We narrowed to 10,645 results for: ESP
-
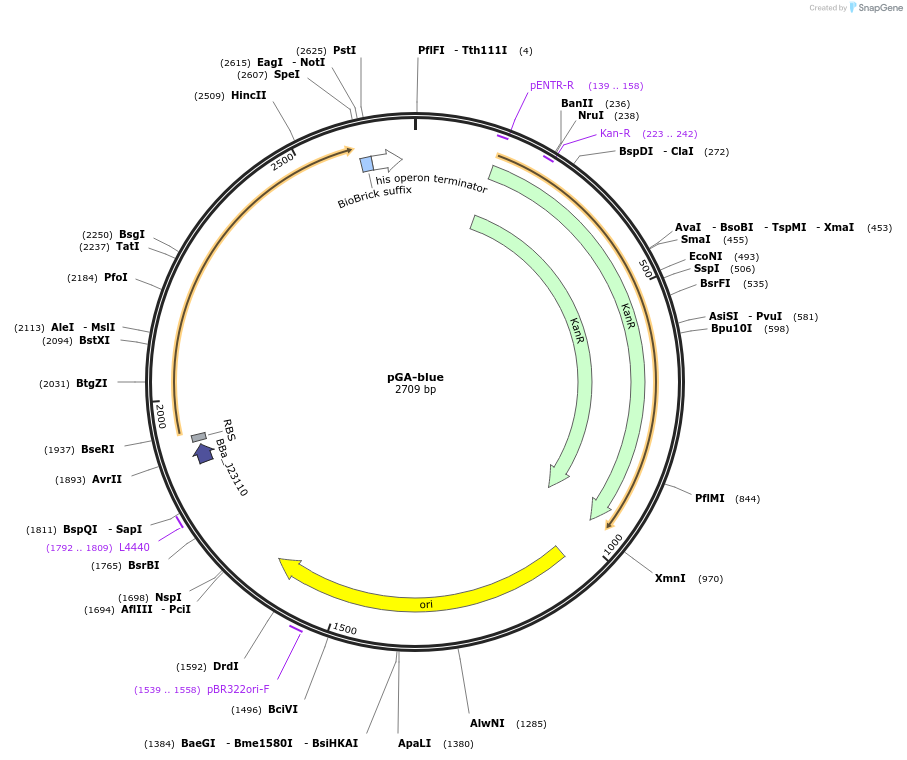
Plasmid#196336PurposeEasy-MISE toolkit Level 0 destination plasmid for parts cloning with Esp3I-based Golden Gate Assembly; allowing fast postitive clones test with AmilCP chromoprotein-based blue/white screeningDepositorTypeEmpty backboneUseSynthetic Biology; Golden gate assembly backboneAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only
-
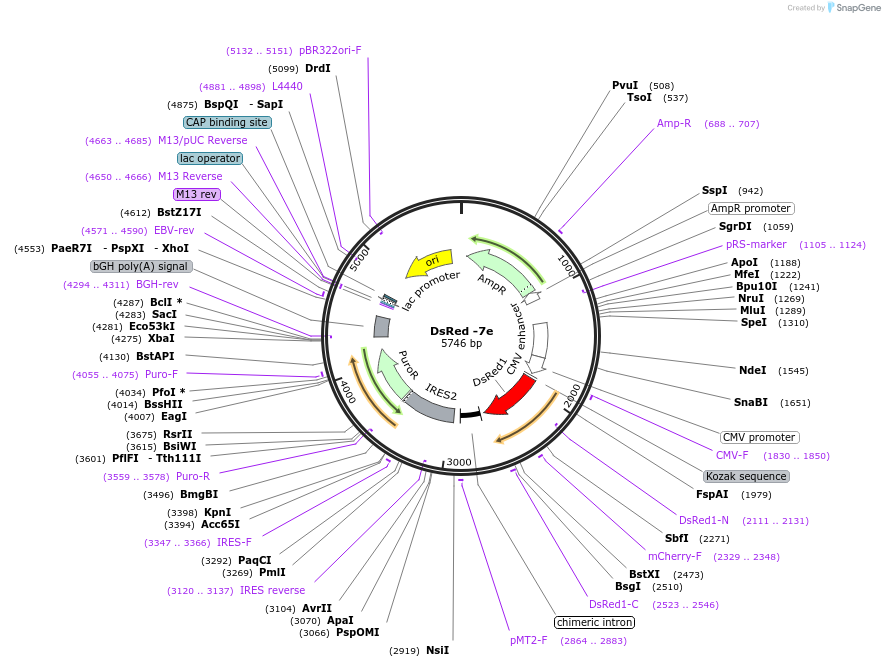
DsRed -7e
Plasmid#191765PurposeExpression of DsRed fused to a negatively charged polypeptide. Overall charge on tetramer: -7eDepositorInsertDsRed
TagsN terminal fusion of negatively charged polypetod…ExpressionBacterial and MammalianPromoterCMVAvailable SinceFeb. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
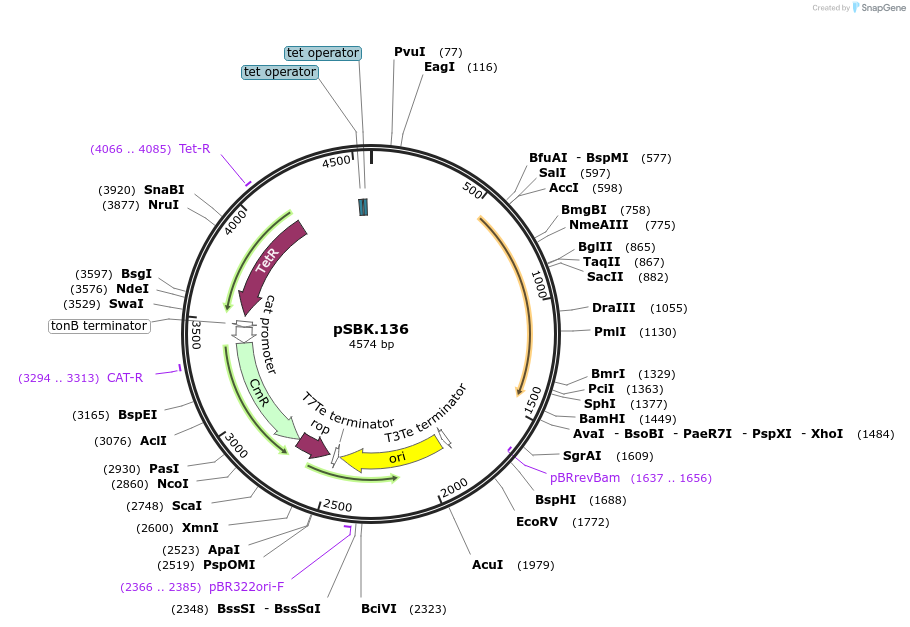
pSBK.136
Plasmid#187220PurposeExpresses Eco1 ncRNA v35 (long a1/a2) and Eco1 ncRNA v35 (long a1/a2, barcode 6) from promoters pSalTTC and pTet*, respectivelyDepositorInsertRetron Eco1 ncRNA v35 (long a1/a2); Retron Eco1 ncRNA v35 (long a1/a2, barcode 6)
ExpressionBacterialPromoterA: pSalTTC; B: pTet*Available SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
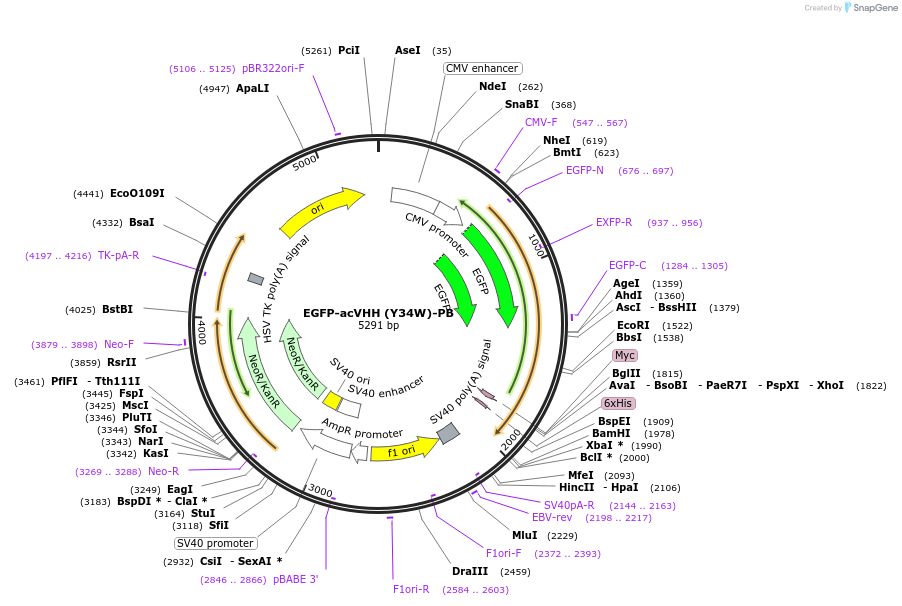
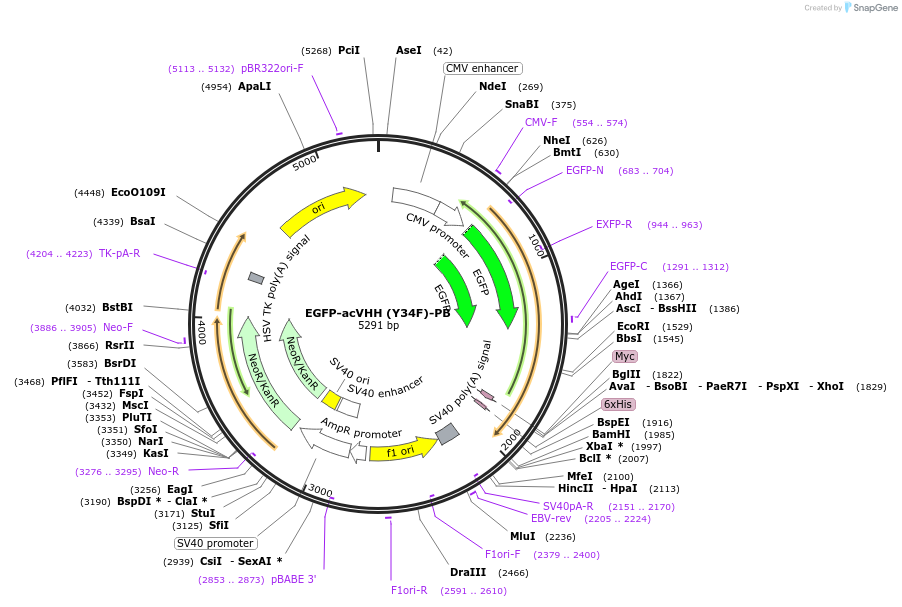
EGFP-acVHH (Y34W)-PB
Plasmid#171072PurposeA COSMO mutant (Y34W) responds to caffeine to induce dimerization of a phospholipid-binding polybasic domain with subsequent plasma membrane translocation.DepositorInsertanti-caffeine nanobody variant
Tagsmodified STIM1–PB tagExpressionMammalianMutationY34WPromoterCMVAvailable SinceJuly 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
EGFP-acVHH (Y34F)-PB
Plasmid#171071PurposeA COSMO mutant (Y34F) responds to caffeine to induce dimerization of a phospholipid-binding polybasic domain with subsequent plasma membrane translocation.DepositorInsertanti-caffeine nanobody variant
Tagsmodified STIM1–PB tagExpressionMammalianMutationY34FPromoterCMVAvailable SinceJuly 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
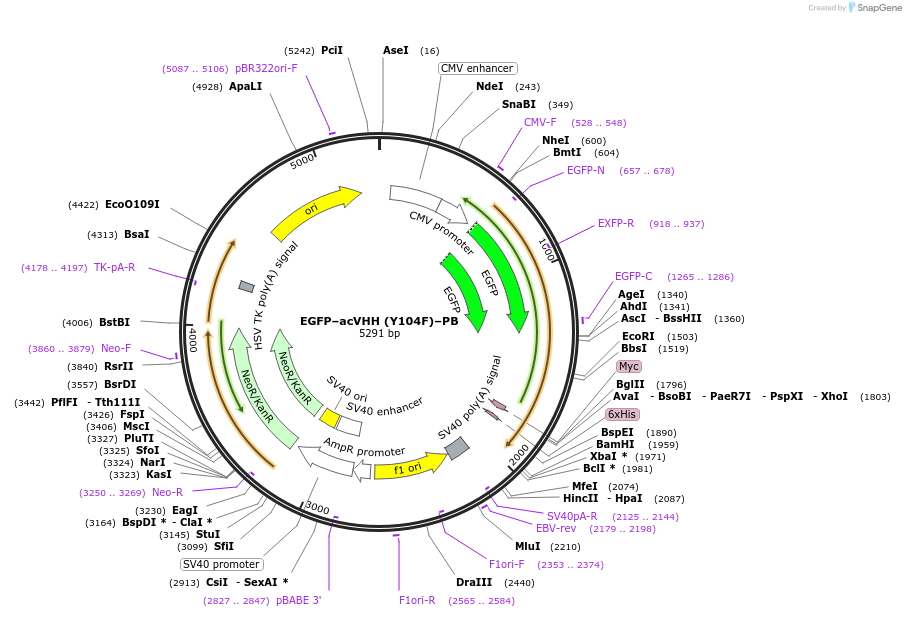
EGFP–acVHH (Y104F)–PB
Plasmid#169683PurposeA COSMO mutant (Y104F) responds to caffeine to induce dimerization of a phospholipid-binding polybasic domain with subsequent plasma membrane translocation.DepositorInsertanti-caffeine nanobody variant
Tagsmodified STIM1–PB tagExpressionMammalianMutationY104FPromoterCMVAvailable SinceJuly 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
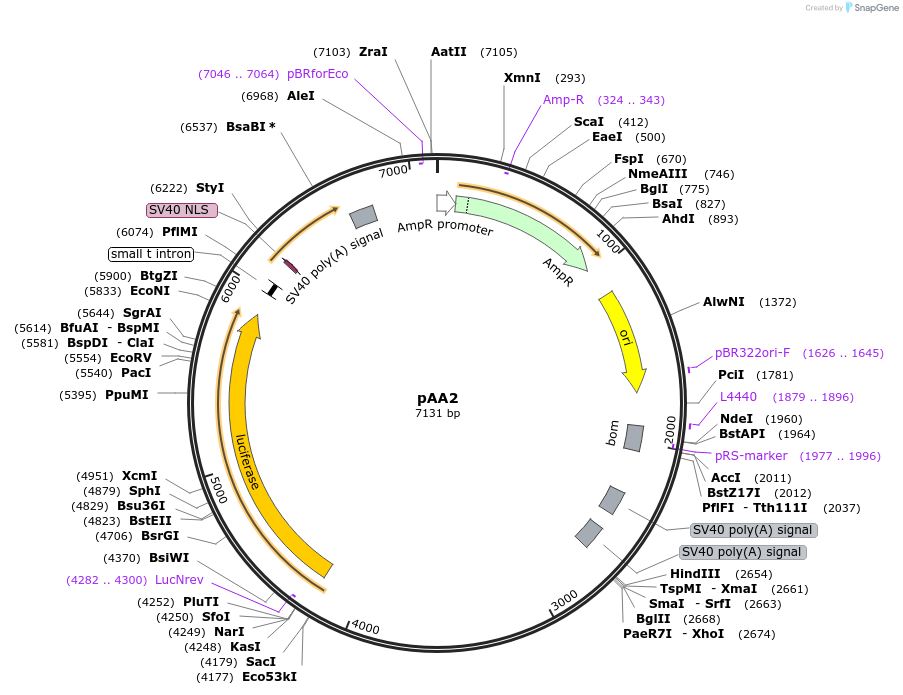
pAA2
Plasmid#154162PurposeFirefly luciferase under the control of 4 copies of: the enhancer for the Drosophila melanogaster Metchnikowin gene, 4 Rel1A-specific NF-_B sites, 3 GATA sites and 3 putative ecdysone-response elements, plus an additional NF-_B site.DepositorInsertFirefly luciferase
UseLuciferaseMutation16 wt NF-κB sites were replaced with Rel1A-specif…Available SinceAug. 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
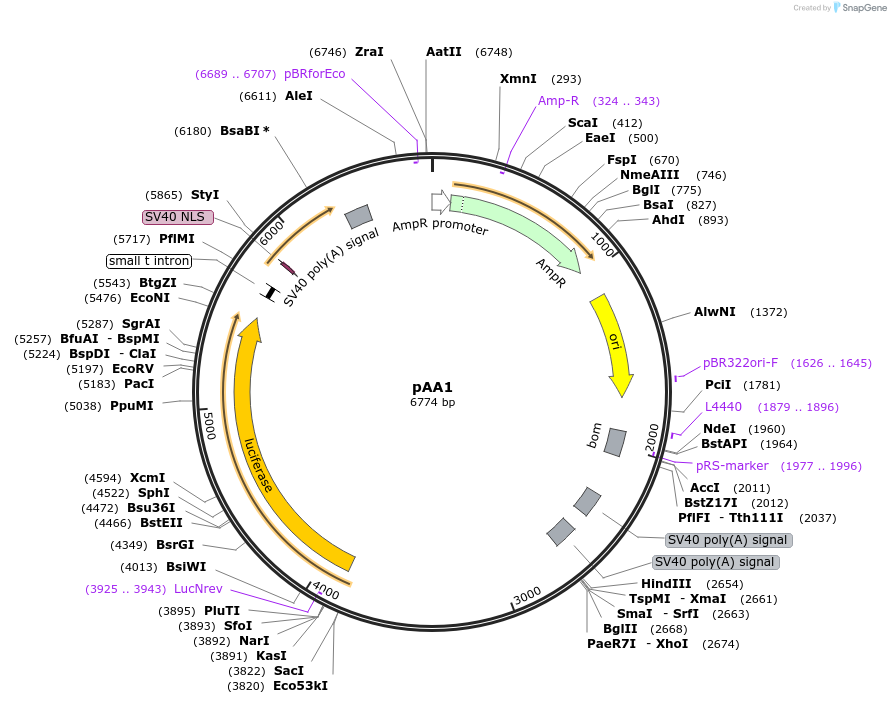
pAA1
Plasmid#154161PurposeFirefly luciferase under the control of 3 copies of: the enhancer for the Drosophila melanogaster Metchnikowin gene, 4 Rel1A-specific NF-_B sites, 3 GATA sites and 3 putative ecdysone-response elements, plus an additional NF-_B site.DepositorInsertFirefly luciferase
UseLuciferaseMutation12 wt NF-κB sites were replaced with Rel1A-specif…Available SinceAug. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
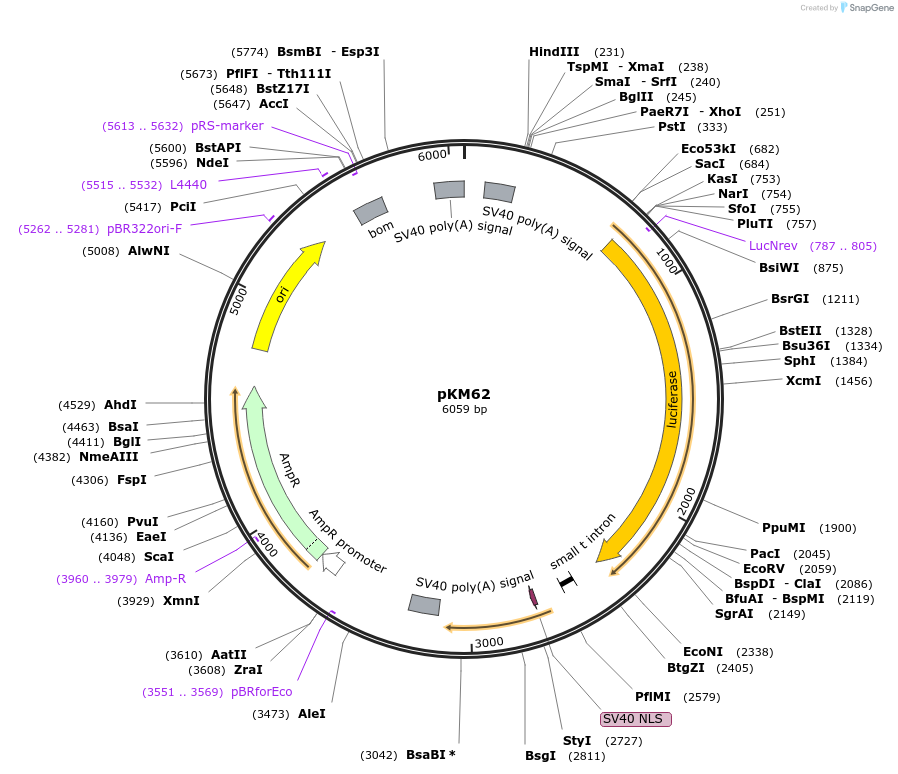
pKM62
Plasmid#154160PurposeFirefly luciferase under the control of the enhancer for the Drosophila melanogaster Metchnikowin gene, with 4 Rel2-specific (VII) NF-_B sites, 3 GATA sites and 3 putative ecdysone-response elements, plus an additional NF-_B site.DepositorInsertFirefly luciferase
UseLuciferaseMutation4 wt NF-κB sites were replaced with Rel2-specific…Available SinceAug. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
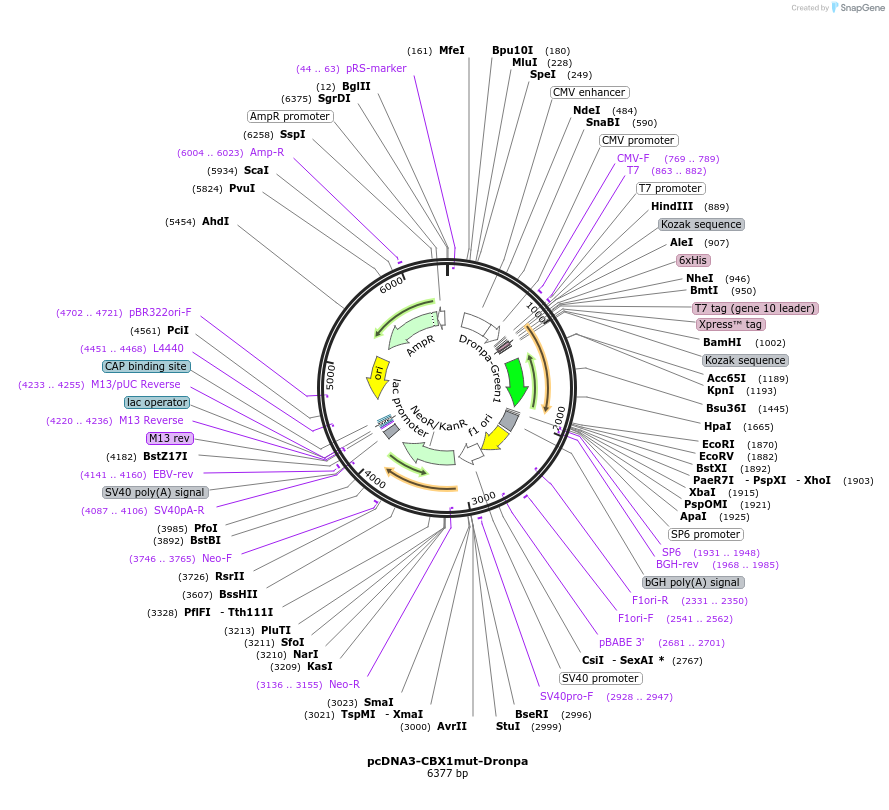
pcDNA3-CBX1mut-Dronpa
Plasmid#138250PurposeTriple-mutant CBX1 chromodomain (residues 20-73) tagged with reversibly switchable FP Dronpa. Mutations V3E/K6E/D40S correspond to positions V22/K25/D59 in full-length CBX1.DepositorInsertCBX1mut-Dronpa
Tags6xHIS - T7 tag (gene 10 leader) - Xpress (TM) tag…ExpressionMammalianPromoterCMVAvailable SinceJuly 30, 2020AvailabilityAcademic Institutions and Nonprofits only -
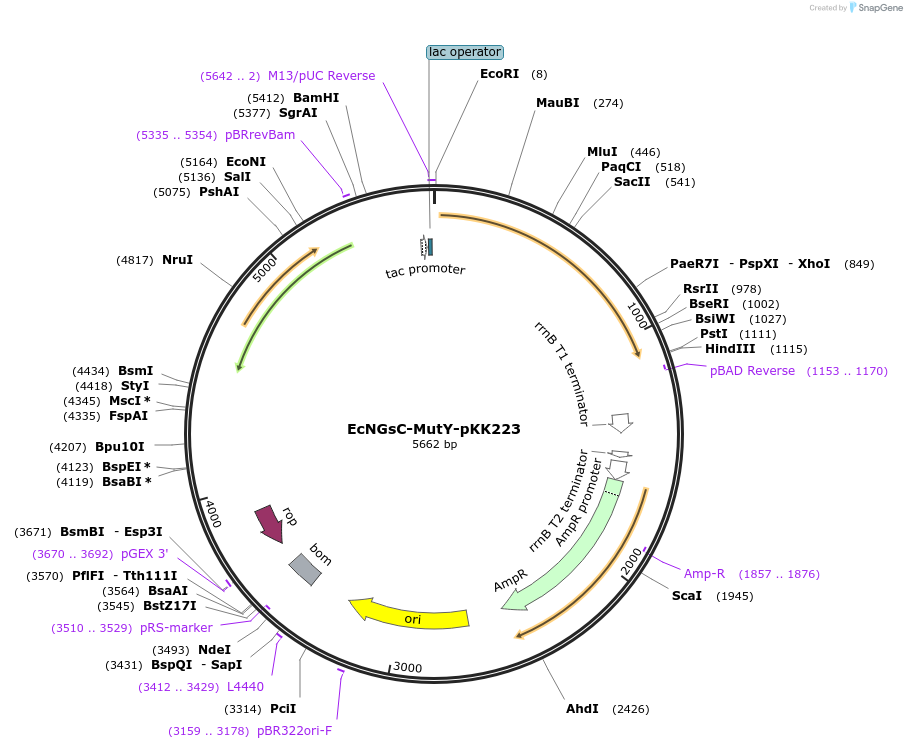
EcNGsC-MutY-pKK223
Plasmid#131376PurposeLow level expression of E.coli G.stearothermophillus MutY chimera proteinDepositorInsertEcNGsC MutY chimera
ExpressionBacterialMutationFusion of two MutY genes. Residues 1-225 are from…PromotertacAvailable SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
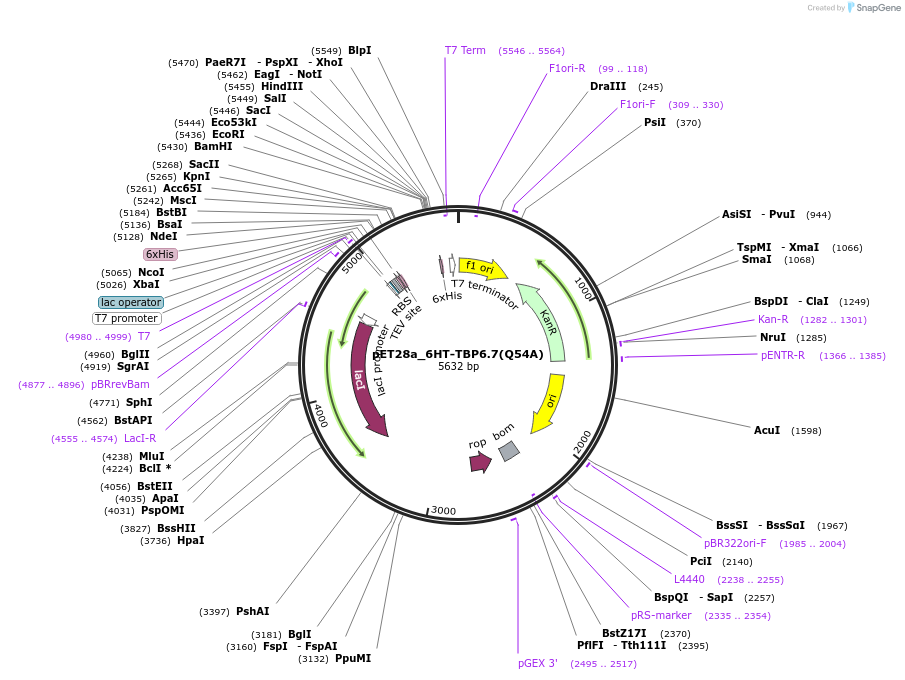
pET28a_6HT-TBP6.7(Q54A)
Plasmid#112730PurposeAlthough not evolved, this residue within TBPs participates in stabilizing the polypeptide loop responsible for RNA recognition.DepositorInsert6His-TEV-TBP6.7(Q54A)
Tags6His-TEVExpressionBacterialMutationQ54APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
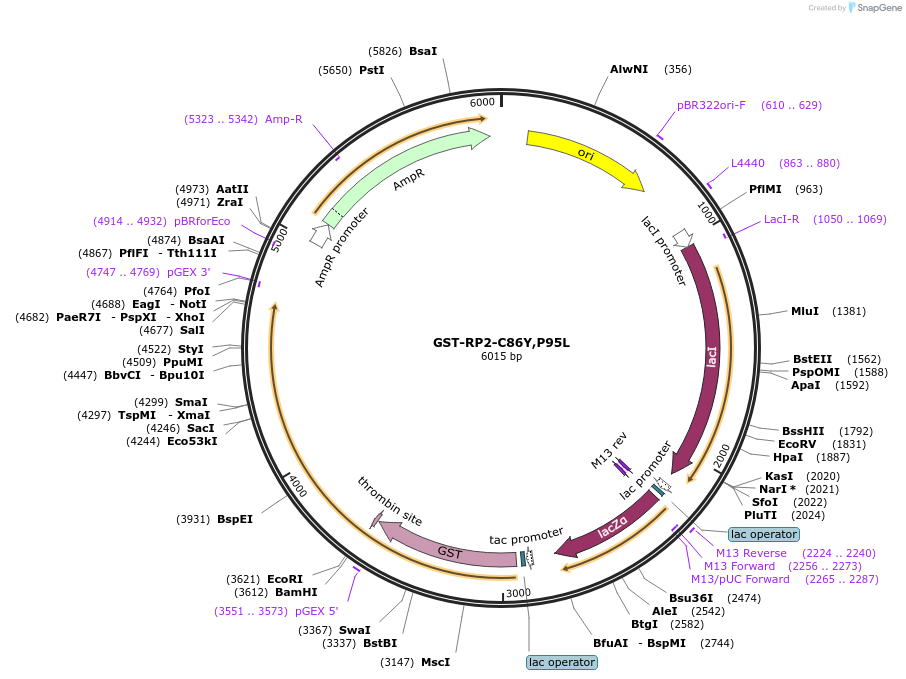
GST-RP2-C86Y,P95L
Plasmid#86074PurposeA bacterial expression plasmid encoding ciliary localization effected RP2 (RP2-C86Y,P95L) with N-terminus GST-tag.DepositorInsertRP2 (RP2 Human)
TagsGSTExpressionBacterialMutationChanged cysteine-86 and proline-95 to tyrosine an…PromotertacAvailable SinceMay 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
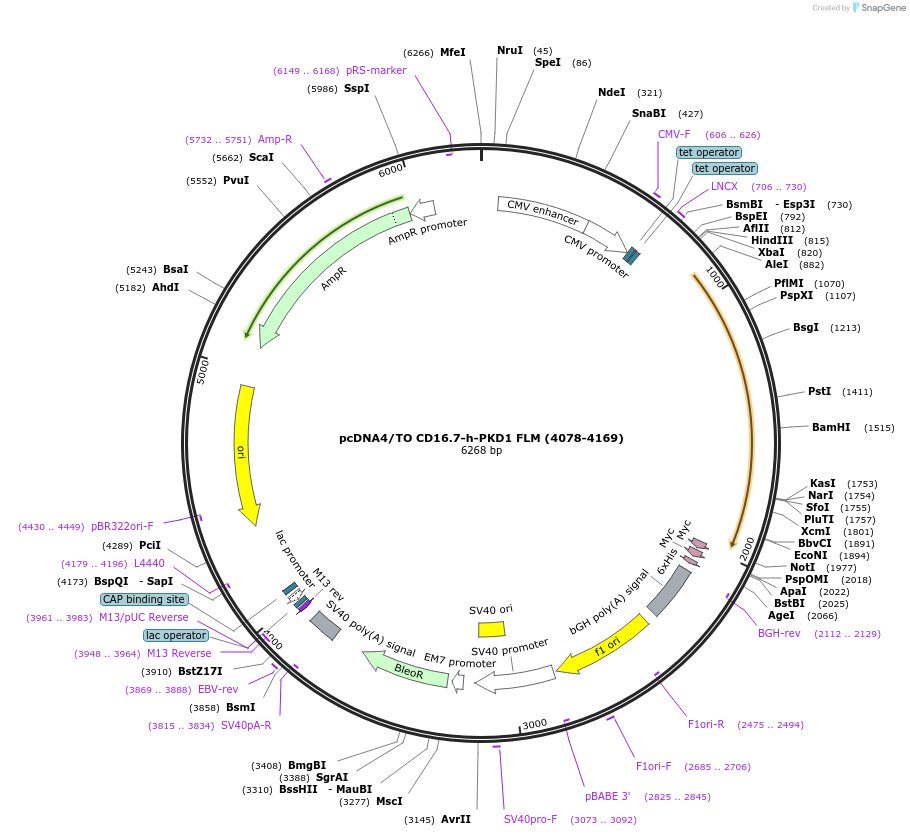
pcDNA4/TO CD16.7-h-PKD1 FLM (4078-4169)
Plasmid#83463PurposeCytoplasmic tail of human PC1 expression cassette with a membrane anchor and PKD1 sequence that corresponds to 4078 to 4169 of PC1 (96)DepositorAvailable SinceNov. 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
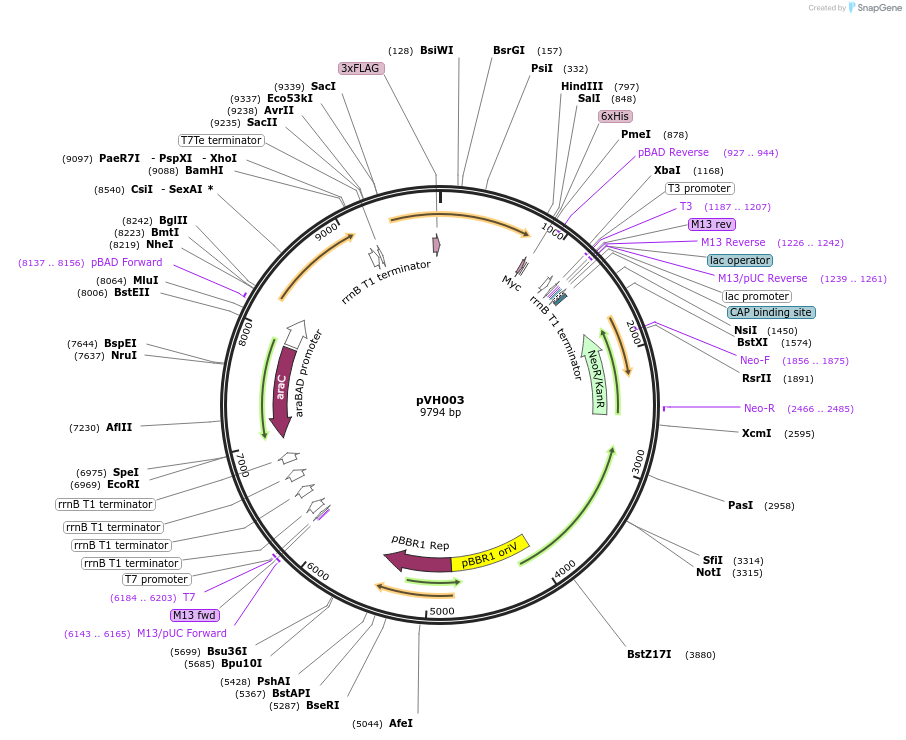
pVH003
Plasmid#80397PurposeContains pBAD-CusR (response regulator) and pCusR-antiscaffold-YFP-AAV. This plasmid was used for microscopy tracking experimentsDepositorUseSynthetic BiologyTagsFused by GS linker to LZx domain and Venus has N-…ExpressionBacterialAvailable SinceSept. 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTH394-SUP45-L34S
Plasmid#29373DepositorInsertSUP45 gene encoding translation release factor 1 from S. cerevisiae (SUP45 Budding Yeast)
ExpressionYeastMutationL34S mutant gene (corresponding to the sup45-36ts…Available SinceMay 23, 2011AvailabilityAcademic Institutions and Nonprofits only -
pTH401-SUP45-R65C
Plasmid#29381DepositorInsertSUP45 gene encoding translation release factor 1 from S. cerevisiae (SUP45 Budding Yeast)
ExpressionYeastMutationR65C mutant gene (corresponding to the sup45-3 al…Available SinceMay 23, 2011AvailabilityAcademic Institutions and Nonprofits only -
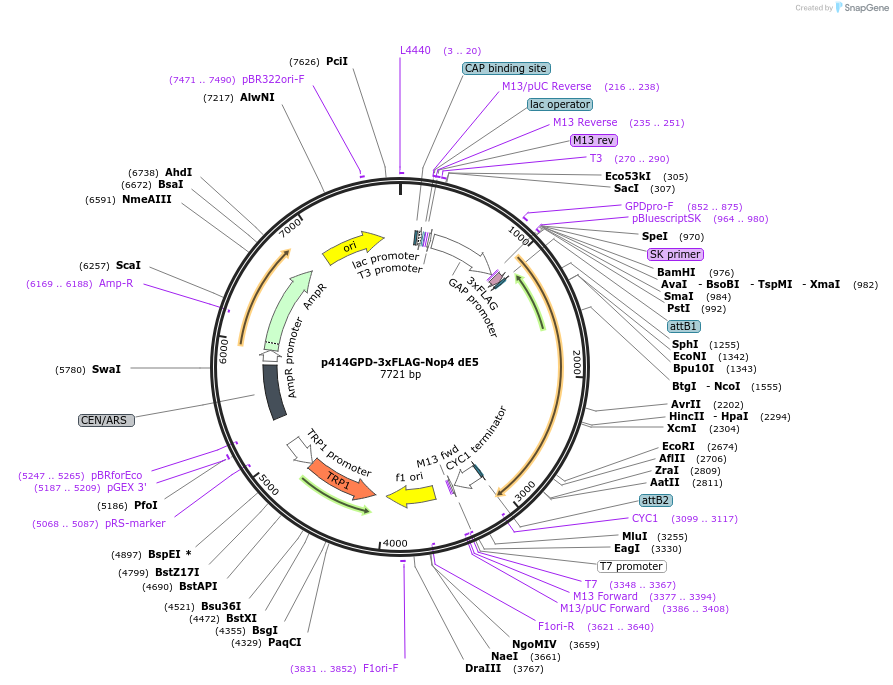
p414GPD-3xFLAG-Nop4 dE5
Plasmid#169263PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-Nop4 delta Exon 5DepositorInsertNop4 delta E5 (NOP4 Budding Yeast)
Tags3xFLAGExpressionYeastMutationdeleted region corresponding to human Exon 5PromoterGPDAvailabilityAcademic Institutions and Nonprofits only -
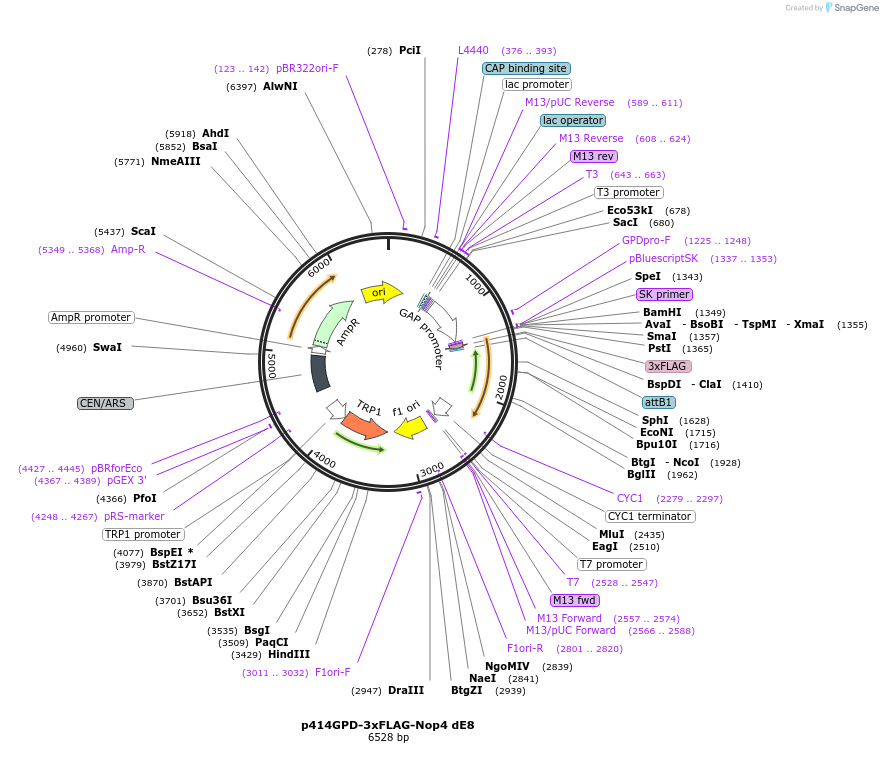
p414GPD-3xFLAG-Nop4 dE8
Plasmid#169264PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-Nop4 delta Exon 8DepositorInsertNop4 delta E8 (NOP4 Budding Yeast)
Tags3xFLAGExpressionYeastMutationdeleted region corresponding to human Exon 8PromoterGPDAvailabilityAcademic Institutions and Nonprofits only -
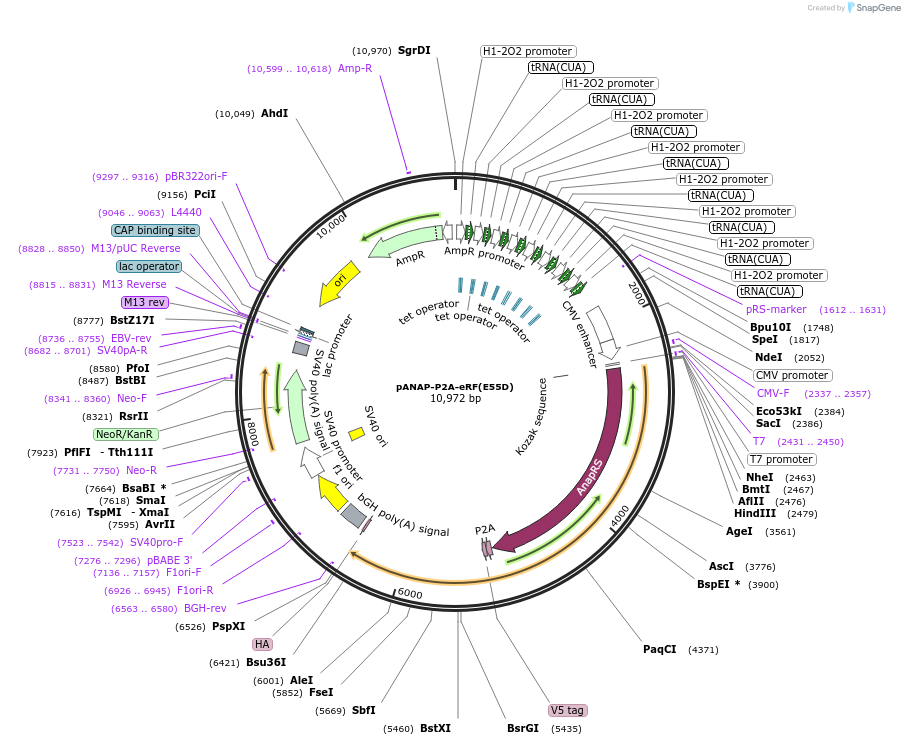
pANAP-P2A-eRF(E55D)
Plasmid#241290PurposeAllows co-translational incorporation of the fluorescent ncAA Anap directly into the target protein; expresses tRNACUA-Leu, AnapRS, and dominant negative ribosomal release factorDepositorInsertthe amber suppressor tRNACUA-Leu
ExpressionMammalianMutationmutant E.coli leucyl synthetase specific for Anap…PromoterCMVAvailable SinceDec. 11, 2025AvailabilityAcademic Institutions and Nonprofits only