We narrowed to 9,586 results for: Coli
-
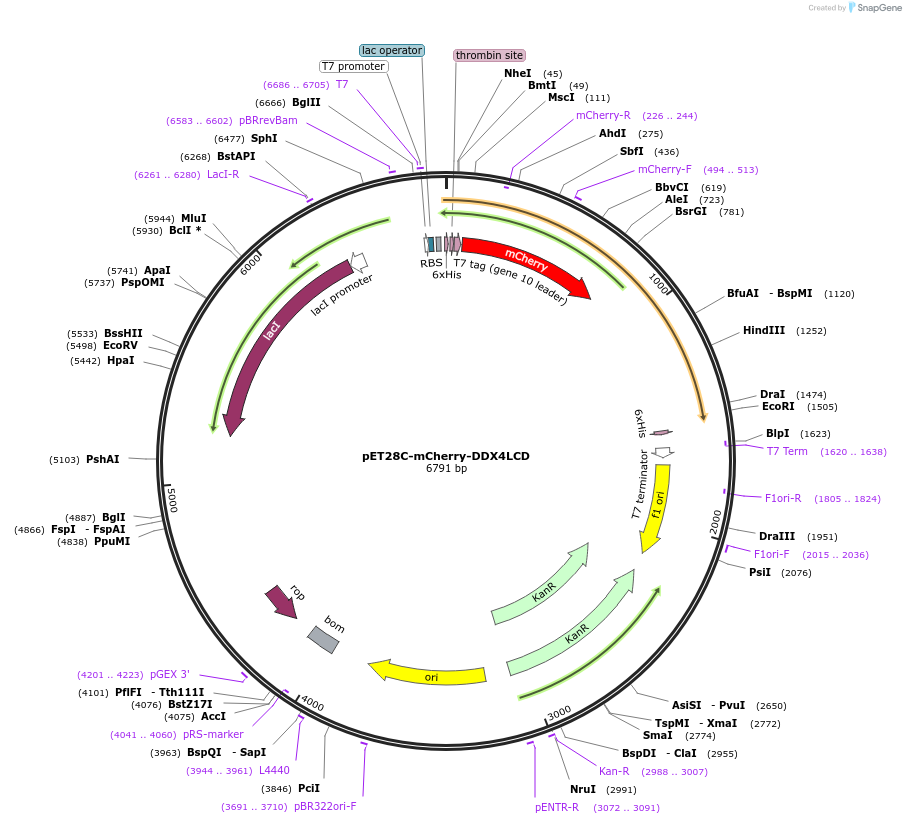
Plasmid#204408PurposeE. coli expression of DDX4 low-complexity domain (1-236) with an N-terminal mCherry tagDepositorAvailable SinceSept. 5, 2023AvailabilityAcademic Institutions and Nonprofits only
-
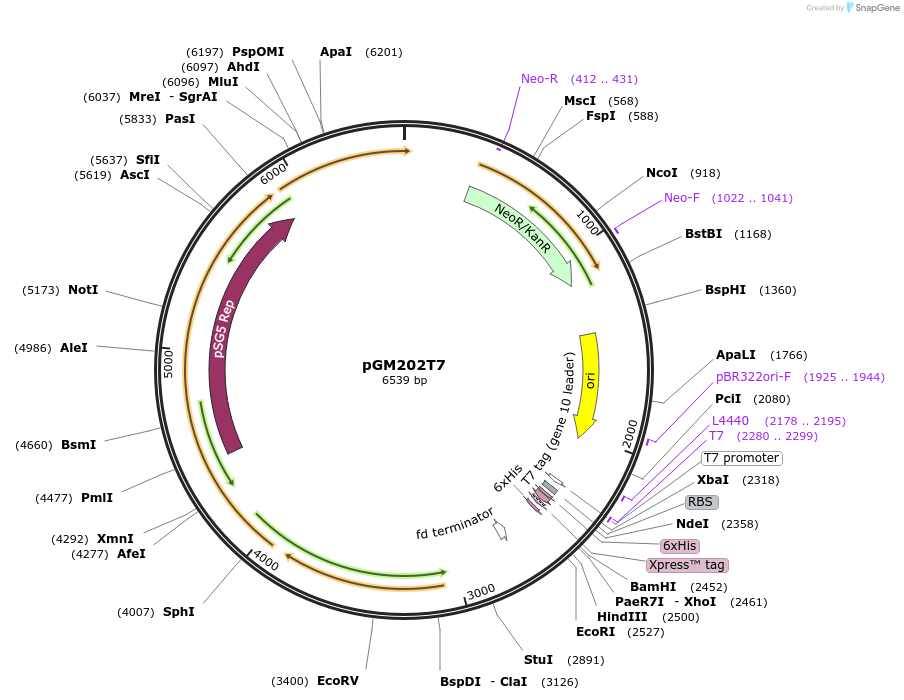
pGM202T7
Plasmid#69993PurposeE. coli/Streptomyces expression vector containing the T7-promoterDepositorInsertsaphII
tsr
rep
sso
T7 promoter
dso
His-Orf
TagsN-terminal and C-terminal His tag encoding sequen…ExpressionBacterialAvailable SinceNov. 13, 2015AvailabilityAcademic Institutions and Nonprofits only -
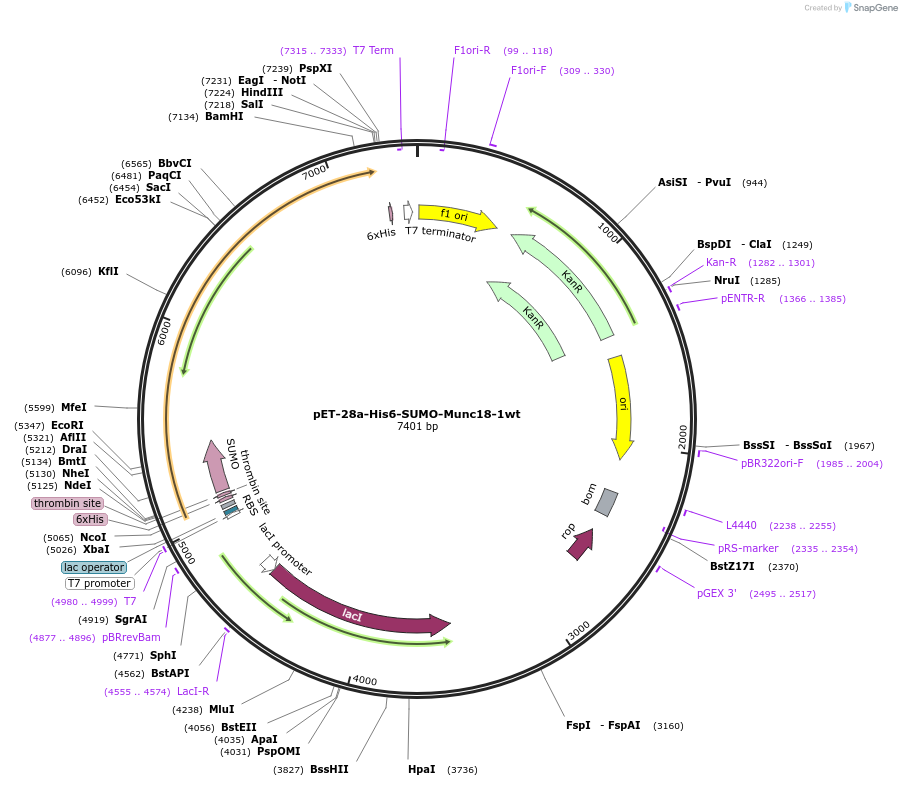
pET-28a-His6-SUMO-Munc18-1wt
Plasmid#135550PurposeExpress rat Munc18-1 in E. coli BL21, purified via HisPur Ni-NTA ResinDepositorInsertMunc18-1wt
Tags6xHis tag, thrombin recognition and cleavage site…ExpressionBacterialMutationNo ATG start codonPromoterT7 promoterAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
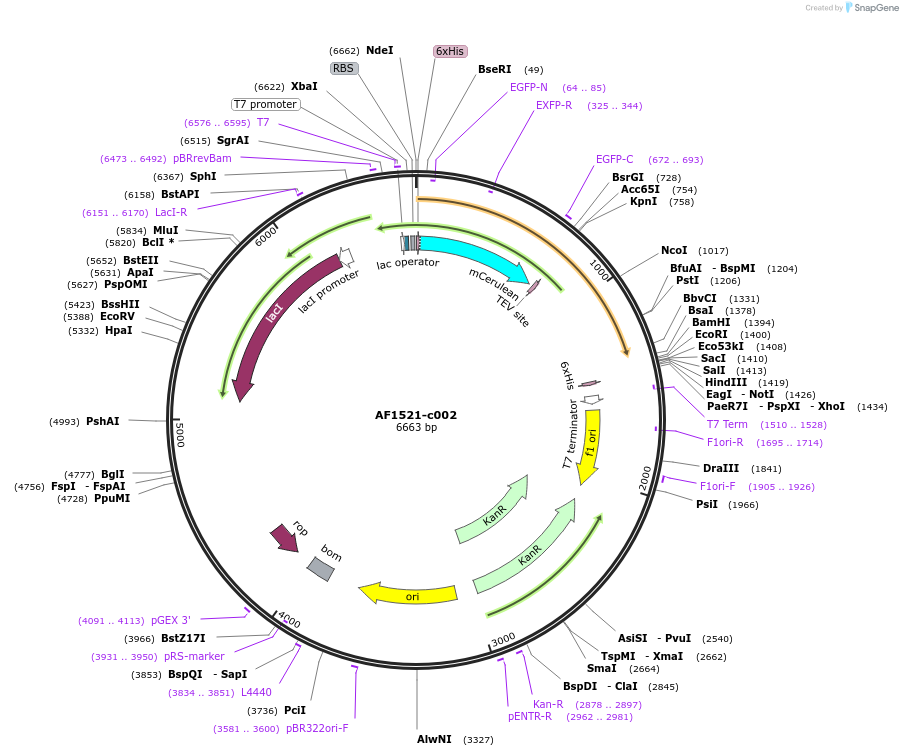
AF1521-c002
Plasmid#173109PurposeExpression of recombinant fusion protein in E. coliDepositorInsertAF_1521 (AF_RS07675 Archaeoglobus fulgidus (strain ATCC 49558 / VC-16 / DSM 4304 / JCM 9628 / NBRC 100126))
TagsmCeruleanExpressionBacterialMutationKH-M1-L192Available SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
HAI
Plasmid#71316PurposeInducible expression of human arginase I in E. coliDepositorAvailable SinceDec. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
pDJSVT86
Plasmid#108965PurposeExpress periplasmic, outer membrane, or secreted proteins in E. coli - OmpA signal and 6His tagDepositorTypeEmpty backboneTagsOmpA 1-27, 6HisExpressionBacterialPromoterT7Available SinceMay 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
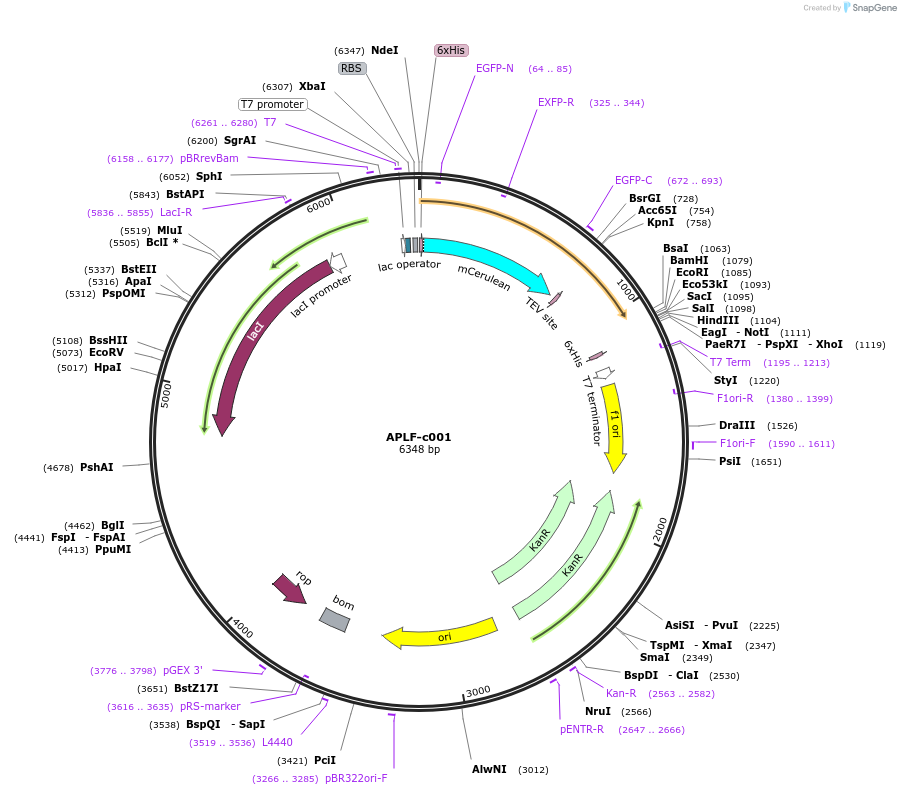
APLF-c001
Plasmid#173077PurposeExpression of recombinant fusion protein in E. coliDepositorAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
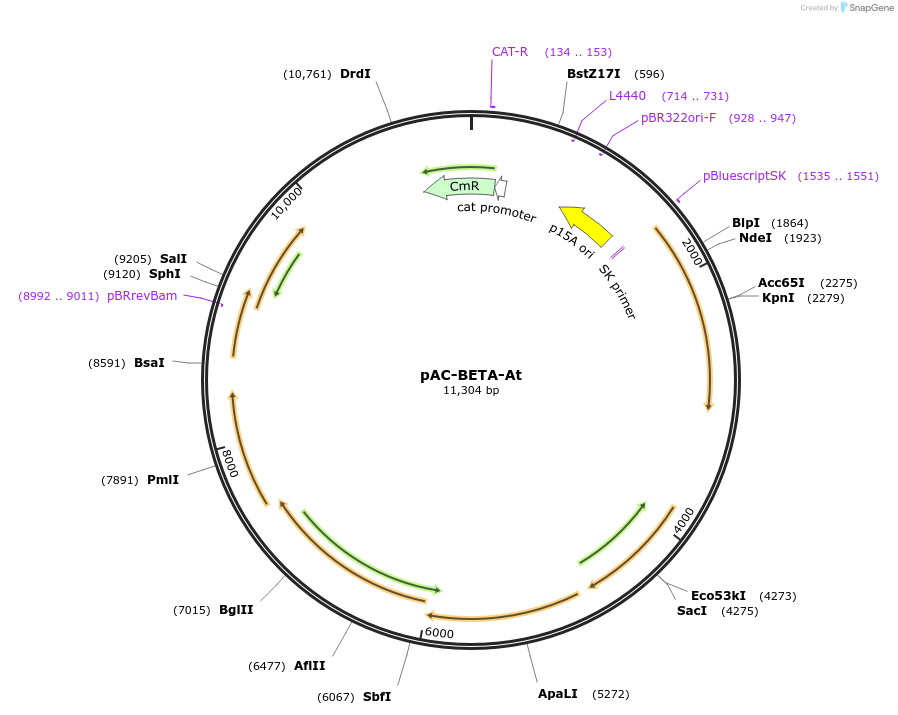
pAC-BETA-At
Plasmid#53288PurposeContains crtE, crtB, and crtI genes of Erwinia herbicola (Pantoea agglomerans) Eho10, and an lcyB cDNA of Arabidopsis thaliana. Use with pY2F to produce alpha-carotene in E. coli.DepositorAvailable SinceJan. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
pGEX6P1-UBE2S-UBD
Plasmid#66713PurposeExpresses the UBE2S-UBD fusion protein in E. coliDepositorAvailable SinceJuly 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
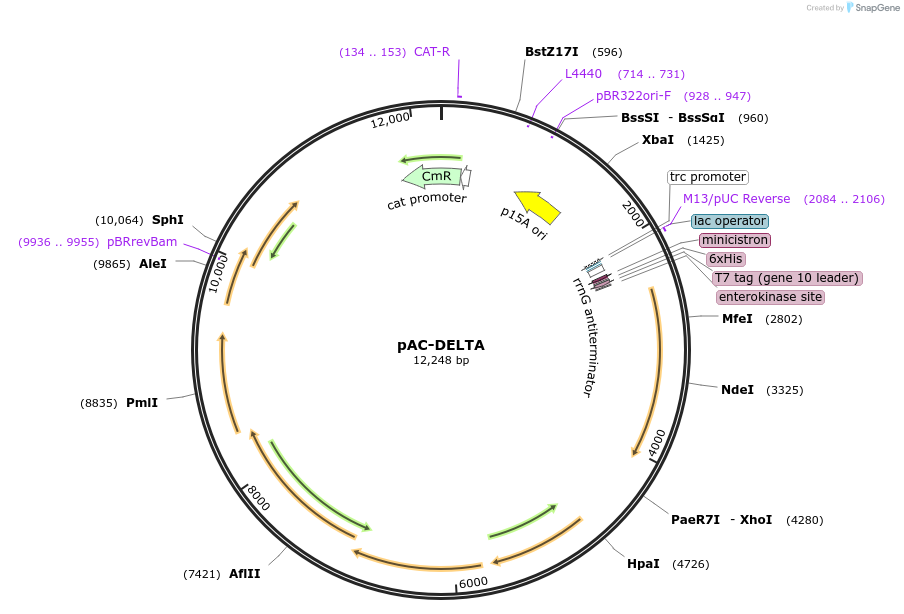
pAC-DELTA
Plasmid#53273PurposeContains crtE, crtB, and crtI genes of Erwinia herbicola (Pantoea agglomerans) Eho10, together with the lcyE gene of Arabidopsis thaliana and thereby produces delta-carotene in E. coliDepositorAvailable SinceJan. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
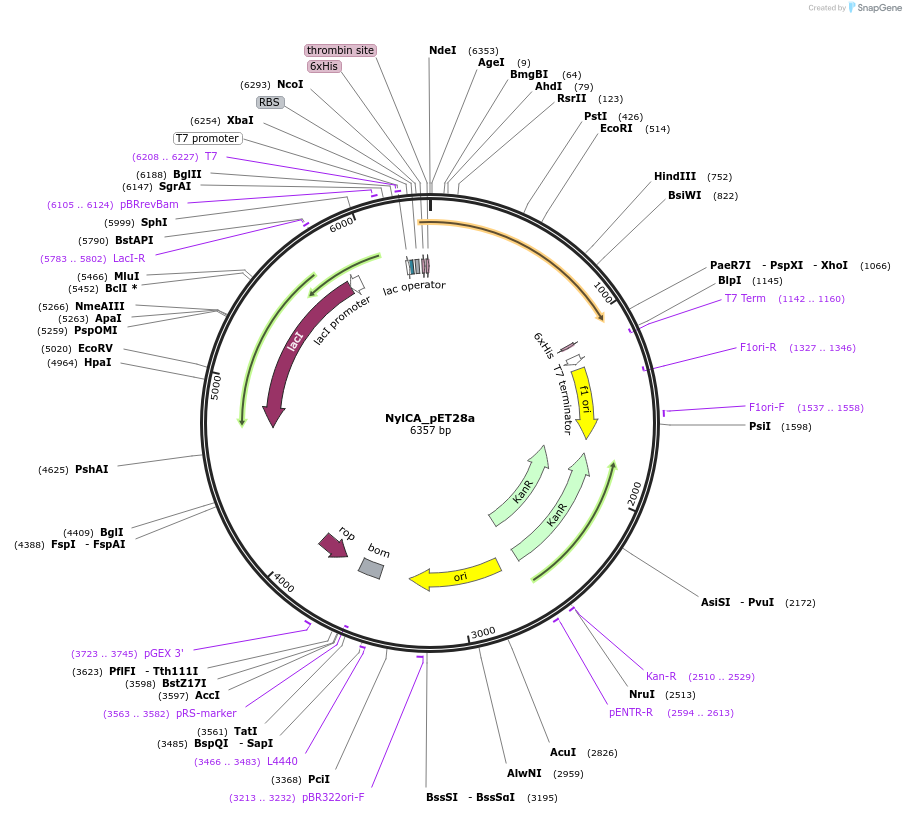
NylCA_pET28a
Plasmid#215418PurposeExpression construct for NylCA gene, codon optimized for expression in E. coliDepositorInsertNylCA
ExpressionBacterialAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
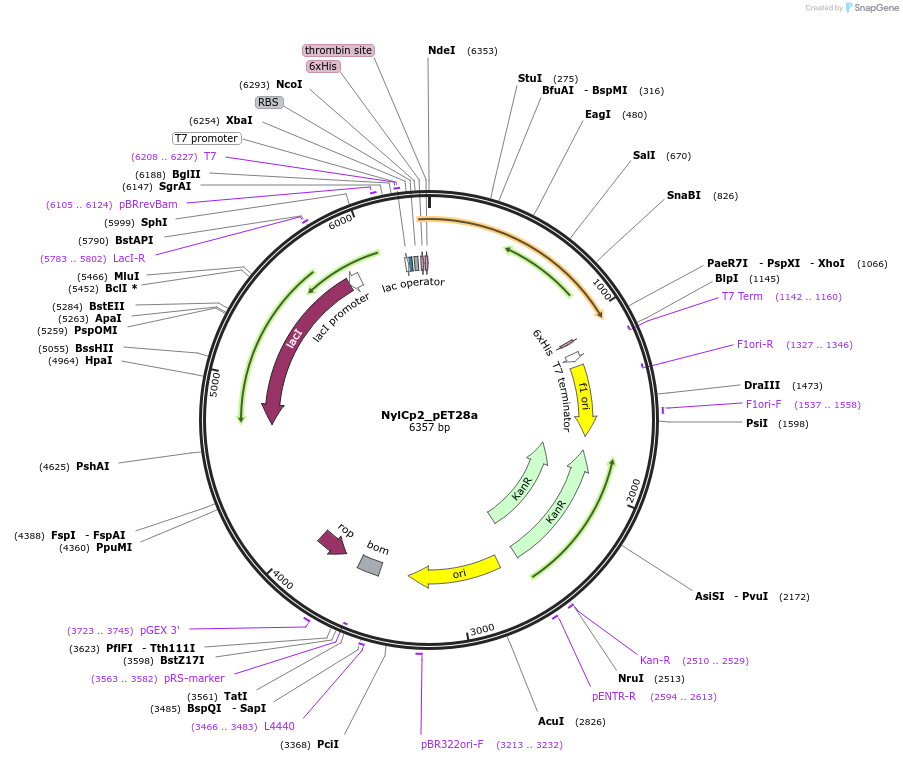
NylCp2_pET28a
Plasmid#215420PurposeExpression construct for NylCp2 gene, codon optimized for expression in E. coliDepositorInsertNylCp2
ExpressionBacterialAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
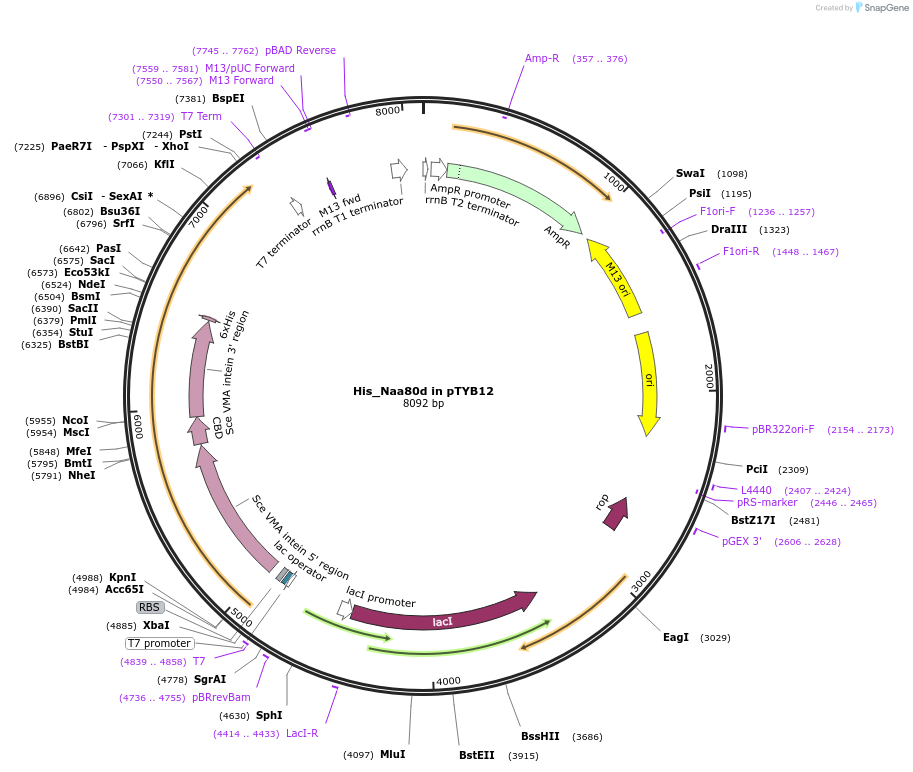
His_Naa80d in pTYB12
Plasmid#188455PurposeExpress in E. coli human Naa80 delta (actin's N-terminal acetyl transferase) with N-terminal intein-chitin binding domain tag and 6xHis tag.DepositorAvailable SinceNov. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
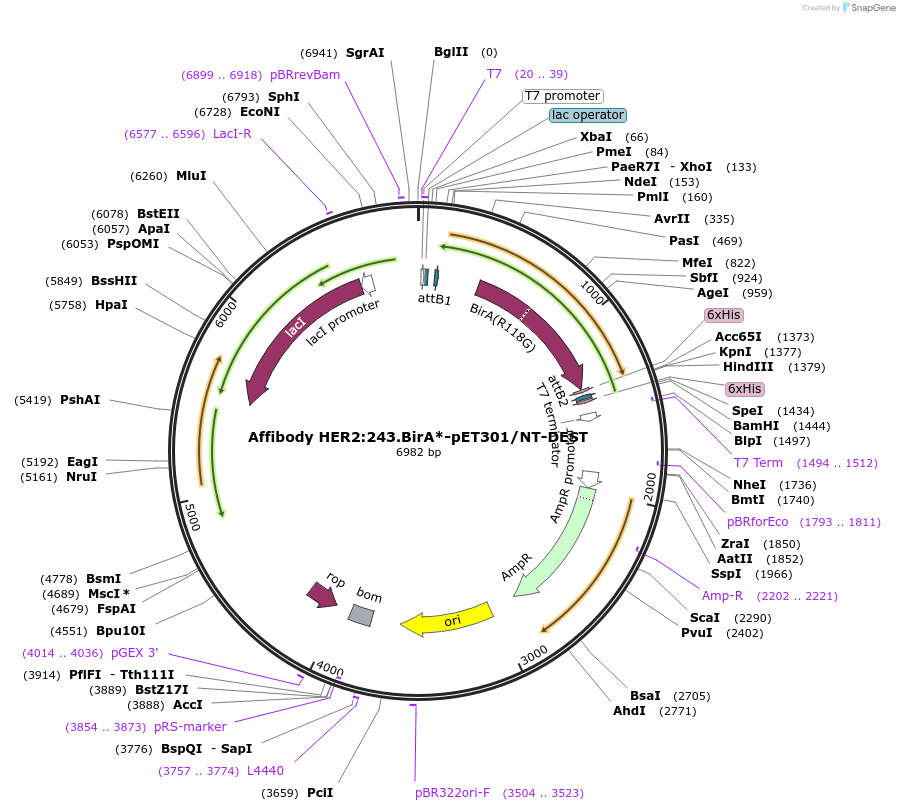
Affibody HER2:243.BirA*-pET301/NT-DEST
Plasmid#174692PurposePlasmid encoding for the bacterial expression of a bispecific coding for the anti-HER2 affibody ZHER2:342 fused to the mutated form of BirA enzyme, BirA*DepositorInsertAnti-HER2 affibody ZHER2:342 fused to mutated form of BirA, BirA*
TagsHIS tagExpressionBacterialMutationR118G mutation in BirAPromoterT7 promoterAvailable SinceJan. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
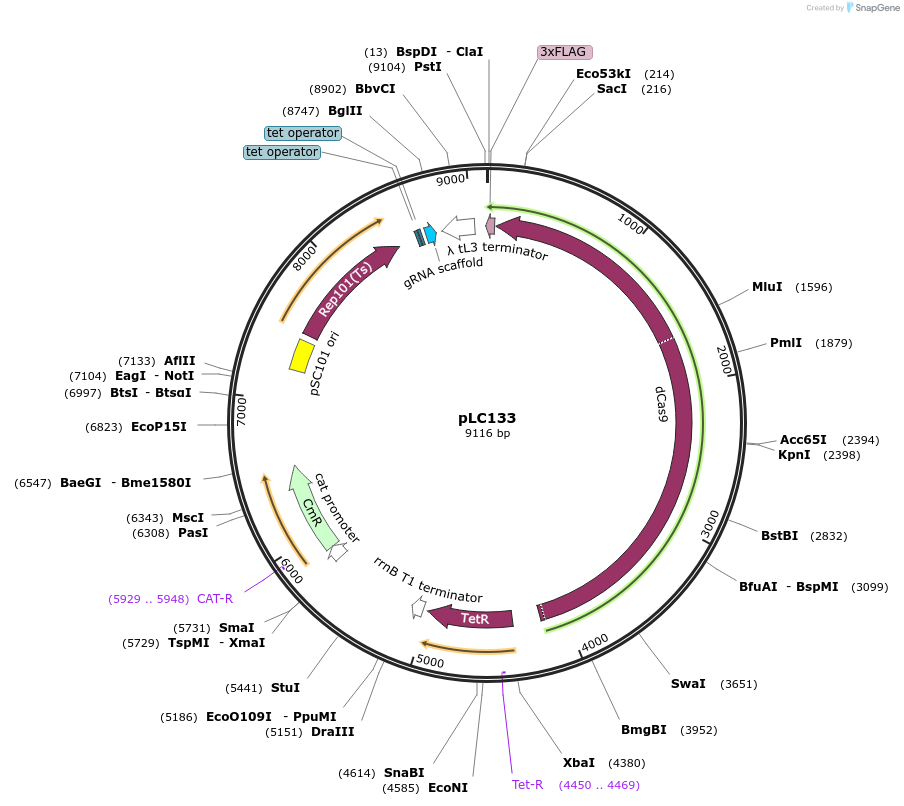
pLC133
Plasmid#198639PurposeExpression plasmid for dCas9 in E. coli used in Chip-seq experiments. dCas9 is tagged with a C-terminal 3x FLAG tag. A guide RNA can be cloned using BsaI restriction sites.DepositorInsertdCas9
Tags3x-FLAGExpressionBacterialPromoterpTetAvailable SinceApril 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
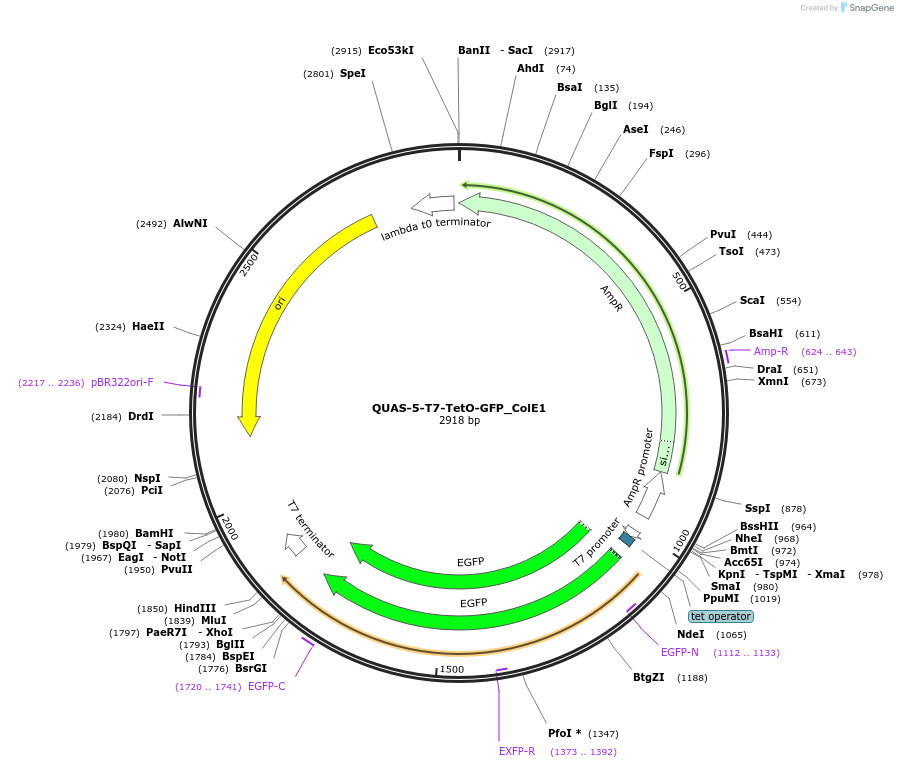
QUAS-5-T7-TetO-GFP_ColE1
Plasmid#171660PurposeQUAS five base pairs upstream of a T7 promoter with a TetO operator. Expresses GFP in BL21(DE3) E. coli according to the Q and Tet systems. Contains the ColE1 origin of replication.DepositorInsertQUAS-5-T7-TetO-GFP ssra-T7 Stop
TagsssrA degradation tag (DAS+4)ExpressionBacterialPromoterT7Available SinceOct. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
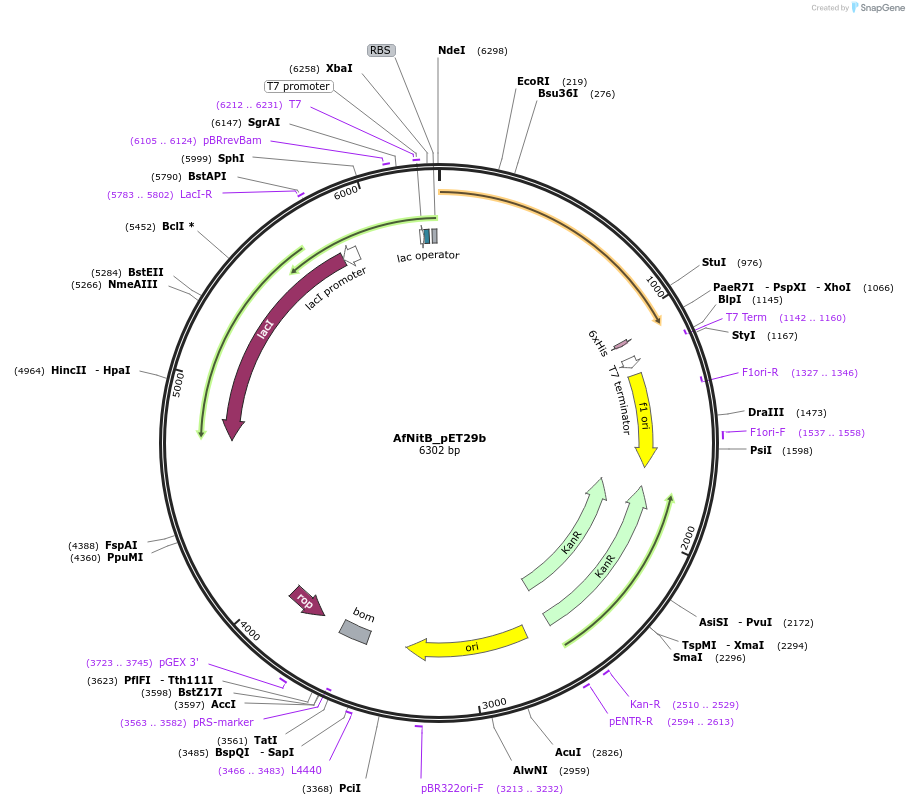
AfNitB_pET29b
Plasmid#215401PurposeExpression construct for AfNitB gene, codon optimized for expression in E. coliDepositorInsertAfNitB
ExpressionBacterialAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
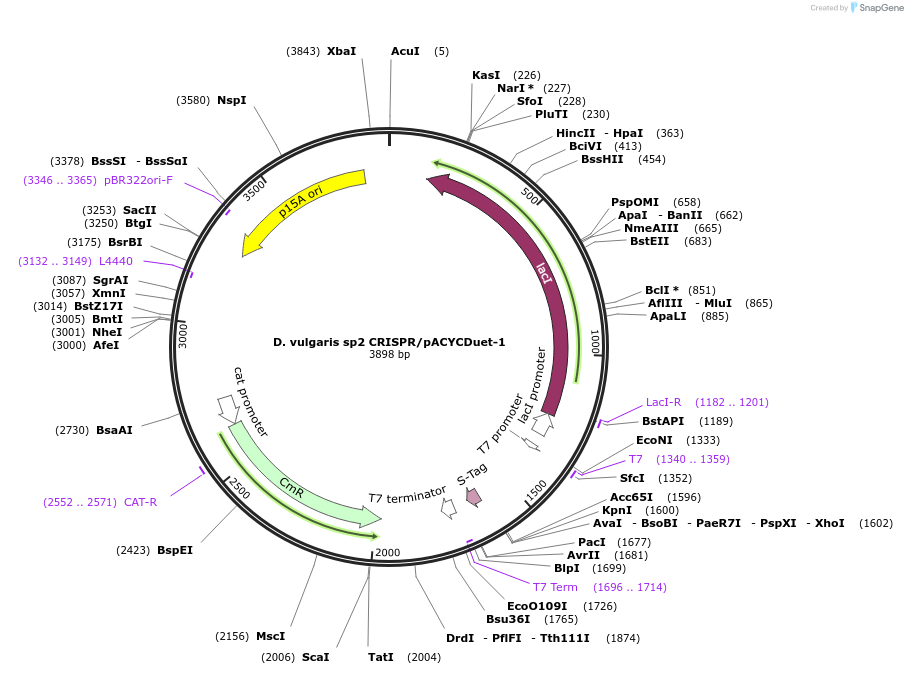
D. vulgaris sp2 CRISPR/pACYCDuet-1
Plasmid#81186PurposeExpresses CRISPR array containing 3 copies of Desulfovibrio vulgaris spacer 2 and flanking repeats in E. coliDepositorInsertSynthetic CRISPR array (3 copies of D. vulgaris spacer #2 flanked by repeats)
UseCRISPRTagsNoneExpressionBacterialPromoterT7Available SinceSept. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
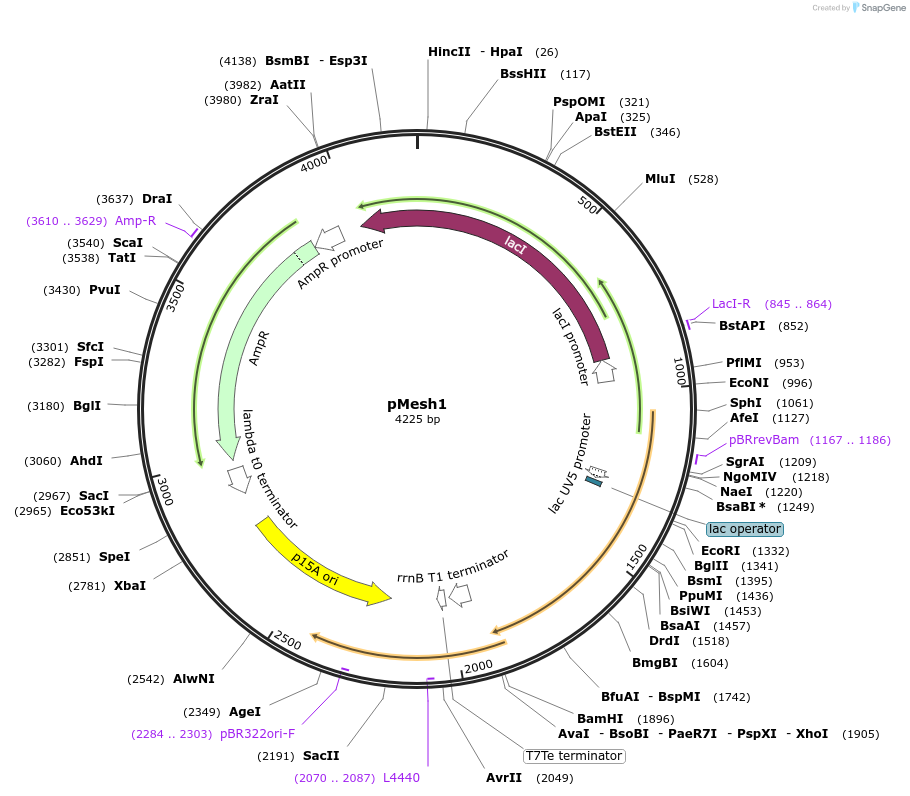
pMesh1
Plasmid#175594PurposeLac inducible, AMP Resistant, non labelled gene mesh1 from D. melanogaster on a low copy backbone. Used to induce controllable hydrolysis of ppGpp in E. coli.DepositorInsertMesh1 (Mesh1 Fly)
ExpressionBacterialMutationCodon optimized for translation in E.coli. AGGAGG…Available SinceNov. 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
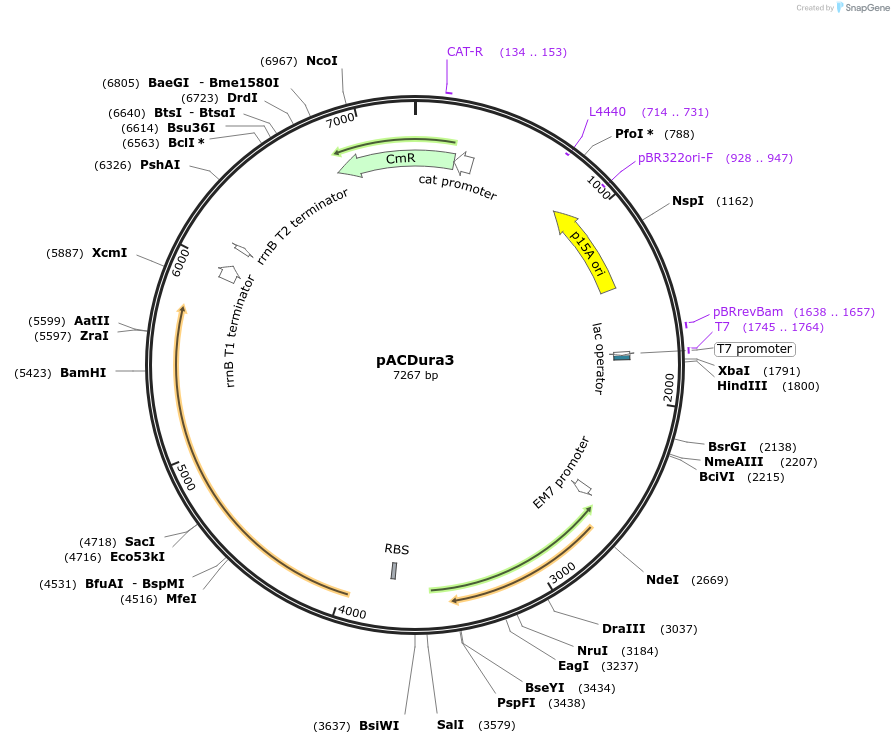
pACDura3
Plasmid#138711PurposeExpresses group II intron Ll.LtrB and IEP (LtrA) from Lactoccus lactis under control of T7 promoter. Ll.LtrB is retargeted to insert into 635s locus of LacZ gene and ura3 gene is cloned inside.DepositorInsertura3 gene (URA3 Budding Yeast)
ExpressionBacterialMutationoptimized ura3 gene for expression in Escherichia…PromoterpEM7 promoterAvailable SinceSept. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
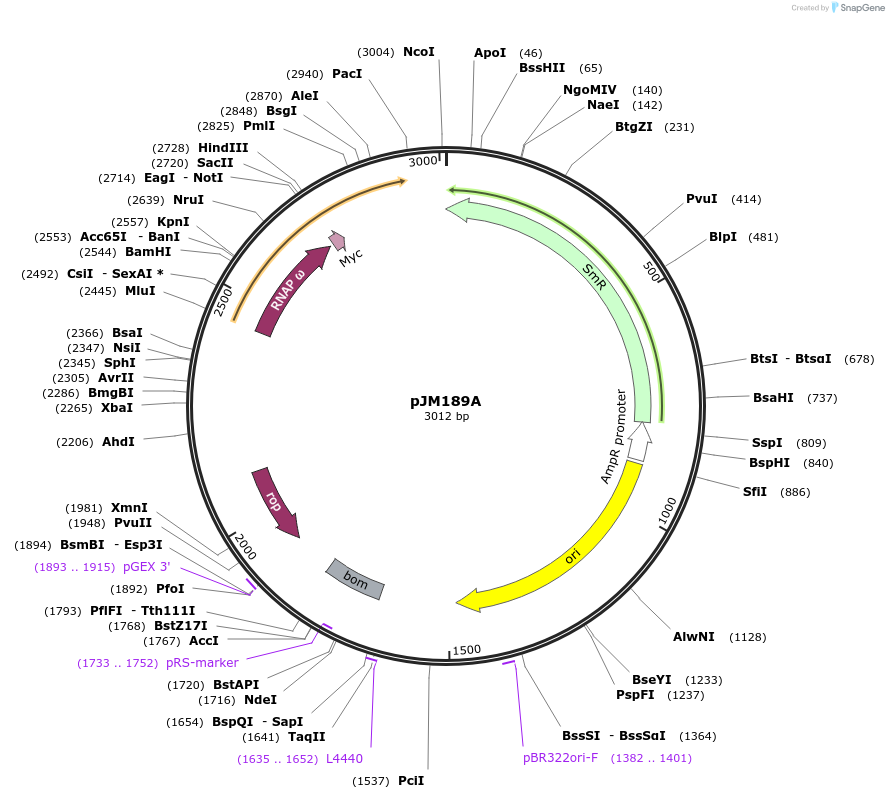
pJM189A
Plasmid#218135PurposeFor hybrid circuit activation. Expresses E. coli RNAP omega subunit fused to ZF degron SD0.DepositorInsertSD0
TagsrpoZAvailable SinceJune 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
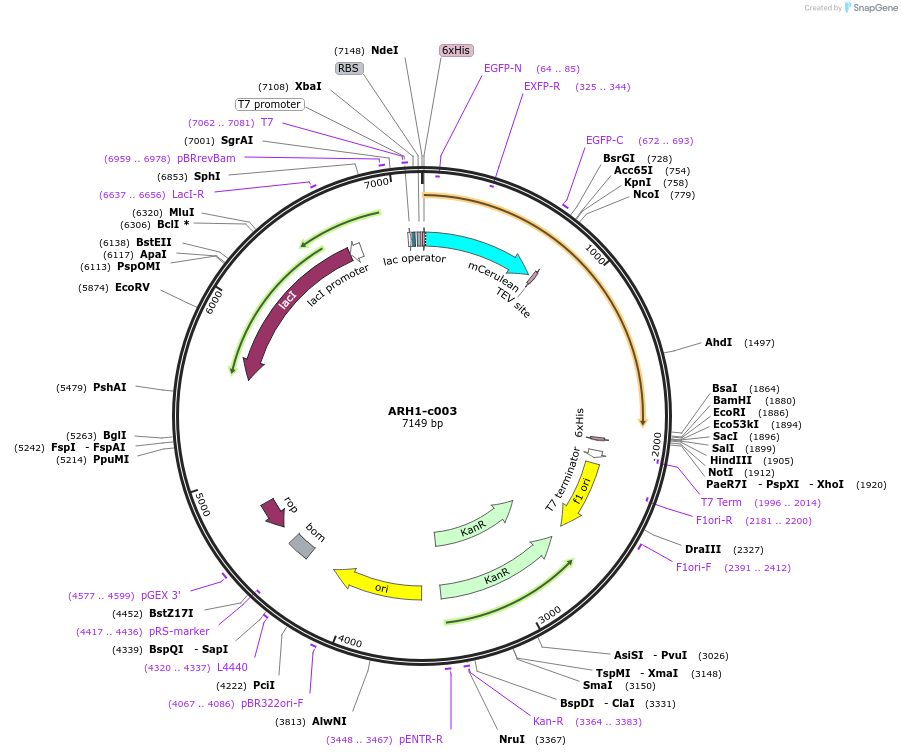
ARH1-c003
Plasmid#173093PurposeExpression of recombinant fusion protein in E. coliDepositorAvailable SinceNov. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
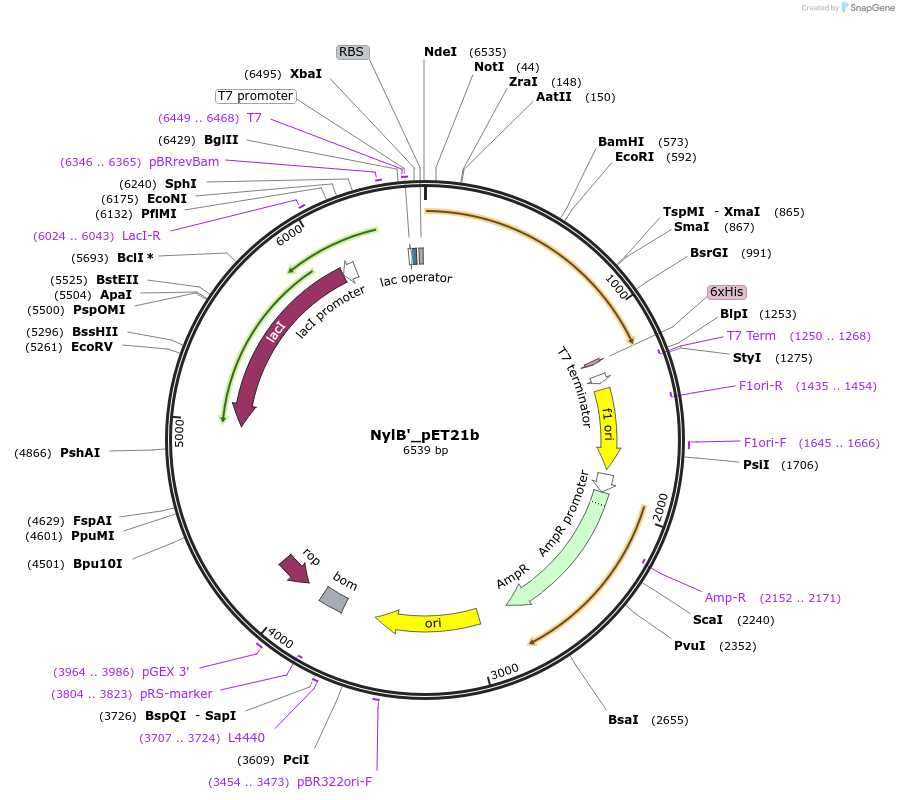
NylB'_pET21b
Plasmid#215416PurposeExpression construct for NylB' gene, codon optimized for expression in E. coliDepositorInsertNylB'
ExpressionBacterialAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
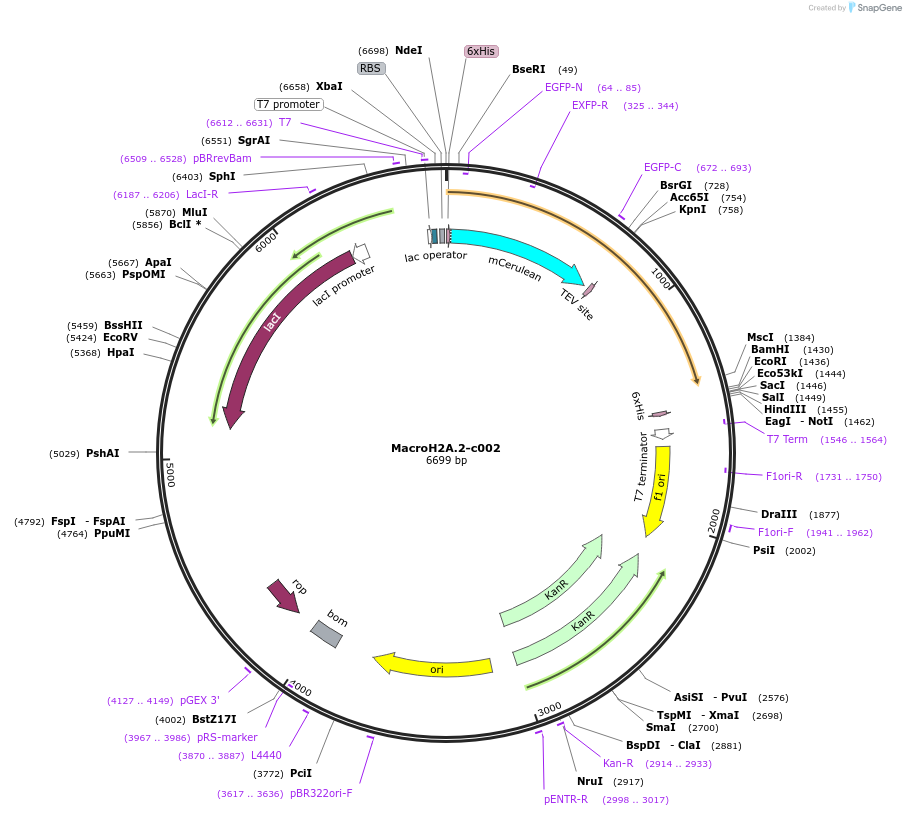
MacroH2A.2-c002
Plasmid#173089PurposeExpression of recombinant fusion protein in E. coliDepositorAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
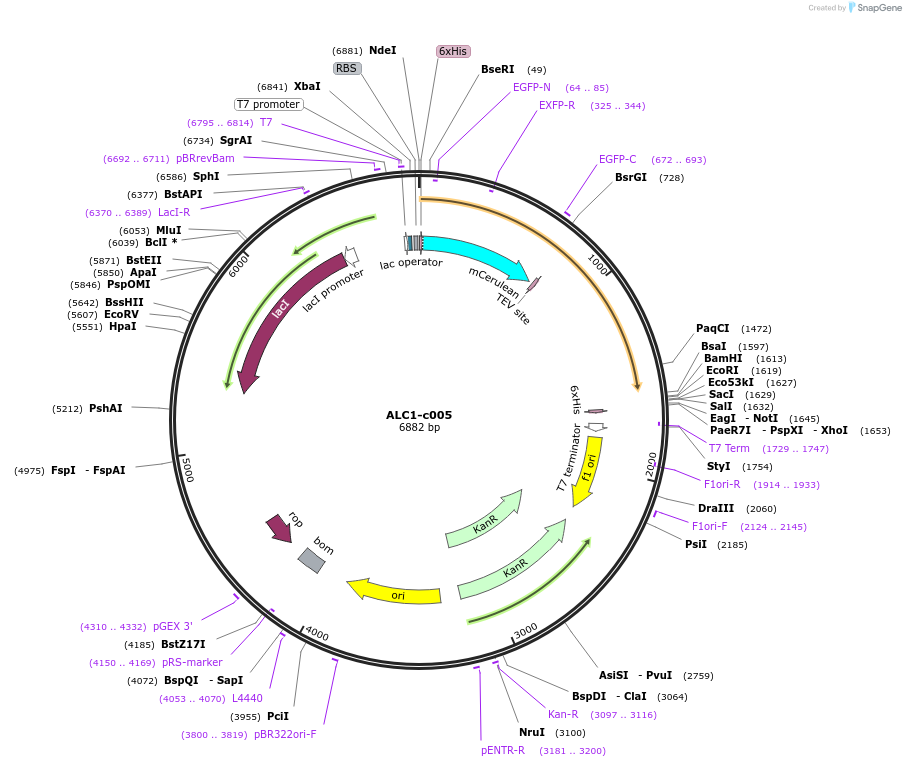
ALC1-c005
Plasmid#173092PurposeExpression of recombinant fusion protein in E. coliDepositorAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
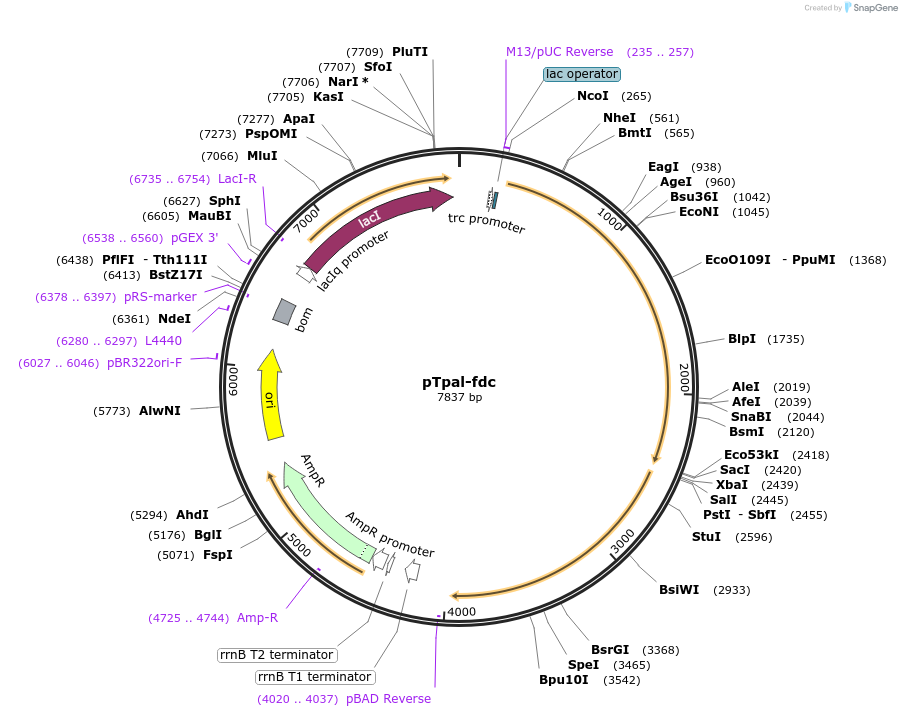
pTpal-fdc
Plasmid#78288PurposeExpresses A. thaliana PAL2 and S. cerevisiae FDC1 in E. coliDepositorExpressionBacterialMutation*see belowPromotertrcAvailable SinceJune 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
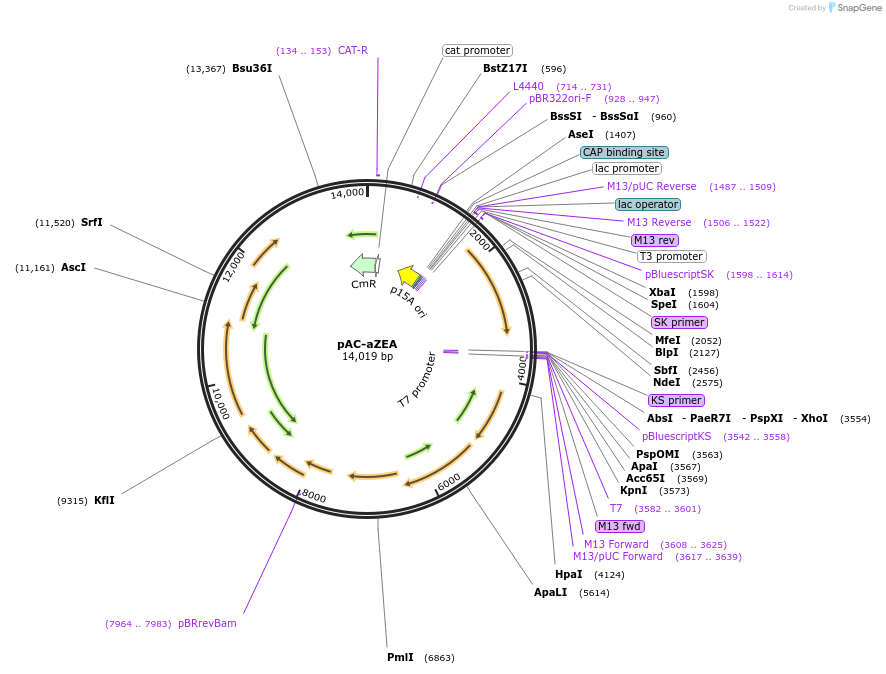
pAC-aZEA
Plasmid#53286PurposeContains crtE, and crtB genes of Erwinia herbicola (Pantoea agglomerans) Eho10, crtI gene of Rhodobacter capsulatus, and lcyE cDNA of Arabidopsis thaliana and produces alpha-zeacarotene in E. coliDepositorAvailable SinceJan. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
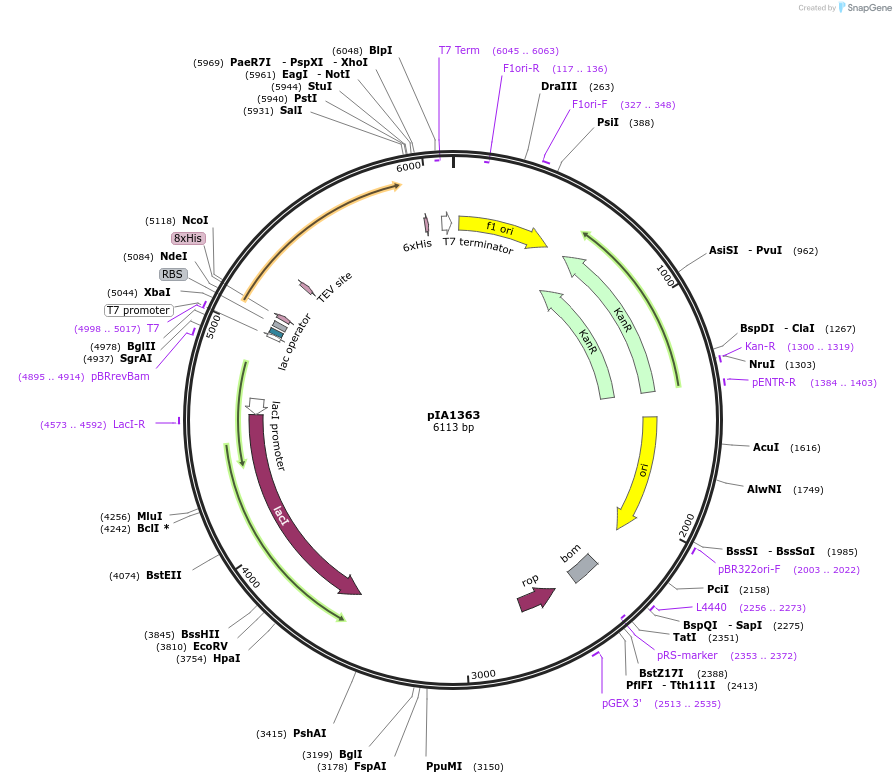
pIA1363
Plasmid#166861PurposeExpression of codon optimized SARS-COV-2 Nsp8 in E. coliDepositorAvailable SinceMarch 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
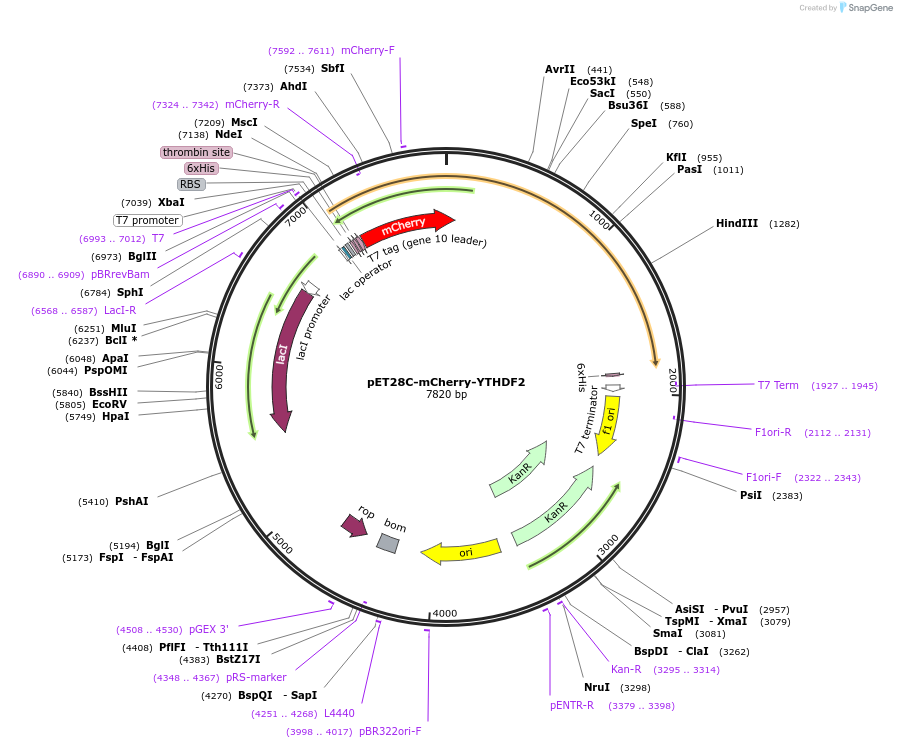
pET28C-mCherry-YTHDF2
Plasmid#204406PurposeE. coli expression of full-length YTHDF2 with an N-terminal mCherry tagDepositorAvailable SinceSept. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
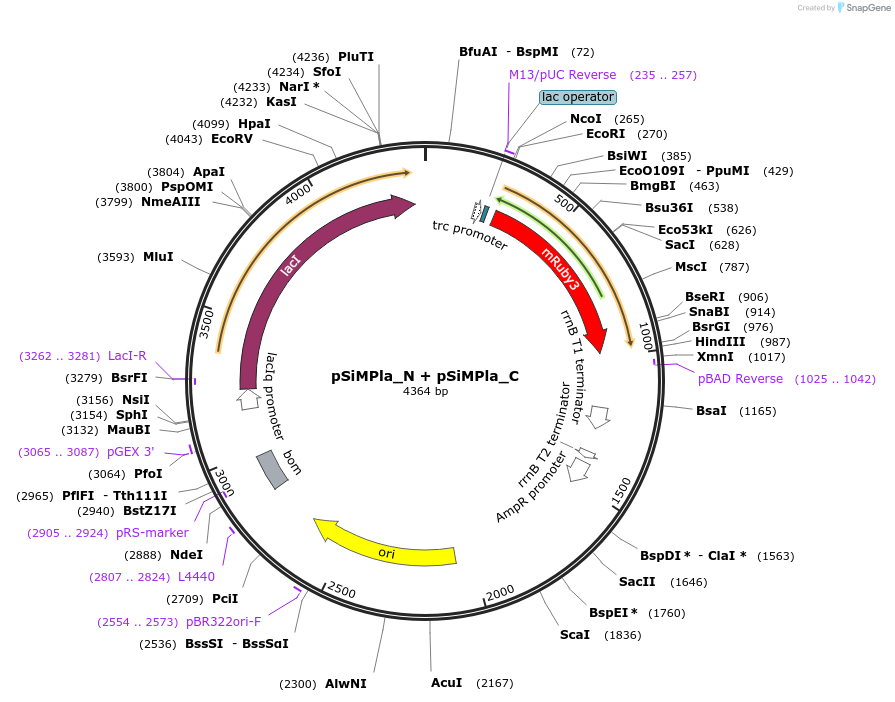
pSiMPlc_N + pSiMPlc_C
Plasmid#134312PurposeMaintenance of 2 plasmids with a single antibiotic in E. coliDepositorInsertcmR_gp41-1 N-intein + cmR_gp41-1 C-intein
ExpressionBacterialAvailable SinceFeb. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
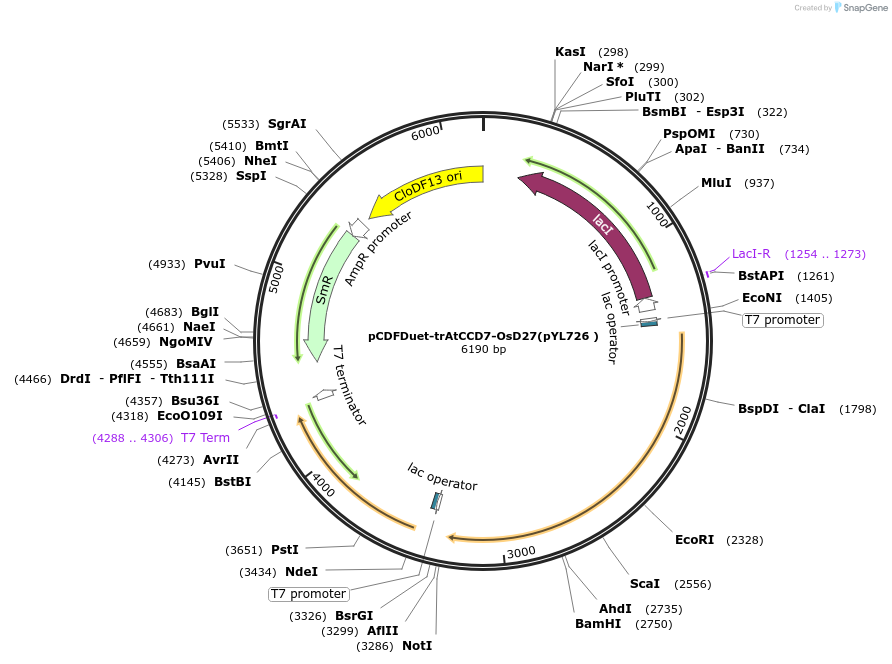
pCDFDuet-trAtCCD7-OsD27(pYL726 )
Plasmid#178285PurposeCo-Expresses OsD27 and trAtCCD7 in E. coli. pCDFDuet-1 carrying D27 from Oryza sativa and N-terminus 31 amino-acid truncated CCD7 from ArabidopsisDepositorInsertsOsD27
trAtCCD7
ExpressionBacterialPromoterT7Available SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
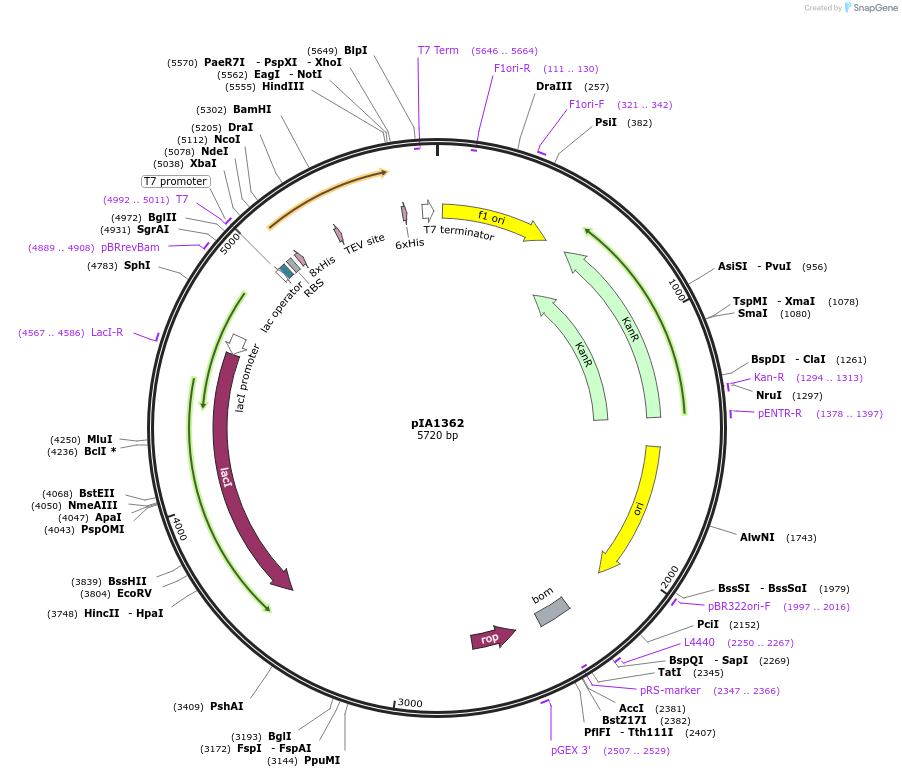
pIA1362
Plasmid#166860PurposeExpression of codon optimized SARS-COV-2 Nsp7 in E. coliDepositorAvailable SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
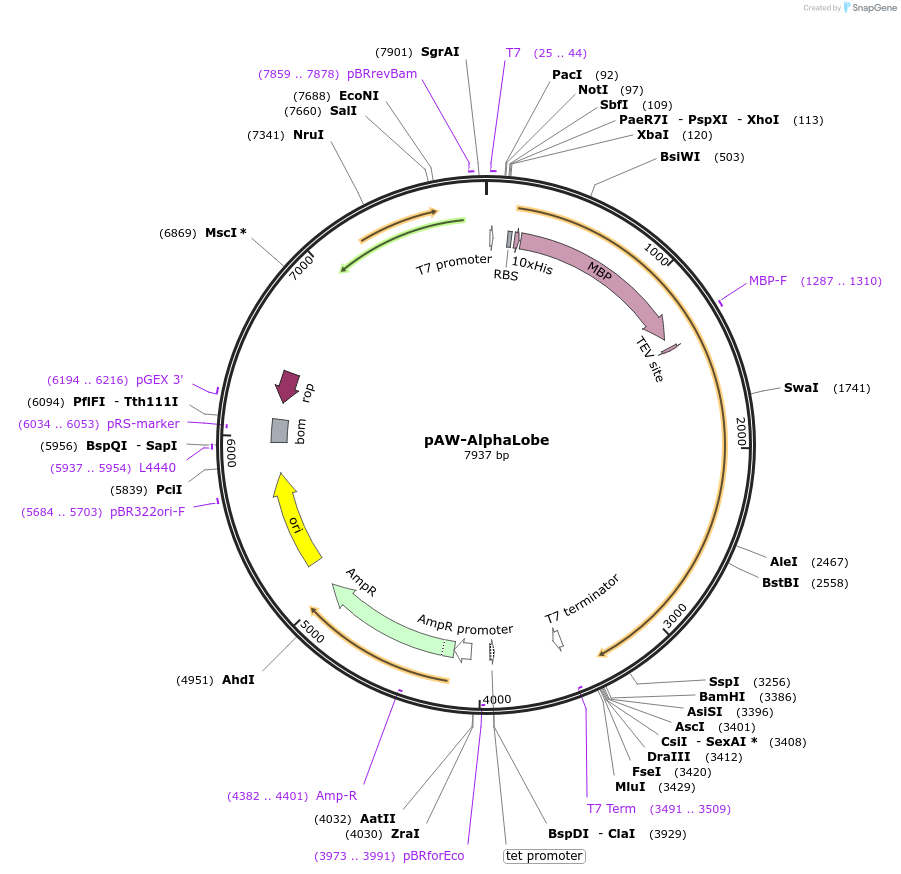
pAW-AlphaLobe
Plasmid#113255PurposeE. coli expression vector for His-MBP-Cas9 Alpha-Helical LobeDepositorInsertS. pyogenes Cas9 alpha-helical lobe
Tags10xHis-MBPExpressionBacterialMutationresidues 56–714; See paper for descriptionPromoterT7Available SinceAug. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
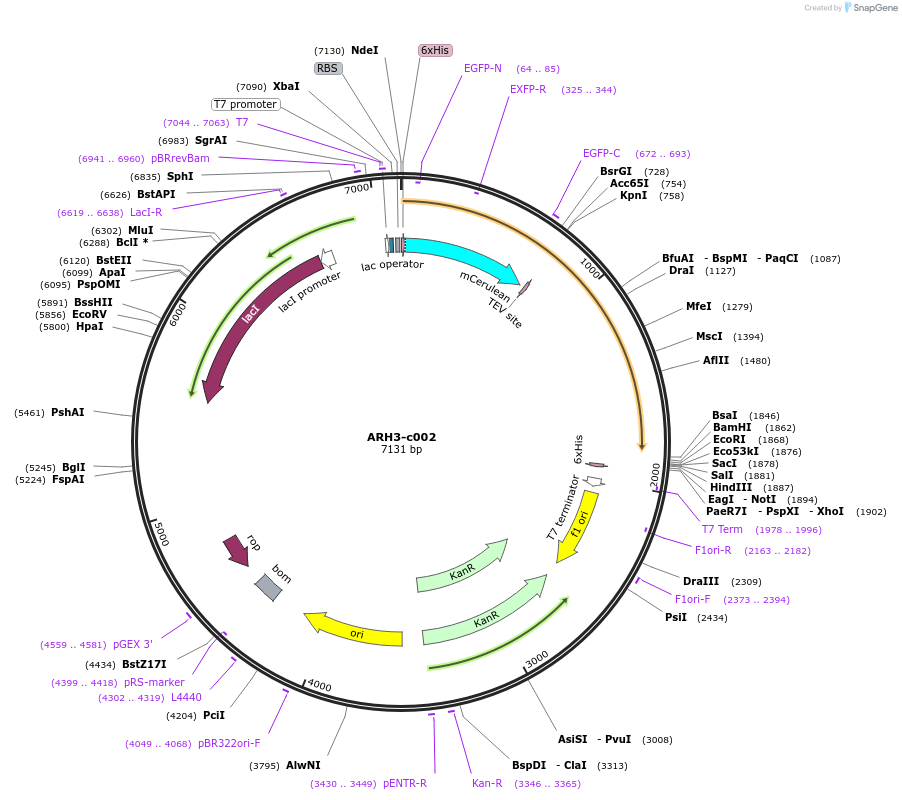
ARH3-c002
Plasmid#173095PurposeExpression of recombinant fusion protein in E. coliDepositorAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
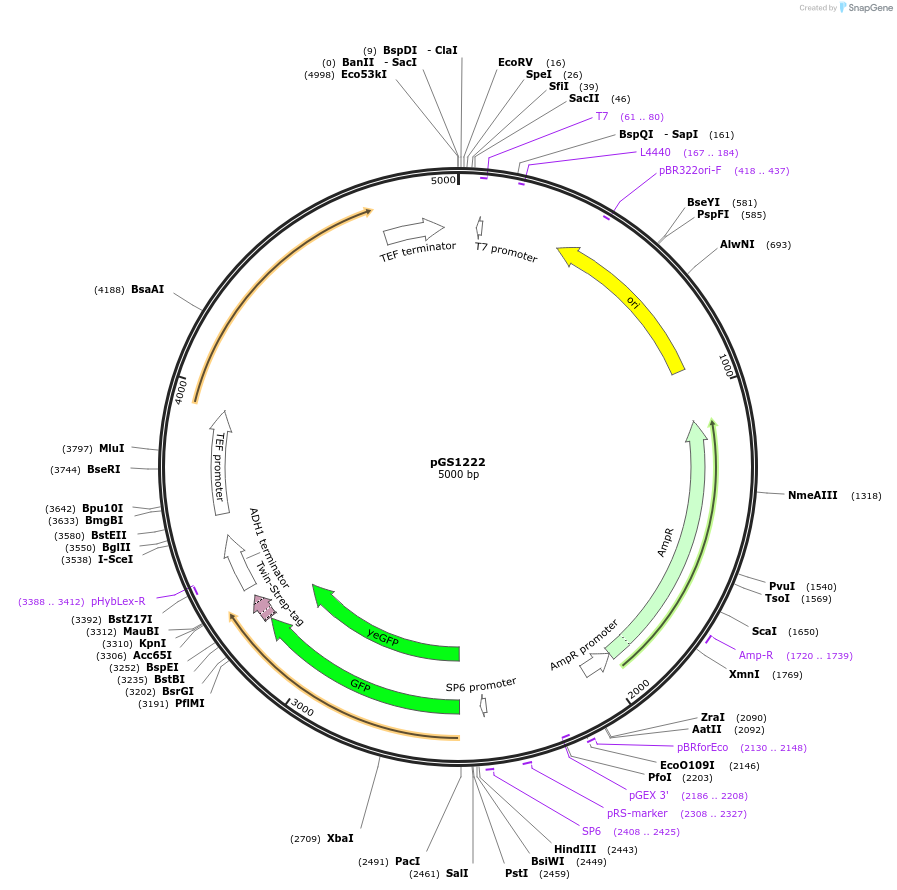
pGS1222
Plasmid#63791PurposeVector to create a cassette for homologous recombination in S. cerevisiae; adds a C-terminal eGFP Twin-Strep tag followed by URA3 selection markerDepositorInserteGFP-Twin-Strep tag followed by URA3
UseE. coli cloning vectorTagseGFP-Twin-Strep tagPromoternoneAvailable SinceApril 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
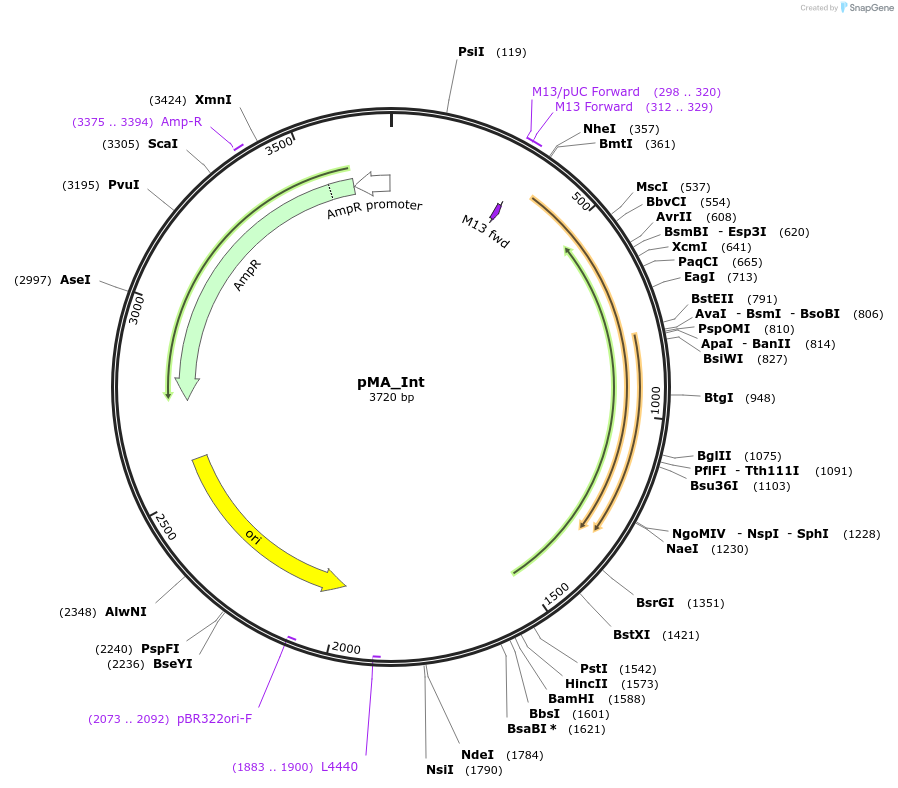
pMA_Int
Plasmid#110096PurposeE. coli replicative vector which contains the mycophage integrase gene required for the attP/attB integration. Is co-transformed in trans together with pFLAG_attP, but cannot replicate in mycobacteria, and is thus lost shortly after electroporation.DepositorInsertmycophage integrase
UseRequired for transformation of attb integrating v…Available SinceJune 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
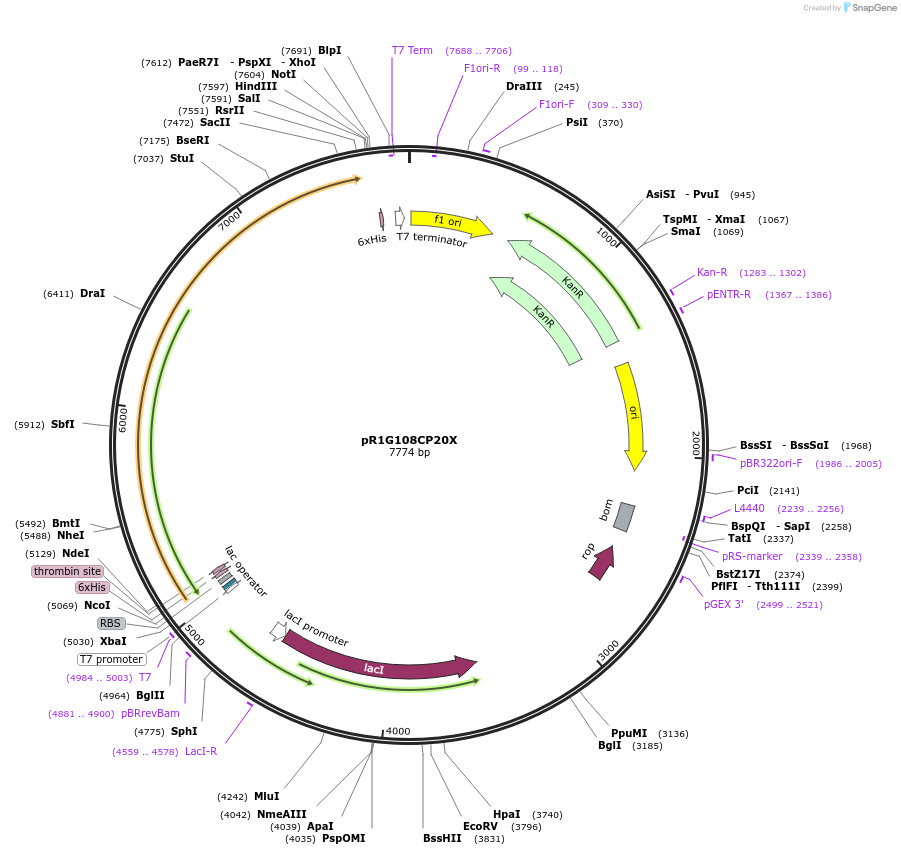
pR1G108CP20X
Plasmid#85086PurposeExpresses PCNA1 variant (G108C) fused to putidaredoxin reductase and putidaredoxin in E. coliDepositorInsertThe G108C variant of PCNA1 fused to putidaredoxin reductase and putidaredoxin
TagsHis6 tagExpressionBacterialMutationG108C in PCNA1PromoterT7 promoterAvailable SinceNov. 28, 2016AvailabilityAcademic Institutions and Nonprofits only -
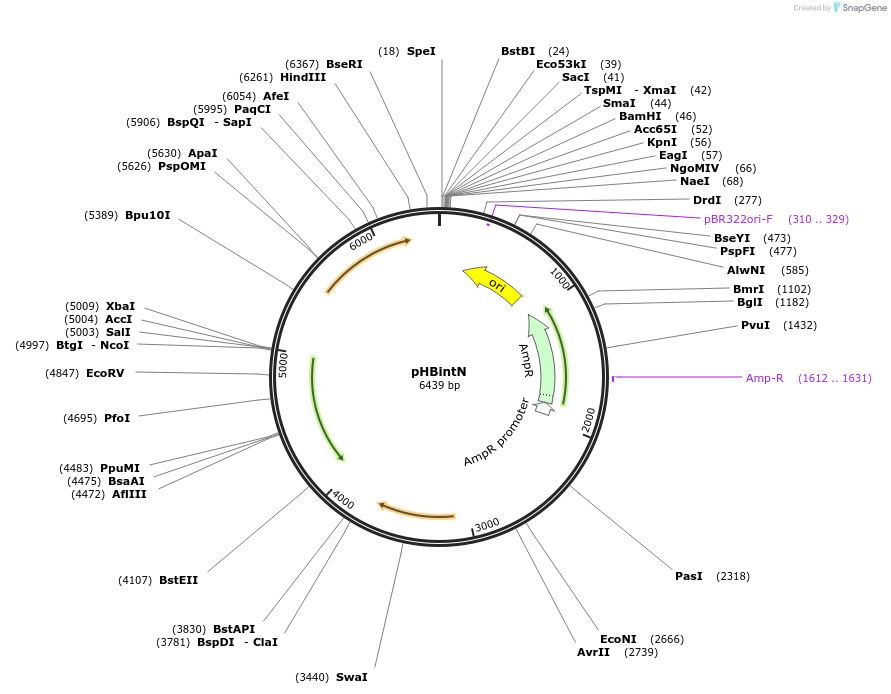
pHBintN
Plasmid#48135PurposeShuttle vector E. coli/B.meg., carrying a temperature sensitive origin of replication for Bacillus megaterium, selection marker neomycine for B. megaterium, integration vectorDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceOct. 11, 2013AvailabilityAcademic Institutions and Nonprofits only -
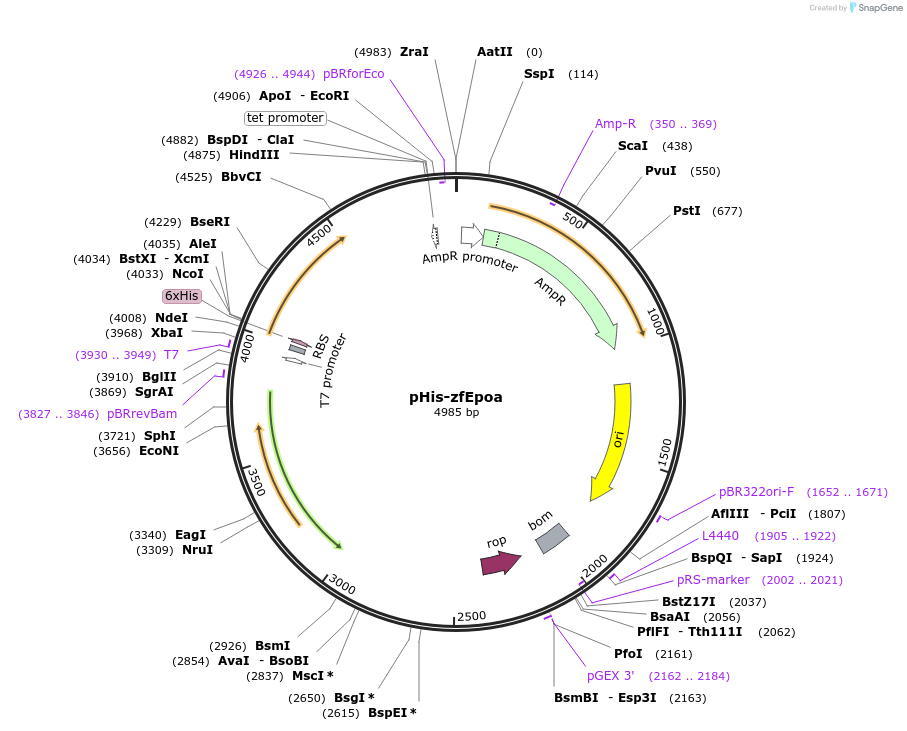
pHis-zfEpoa
Plasmid#65611PurposeExpress soluble His-tagged zebrafish erythropoietin in E. coliDepositorAvailable SinceSept. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
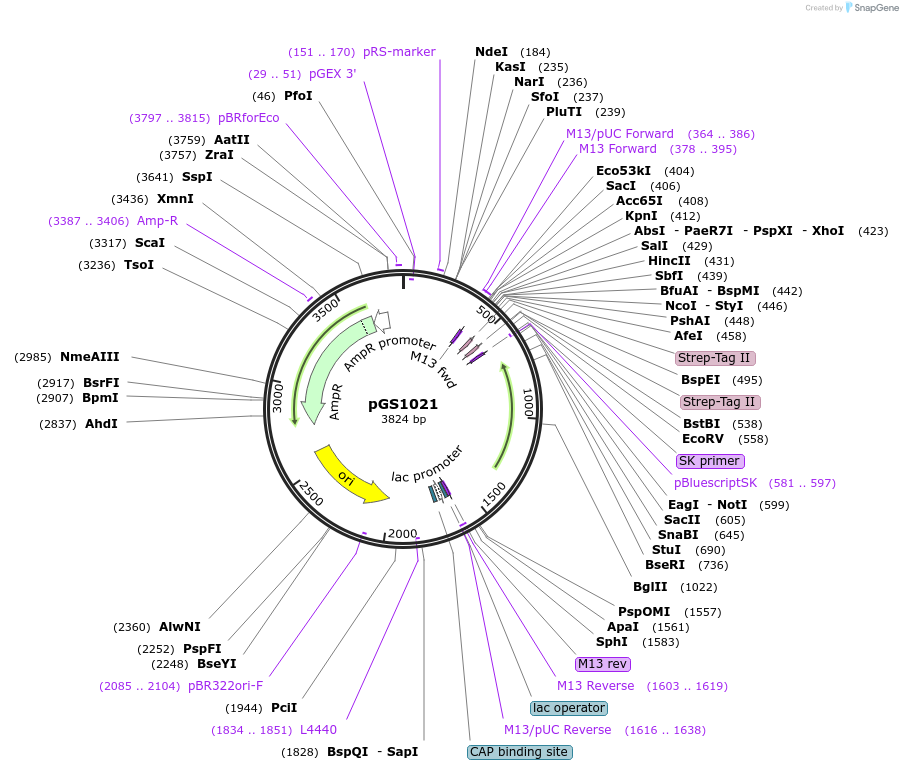
pGS1021
Plasmid#59575PurposeVector to create a cassette for homologous recombination in S. cerevisiae; adds a C-terminal Twin-Strep tag and a TRP1 selection markerDepositorInsertTwin-Strep tag optimized for PCR amplification followed by TRP1 selection marker
UseE. coli cloning vectorTagsTwin-Strep tagPromoterNoneAvailable SinceAug. 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
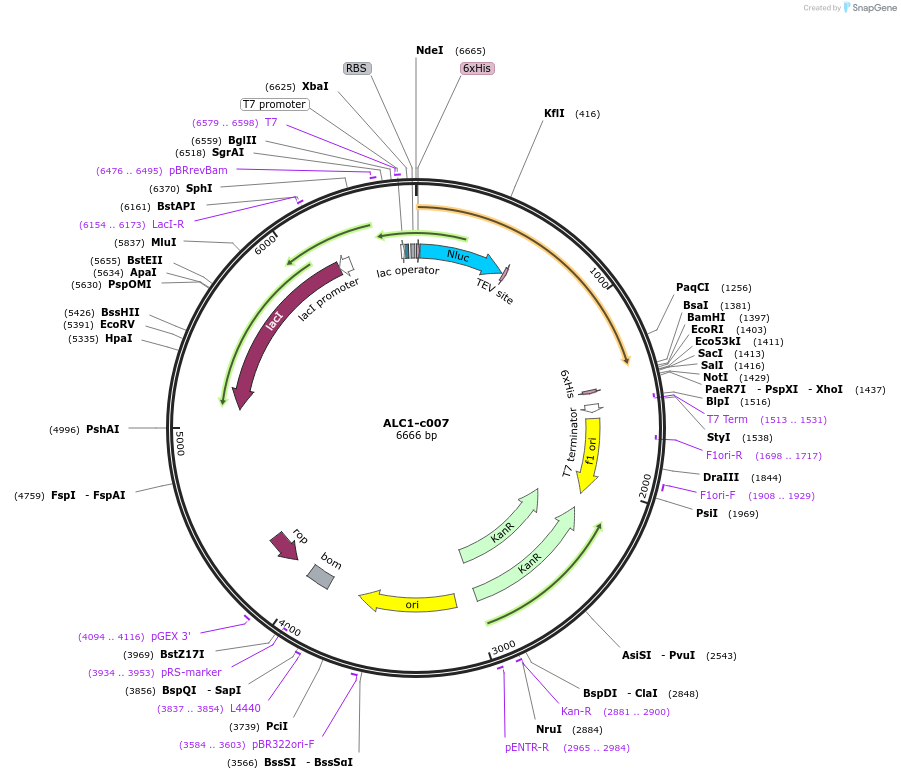
ALC1-c007
Plasmid#173112PurposeExpression of recombinant fusion protein in E. coliDepositorAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
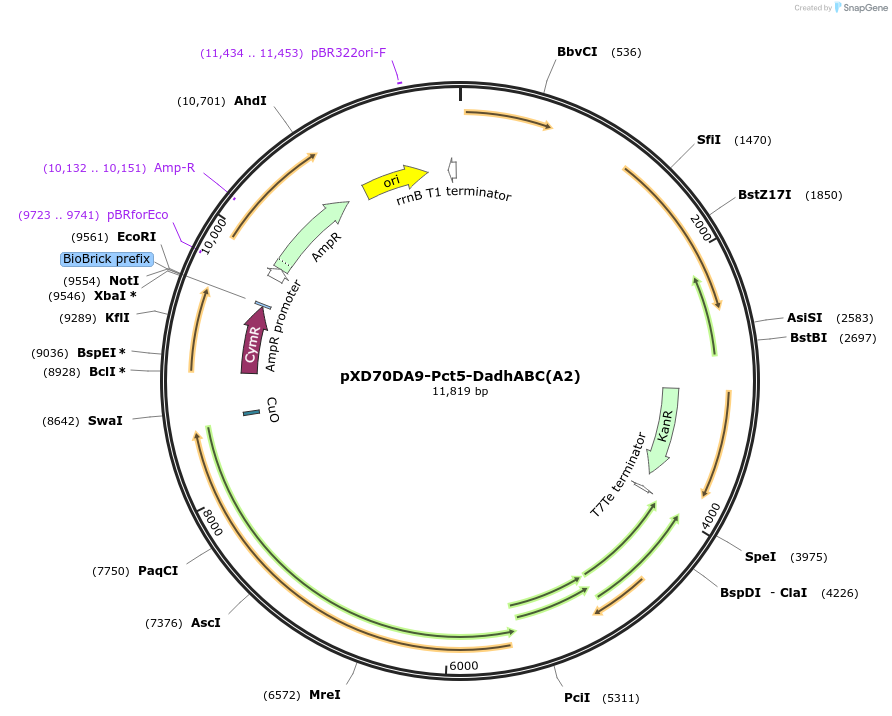
pXD70DA9-Pct5-DadhABC(A2)
Plasmid#191632PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), cumate-inducible promoter Pct5-dopamine dehydroxylase from dopamine-metabolizing E. lenta A2 strainDepositorInsertDopamine dehydroxylase
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
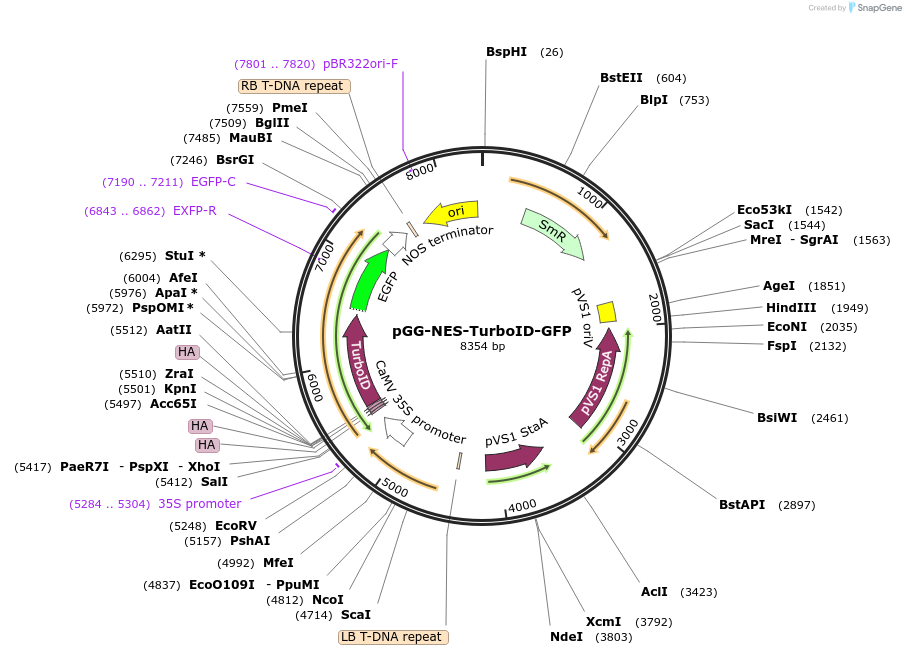
pGG-NES-TurboID-GFP
Plasmid#209401PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a nuclear export signal (NES) in plantaDepositorInsertNES-3XHA-TurboID
TagsGFPExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
pSiMPla_N + pSiMPla_C
Plasmid#134314PurposeMaintenance of 2 plasmids with a single antibiotic in E. coliDepositorInsertampR_gp41-1 N-intein + ampR_gp41-1 C-intein
ExpressionBacterialAvailable SinceMarch 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
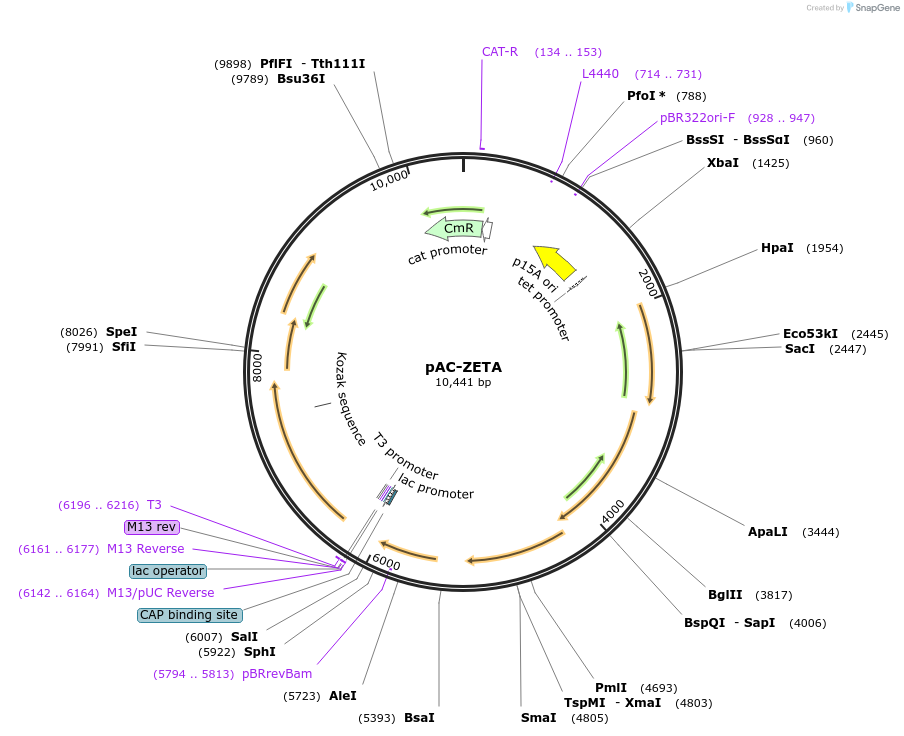
pAC-ZETA
Plasmid#53316PurposeContains Erwinia herbicola Eho10 genes crtE and crtB, together with the pds (crtP) gene of Synechococcus PCC7942 fused to the lacZ gene at the N terminus. Produces zeta-carotene in E. coli.DepositorInsertpds
UseLow copy number bacterial cloning vectorPromoterlazZAvailable SinceJan. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
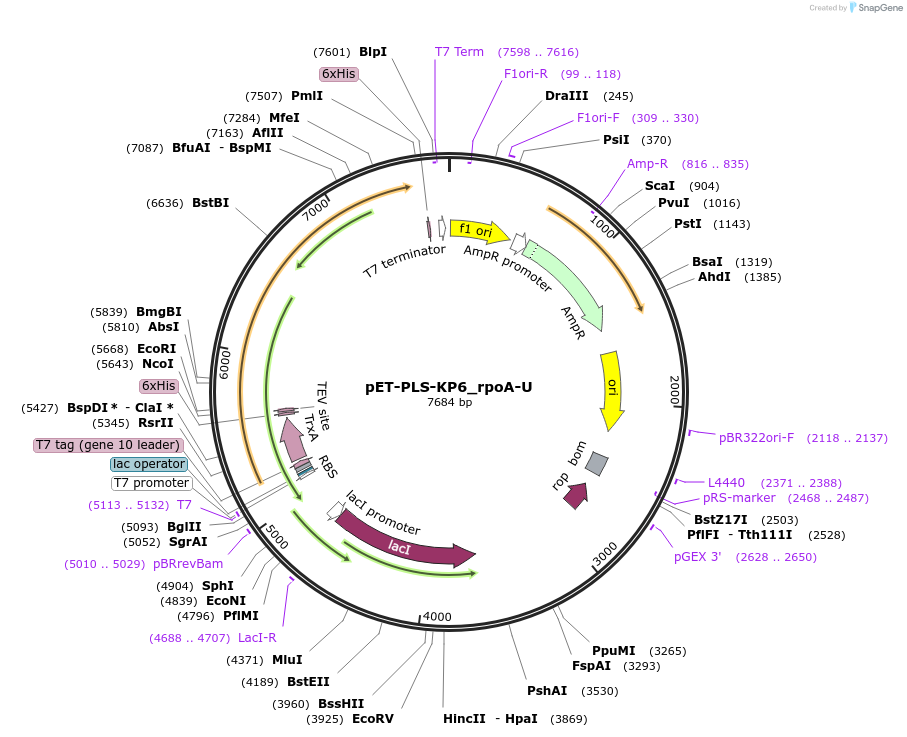
pET-PLS-KP6_rpoA-U
Plasmid#190967PurposeTest the U-to-C editing activity of KP6 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP6
TagsHisExpressionBacterialPromoterT7Available SinceOct. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
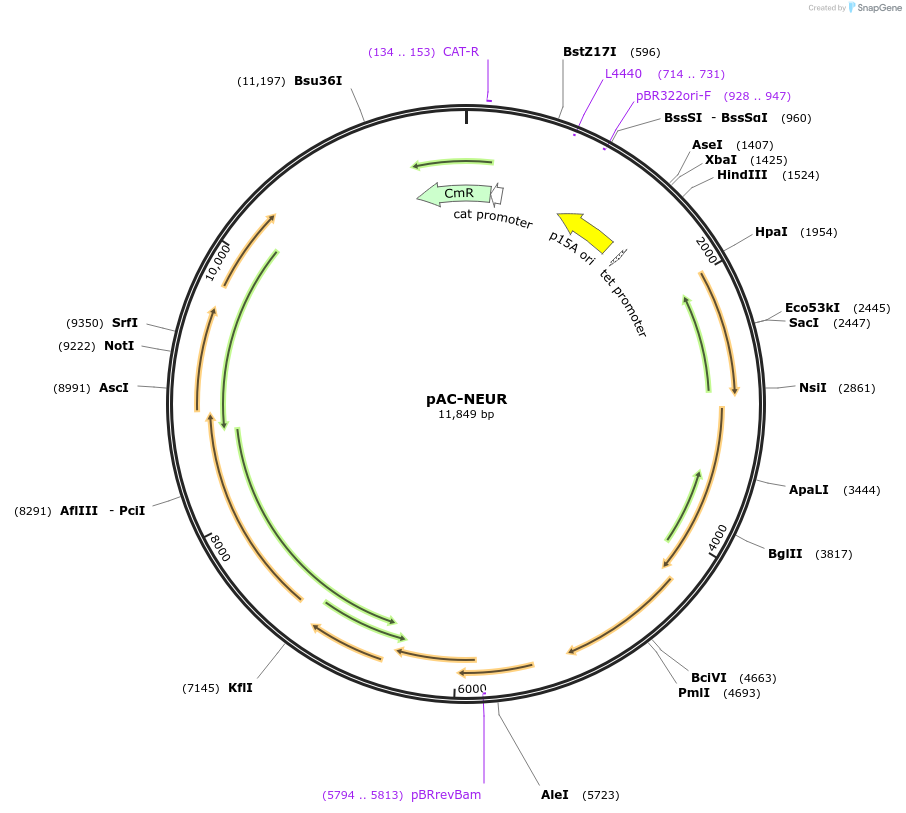
pAC-NEUR
Plasmid#53271PurposeContains the Erwinia herbicola (Pantoea agglomerans) Eho10 genes crtE and crtB, and the Rhodobacter capsulatus crtI gene and thereby produces neurosporene in Escherichia coliDepositorInsertcrtI
UseLow copy number bacterial cloning vectorPromoterendogenous promotersAvailable SinceJan. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
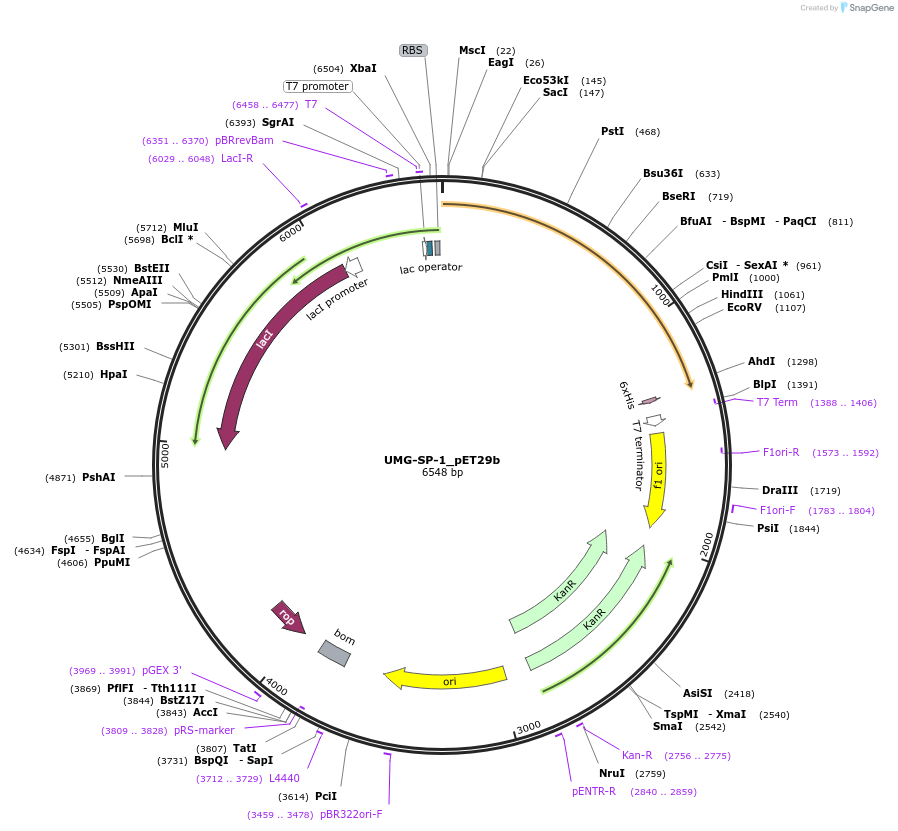
UMG-SP-1_pET29b
Plasmid#215433PurposeExpression construct for UMG-SP-1 gene, codon optimized for expression in E. coliDepositorInsertUMG-SP-1
ExpressionBacterialAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
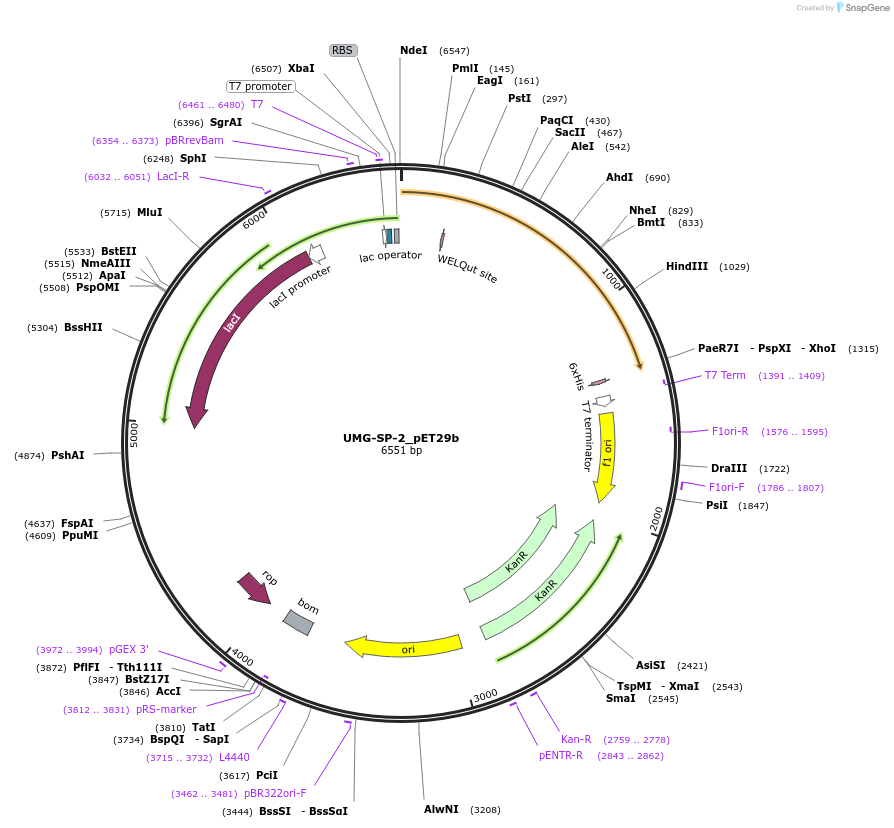
UMG-SP-2_pET29b
Plasmid#215434PurposeExpression construct for UMG-SP-2 gene, codon optimized for expression in E. coliDepositorInsertUMG-SP-2
ExpressionBacterialAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only