We narrowed to 27,413 results for: CAT;
-
Plasmid#47024PurposeEncodes a template from which gRNAs can be made via InFusion cloning. The Medicago truncatula U6.6 promoter drives the gRNA. For use in plants.DepositorInsertgRNA Shuttle
UseCRISPR; Cas9ExpressionPlantMutationG to T cloning mutation at position 323PromoterMedicago truncatula U6.6Available SinceSept. 3, 2013AvailabilityIndustry, Academic Institutions, and Nonprofits -
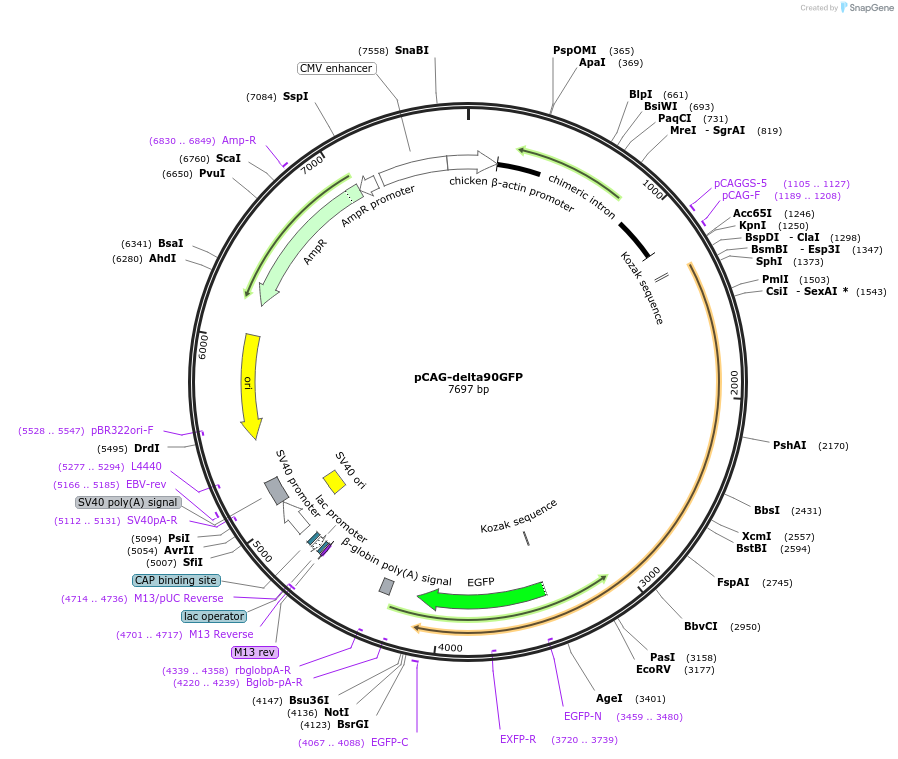
pCAG-delta90GFP
Plasmid#26645DepositorInsertdelta90 beta-catenin (Ctnnb1 Mouse)
TagsGFPExpressionMammalianMutationDeletion of first 90 aa of beta cateninAvailable SinceNov. 8, 2010AvailabilityAcademic Institutions and Nonprofits only -
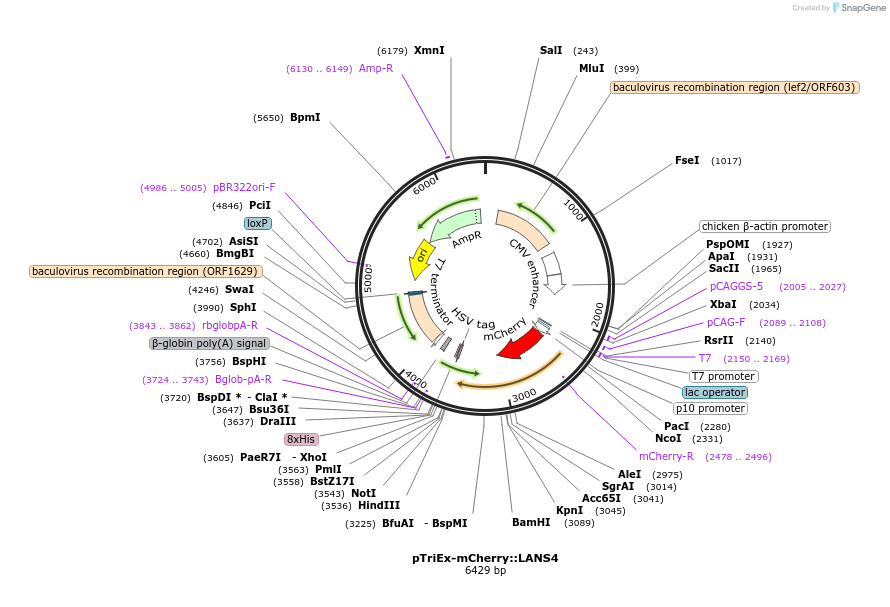
pTriEx-mCherry::LANS4
Plasmid#60785PurposeControl of Protein Activity and Cell Fate Specification via Light-Mediated Nuclear TranslocationDepositorInsertLANS4
TagsmCherryExpressionBacterial, Insect, and Mamm…PromoterCMVAvailable SinceAug. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
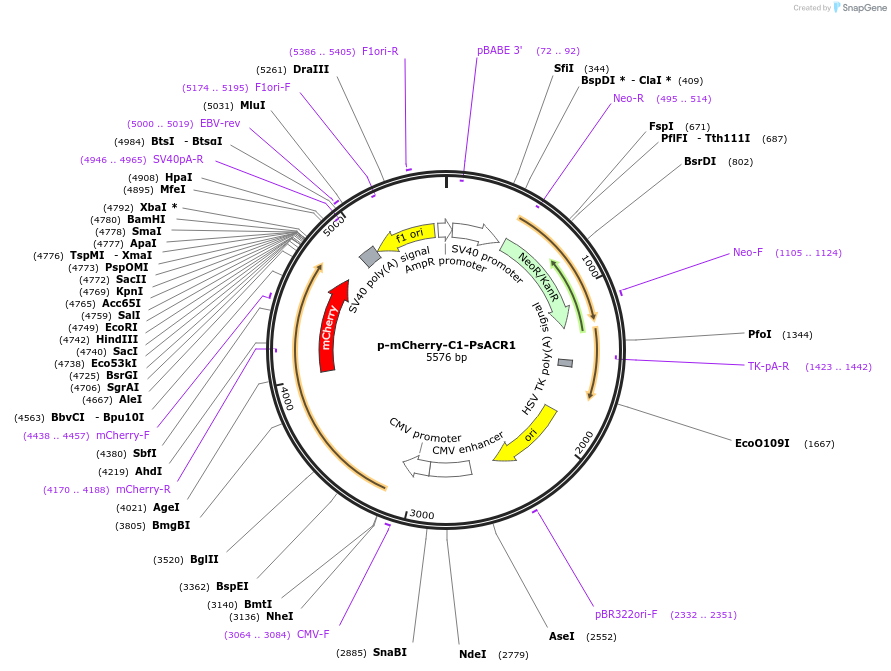
p-mCherry-C1-PsACR1
Plasmid#85465PurposeNatural anion conducting Channelrhodopsin with red shifted absorption. Closing kinetics 80ms. Codon optimized for mammalian expression (human/mouse)DepositorInsertSynthetic construct PsChR1 gene
TagsmCherryExpressionMammalianPromoterCMV (+enhancer)Available SinceJan. 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
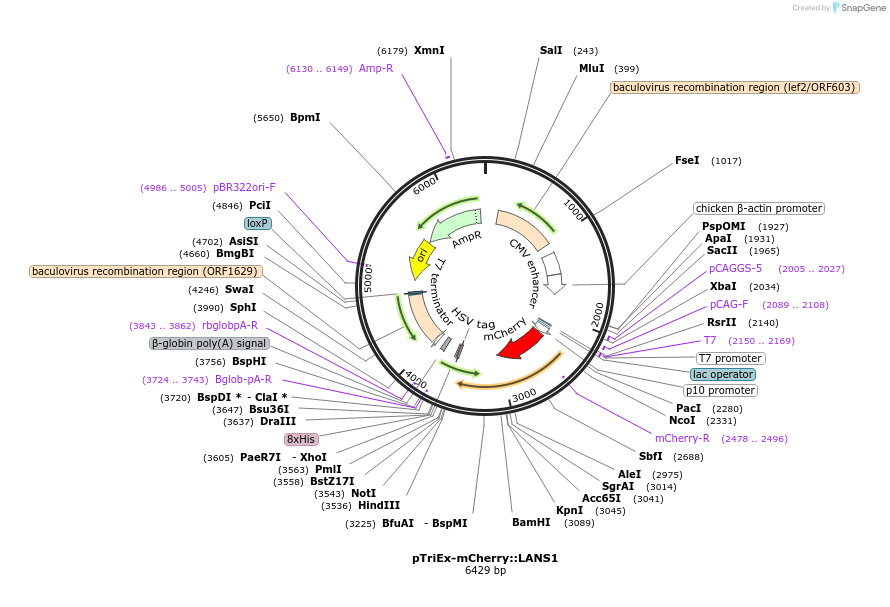
pTriEx-mCherry::LANS1
Plasmid#60784PurposeControl of Protein Activity and Cell Fate Specification via Light-Mediated Nuclear TranslocationDepositorInsertLANS1
TagsmCherryExpressionBacterial, Insect, and Mamm…PromoterCMVAvailable SinceAug. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
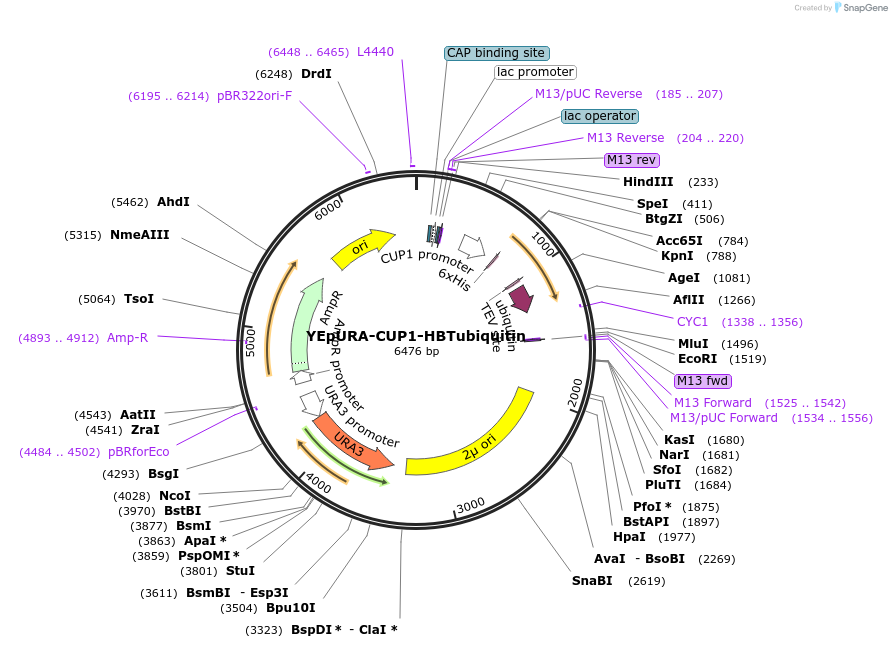
YEpURA-CUP1-HBTubiquitin
Plasmid#26900DepositorInsertHBT-tagged ubiquitin
ExpressionYeastAvailable SinceJan. 24, 2011AvailabilityAcademic Institutions and Nonprofits only -
pcDNA-NES-ZapCY2
Plasmid#59016PurposeCytosolic zinc-binding FRET biosensorDepositorInsertNES-ZapCY2 genetically encoded cytosolic Zn(II) sensor
TagsNuclear Exclusion SignalExpressionMammalianMutationSee commentPromoterCMVAvailable SinceAug. 21, 2014AvailabilityAcademic Institutions and Nonprofits only -
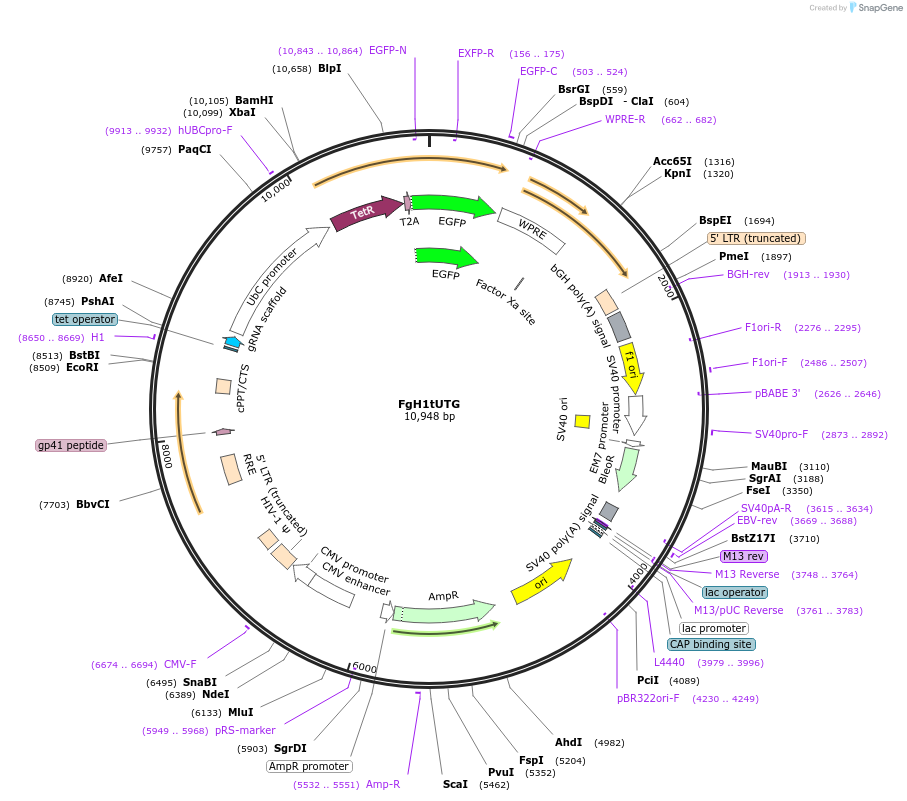
FgH1tUTG
Plasmid#70183PurposeInducible expression of guide RNA with fluorescent GFP reporterDepositorInsertH1-Tet-sgrna cassette
UseCRISPR and LentiviralExpressionMammalianAvailable SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
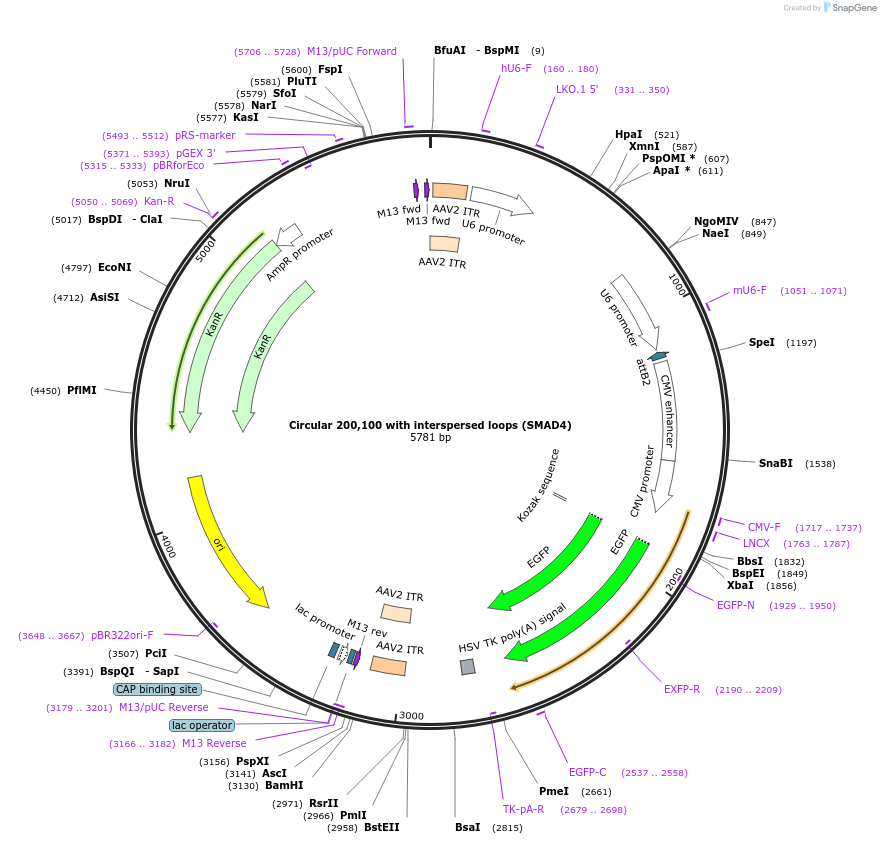
Circular 200,100 with interspersed loops (SMAD4)
Plasmid#180191PurposeAAV vector carrying a guide RNA targeting the human SMAD4 mRNADepositorInsertCircular 200,100 guide RNA with interspersed loops
UseAAVExpressionMammalianPromoterHuman U6Available SinceApril 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
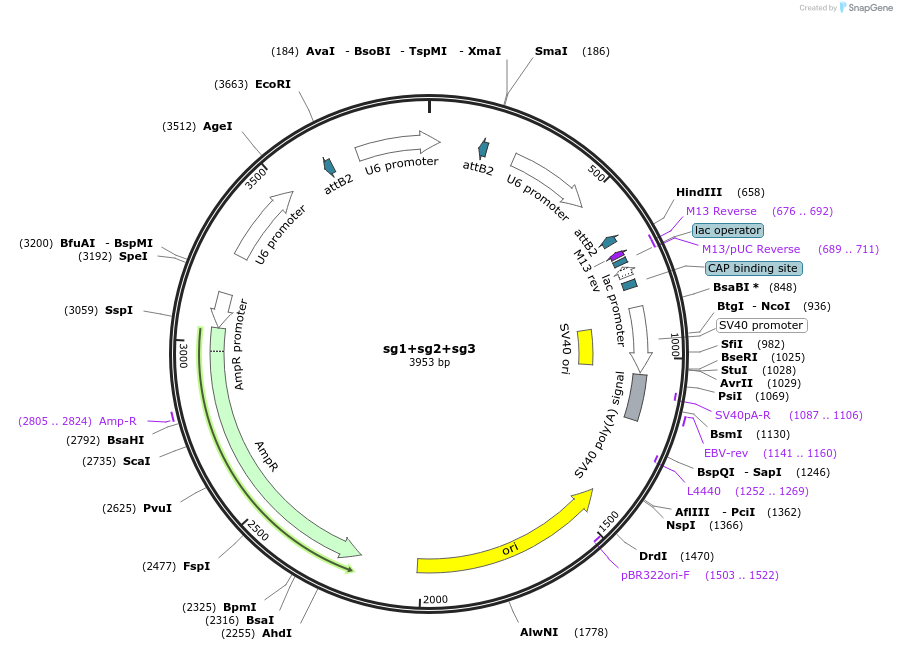
sg1+sg2+sg3
Plasmid#113969PurposeTriple short guide RNA targeting GTATAGCATACATTATACG, TACCACATTTGTAGAGGTT & CAATGTATCTTATCATGTCDepositorInsertsg1+sg2+sg3
ExpressionMammalianPromoterU6Available SinceMay 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
pSLQ7535 pHR (hU6-crSURF2-EFS-PuroR-WPRE)
Plasmid#214879PurposeLentiviral vector encoding RfxCas13d targeting SURF2 guide arrayDepositorInserthU6-crSURF2-EFS-PuroR-WPRE
UseCRISPR and LentiviralExpressionMammalianPromoterhU6Available SinceApril 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
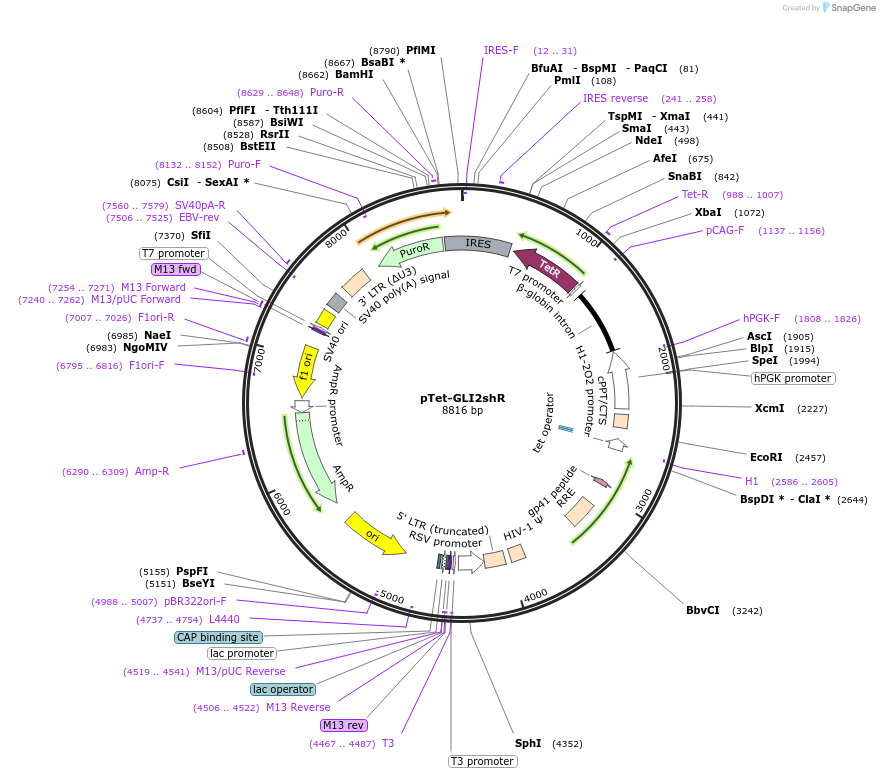
pTet-GLI2shR
Plasmid#136691PurposeInducible Gli2 shRNA lentiviral plasmid (target sequence: human GLI2 5′CCGGCCTGGCATGACTACCACTATGCTCGAGCATAGTGGTAGTCATGCCAGGTTTTTG 3′)DepositorInsertshRNA_humanGli2 (GLI2 Human)
UseLentiviralAvailable SinceJune 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
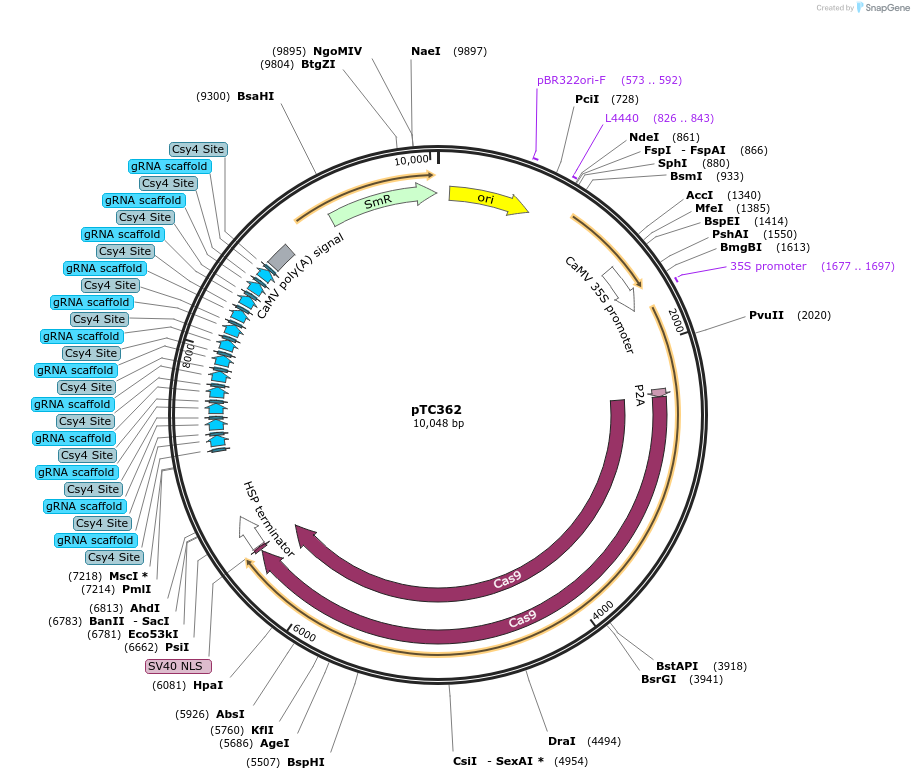
pTC362
Plasmid#91216Purposeprotoplast vector for targeted deletion of 6 genes in tomatoDepositorInsertgRNAs targeting 6 tomato genes
UseCRISPRExpressionPlantAvailable SinceJuly 14, 2017AvailabilityAcademic Institutions and Nonprofits only -
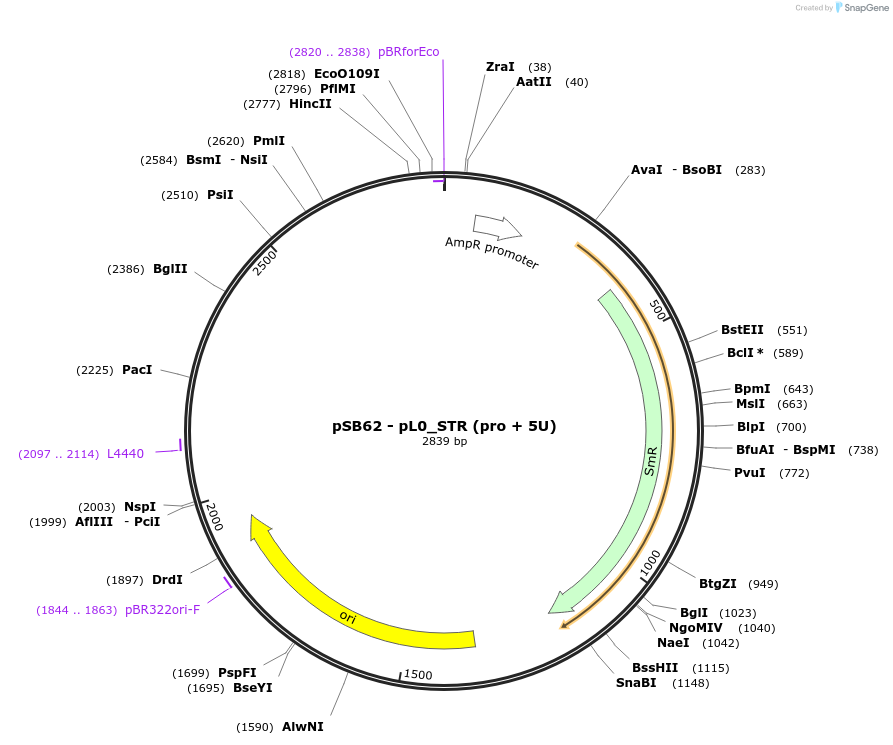
pSB62 - pL0_STR (pro + 5U)
Plasmid#123186PurposeGolden Gate (MoClo; PRO + 5U) compatible STR1 promoter from Catharanthus roseusDepositorInsertCatharanthus roseus STR1 promoter and 5'UTR
UsePart for plant expressionExpressionPlantMutationNo BpiI and BsaI sitesAvailable SinceMay 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
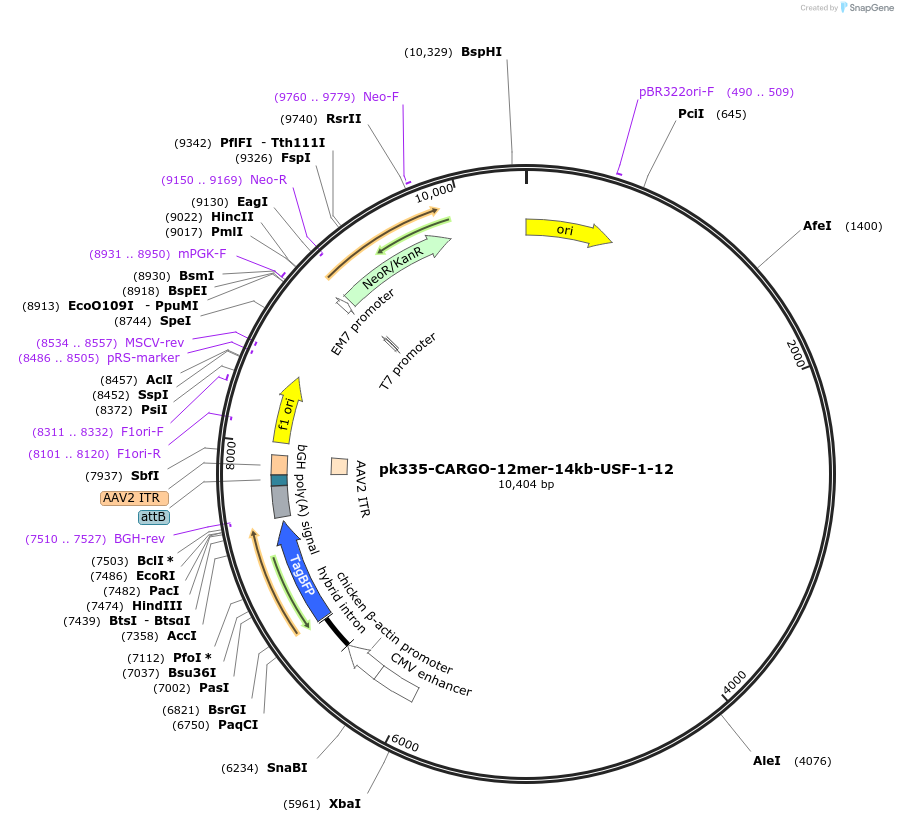
pk335-CARGO-12mer-14kb-USF-1-12
Plasmid#227472Purpose12-mer gRNA array targeting Sp dCas9/Cas9 to the Prdm8-Fgf5 locusDepositorInsertCARGO to 14kb Upstream Fgf5
UseCRISPRAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
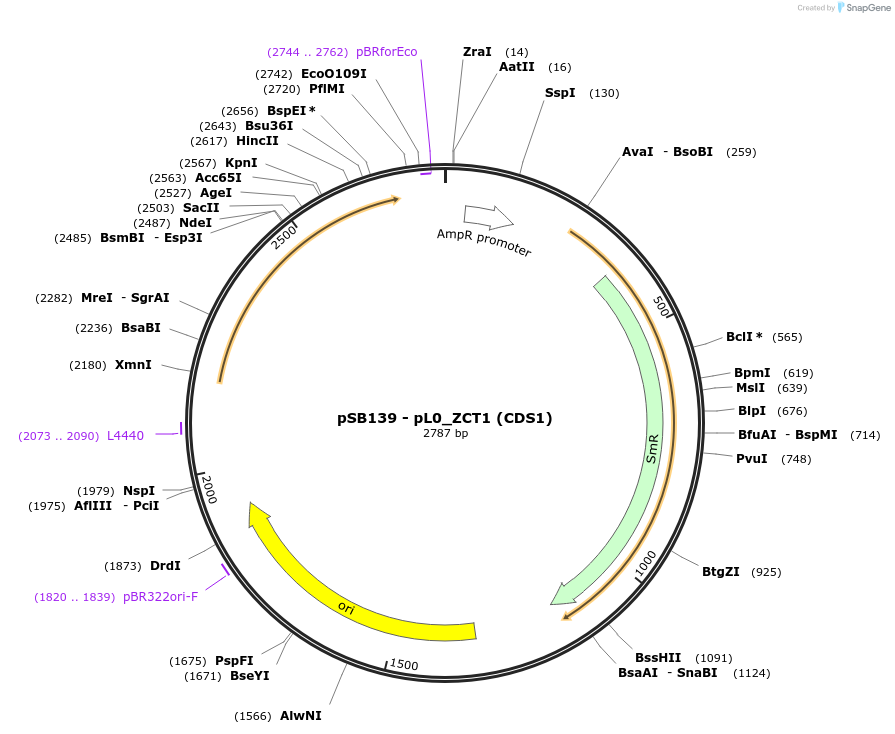
pSB139 - pL0_ZCT1 (CDS1)
Plasmid#123184PurposeGolden Gate (MoClo; CDS1) compatible ZCT1 gene from Catharanthus roseus; a repressor of the STR1 promoter from C. roseusDepositorInsertZCT1 from Catharanthus roseus
UsePart for plant expressionExpressionPlantMutationNo BpiI and BsaI sitesAvailable SinceMay 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
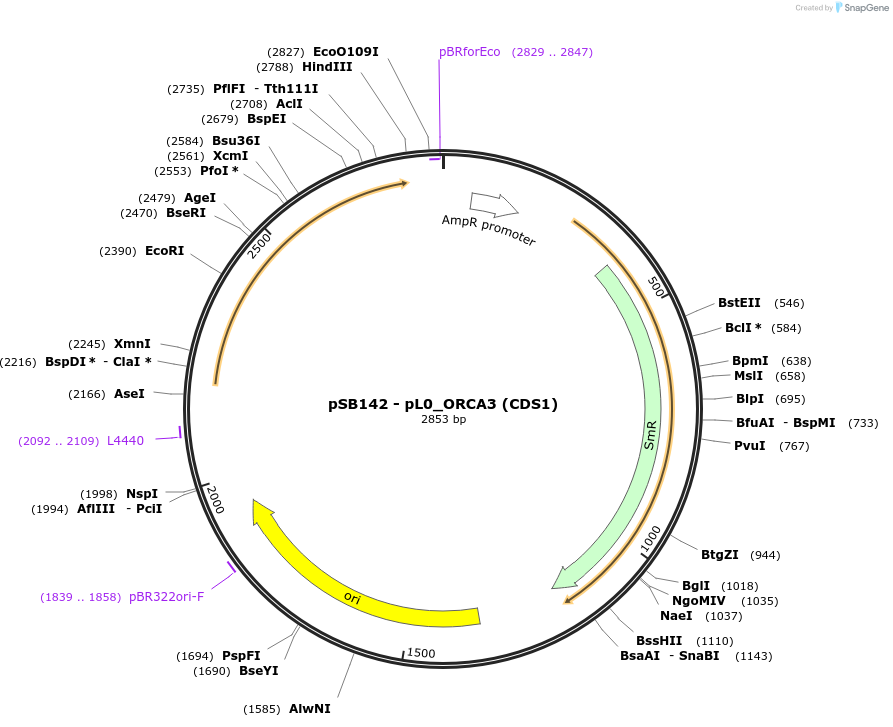
pSB142 - pL0_ORCA3 (CDS1)
Plasmid#123185PurposeGolden Gate (MoClo; CDS1) compatible ORCA3 gene from Catharanthus roseus; an activator of the STR1 promoter from C. roseusDepositorInsertORCA3 from Catharanthus roseus
UsePart for plant expressionExpressionPlantMutationNo BpiI and BsaI sitesAvailable SinceMay 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
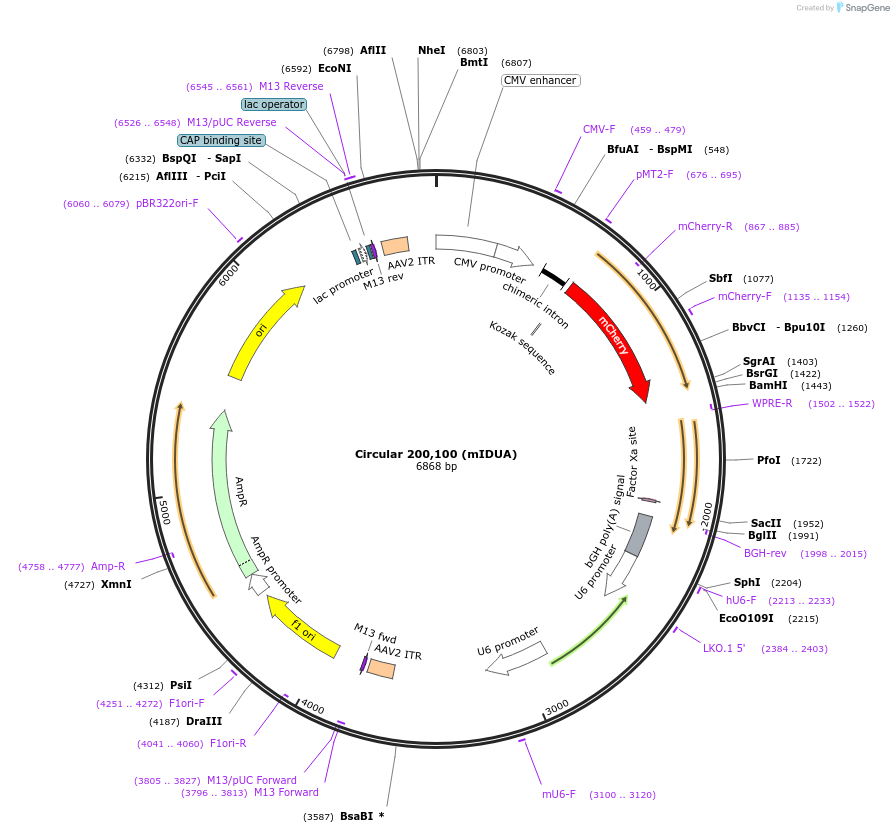
Circular 200,100 (mIDUA)
Plasmid#170123PurposeAAV vector carrying 2 copies of a guide RNA targeting the mouse IDUA mRNA, expressed from human and mouse U6 promotersDepositorInsertCircular 200,100 guide RNA (Idua Mouse)
UseAAVExpressionMammalianPromoterHuman U6 and mouse U6Available SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
pEEF1D-FLAG (pcDNA 3.1+) LG6-1
Plasmid#37365DepositorAvailable SinceJuly 17, 2012AvailabilityAcademic Institutions and Nonprofits only -
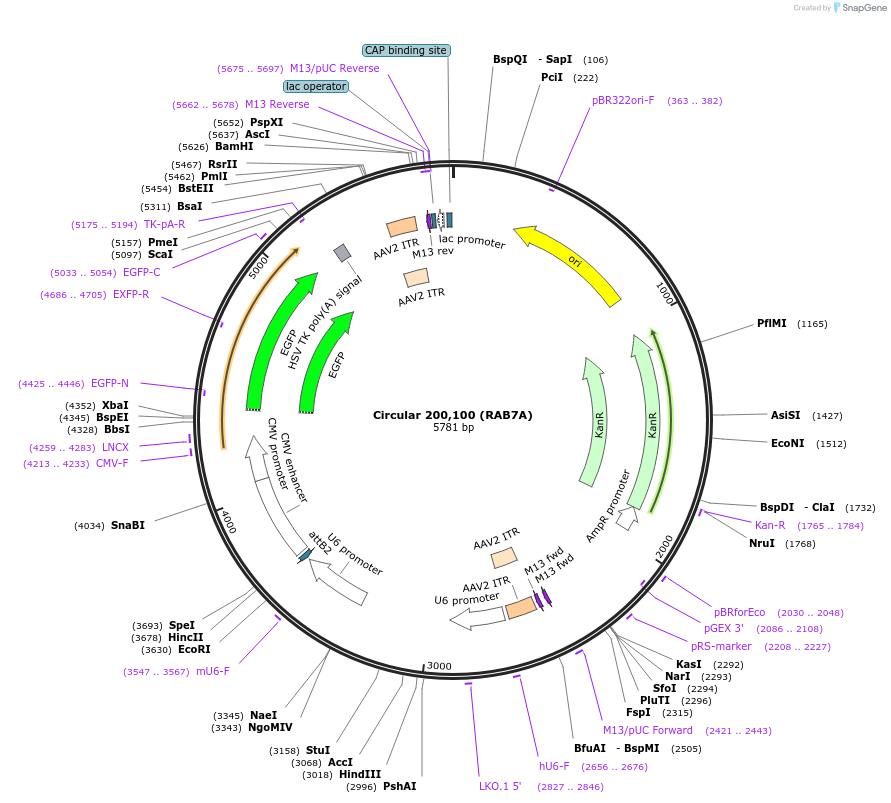
Circular 200,100 (RAB7A)
Plasmid#170118PurposeAAV vector carrying a guide RNA targeting the human RAB7A mRNADepositorAvailable SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only