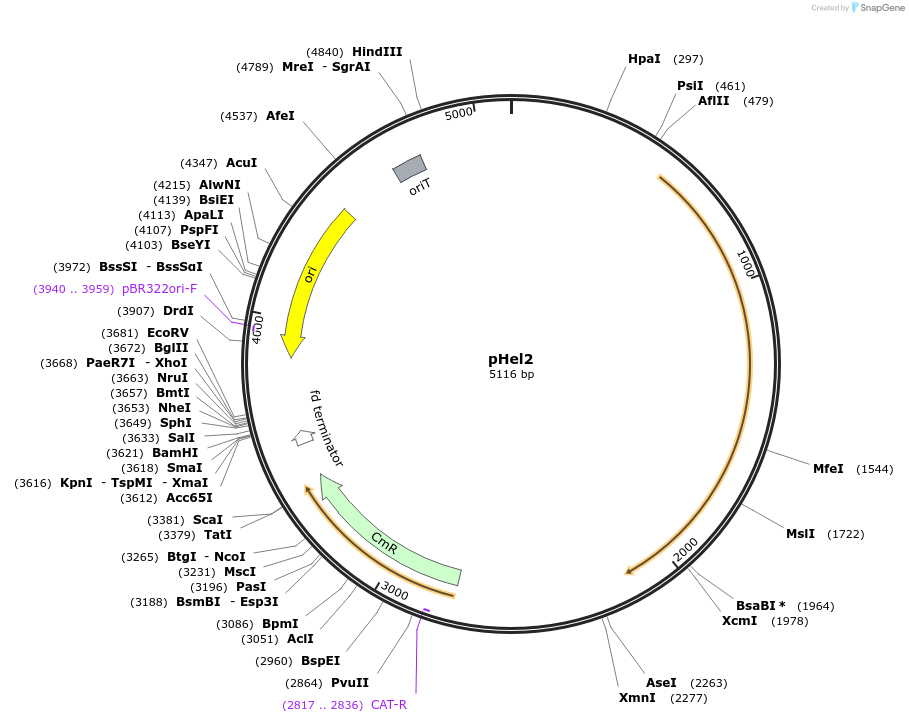
pHel2
Plasmid#102960PurposeE.coli/H.Pylori shuttle vectorDepositorTypeEmpty backboneUsePlasmid shuttle vector system for genetic complem…Available SinceNov. 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
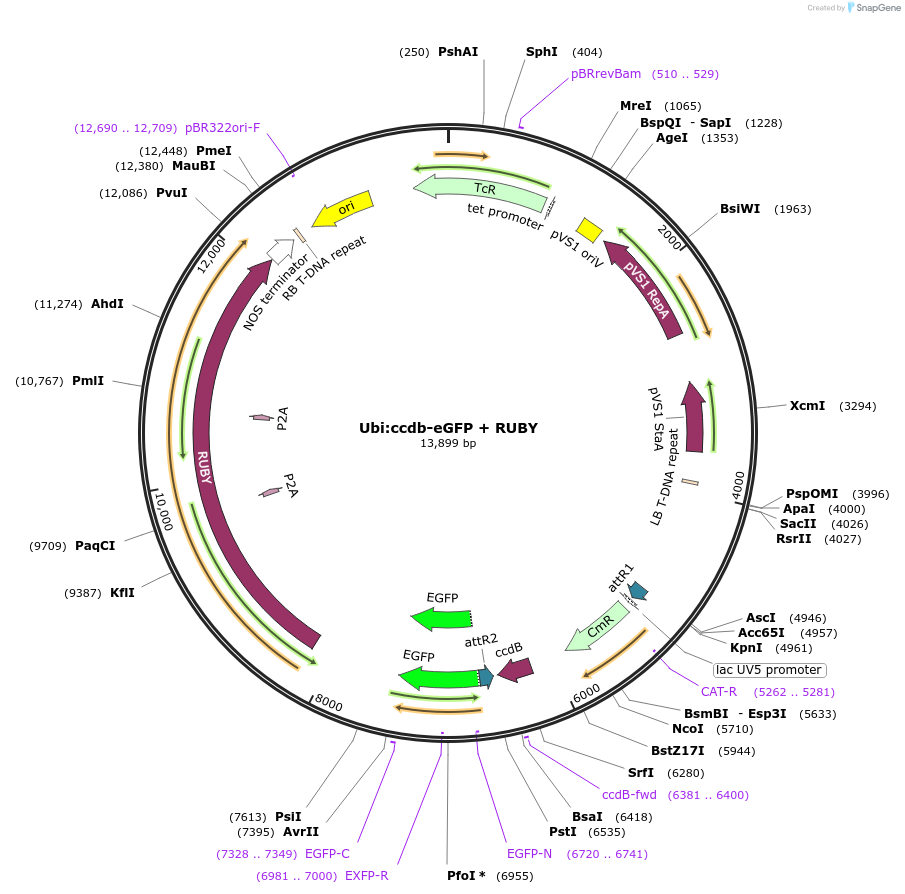
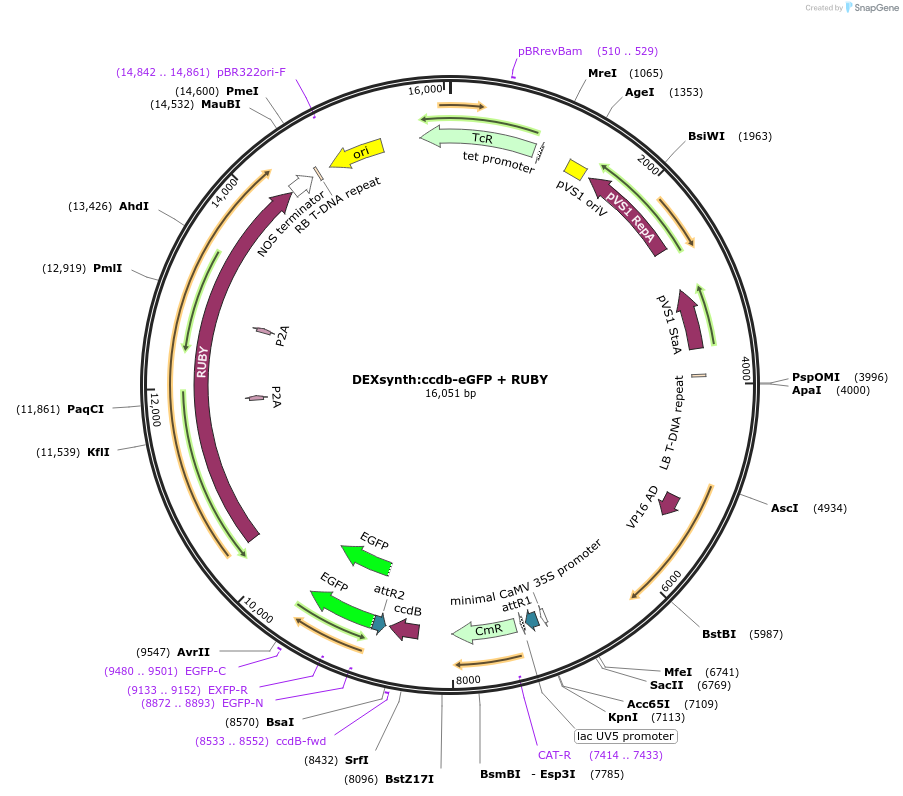
Ubi:ccdb-eGFP + RUBY
Plasmid#233310PurposeGATEWAY compatible vector with a soybean Ubiquitin (GmUbi) promoter driving in frame epitope fusion of eGFP paired with ccdb selection toxin and RUBY markerDepositorInsertUbi:ccdb-eGFP + RUBY
UseGateway destinationTagseGFPAvailable SinceMarch 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
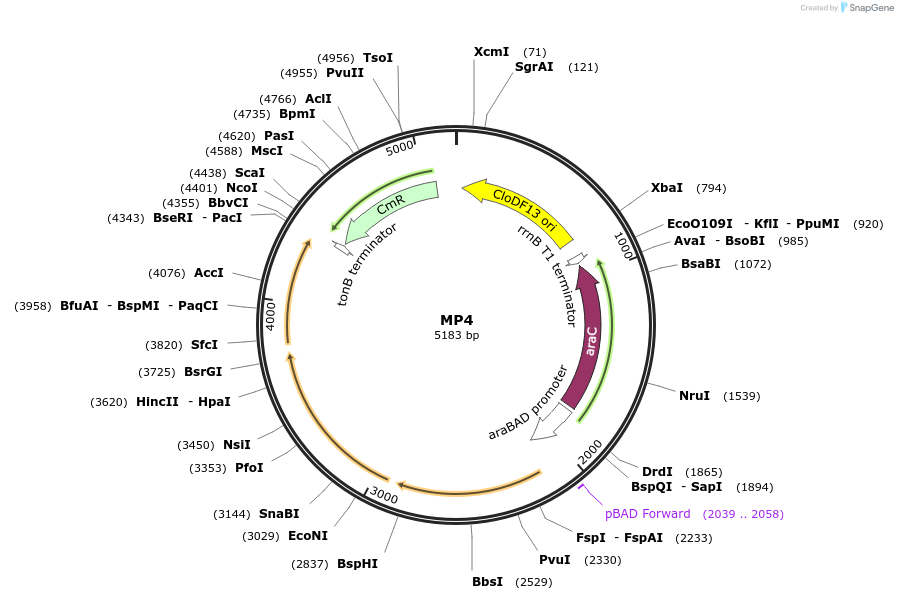
MP4
Plasmid#69652PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
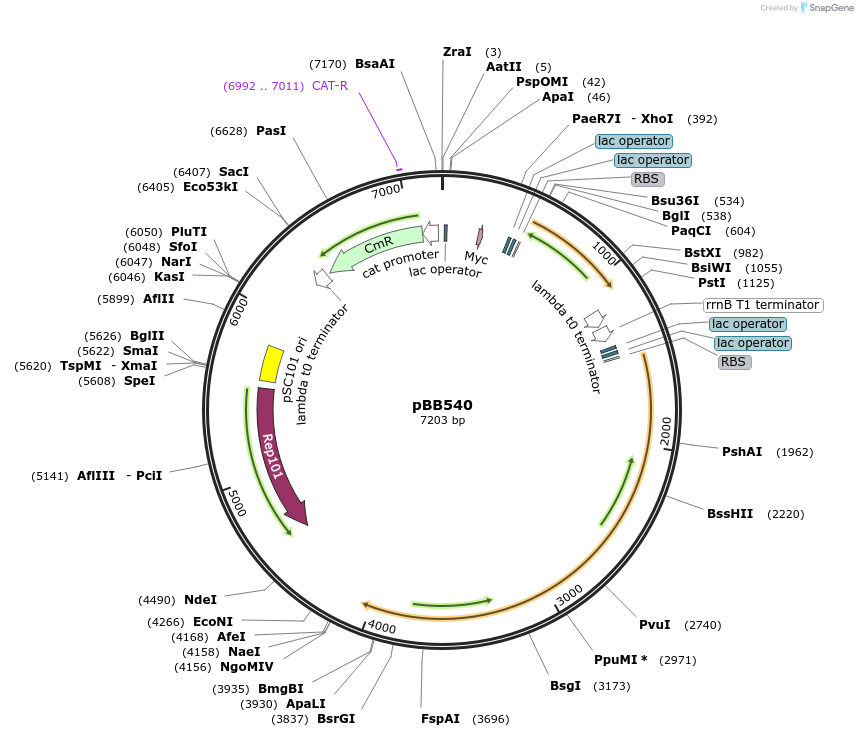
pBB540
Plasmid#27393DepositorInsertgrpE, clpB
ExpressionBacterialAvailable SinceApril 26, 2012AvailabilityAcademic Institutions and Nonprofits only -
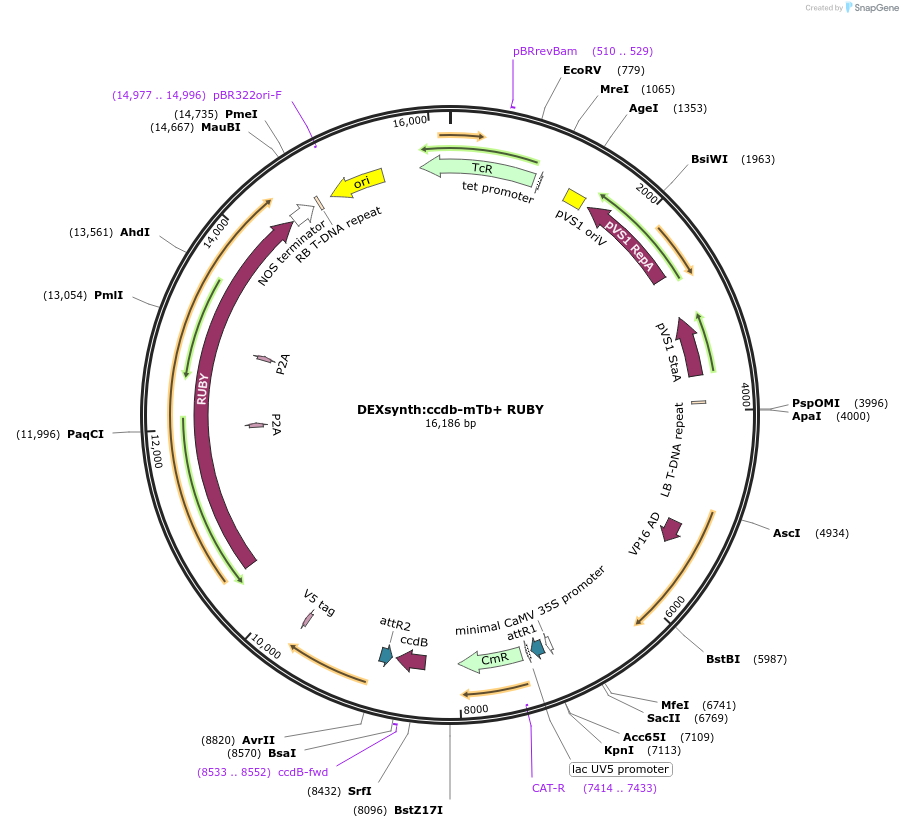
DEXsynth:ccdb-mTb+ RUBY
Plasmid#233322PurposeGATEWAY compatible vector with a dexamethasone (DEX)-inducible promoter driving in frame epitope fusion miniTurbo-ID paired with ccdb selection toxin and RUBY markerDepositorInsertDEXsynth:ccdb-mTb+ RUBY
UseGateway destinationTagsminiTurbo-IDAvailable SinceMarch 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
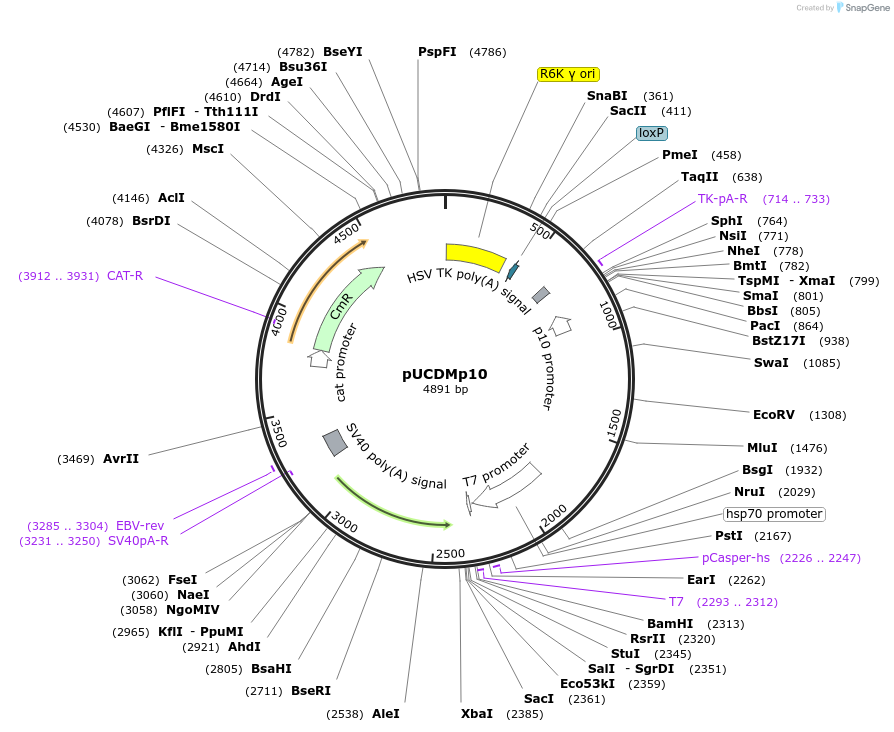
pUCDMp10
Plasmid#170461PurposeEarlyBac insect cell expression systemDepositorInsertNew vector
ExpressionInsectAvailable SinceMarch 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
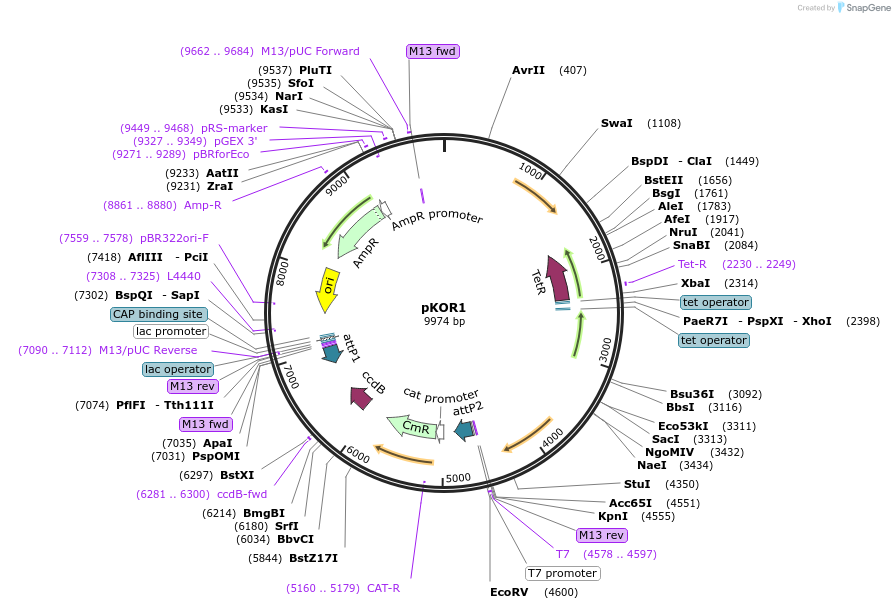
pKOR1
Plasmid#133446PurposeEscherichia coli/S. aureus shuttle vector, permits rapid cloning via lambda recombination and ccdB selectionDepositorTypeEmpty backboneUseE.coli/s.aureus shuttle vectorAvailable SinceOct. 30, 2019AvailabilityAcademic Institutions and Nonprofits only -
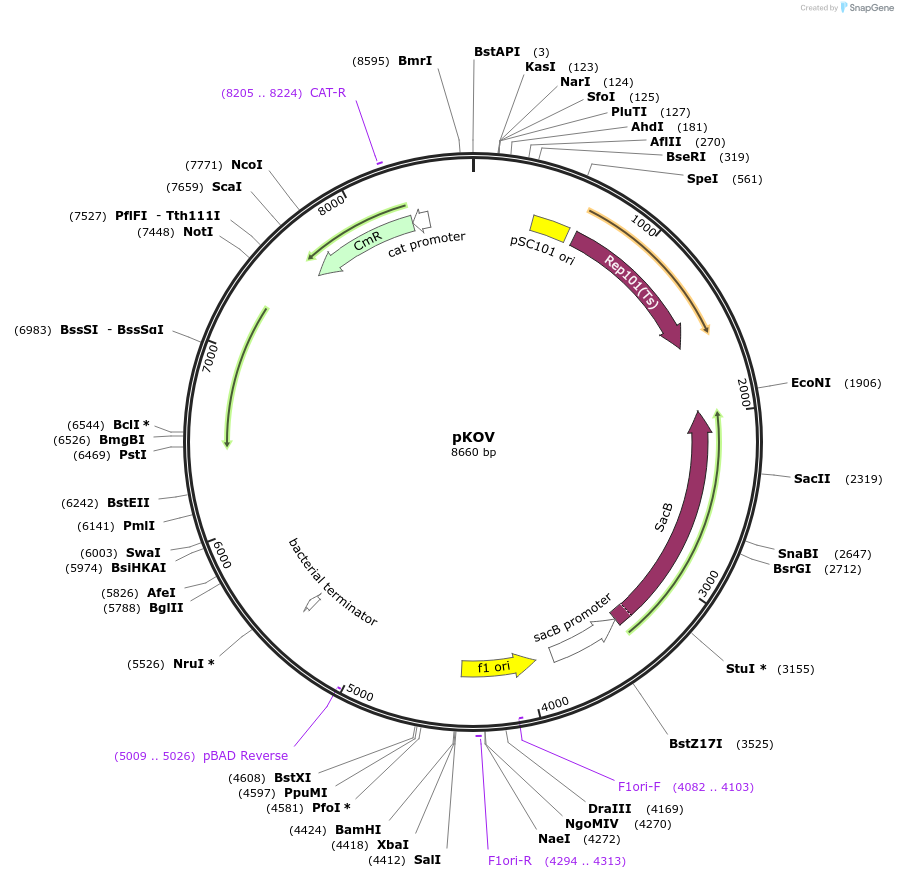
pKOV
Plasmid#25769DepositorTypeEmpty backboneExpressionBacterialAvailable SinceAug. 3, 2010AvailabilityAcademic Institutions and Nonprofits only -
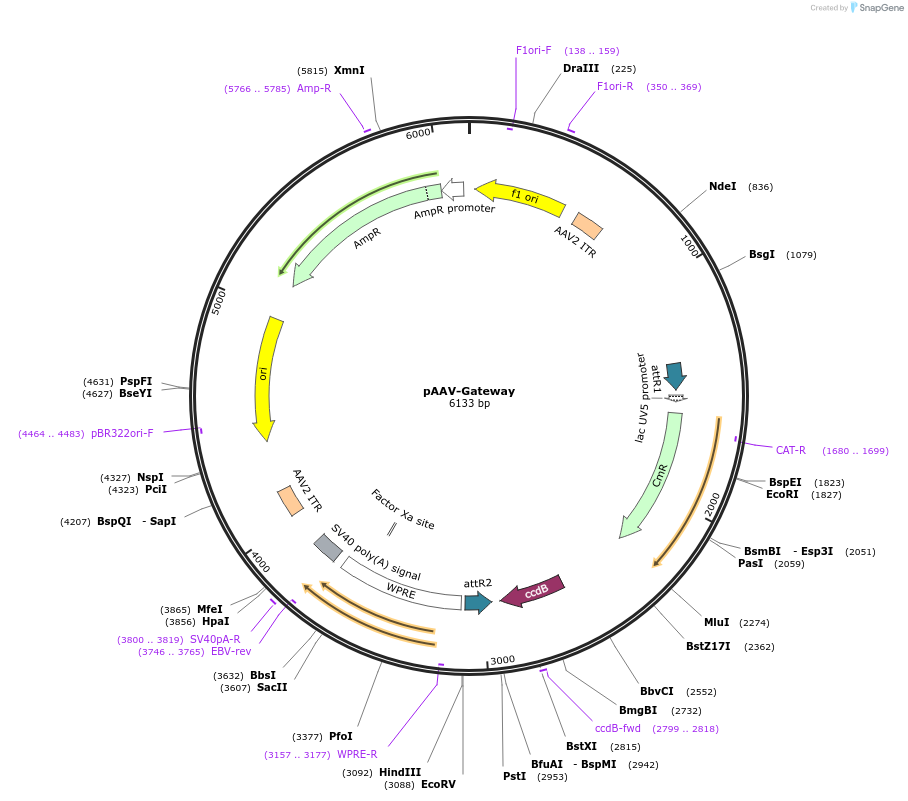
pAAV-Gateway
Plasmid#32671DepositorTypeEmpty backboneUseAAV; AavAvailable SinceJune 29, 2012AvailabilityAcademic Institutions and Nonprofits only -
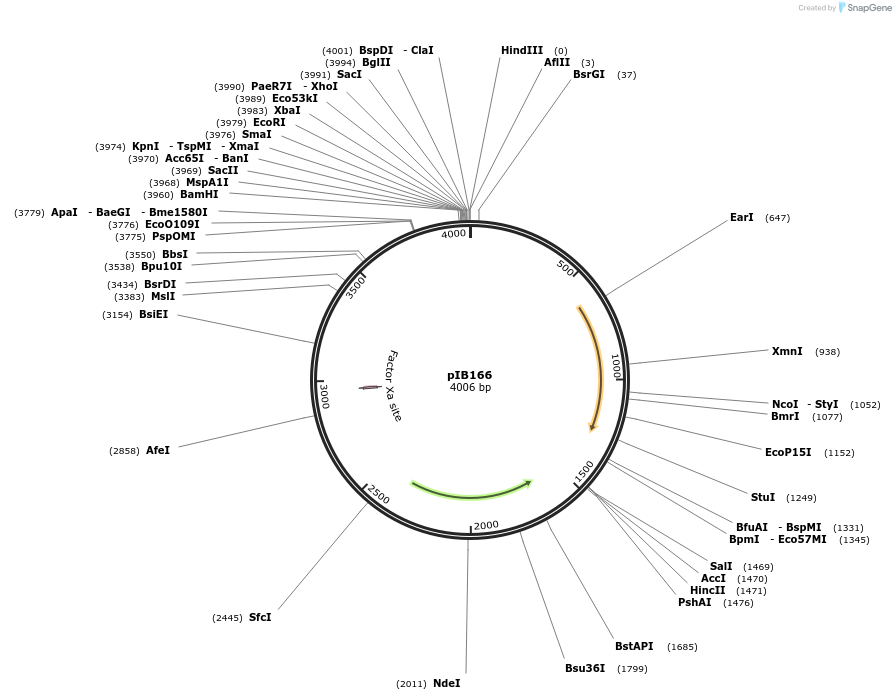
pIB166
Plasmid#90189PurposeE. coli - Streptococci shuttle plasmid for gene expression in streptococci with P23 promoterDepositorTypeEmpty backboneTagsNoneExpressionBacterialAvailable SinceApril 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
lvl0 nluc CDS
Plasmid#153404PurposeNanoluciferase coding sequenceDepositorInsertnanoluciferase CDS
ExpressionPlantAvailable SinceOct. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
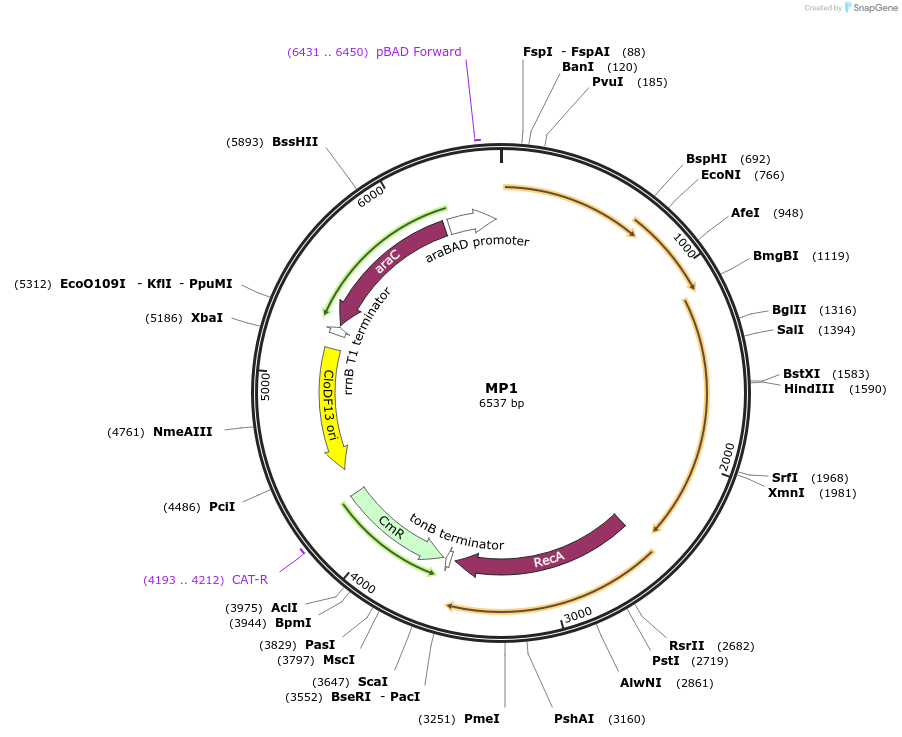
MP1
Plasmid#69627PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 umuD' umuC recA730
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
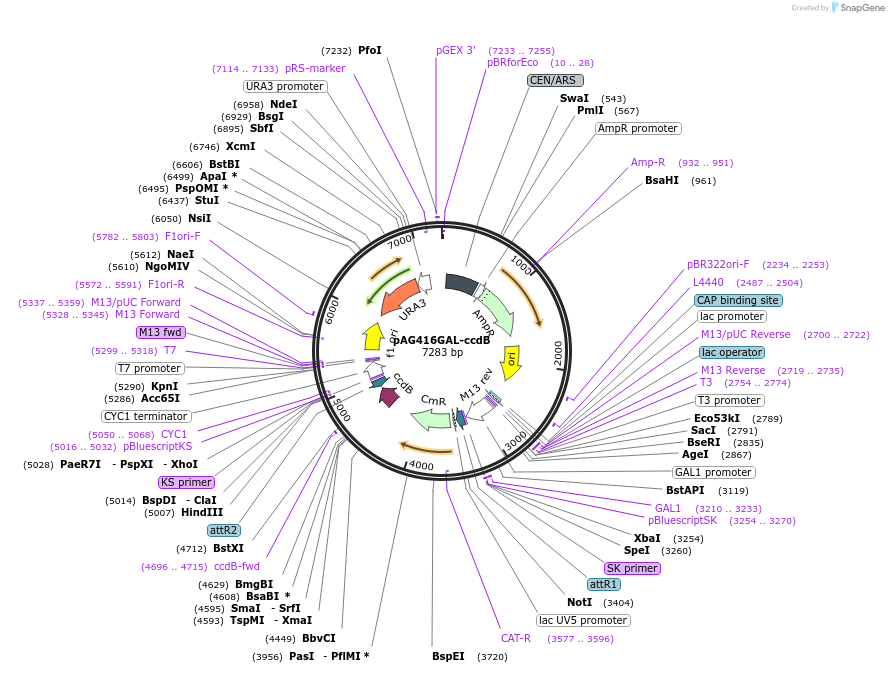
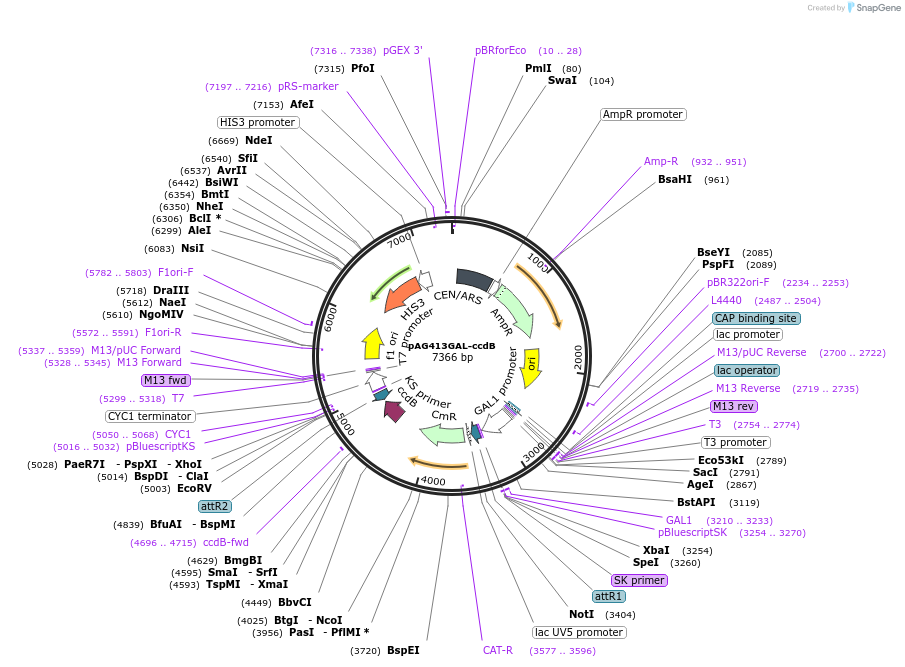
pAG416GAL-ccdB
Plasmid#14147DepositorHas ServiceCloning Grade DNATypeEmpty backboneUseGateway destinationExpressionYeastAvailable SinceMarch 16, 2007AvailabilityAcademic Institutions and Nonprofits only -
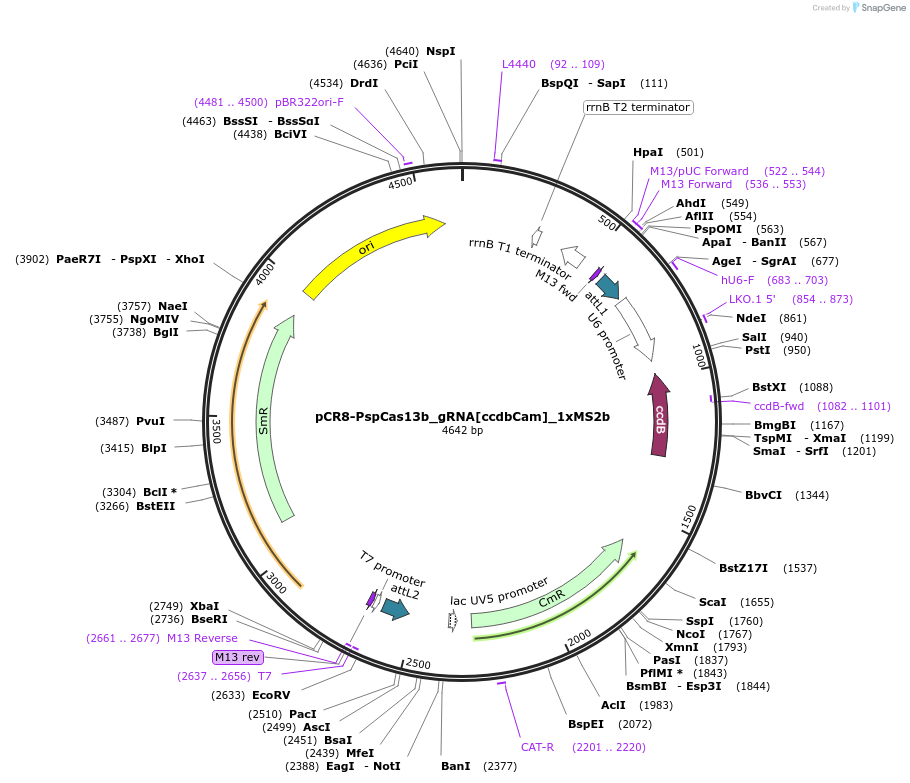
pCR8-PspCas13b_gRNA[ccdbCam]_1xMS2b
Plasmid#196847Purposebackbone for gRNA cloning of dpspCas13b tagged by 1xMS2DepositorTypeEmpty backboneUseCRISPRAvailable SinceMarch 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
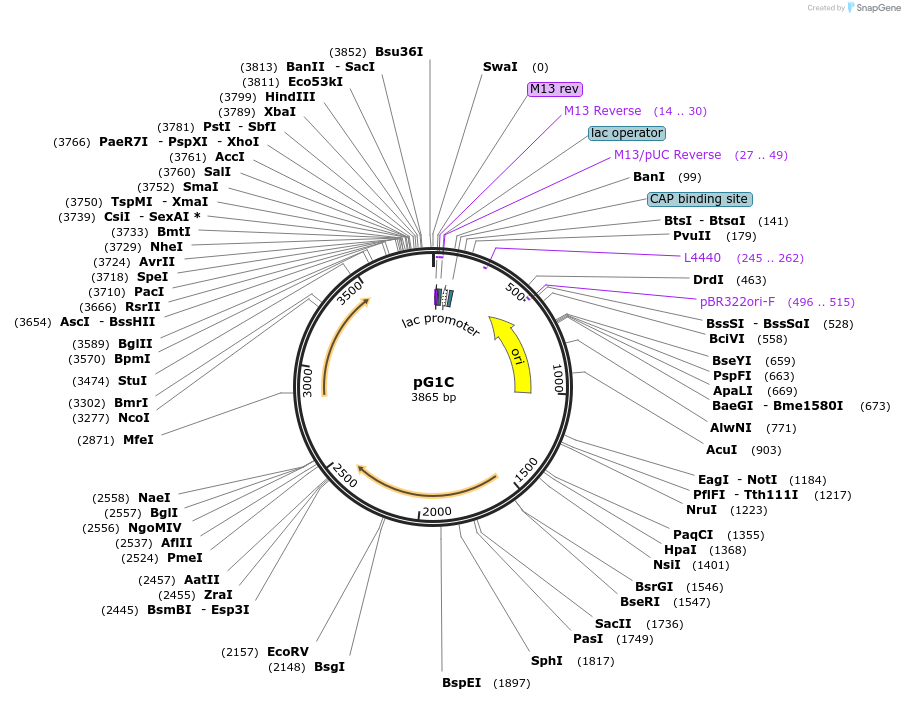
pG1C
Plasmid#71740PurposeModular shuttle vector for Geobacillus and E. coliDepositorTypeEmpty backboneUseSynthetic Biology; Shuttle vectorExpressionBacterialAvailable SinceJuly 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
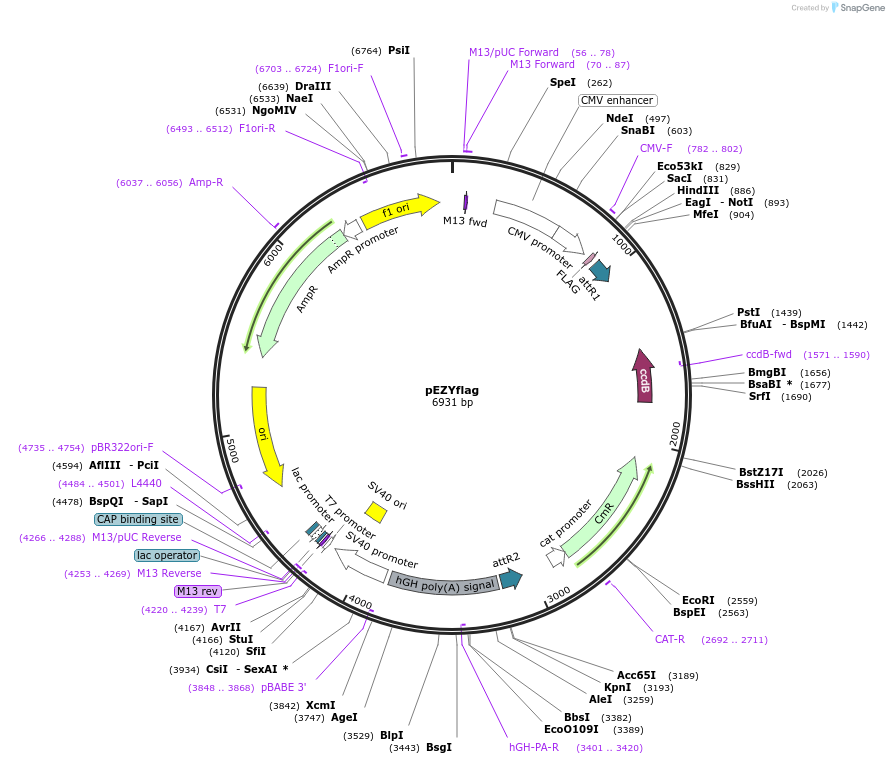
pEZYflag
Plasmid#18700PurposepFlag-CMV-2 converted Gateway Destination vector for expressing Flag tagged fusion proteins in mammalian cellsDepositorTypeEmpty backboneTagsFlagExpressionMammalianPromoterCMVAvailable SinceJuly 26, 2008AvailabilityAcademic Institutions and Nonprofits only -
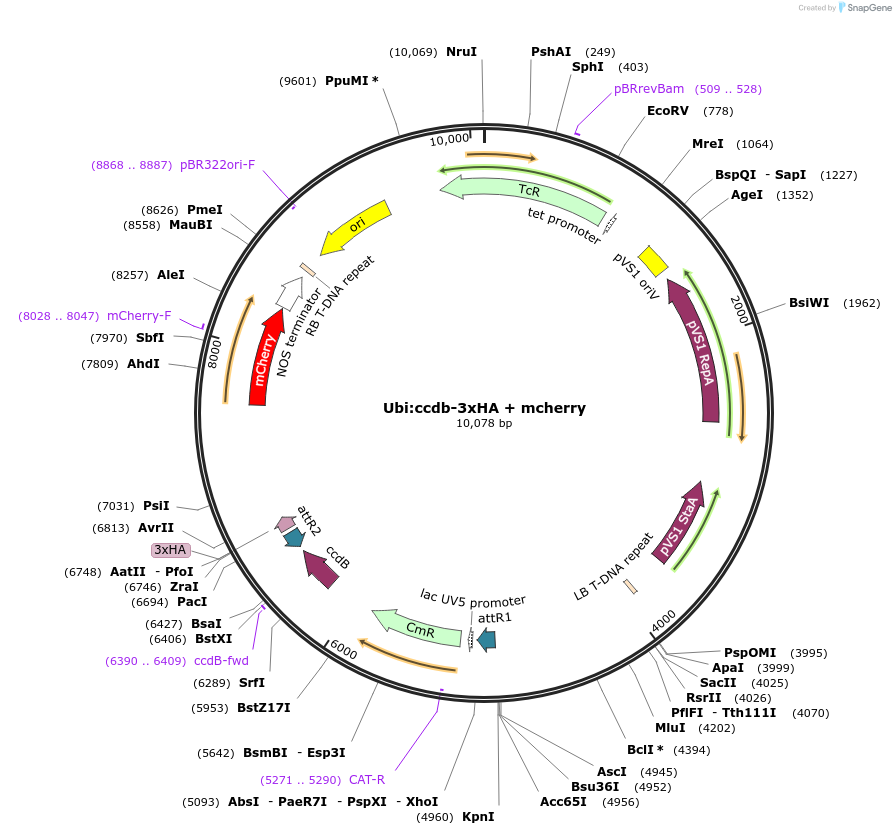
Ubi:ccdb-3xHA + mcherry
Plasmid#233302PurposeGATEWAY compatible vector with a soybean Ubiquitin (GmUbi) promoter driving in frame epitope fusion of 3xHA tag paired with ccdb selection toxin and mCherry markerDepositorInsertUbi:ccdb-3xHA + mcherry
UseGateway destinationTags3xHAAvailable SinceFeb. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
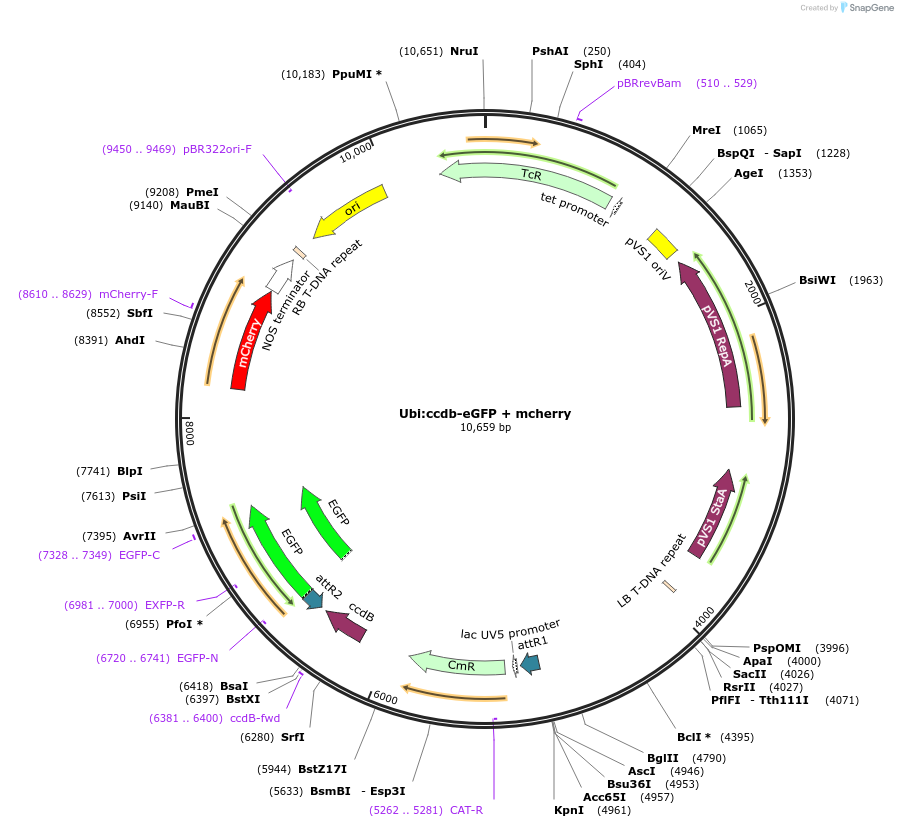
Ubi:ccdb-eGFP + mcherry
Plasmid#233300PurposeGATEWAY compatible vector with a soybean Ubiquitin (GmUbi) promoter driving in frame epitope fusion of eGFP paired with ccdb selection toxin and mCherry markerDepositorInsertUbi:ccdb-eGFP + mcherry
UseGateway destinationTagseGFPAvailable SinceFeb. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
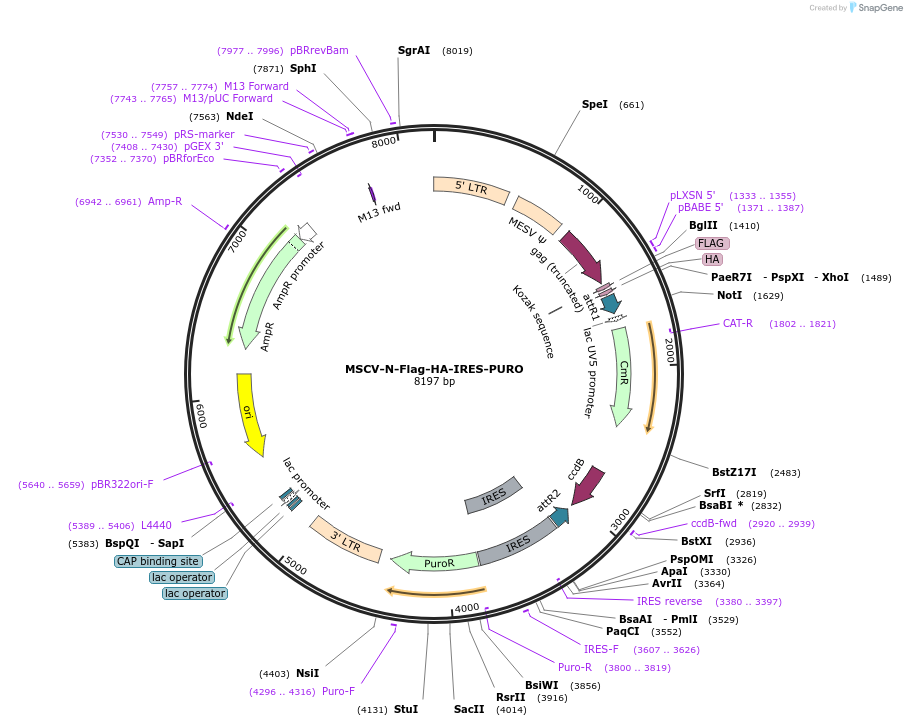
MSCV-N-Flag-HA-IRES-PURO
Plasmid#41033DepositorTypeEmpty backboneUseRetroviral; Gateway destination vectorTagsFlag, HA, and IRES PuroExpressionMammalianPromoterLTR-driven expressionAvailable SinceOct. 26, 2012AvailabilityAcademic Institutions and Nonprofits only -
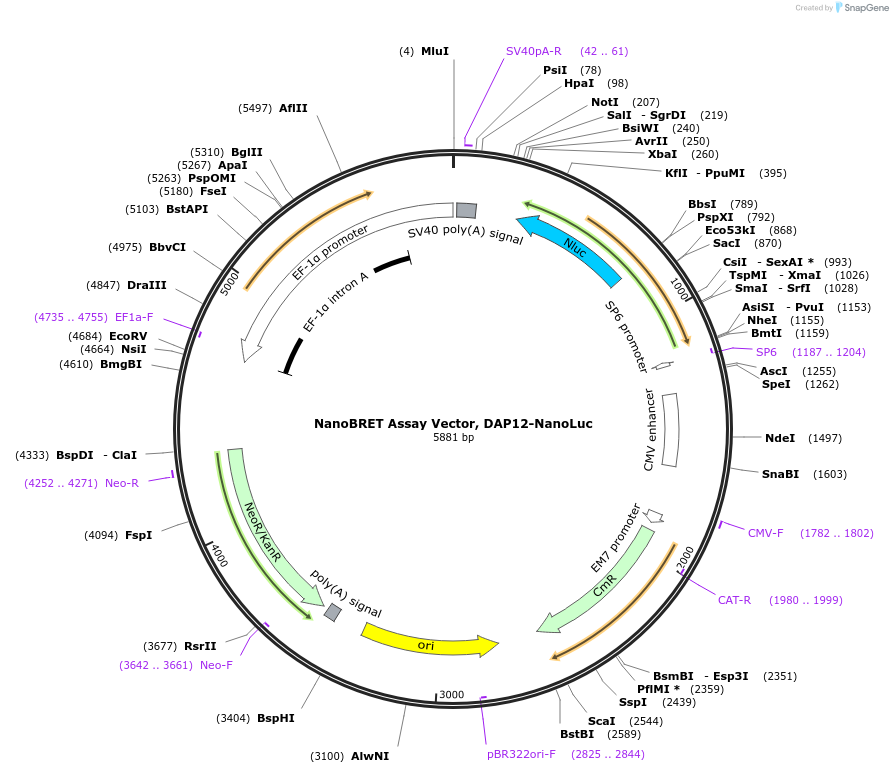
NanoBRET Assay Vector, DAP12-NanoLuc
Plasmid#238723PurposeExpress DAP12-NanoLuc Fusion Protein in Mammalian Cells under a CMV promoterDepositorHas ServiceDNAInsertDAP12 (TYROBP Human)
TagsNanoLuc (R)ExpressionMammalianMutationNonePromoterCMVAvailable SinceSept. 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
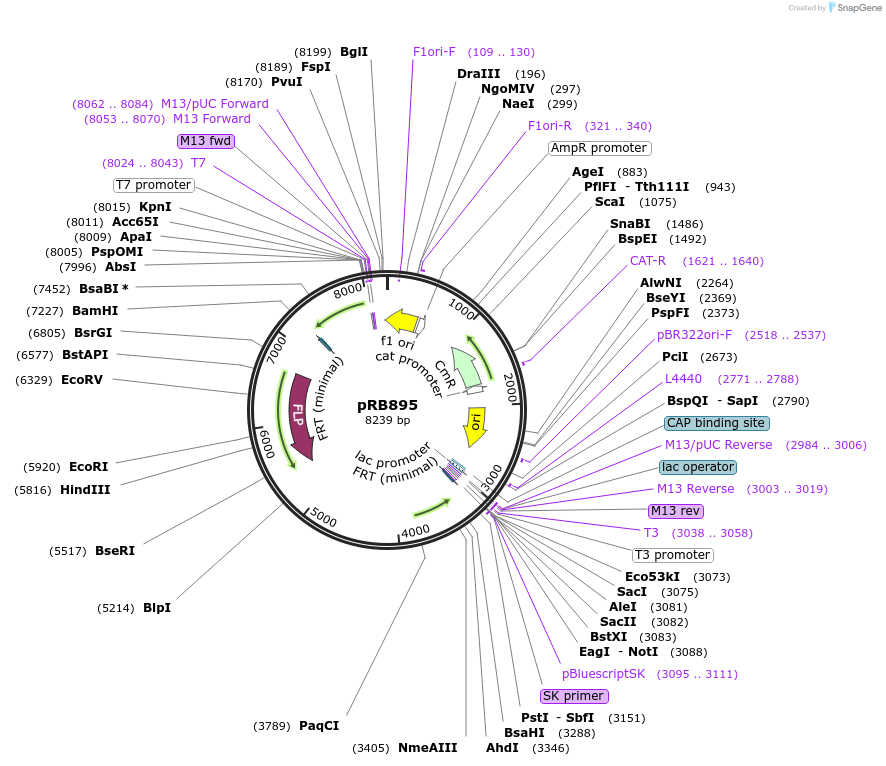
pRB895
Plasmid#124586PurposeC-terminal fusions of C. albicans optimized mNeonGreen fluorescent protein.DepositorTypeEmpty backboneUseSynthetic Biology; Candida albicans expressionTagsmNeonGreenAvailable SinceMay 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
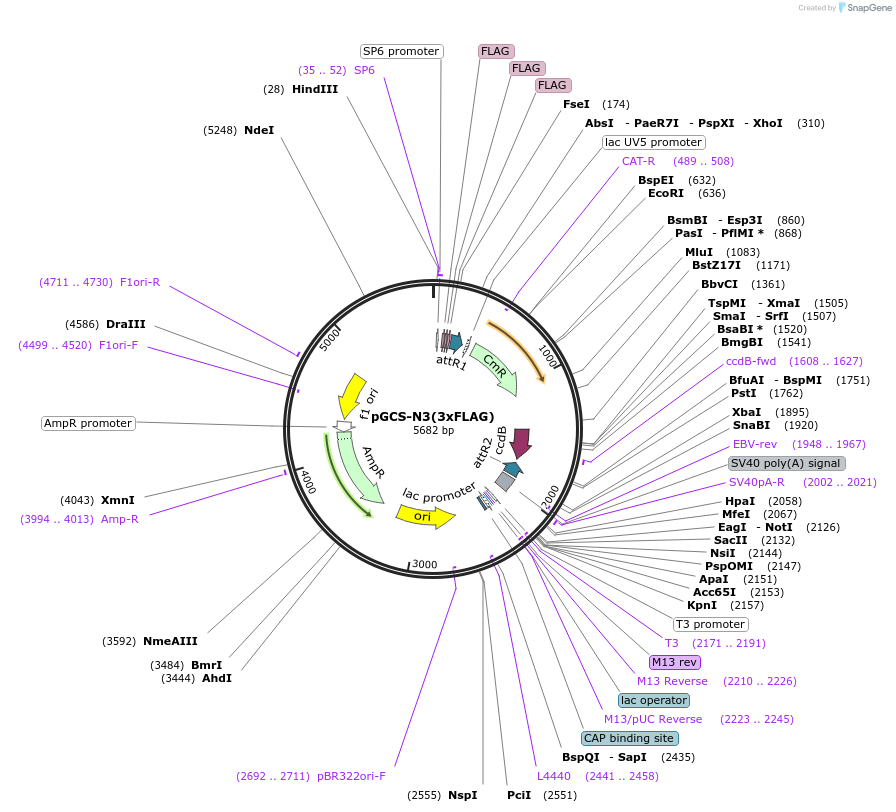
pGCS-N3(3xFLAG)
Plasmid#85720PurposeGateway destination vectorDepositorTypeEmpty backboneUseMammalian/avian/xenopus/zebrafishTags3xFLAGAvailable SinceFeb. 16, 2017AvailabilityAcademic Institutions and Nonprofits only -
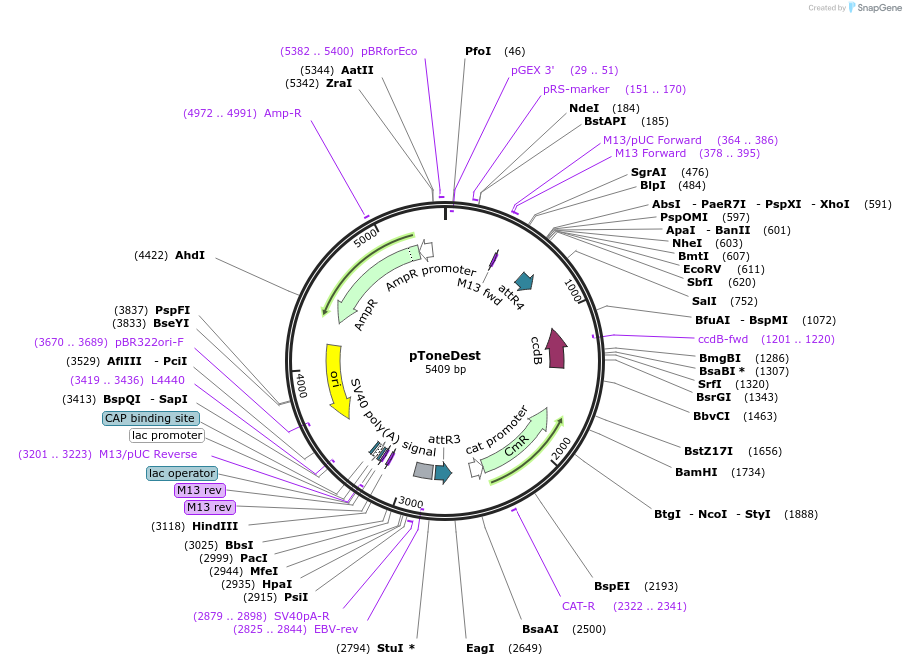
pToneDest
Plasmid#67691PurposeattR4-attR3 Destination cassette in Tol1 backboneDepositorTypeEmpty backboneAvailable SinceAug. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
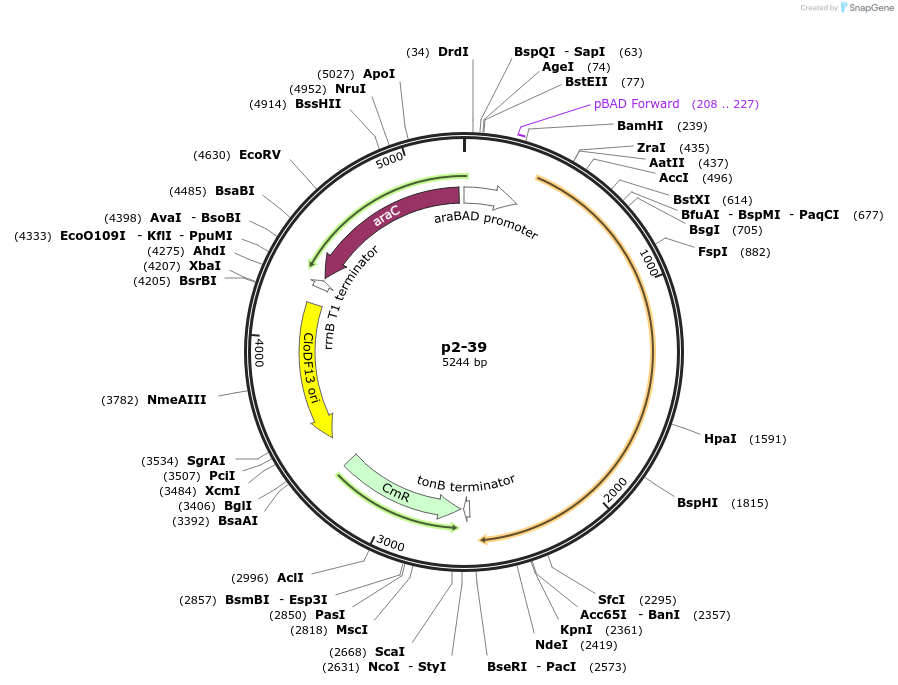
p2-39
Plasmid#118086PurposeZB-linker-split C-terminal T7 RNAP expression plasmidDepositorInsertZB-linker-split C-terminal T7 RNAP expression plasmid
UseSynthetic BiologyAvailable SinceNov. 7, 2018AvailabilityAcademic Institutions and Nonprofits only -
DEXsynth:ccdb-eGFP + RUBY
Plasmid#233317PurposeGATEWAY compatible vector with a dexamethasone (DEX)-inducible promoter driving in frame epitope fusion of eGFP paired with ccdb selection toxin and RUBY markerDepositorInsertDEXsynth:ccdb-eGFP + RUBY
UseGateway destinationTagseGFPAvailable SinceMarch 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
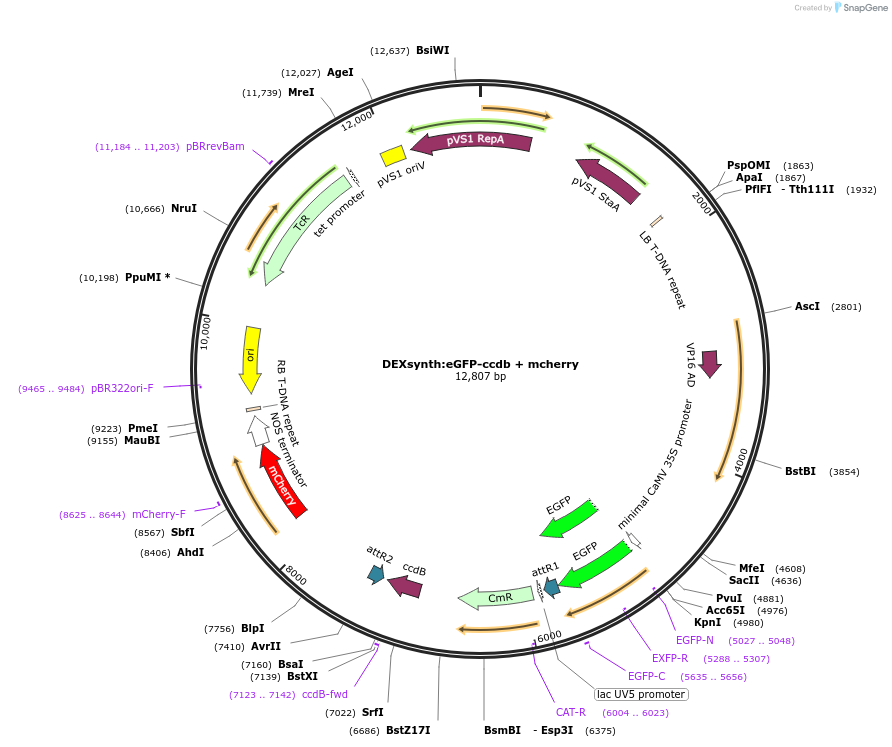
DEXsynth:eGFP-ccdb + mcherry
Plasmid#233311PurposeGATEWAY compatible vector with a dexamethasone (DEX)-inducible promoter driving in frame epitope fusion of eGFP paired with ccdb selection toxin and mCherry markerDepositorInsertDEXsynth:eGFP-ccdb + mcherry
UseGateway destinationTagseGFPAvailable SinceFeb. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
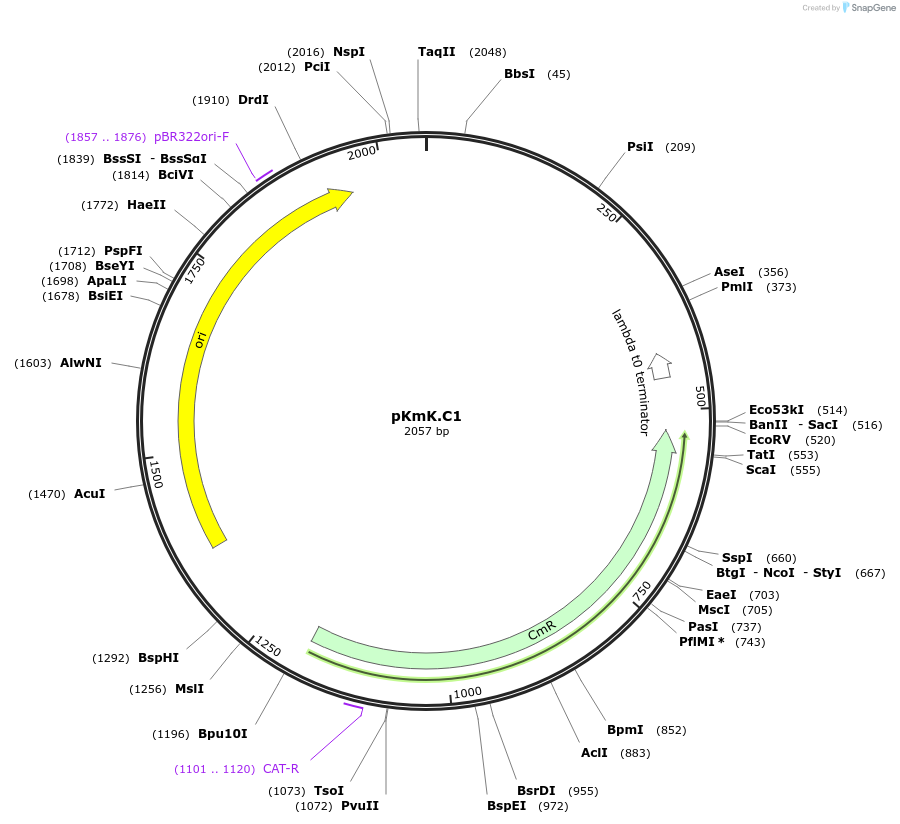
pKmK.C1
Plasmid#125059PurposeStorage plasmid for K.marxianus centromereDepositorInsertKmARS7/CenD
UseSynthetic BiologyExpressionYeastAvailable SinceMay 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
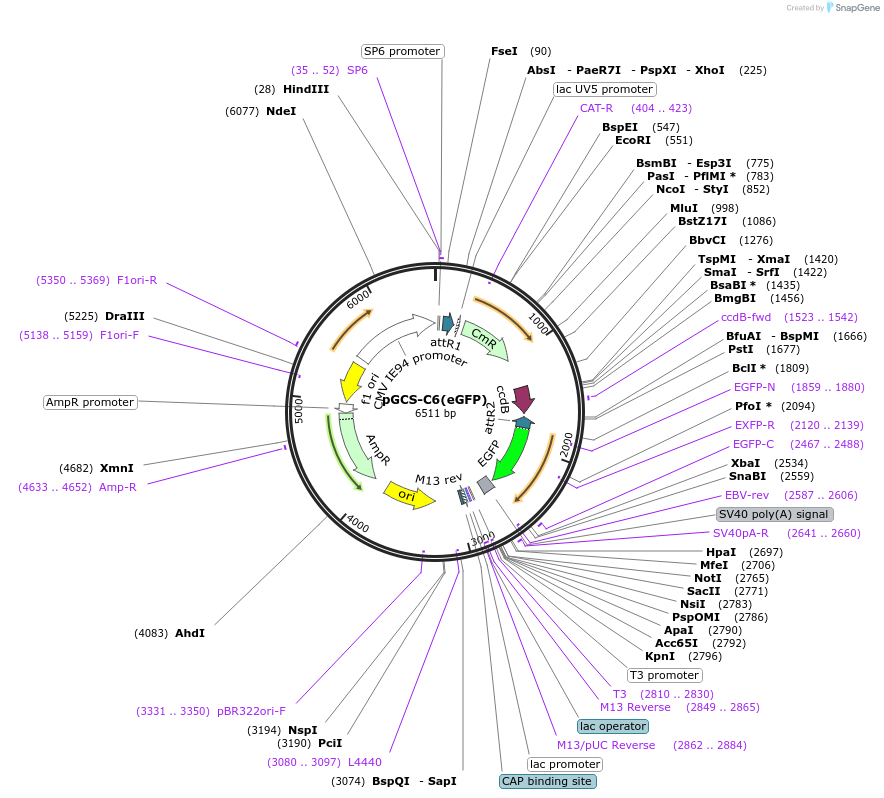
pGCS-C6(eGFP)
Plasmid#85729PurposeGateway destination vectorDepositorTypeEmpty backboneUseMammalian/avian/xenopus/zebrafishTagsEGFPAvailable SinceJan. 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
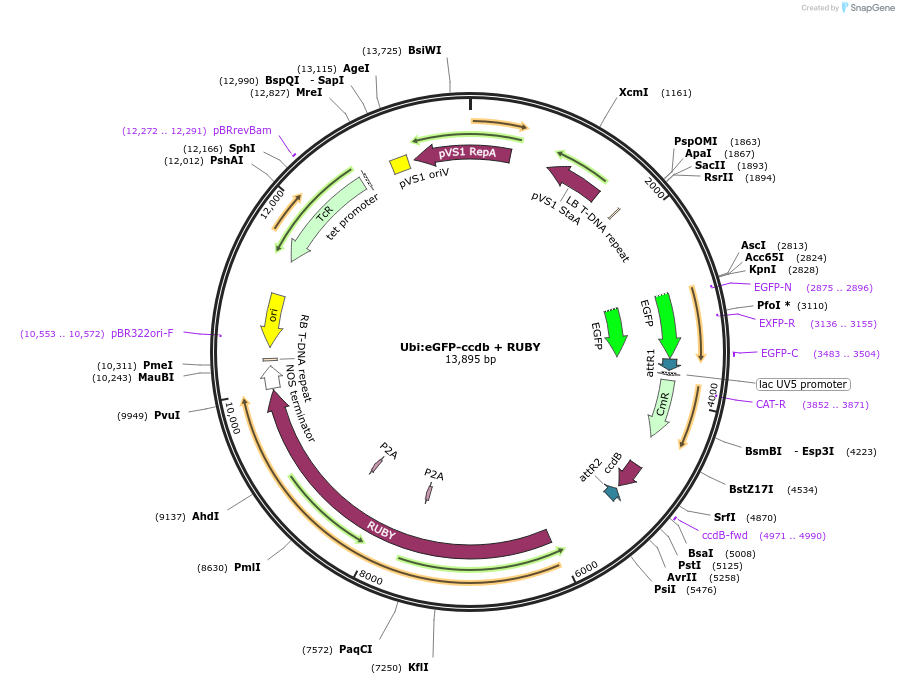
Ubi:eGFP-ccdb + RUBY
Plasmid#233309PurposeGATEWAY compatible vector with a soybean Ubiquitin (GmUbi) promoter driving in frame epitope fusion of eGFP paired with ccdb selection toxin and RUBY markerDepositorInsertUbi:eGFP-ccdb + RUBY
UseGateway destinationTagseGFPAvailable SinceMarch 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
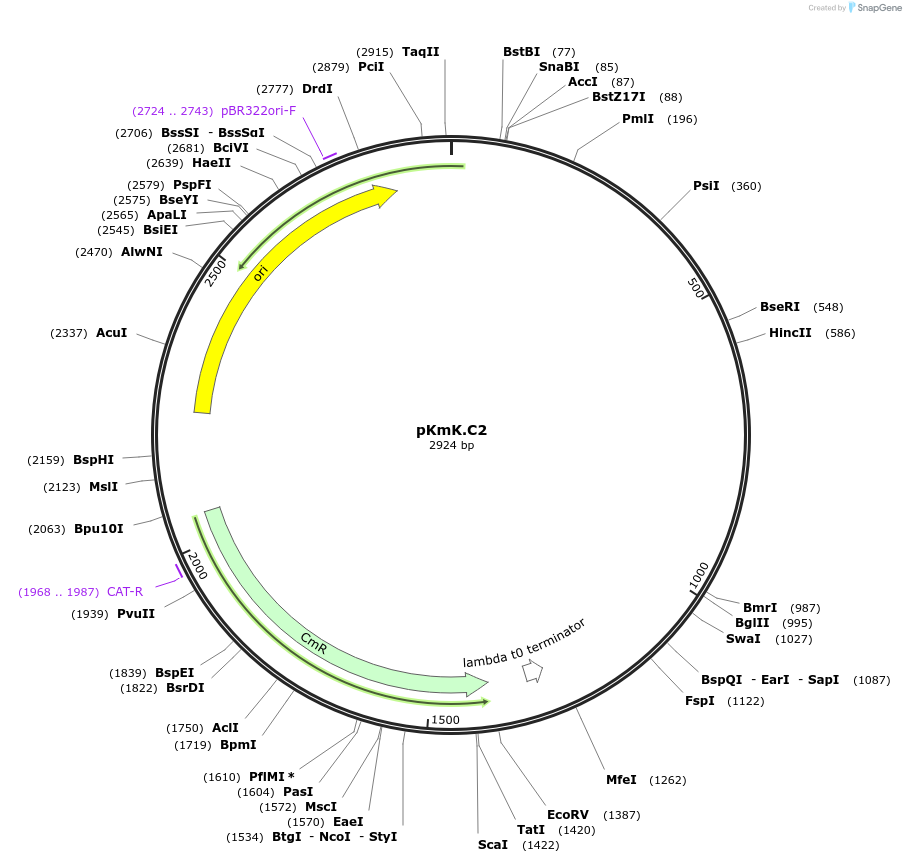
pKmK.C2
Plasmid#125060PurposeStorage plasmid for K.marxianus centromereDepositorInsertKmARS/CEN5
UseSynthetic BiologyExpressionYeastAvailable SinceMay 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
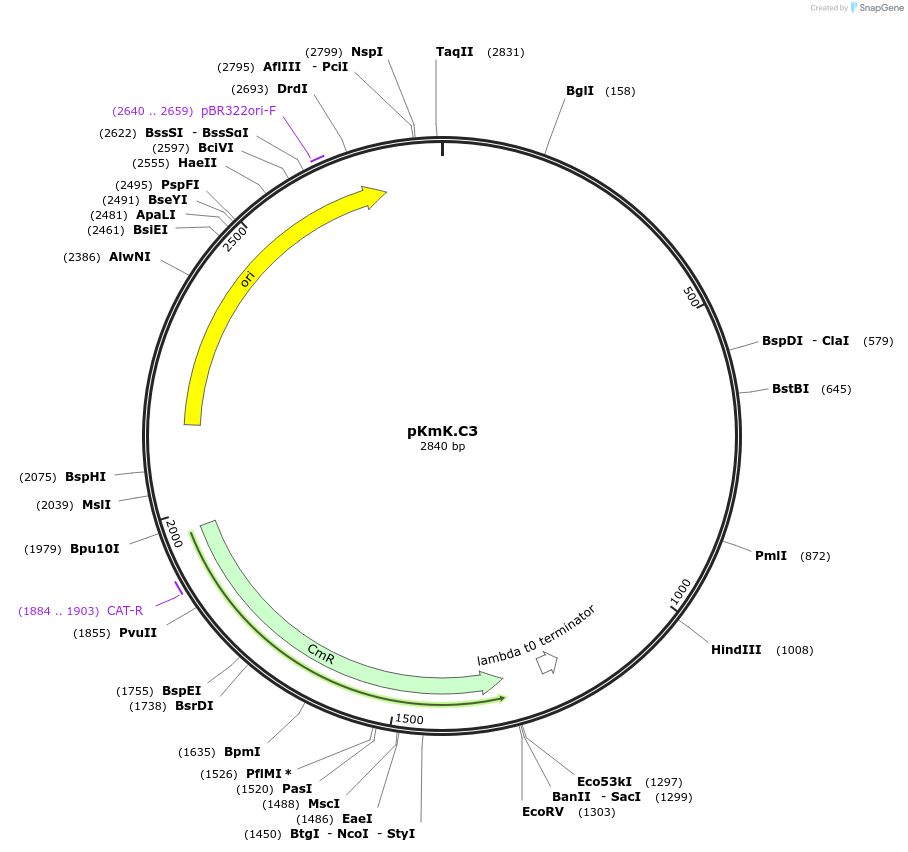
pKmK.C3
Plasmid#125061PurposeStorage plasmid for K.marxianus centromereDepositorInsertKmARS/CEN6
UseSynthetic BiologyExpressionYeastAvailable SinceMay 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
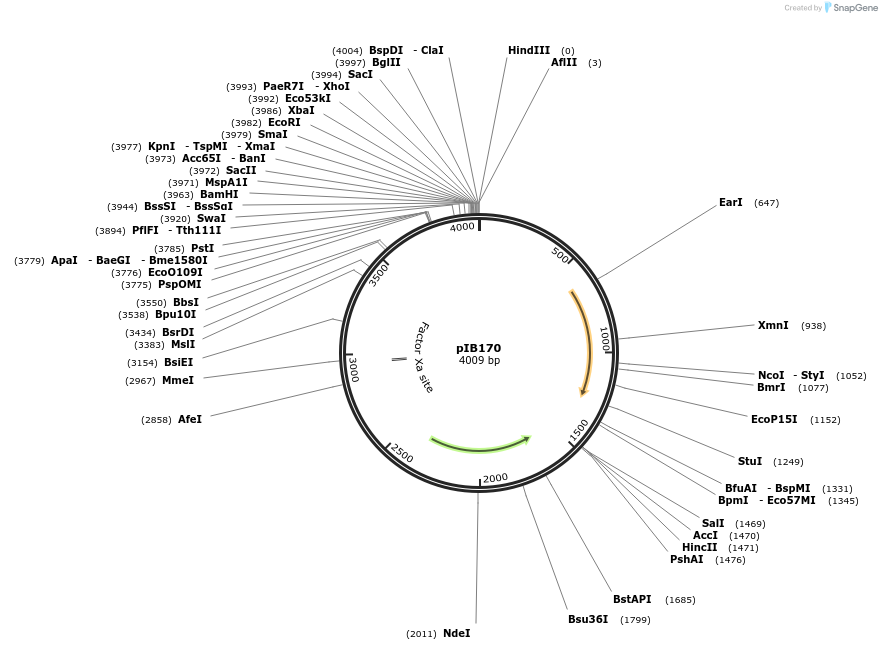
pIB170
Plasmid#90193PurposeE. coli - Streptococci shuttle plasmid for gene expression in streptococci with Pveg promoterDepositorTypeEmpty backboneTagsNoneExpressionBacterialAvailable SinceApril 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
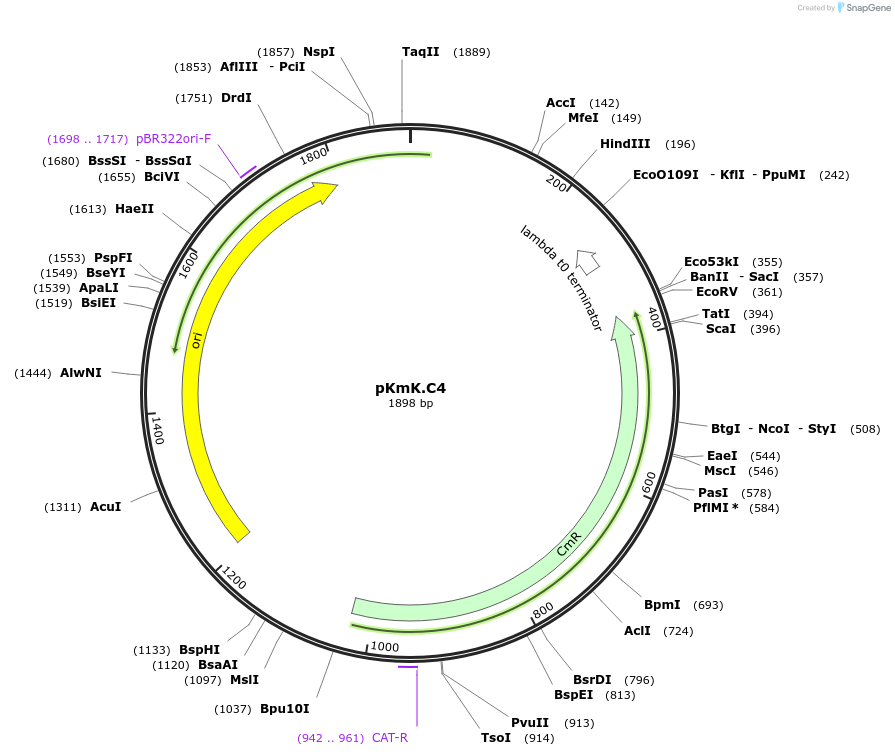
pKmK.C4
Plasmid#125062PurposeStorage plasmid for K.marxianus ARSDepositorInsertKmARS2
UseSynthetic BiologyExpressionYeastAvailable SinceMay 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
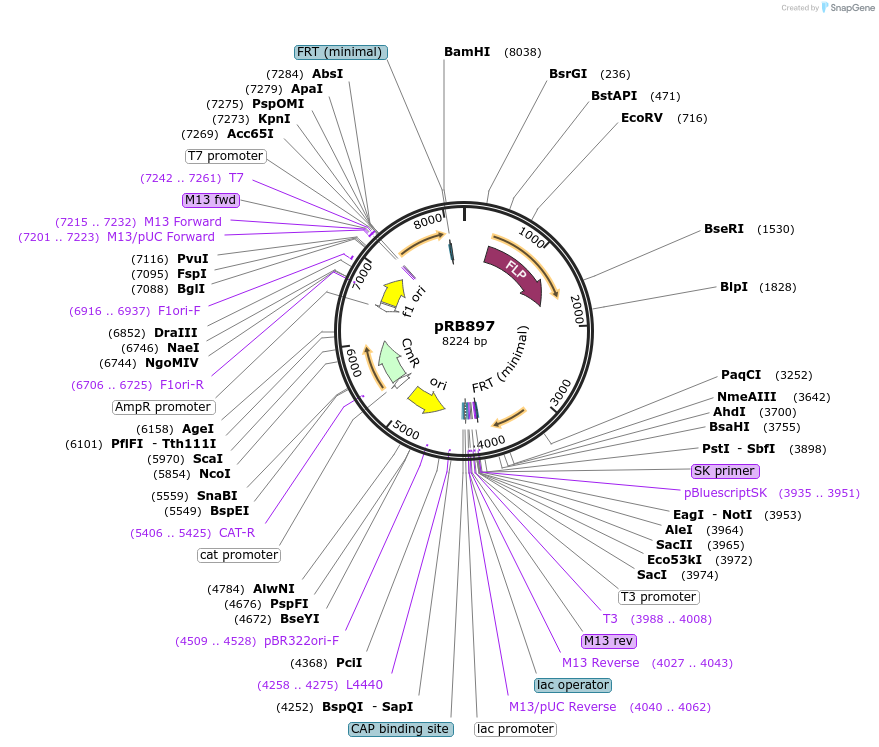
pRB897
Plasmid#124587PurposeC-terminal fusions of C. albicans optimized mScarlet fluorescent protein.DepositorTypeEmpty backboneUseSynthetic Biology; Candida albicans expressionTagsmScarletAvailable SinceApril 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
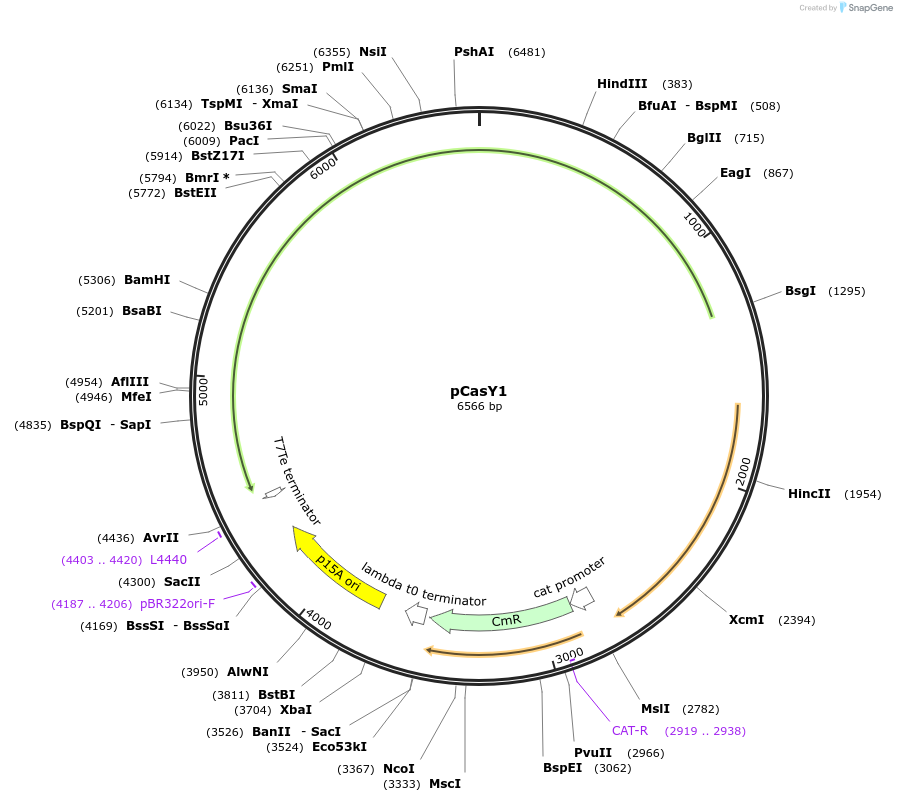
pCasY1
Plasmid#87687Purposep15A plasmid expressing CasY1DepositorInsertCasY1
ExpressionBacterialAvailable SinceApril 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
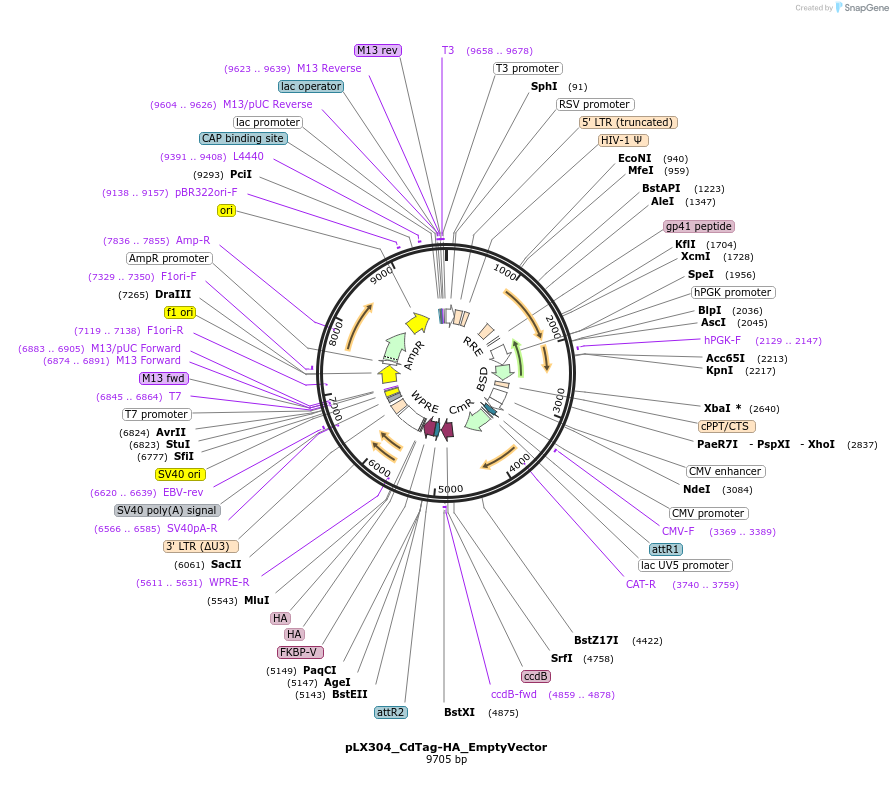
pLX304_CdTag-HA_EmptyVector
Plasmid#221466PurposeGateway backbone for C-terminal dTag cloningDepositorTypeEmpty backboneUseLentiviralTagsFKBP F36VExpressionMammalianAvailable SinceJuly 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
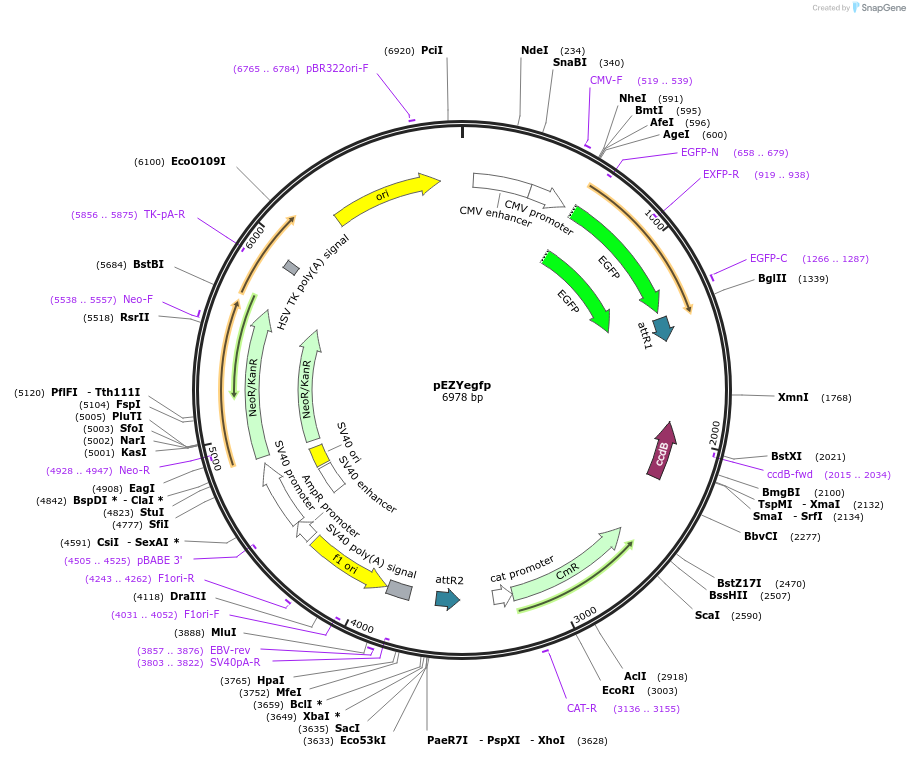
pEZYegfp
Plasmid#18671PurposepEGFPC4 converted Gateway Destination vector for expressing EGFP tagged fusion proteins in mammalian cellsDepositorTypeEmpty backboneTagsGFPExpressionMammalianPromoterCMVAvailable SinceJuly 26, 2008AvailabilityAcademic Institutions and Nonprofits only -
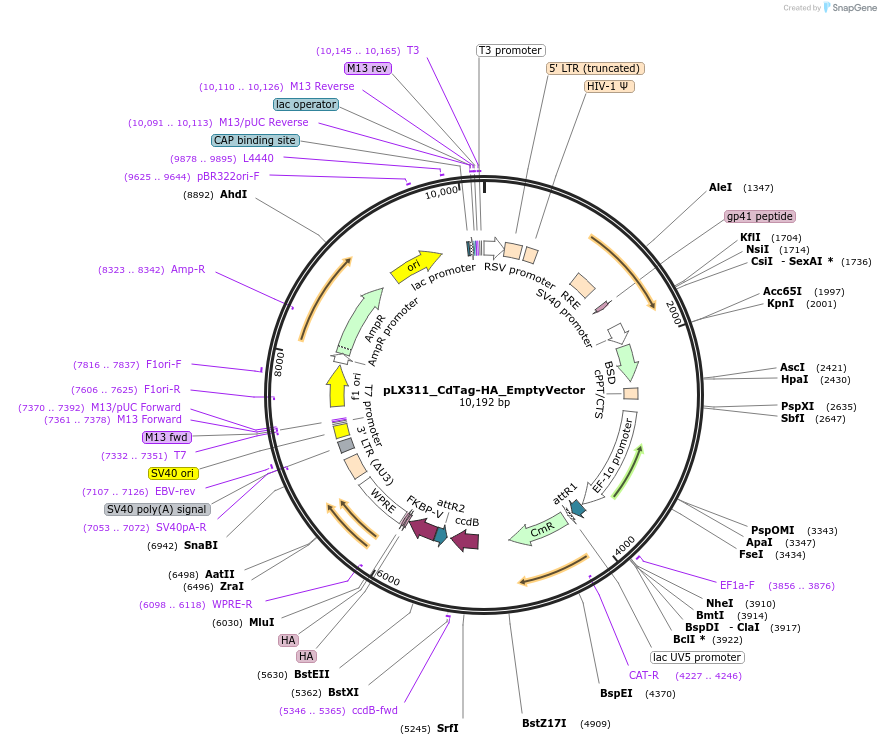
pLX311_CdTag-HA_EmptyVector
Plasmid#221468PurposeGateway backbone for C-terminal dTag cloningDepositorTypeEmpty backboneUseLentiviralTagsFKBP F36VExpressionMammalianAvailable SinceAug. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
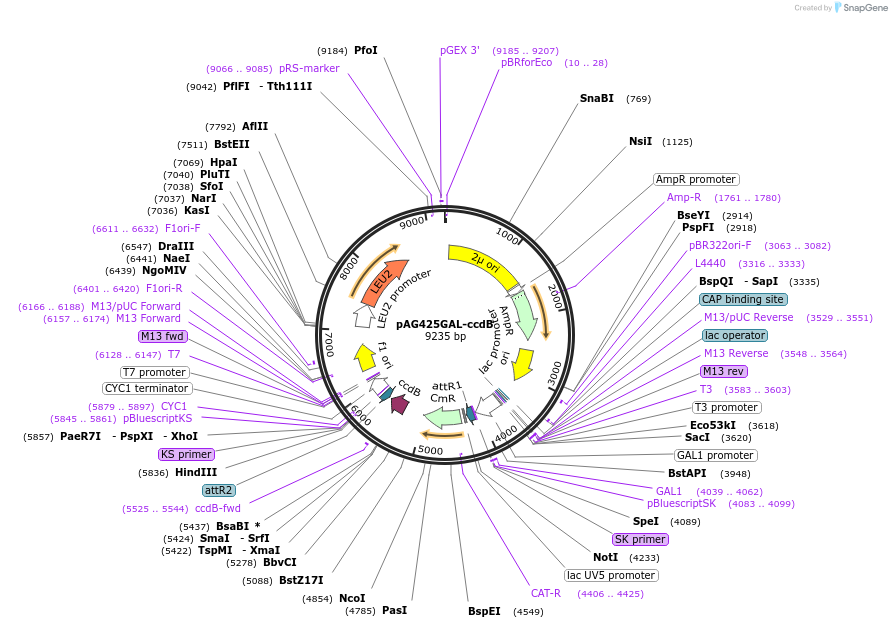
pAG425GAL-ccdB
Plasmid#14153DepositorHas ServiceCloning Grade DNATypeEmpty backboneUseGateway destinationExpressionYeastAvailable SinceMarch 16, 2007AvailabilityAcademic Institutions and Nonprofits only -
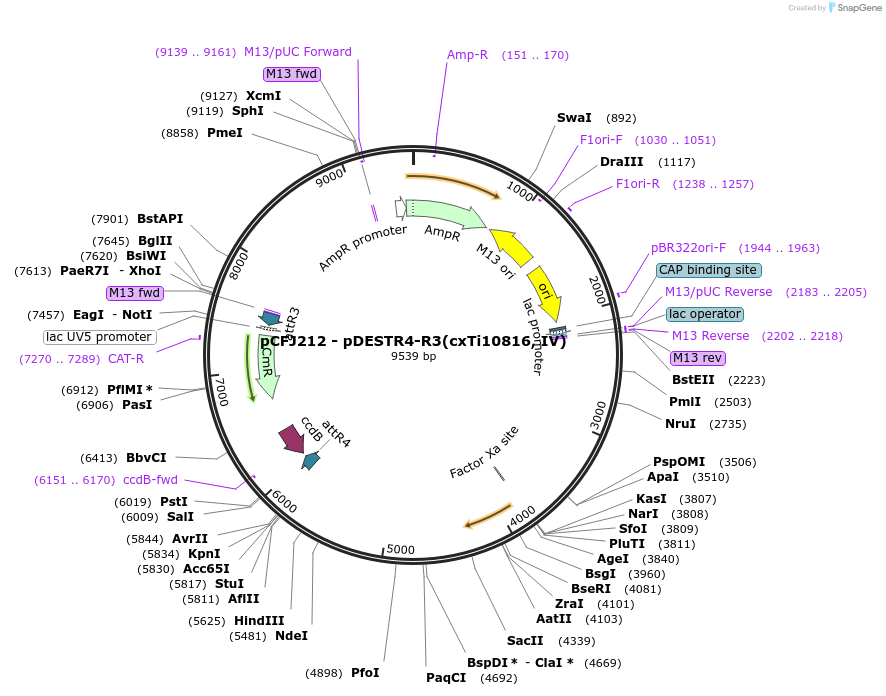
pCFJ212 - pDESTR4-R3(cxTi10816, IV)
Plasmid#34864DepositorInsertcxTi10816 targeting
UseWorm targetingAvailable SinceJune 25, 2012AvailabilityAcademic Institutions and Nonprofits only -
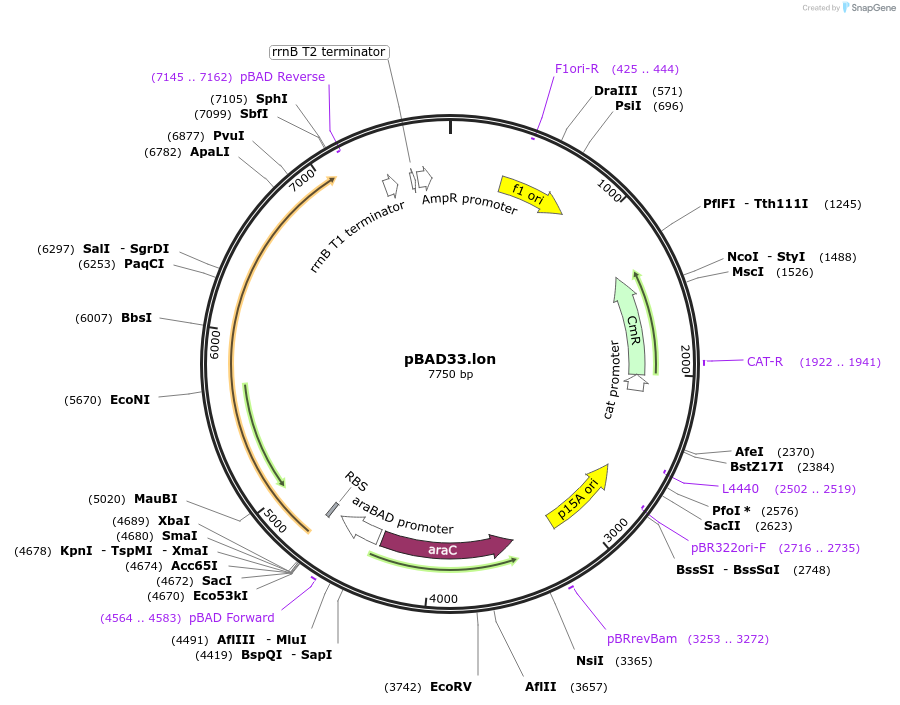
pBAD33.lon
Plasmid#22145DepositorAvailable SinceSept. 28, 2009AvailabilityAcademic Institutions and Nonprofits only -
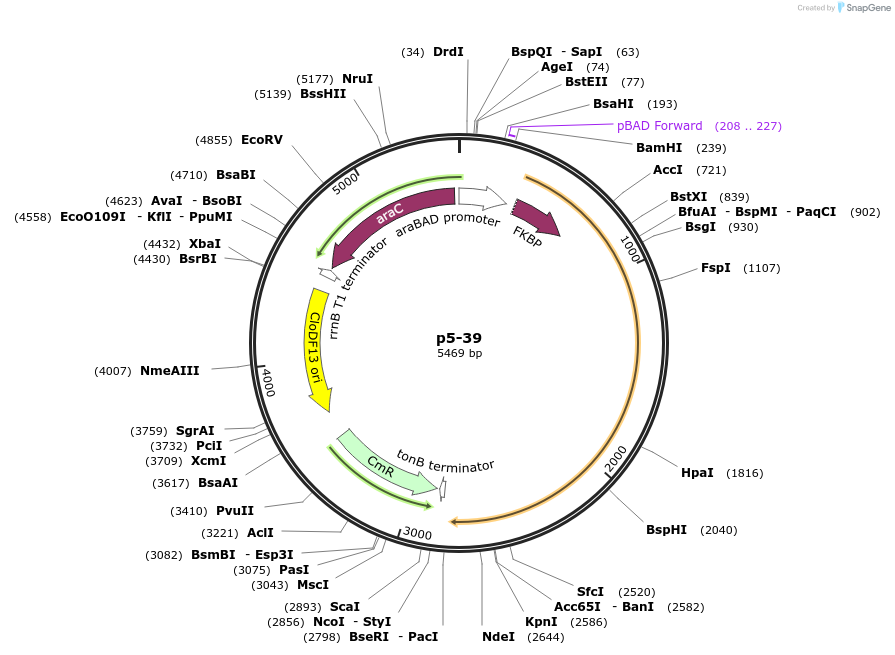
p5-39
Plasmid#118088PurposeFKBP-TSGGSG-Split C-terminal T7 RNAP expression plasmidDepositorInsertFKBP-TSGGSG-Split C-terminal T7 RNAP expression plasmid
UseSynthetic BiologyAvailable SinceNov. 7, 2018AvailabilityAcademic Institutions and Nonprofits only -
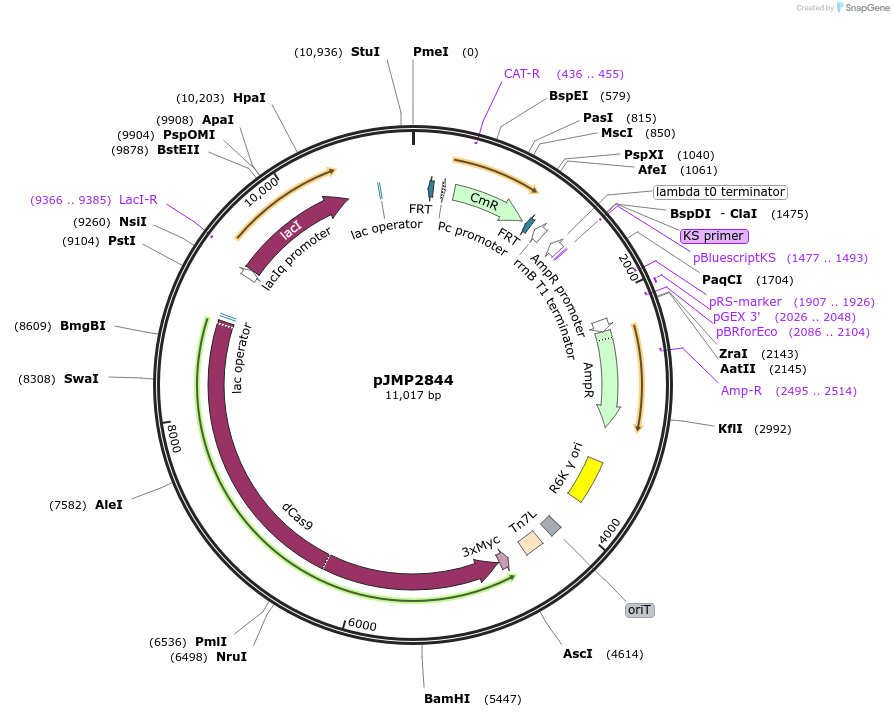
pJMP2844
Plasmid#160675PurposeMobile CRISPRi empty sgRNA (BsaI) for cloning new sgRNAsDepositorTypeEmpty backboneExpressionBacterialAvailable SinceNov. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
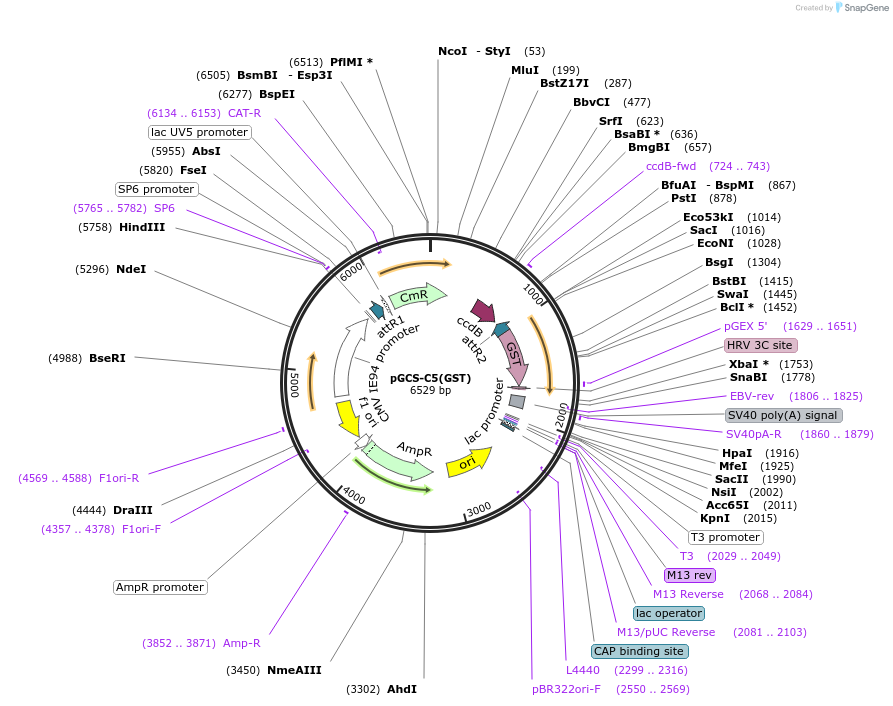
pGCS-C5(GST)
Plasmid#85728PurposeGateway destination vectorDepositorTypeEmpty backboneUseMammalian/avian/xenopus/zebrafishTagsGSTAvailable SinceJan. 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
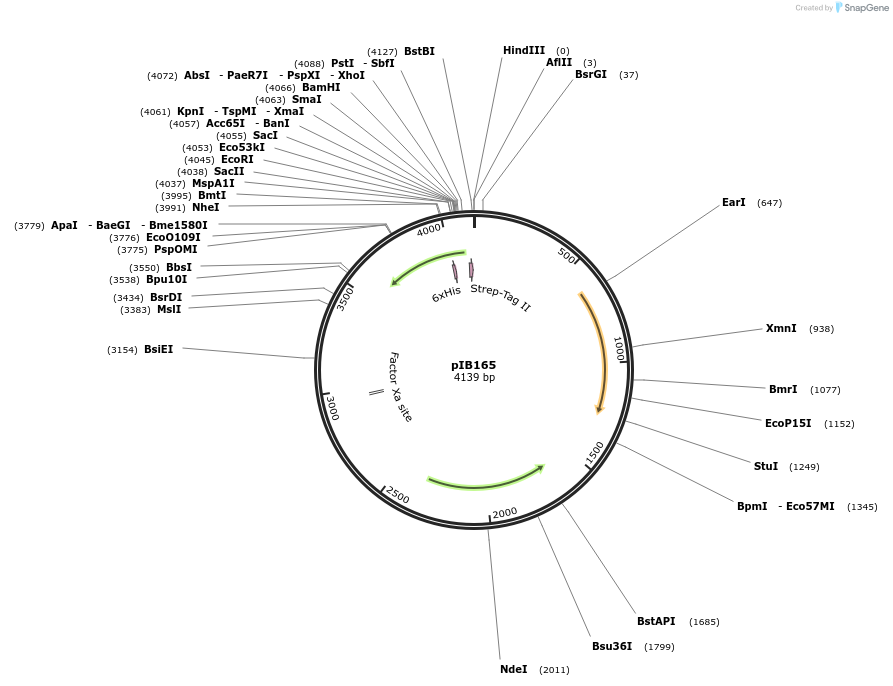
pIB165
Plasmid#90188PurposeE. coli - Streptococci shuttle plasmid for gene expression in streptococci with P23 promoterDepositorTypeEmpty backboneTagsHis-tag and Strep-tagExpressionBacterialAvailable SinceApril 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
pAG413GAL-ccdB
Plasmid#14141DepositorHas ServiceCloning Grade DNATypeEmpty backboneUseGateway destinationExpressionYeastAvailable SinceMarch 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
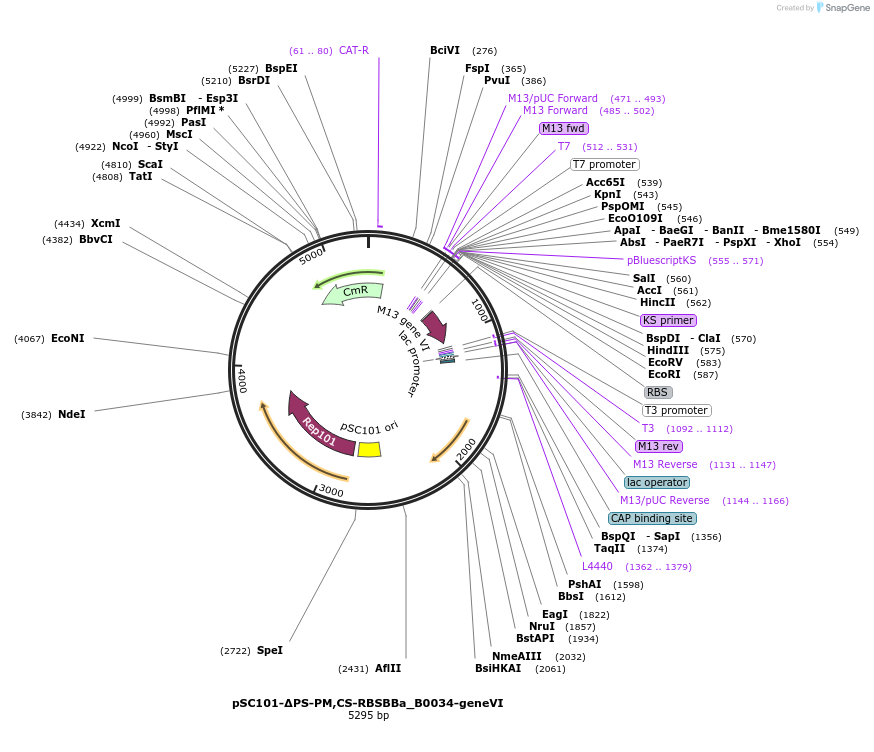
pSC101-ΔPS-PM,CS-RBSBBa_B0034-geneVI
Plasmid#134356PurposeAccessory plasmidDepositorInsertGene VI
ExpressionBacterialAvailable SinceJune 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
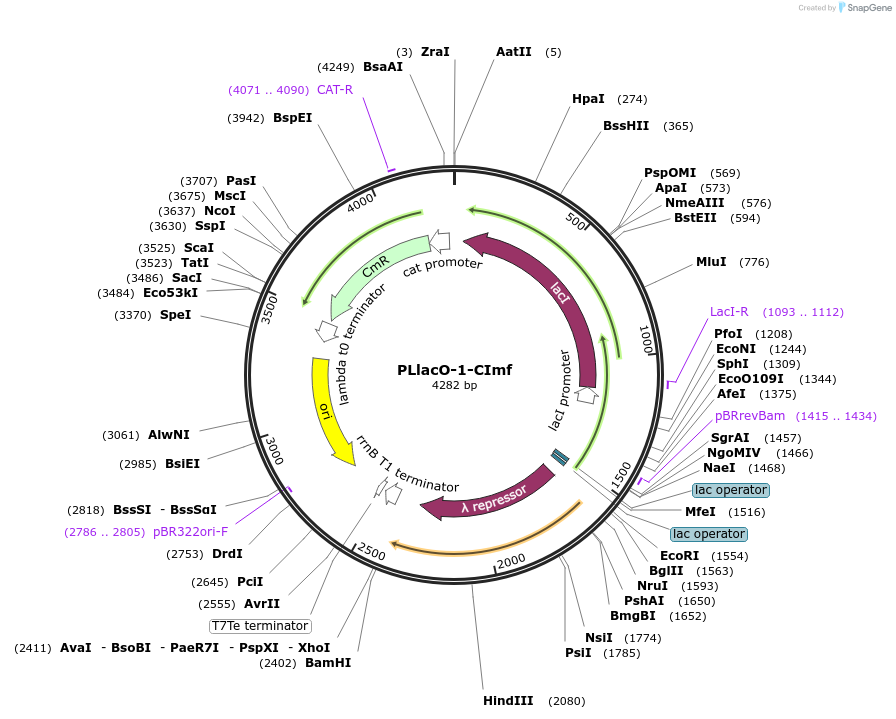
PLlacO-1-CImf
Plasmid#40126DepositorInsertLac inducible lambda repressor
UseSynthetic BiologyTagsmf-lon ssrA tagExpressionBacterialAvailable SinceSept. 12, 2012AvailabilityAcademic Institutions and Nonprofits only