We narrowed to 17,883 results for: Ret;
-
Plasmid#24520DepositorInsertECFP, EYFP
TagsECFP and EYFPExpressionMammalianAvailable SinceJune 3, 2011AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1 mSTIM1 myc
Plasmid#17732DepositorAvailable SinceApril 14, 2008AvailabilityAcademic Institutions and Nonprofits only -
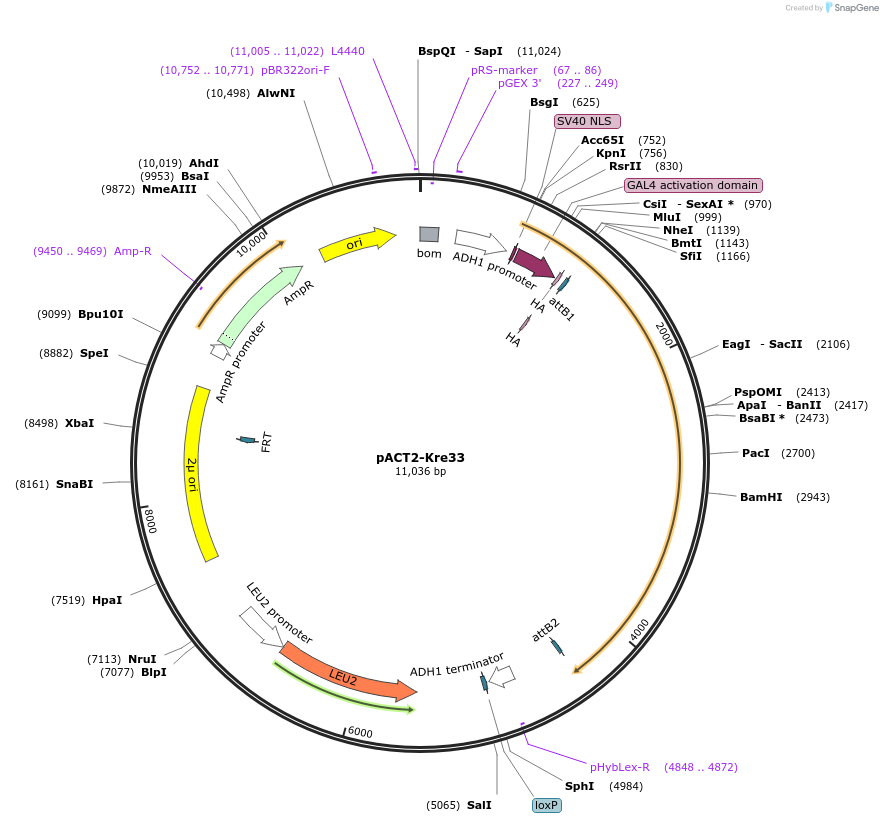
pACT2-Kre33
Plasmid#99990PurposeExpress yeast Kre33 with Y2H activation domain fusionDepositorAvailable SinceNov. 20, 2017AvailabilityAcademic Institutions and Nonprofits only -
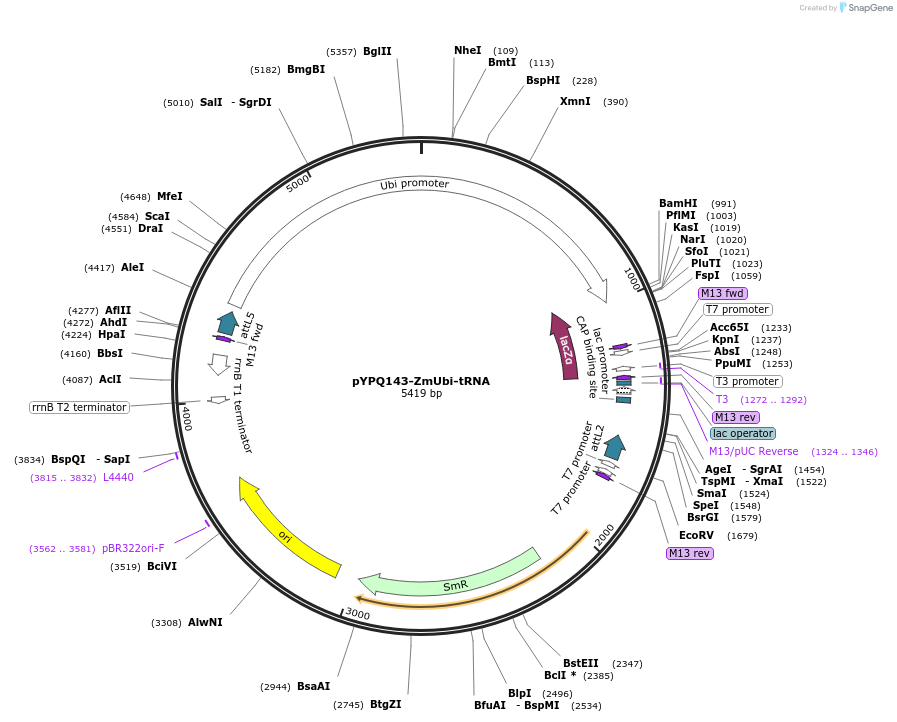
pYPQ143-ZmUbi-tRNA
Plasmid#158400PurposeGateway entry clone and Golden Gate recipient for pYPQ131-tRNA2.0 to pYPQ133-tRNA2.0; assembly of 3 gRNAsDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
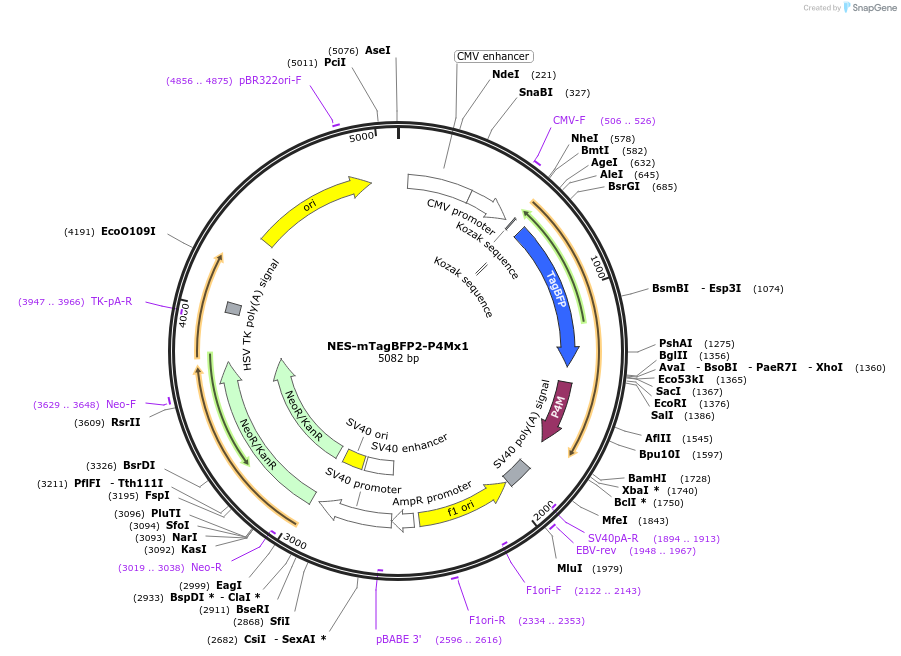
NES-mTagBFP2-P4Mx1
Plasmid#108142PurposePI4P biosensorDepositorInsertX.leavis map2k1.L(32-44):mTagBFP2:L. penomophila SidM(546-647)
ExpressionMammalianPromoterCMVAvailable SinceApril 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
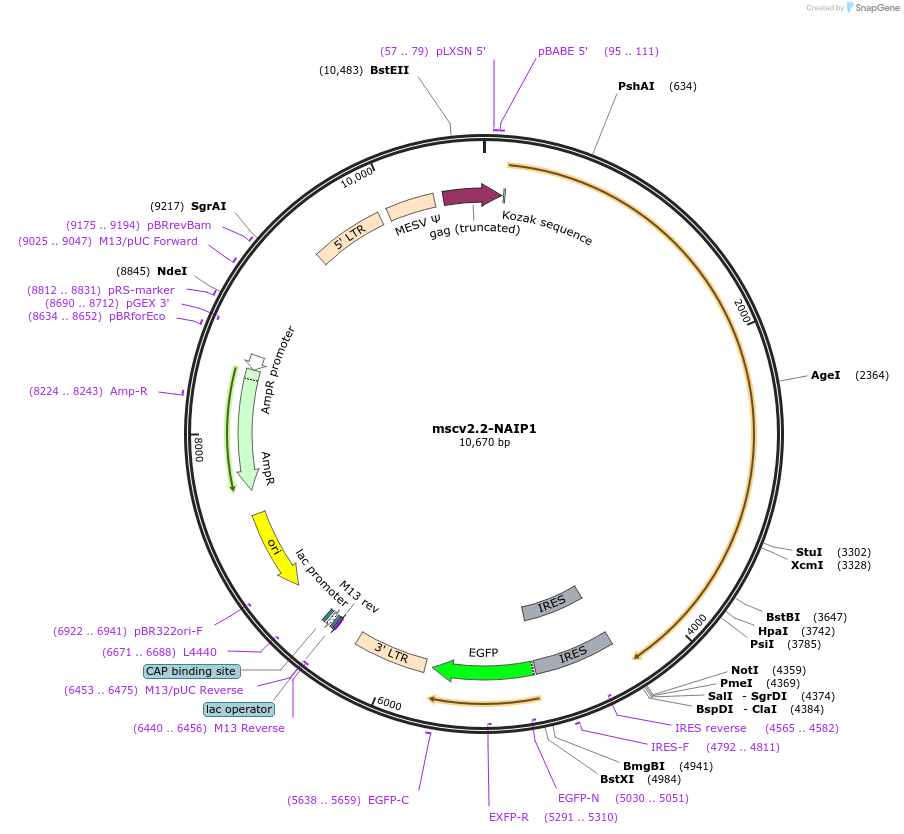
mscv2.2-NAIP1
Plasmid#60200PurposeMouse NAIP1 expressed under the constitutive, moderate-level retroviral LTR promoter, expresses in mammalian cells; IRES-GFP reporter downstream of NAIP1DepositorInsertNAIP1
UseRetroviralExpressionMammalianPromoterLTRAvailable SinceFeb. 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
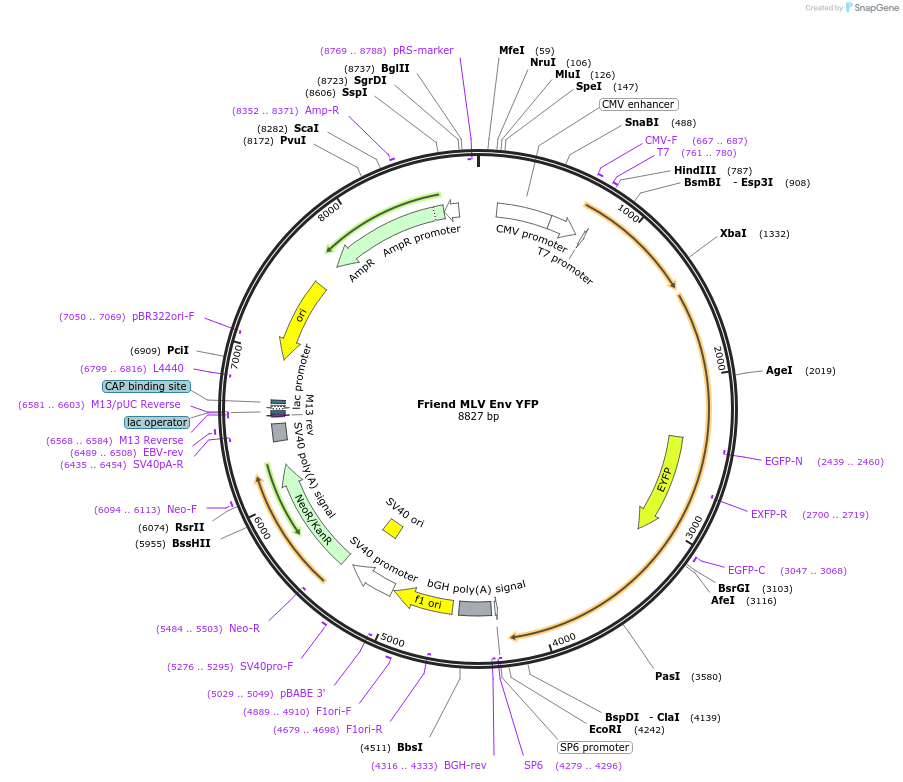
Friend MLV Env YFP
Plasmid#1820DepositorAvailable SinceMay 19, 2005AvailabilityAcademic Institutions and Nonprofits only -
YFP-DBD
Plasmid#14874DepositorAvailable SinceAug. 9, 2007AvailabilityAcademic Institutions and Nonprofits only -
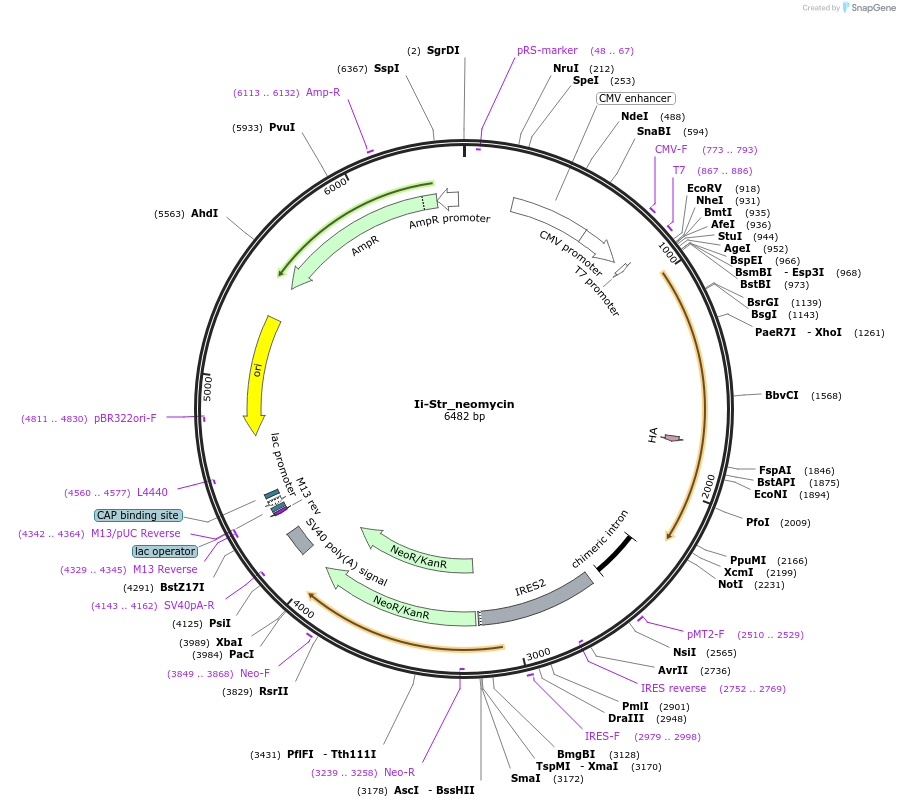
Ii-Str_neomycin
Plasmid#65308PurposeLuminal ER hook only (RUSH system)DepositorInsertInvariant chain (p33)
ExpressionMammalianMutationStreptavidin in its lumenal partPromoterCMVAvailable SinceAug. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
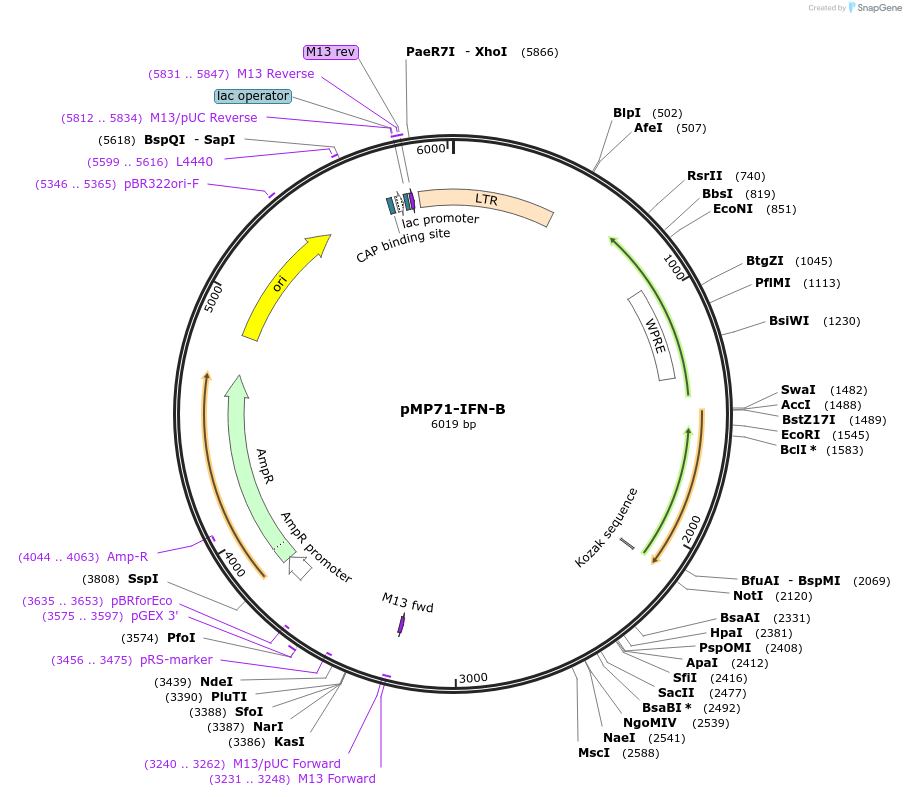
pMP71-IFN-B
Plasmid#174597PurposeExpresses secreted murine interferon betaDepositorInsertsecreted IFN-beta
ExpressionMammalianAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
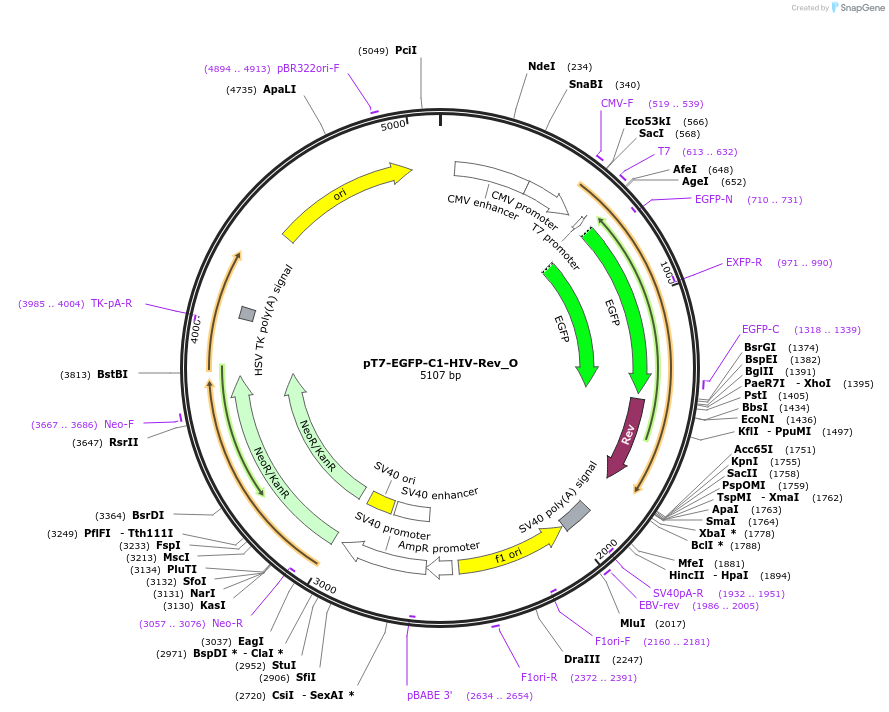
pT7-EGFP-C1-HIV-Rev_O
Plasmid#147113PurposeMammalian Expression of HIV-RevDepositorInsertHIV-Rev
ExpressionMammalianAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
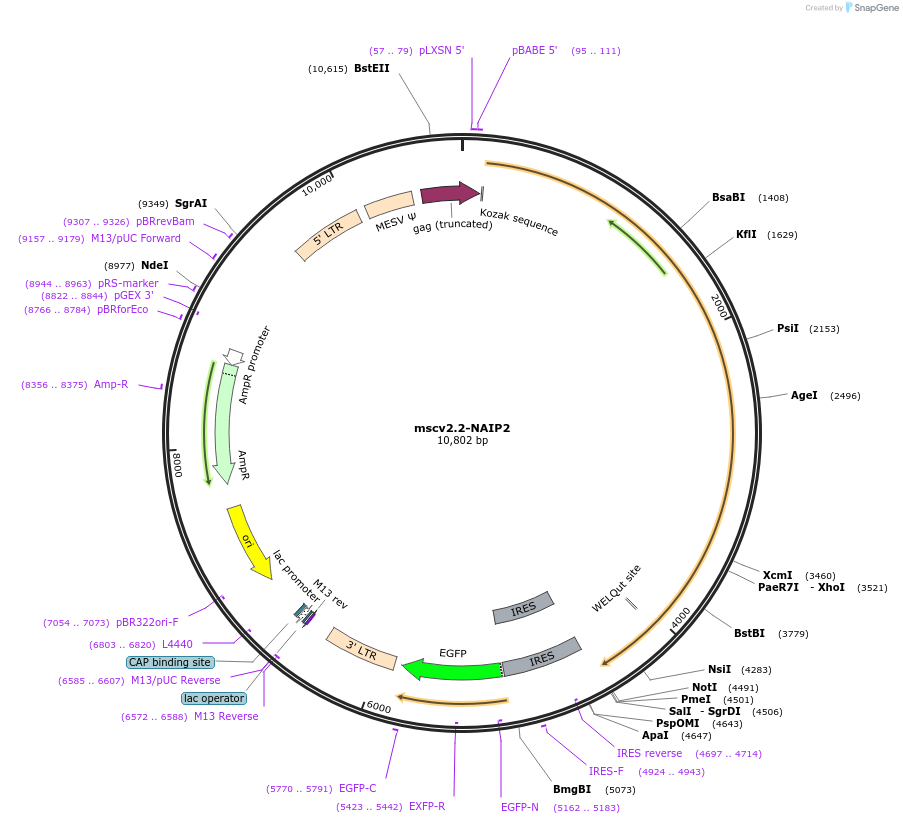
mscv2.2-NAIP2
Plasmid#60201PurposeMouse NAIP2 expressed under the constitutive, moderate-level retroviral LTR promoter, expresses in mammalian cells; IRES-GFP reporter downstream of NAIP2DepositorAvailable SinceFeb. 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
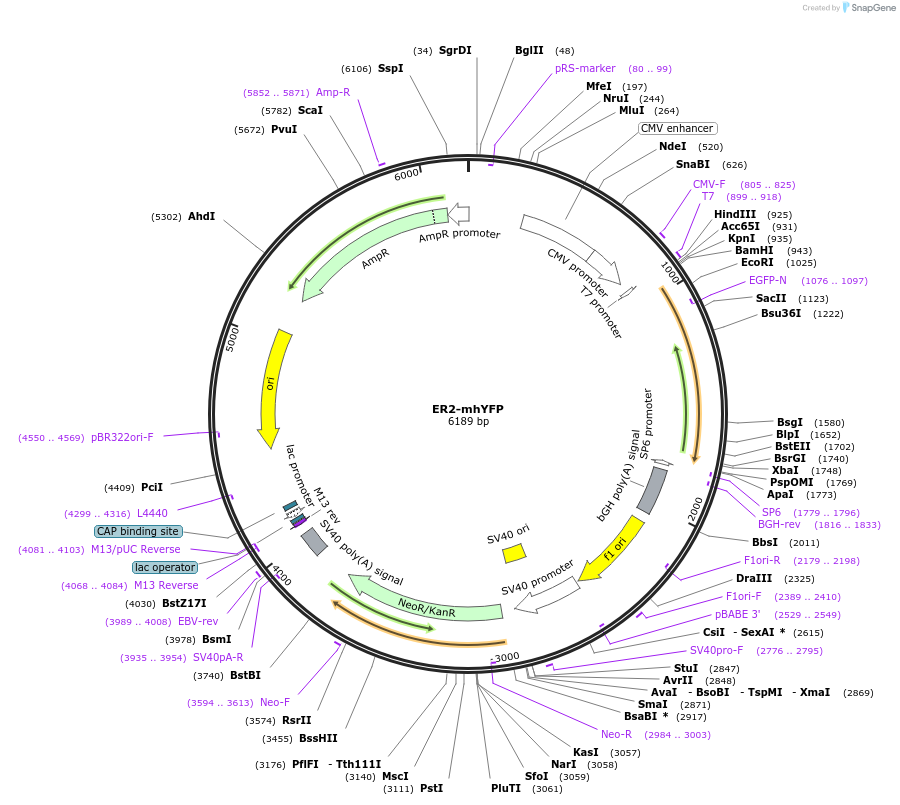
ER2-mhYFP
Plasmid#186528PurposeLocalization and retention of monomeric hyperfolder YFP fluorescent protein in the ER; improved targeting efficiencyDepositorInsertMonomeric hyperfolder YFP
TagsCalreticulin and KDELExpressionMammalianMutationhfYFP-S147P/L195M/V206KPromoterCMVAvailable SinceNov. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
pMXs-MYT1L
Plasmid#32926PurposeRetroviral expression of human Myt1LDepositorAvailable SinceNov. 30, 2011AvailabilityAcademic Institutions and Nonprofits only -
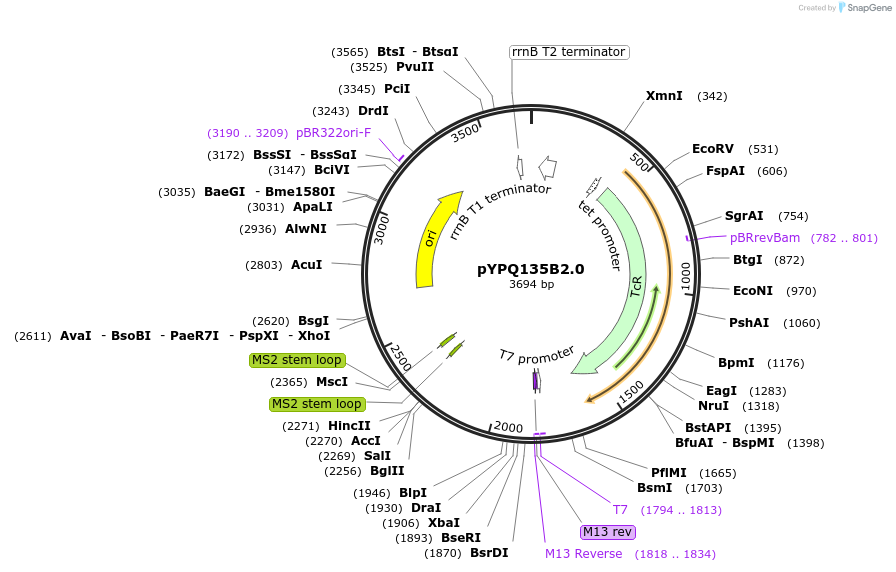
pYPQ135B2.0
Plasmid#167159PurposeGolden Gate entry vector to express the 5th gRNA with gRNA2.0 scaffold (with four MS2 binding sites) under AtU3 promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterAtU3Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
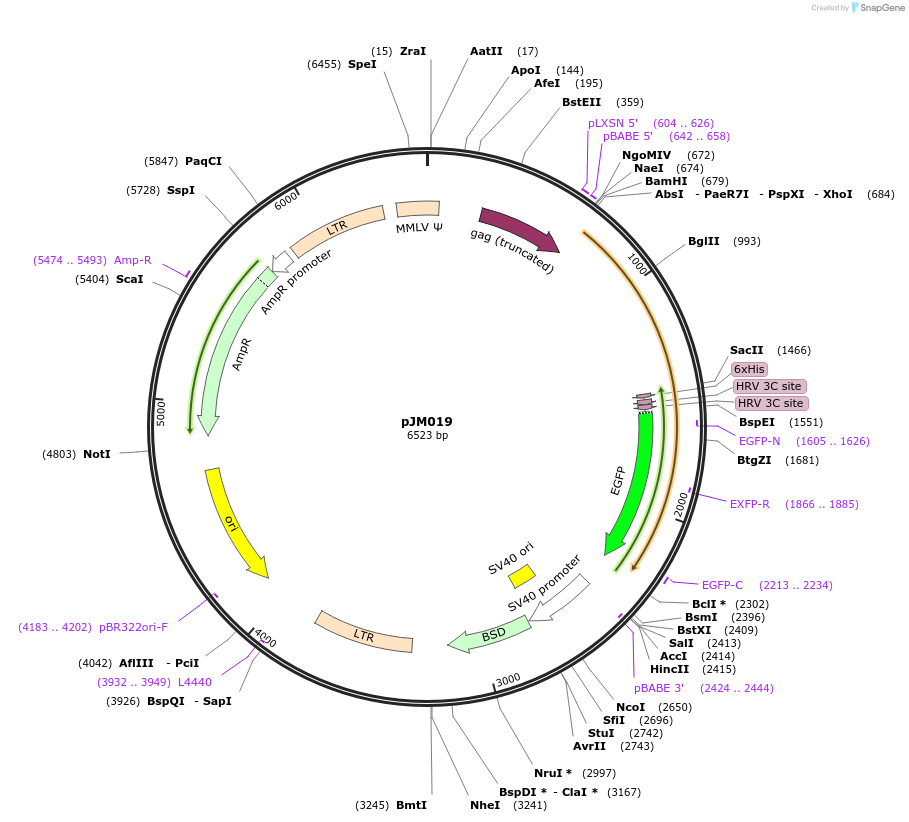
pJM019
Plasmid#134158PurposeRetroviral plasmid expressing GFP PSMA3DepositorAvailable SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
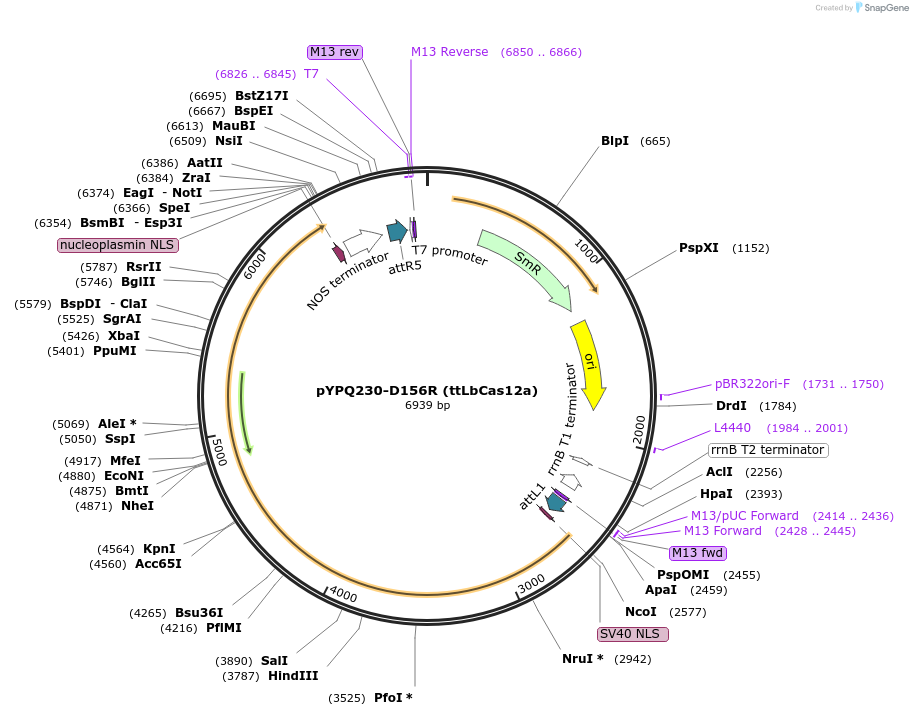
pYPQ230-D156R (ttLbCas12a)
Plasmid#195366PurposeA temperature-tolerant LbCas12a mutant (D156R), also known as ttLbCas12aDepositorInsertLbCas12a-D156R (ttLbCas12a)
UseCRISPR; Gateway compatible lbcas12a-d156r (ttlbca…ExpressionPlantPromoterZmUBI1Available SinceMay 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
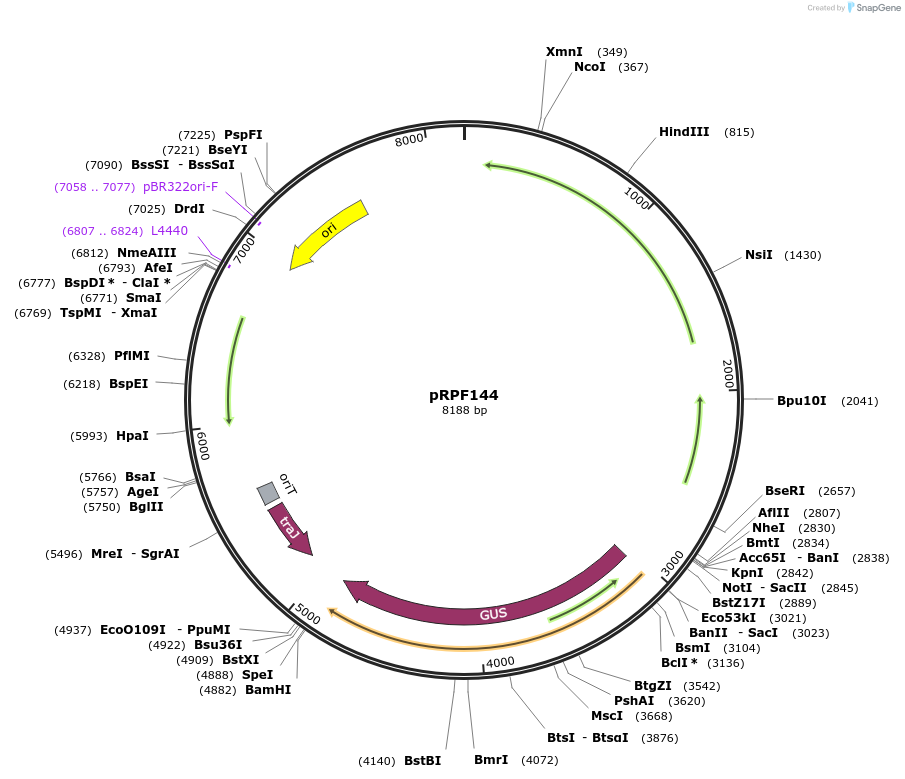
pRPF144
Plasmid#106372PurposeC. difficile constitutive expression system. Shuttle plasmid containing constitutive Pcwp2 -gusADepositorInsertPcwp2-gusA (gusA )
UseE. coli - c. difficile shuttle vectorMutationcodon optimised for C. difficilePromoterPcwp2Available SinceMarch 7, 2018AvailabilityAcademic Institutions and Nonprofits only -
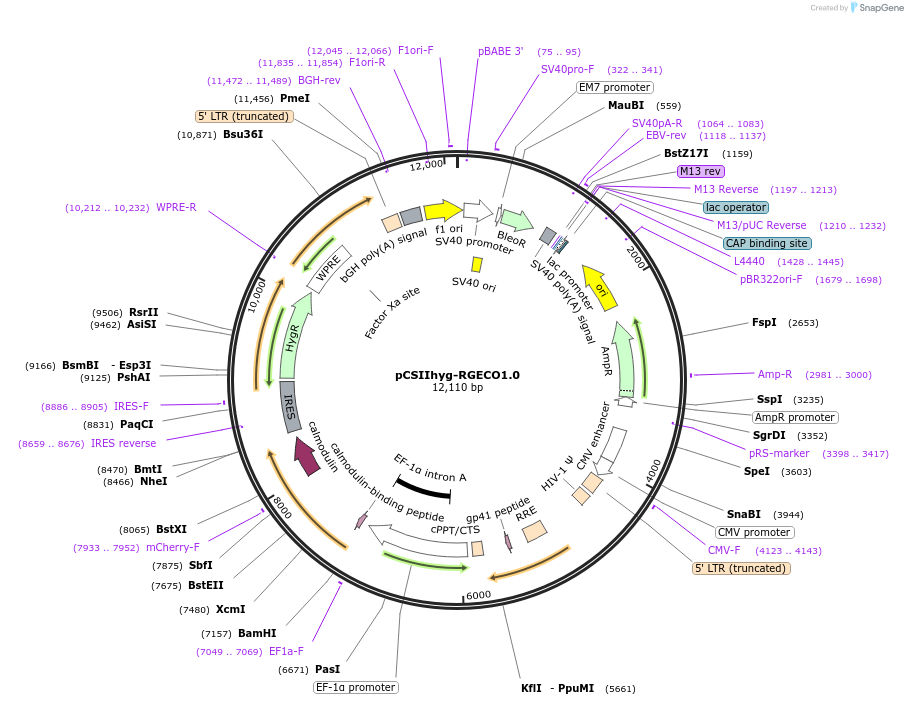
pCSIIhyg-RGECO1.0
Plasmid#175813PurposeThe lentiviral vector for R-GECO1.0, a genetically encoded red fluorescent Ca2+ sensor.DepositorInsertRGECO1.0
UseLentiviralExpressionMammalianAvailable SinceFeb. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
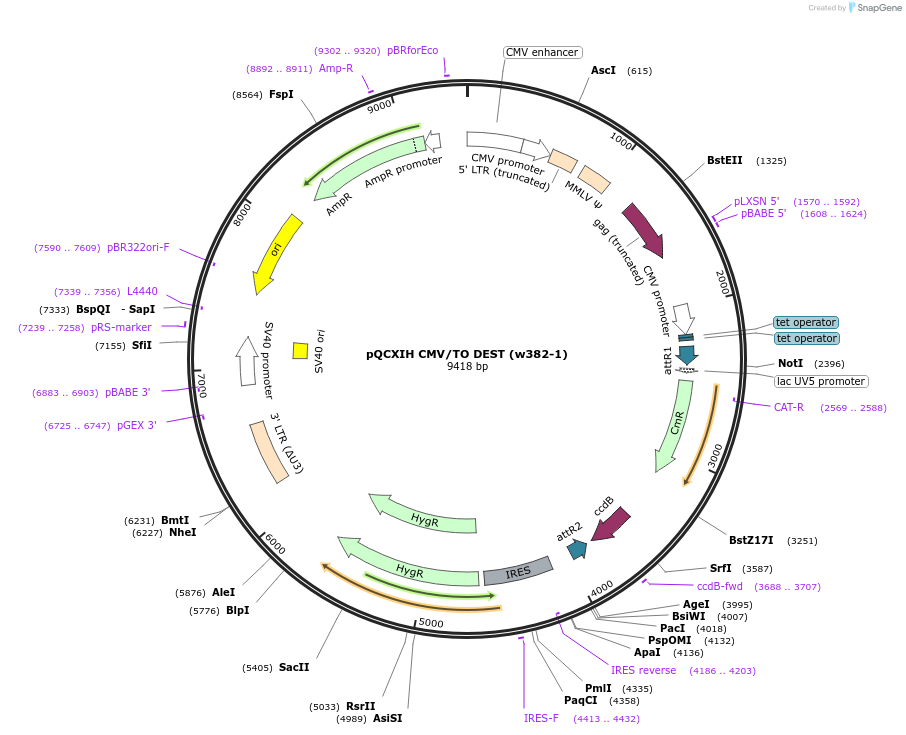
pQCXIH CMV/TO DEST (w382-1)
Plasmid#17394PurposeRetroviral Gateway destination vector, Expression, CMV/TO promoter, HygroDepositorTypeEmpty backboneUseRetroviral; Destination vectorExpressionMammalianAvailable SinceAug. 12, 2009AvailabilityAcademic Institutions and Nonprofits only -
pCAGGS-N2c(G)
Plasmid#73481PurposeN2c Glycoprotein expression vectorDepositorInsertN2c(G)
ExpressionMammalianAvailable SinceMarch 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
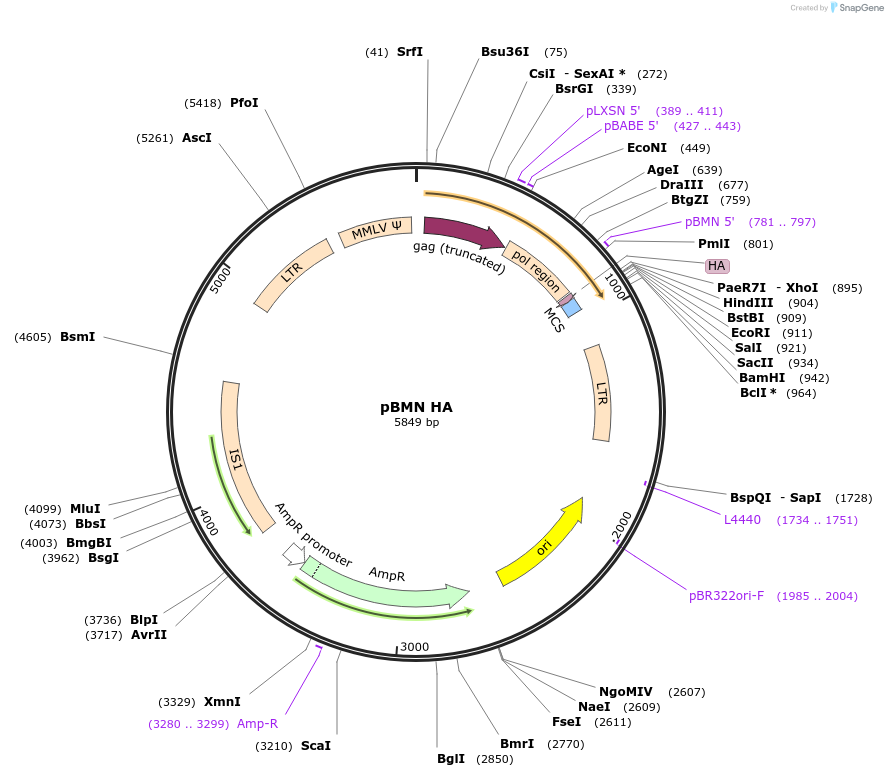
pBMN HA
Plasmid#188645PurposeThis is the Lazarou lab's standard retrovirus vector backbone for expression of HA-tag fusions onto the N-terminus of cloned genesDepositorTypeEmpty backboneUseRetroviralTagsHAExpressionMammalianAvailable SinceMay 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
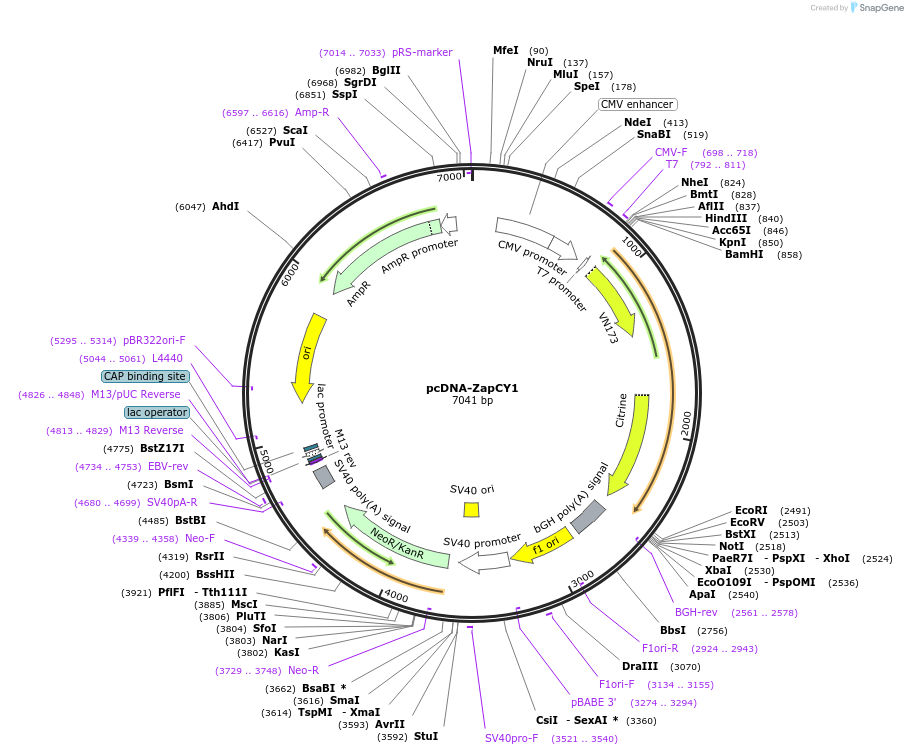
pcDNA-ZapCY1
Plasmid#36319DepositorInsertZapCY1 high affinity Zn(II) sensor
ExpressionMammalianPromoterCMVAvailable SinceMay 18, 2012AvailabilityAcademic Institutions and Nonprofits only -
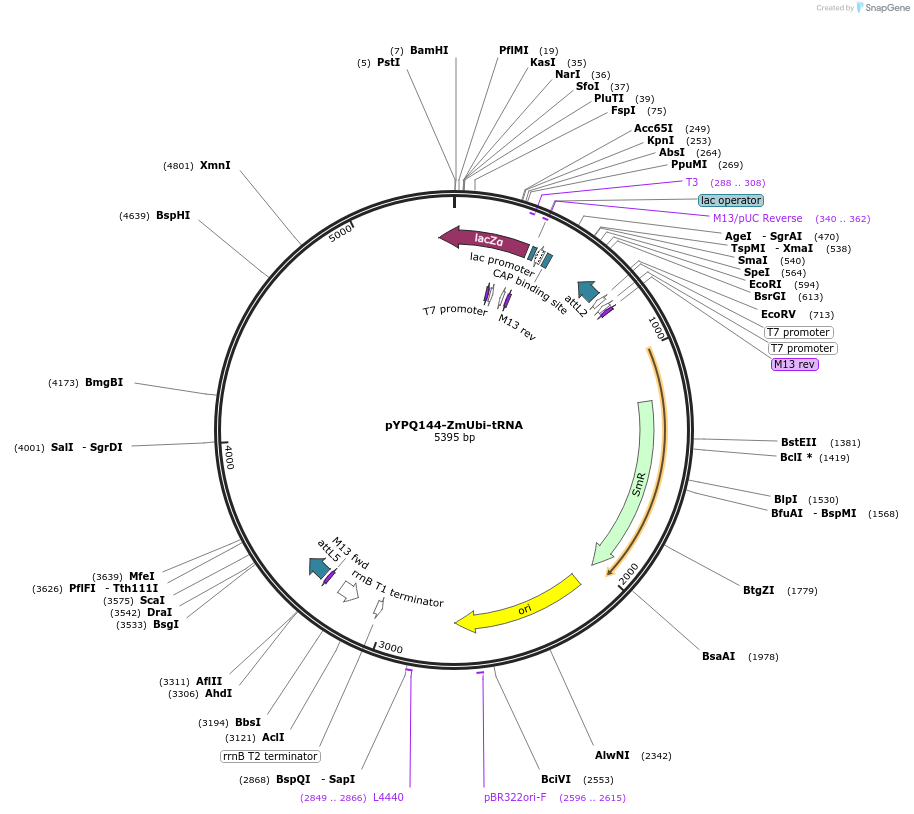
pYPQ144-ZmUbi-tRNA
Plasmid#158402PurposeGateway entry clone and Golden Gate recipient for pYPQ131-tRNA2.0 to pYPQ134-tRNA2.0; assembly of 4 gRNAsDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
Wnt7A STOP
Plasmid#35877DepositorAvailable SinceMay 7, 2013AvailabilityAcademic Institutions and Nonprofits only -
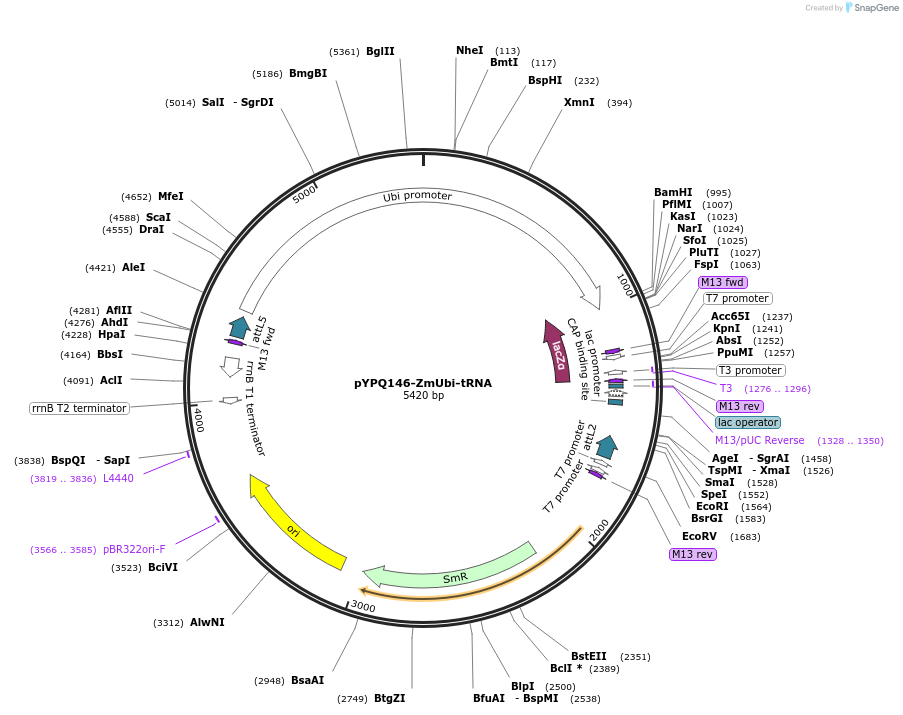
pYPQ146-ZmUbi-tRNA
Plasmid#158404PurposeGateway entry clone and Golden Gate recipient for pYPQ131-tRNA2.0 to pYPQ136-tRNA2.0; assembly of 6 gRNAsDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJuly 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
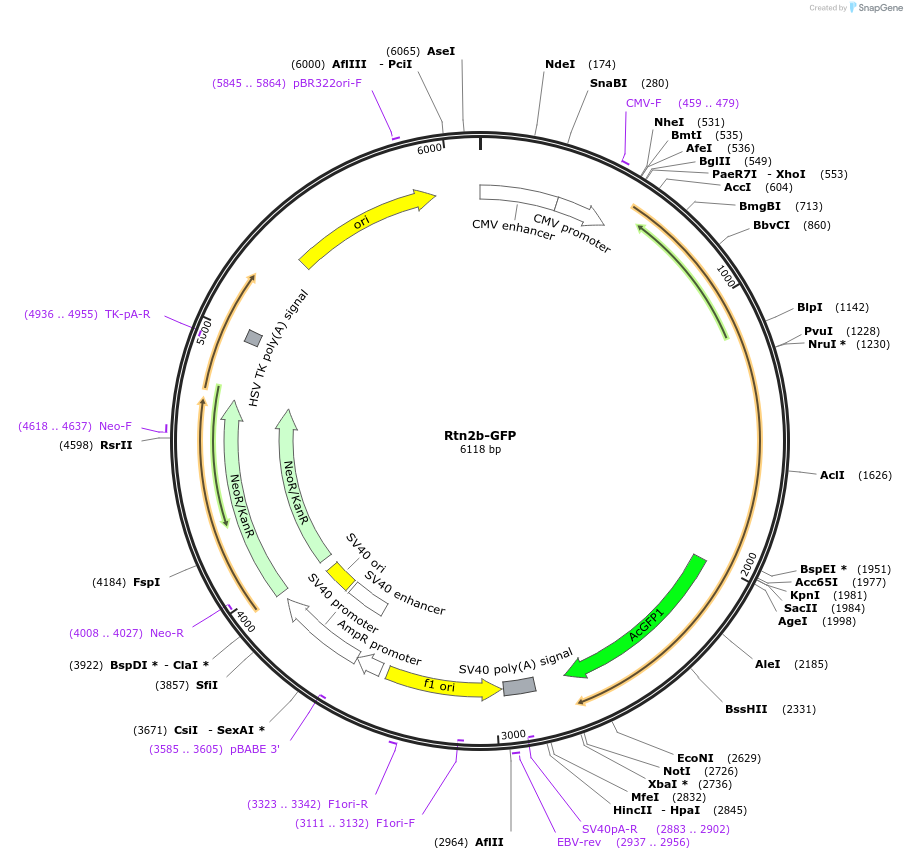
Rtn2b-GFP
Plasmid#186599PurposeEncodes human full-length Reticulon 2 isoform B fused to GFPDepositorAvailable SinceFeb. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
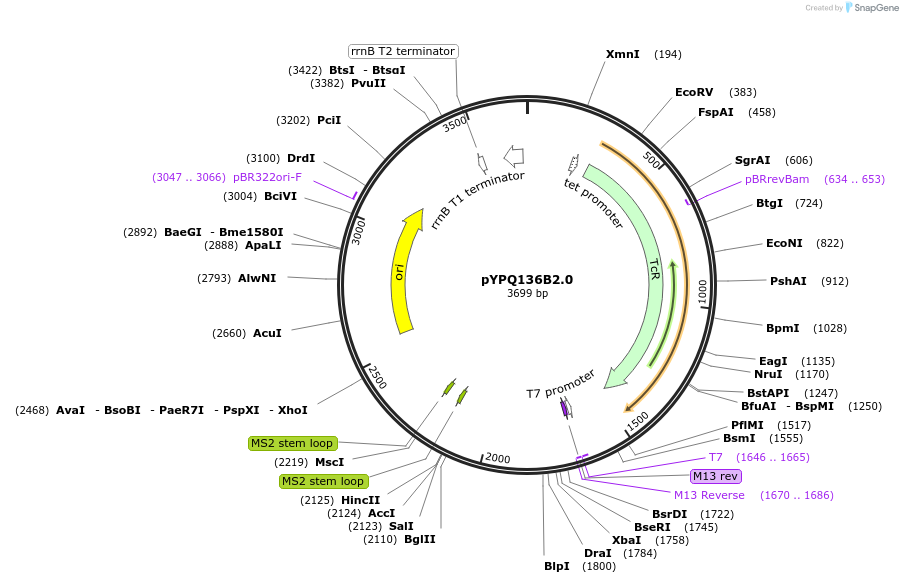
pYPQ136B2.0
Plasmid#167160PurposeGolden Gate entry vector to express the 6th gRNA with gRNA2.0 scaffold (with four MS2 binding sites) under AtU3 promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterAtU3Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
mCitrine-USP19
Plasmid#78593PurposeExpress mCitrine-USP19 in mammalian cellsDepositorAvailable SinceJune 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
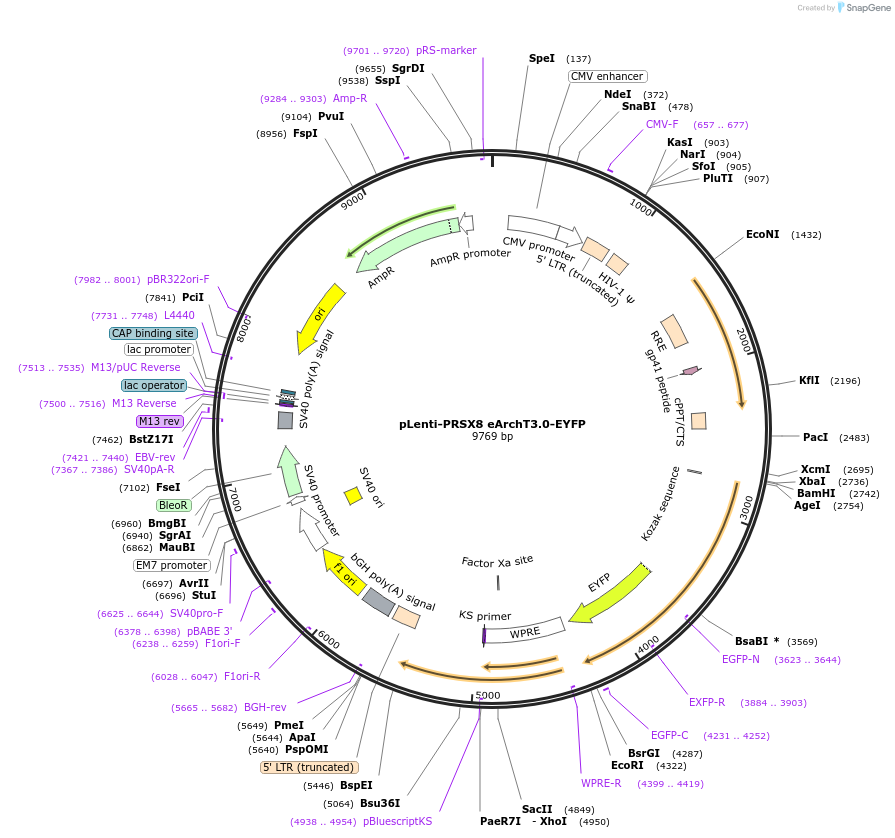
pLenti-PRSX8 eArchT3.0-EYFP
Plasmid#89538PurposeExpresses eArchT 3.0-eYFP in Phox2 (including catecholamine) neuronsDepositorInserteArchT 3.0
UseLentiviralTagsEYFPExpressionMammalianPromoterPRSx8Available SinceAug. 8, 2017AvailabilityAcademic Institutions and Nonprofits only