We narrowed to 23,118 results for: Sis
-
Plasmid#69558PurposeIn vitro transcription/translation (T7)DepositorInsertUBB (Ubb Mouse)
ExpressionBacterialMutationThe two last codons (TATTAA; encoding the last ty…PromoterT7Available SinceDec. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pET23a-MmUBB(Y305P)
Plasmid#69557PurposeBacterial expression; in vitro transcription/translation (T7)DepositorInsertUBB (Ubb Mouse)
ExpressionBacterialMutationCodon TAT encoding tyrosine 305 was changed to CC…PromoterT7Available SinceDec. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pPAt2S3 (GB0029)
Plasmid#68162PurposeProvides the Arabidopsis thaliana Seed-specific promoter 2S3 as a level 0 GoldenBraid partDepositorInsertAt2S3 promoter
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceOct. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
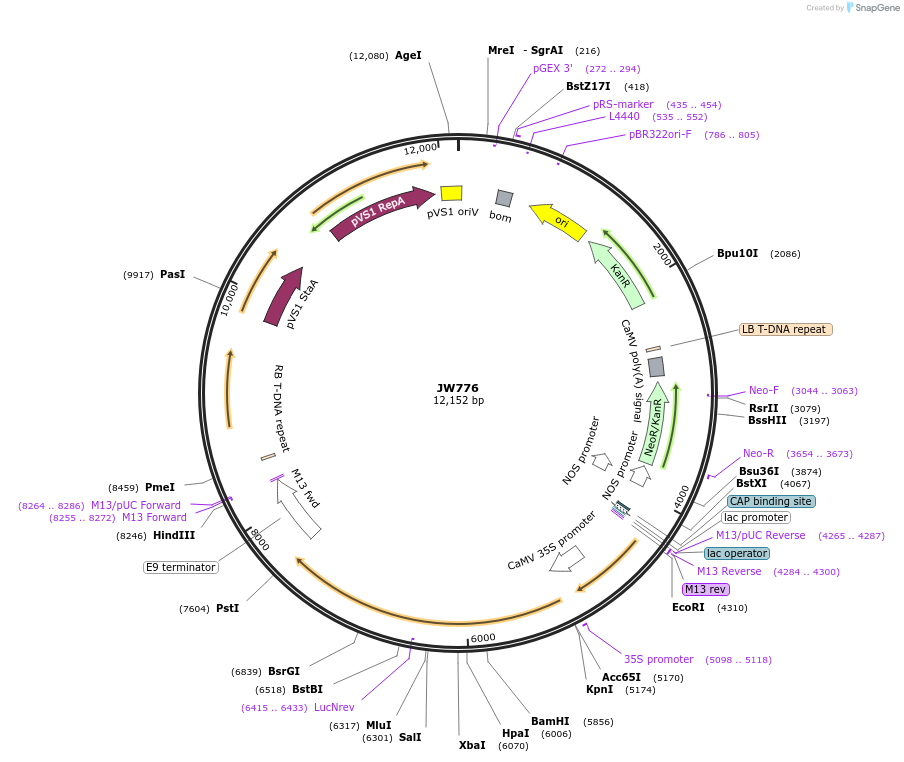
JW776
Plasmid#64817PurposeBiFC rCUC2DepositorAvailable SinceSept. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
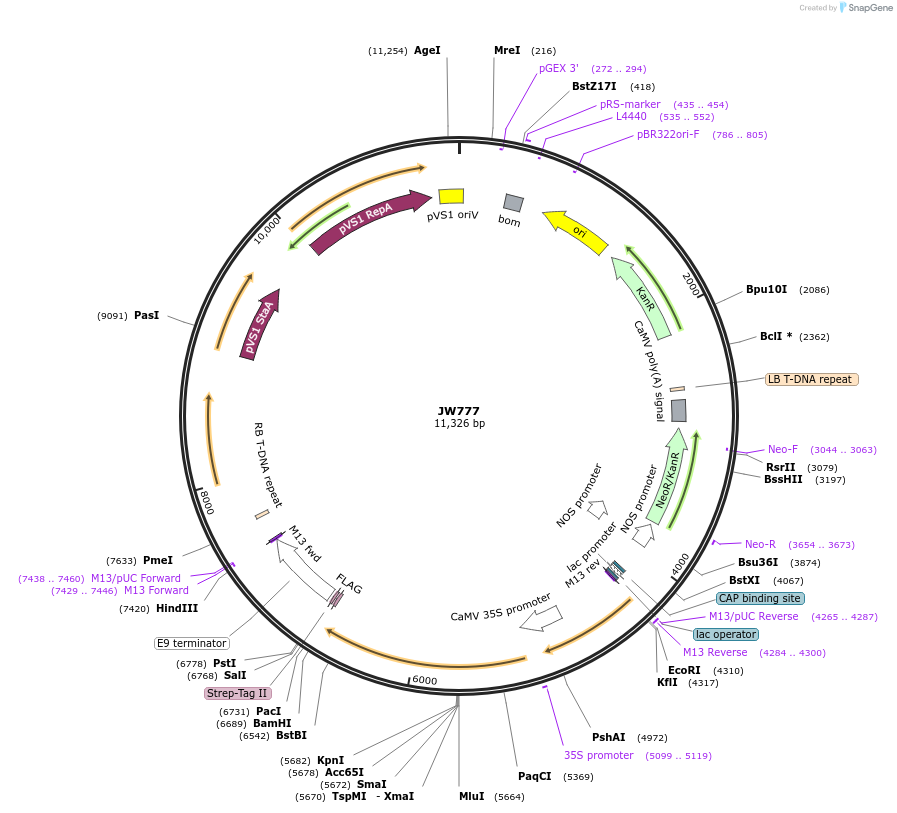
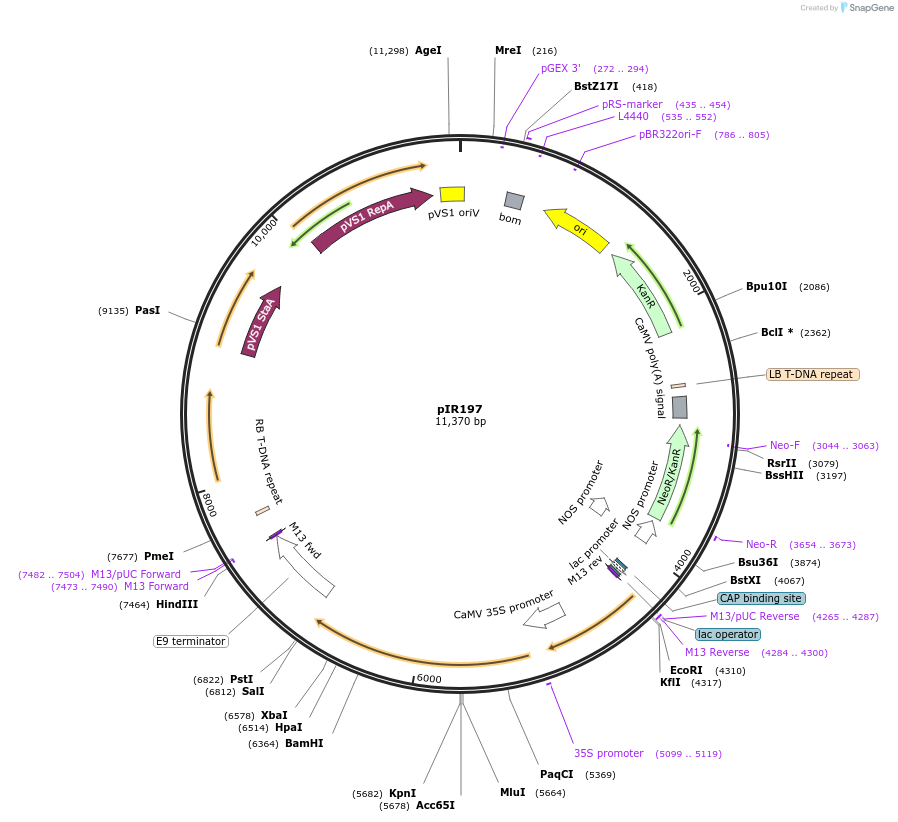
pIR197
Plasmid#64819PurposeBiFC rCUC2DepositorAvailable SinceSept. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
MT583
Plasmid#63114PurposeThe gDNA construct is designed for arabidopsis ecotype Est-1 ACD6 expression in other genetic backgrounds.DepositorInsertACD6 (Accelerated Cell Death 6)
ExpressionPlantAvailable SinceSept. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
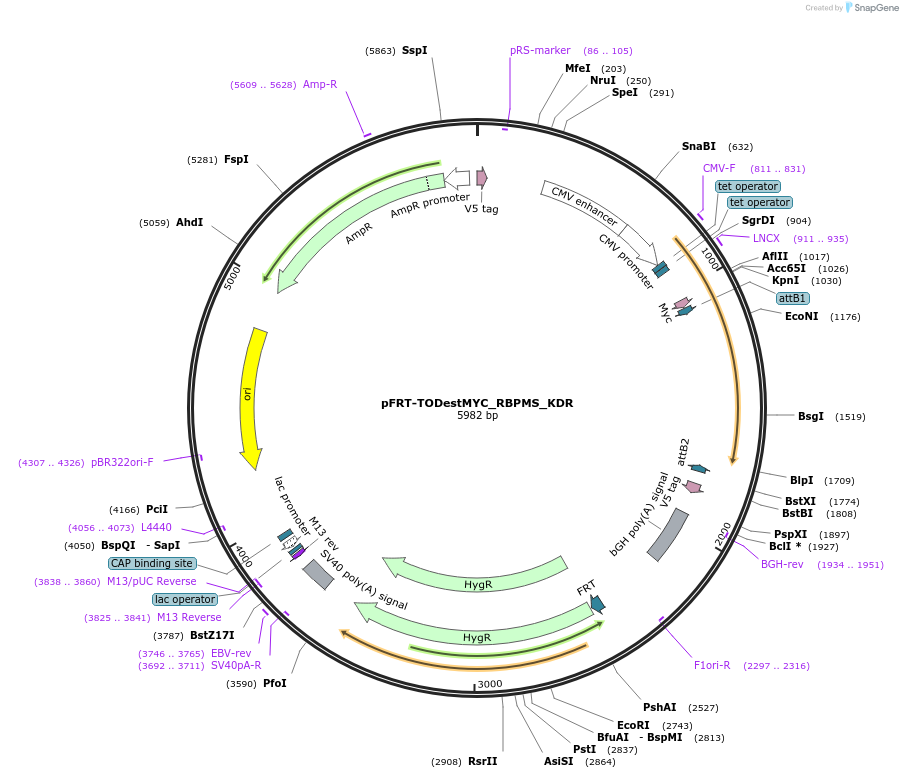
pFRT-TODestMYC_RBPMS_KDR
Plasmid#65667PurposeMYC expression vector with RBPMS encoding silent mutation to render resistant to siRNA R2 (Farazi et al. 2014)DepositorInsertRBPMS (RBPMS Human)
TagsMYCExpressionMammalianMutationintroduced HindIII site to exchange FLAG-HA tag w…PromoterCMV, 2x TetAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
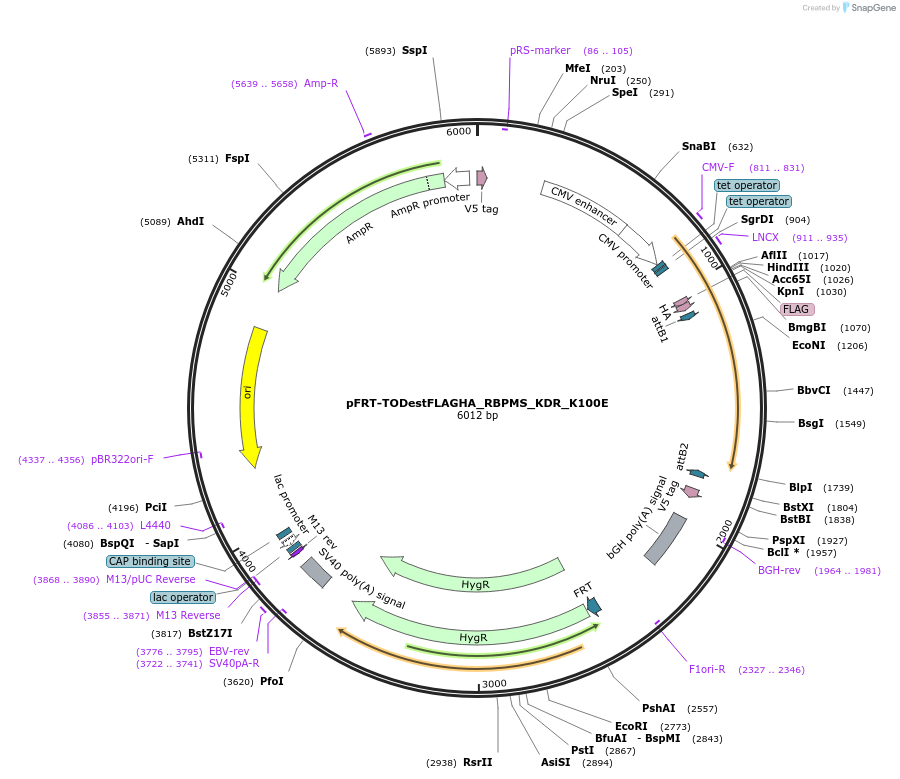
pFRT-TODestFLAGHA_RBPMS_KDR_K100E
Plasmid#65093PurposeDestination vector with RBPMS resistant to siRNA treatment by s21729 (Applied Biosystems) with K100E mutation for stable cell line generationDepositorInsertRBPMS (RBPMS Human)
TagsFLAG HAExpressionMammalianMutationchanged lysine 100 to glutamic acid (KDR backgrou…PromoterCMV, 2x TetAvailable SinceJune 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
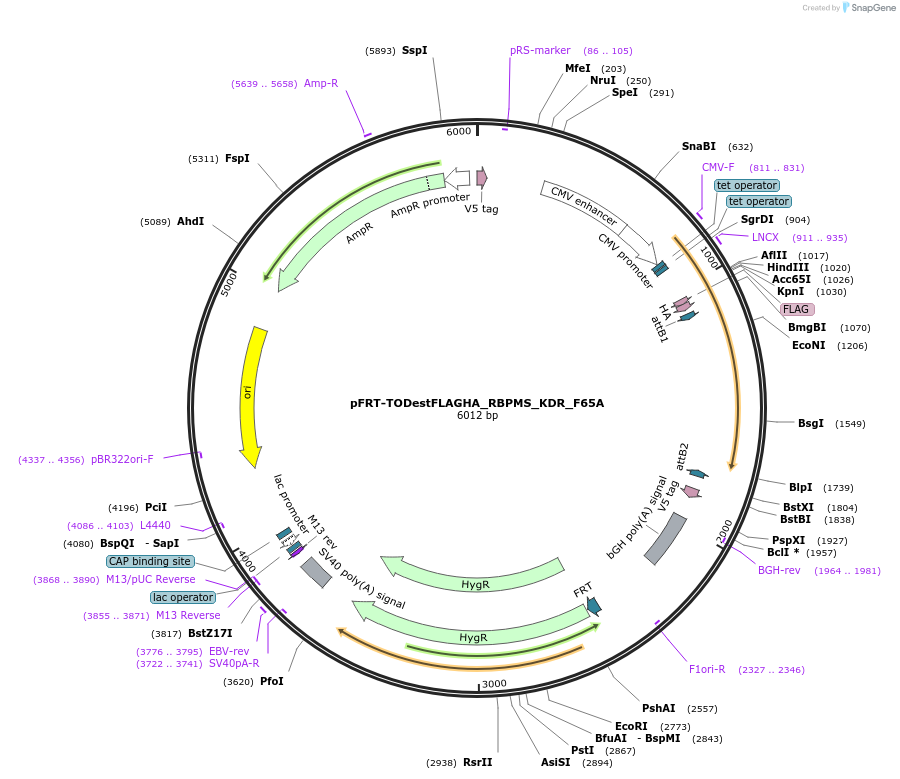
pFRT-TODestFLAGHA_RBPMS_KDR_F65A
Plasmid#65092PurposeDestination vector with RBPMS resistant to siRNA treatment by s21729 (Applied Biosystems) with F65A mutation for stable cell line generationDepositorInsertRBPMS (RBPMS Human)
TagsFLAG HAExpressionMammalianMutationchange phenylalanine 65 to alanine (KDR backbone)PromoterCMV, 2x TetAvailable SinceJune 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
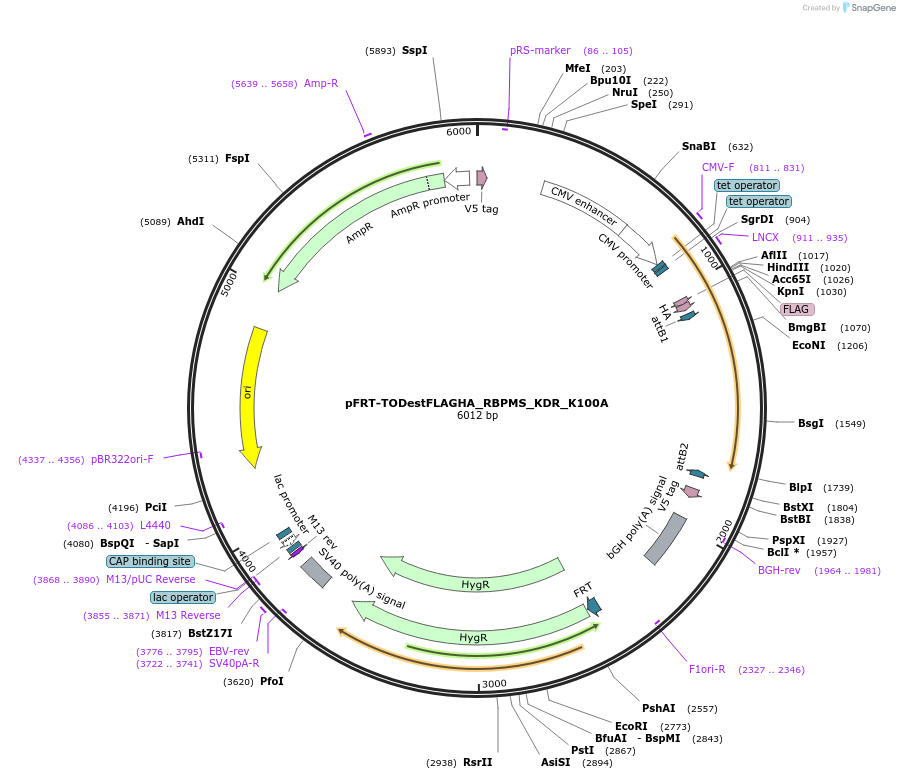
pFRT-TODestFLAGHA_RBPMS_KDR_K100A
Plasmid#65663PurposeFLAG-HA expression vector with RBPMS carrying mutation K100A and encoding silent mutation to render resistant to siRNA R2 (Farazi et al. 2014)DepositorInsertRBPMS (RBPMS Human)
TagsFLAG HAExpressionMammalianMutationchanged lysine 100 to alanine (KDR background)PromoterCMV, 2x TetAvailable SinceJune 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
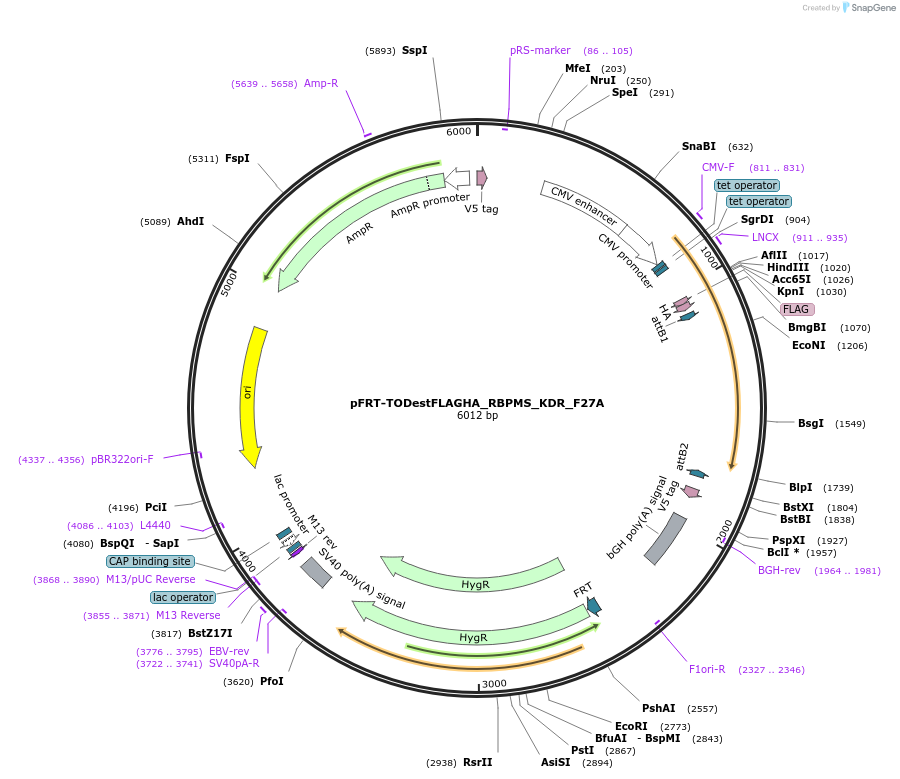
pFRT-TODestFLAGHA_RBPMS_KDR_F27A
Plasmid#65661PurposeFLAG-HA expression vector with RBPMS carrying mutation F27A and encoding silent mutation to render resistant to siRNA R2 (Farazi et al. 2014)DepositorInsertRBPMS (RBPMS Human)
TagsFLAG HAExpressionMammalianMutationchanged phenylalanine 27 to alanine (KDR backgrou…PromoterCMV, 2x TetAvailable SinceJune 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
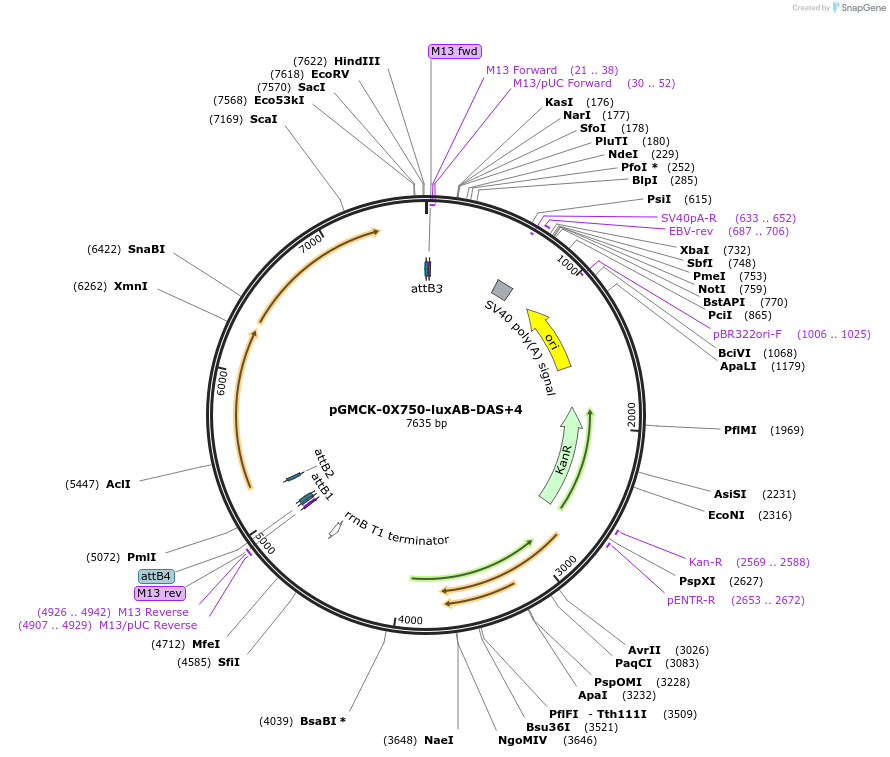
pGMCK-0X750-luxAB-DAS+4
Plasmid#49517PurposeKanamycin resistant; expresses luxAB-DAS+4 under the control of a promoter containing PtetO-4C5G; integrates into the mycobacteriophage att-L5 siteDepositorInsertP750-luxAB-DAS+4
TagsDAS+4 (AANDENYSENYADAS)ExpressionBacterialPromoterP750Available SinceJan. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
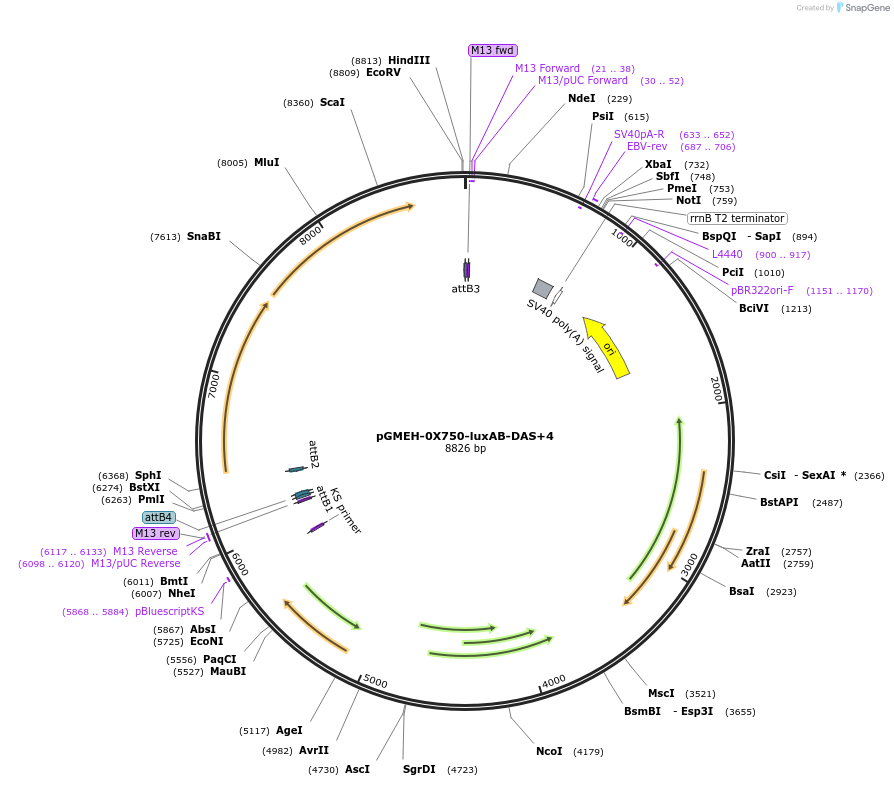
pGMEH-0X750-luxAB-DAS+4
Plasmid#49519PurposeHygromycin resistant; expresses luxAB-DAS+4 under the control of a promoter containing PtetO-4C5G; replicates episomallyDepositorInsertP750-luxAB-DAS+4
TagsDAS+4 (AANDENYSENYADAS)ExpressionBacterialPromoterP750Available SinceJan. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
-
-
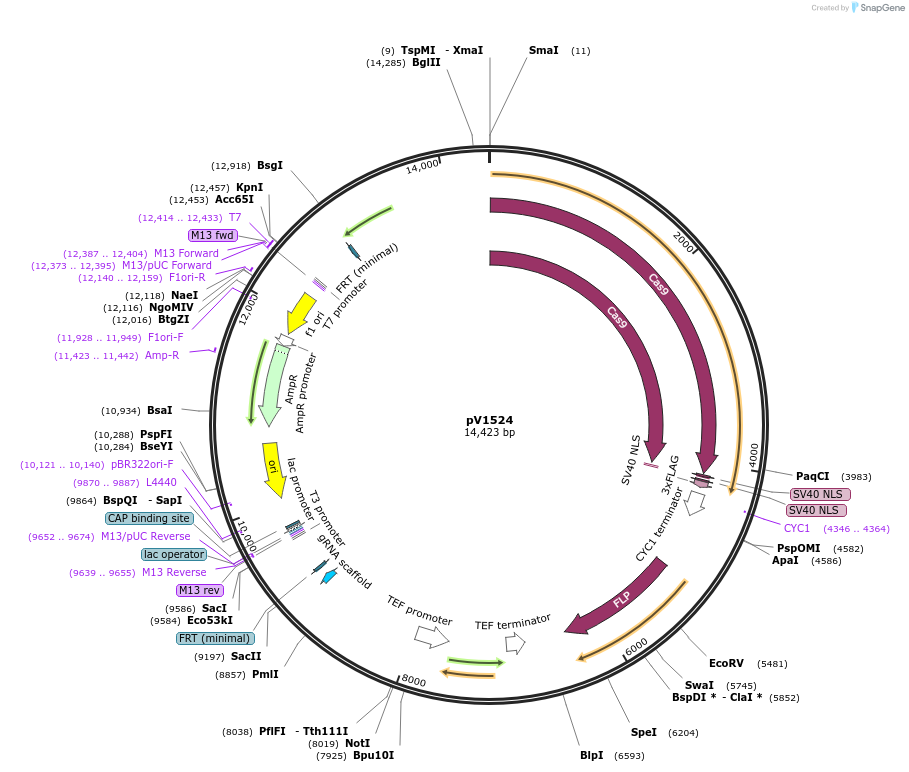
pV1524
Plasmid#111431PurposeCaCas9/gRNA Solo entry plasmid for cloning guides - contains stuffer with BsmBI sites - inserts at Neut5L - Recyclable for serial mutagenesis (Mal2-FLP)DepositorInsertCaCas9/sgRNA-BsmBI stuffer
UseCRISPRExpressionYeastAvailable SinceAug. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
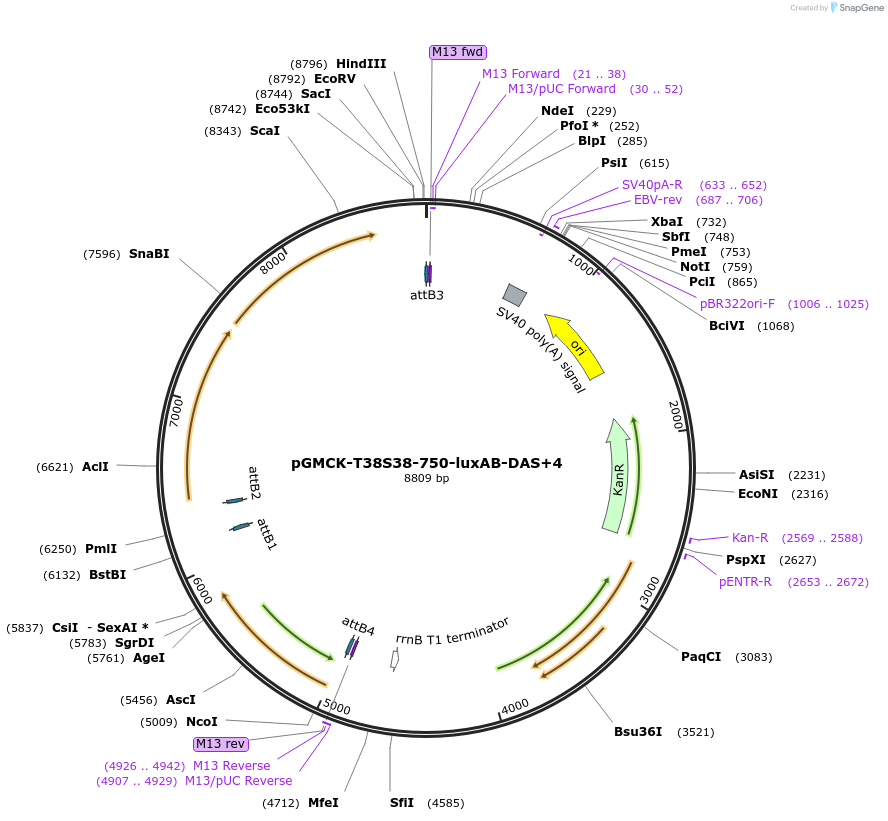
pGMCK-T38S38-750-luxAB-DAS+4
Plasmid#49518PurposeKanamycin resistant; expresses luxAB-DAS+4 under the control of a promoter containing PtetO-4C5G; constitutively expresses reverse TetR (tetR38); integrates into the mycobacteriophage att-L5 siteDepositorInsertT38S8-P750-luxAB-DAS+4
TagsDAS+4 (AANDENYSENYADAS)ExpressionBacterialPromoterP750Available SinceJan. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
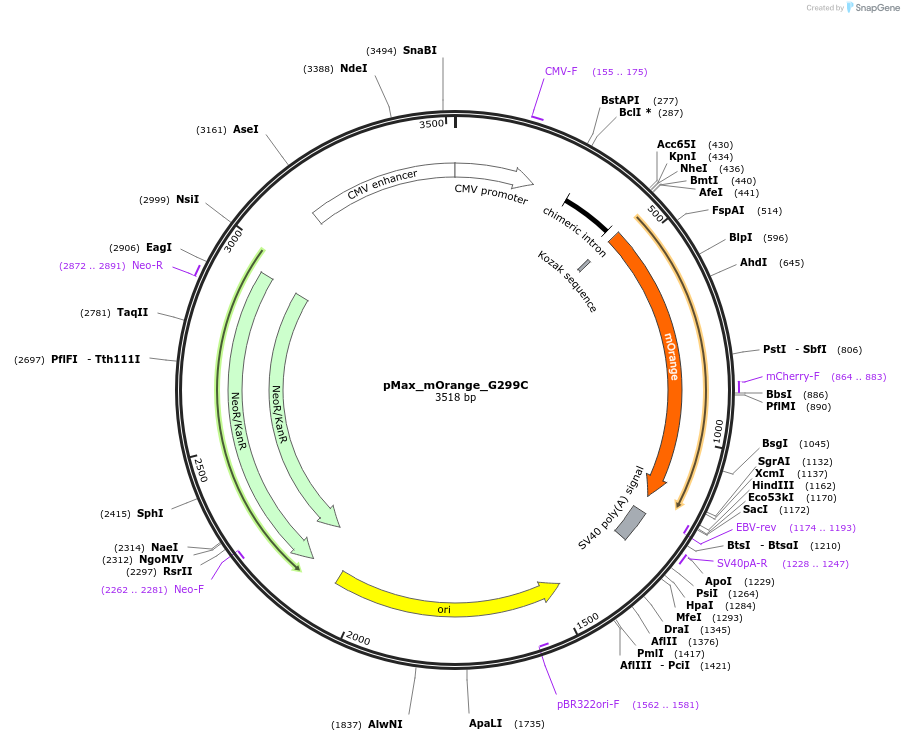
pMax_mOrange_G299C
Plasmid#177829PurposeSite-specific mutagenesis of mOrange for the purposes of creating a site-specific G:G mispair for the purposes of measuring MMR activity via Host Cell Reactivation (HCR).DepositorInsertmOrange_G299C
ExpressionMammalianMutationG299CPromoterCMVAvailable SinceNov. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
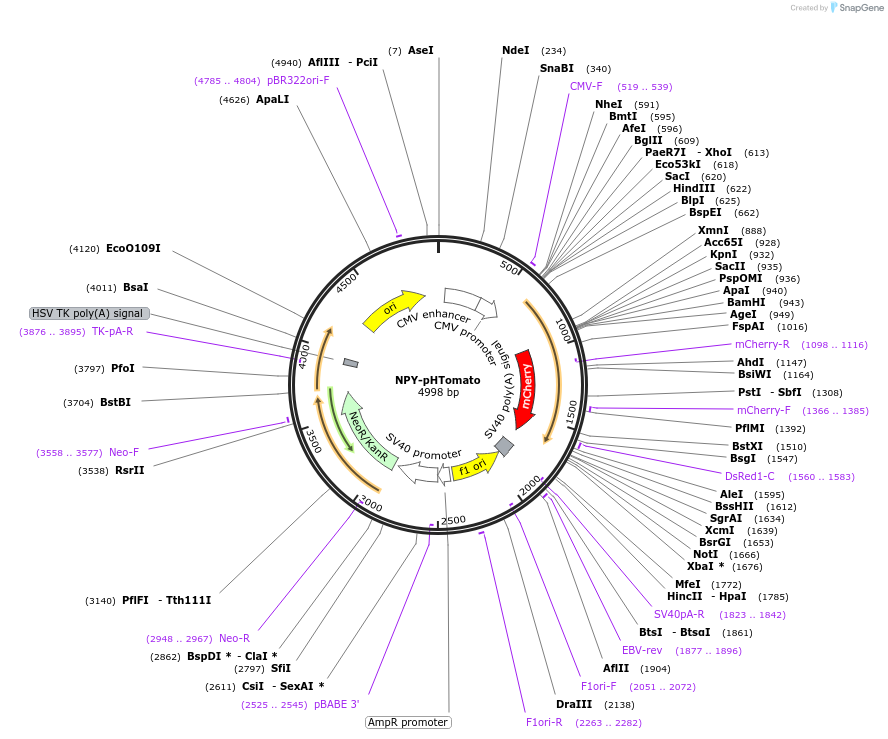
NPY-pHTomato
Plasmid#83501PurposeExpression of a fluorescent granule marker, neuropeptide-Y, for localization and exocytosis studies.DepositorAvailable SinceNov. 8, 2016AvailabilityAcademic Institutions and Nonprofits only -
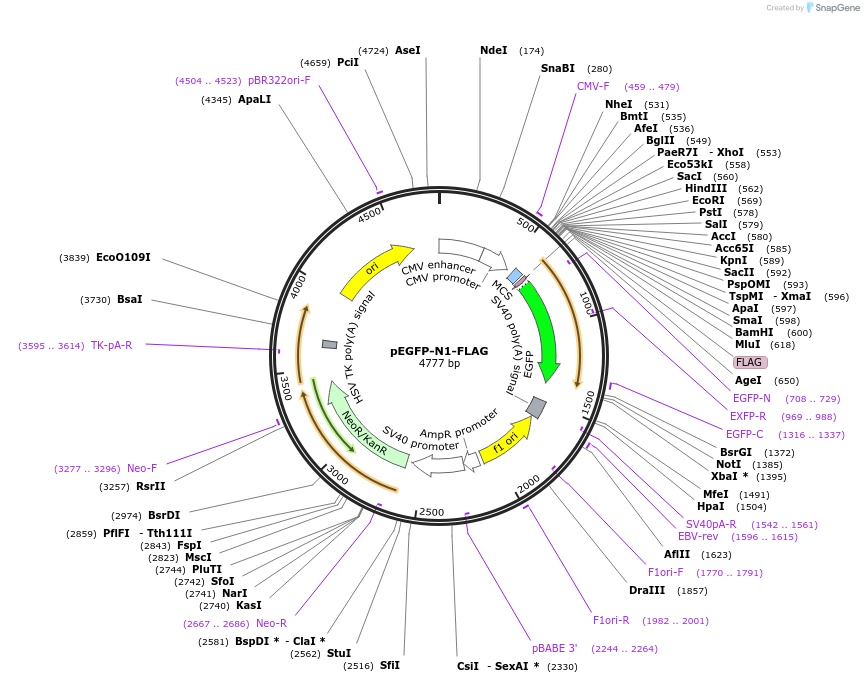
pEGFP-N1-FLAG
Plasmid#60360PurposePlasmid for cloning and expression of FLAG-GFP tagged proteins (C-terminal tag). Confers resistance to G418.DepositorHas ServiceCloning Grade DNATypeEmpty backboneTagsEGFP and FLAGExpressionMammalianPromoterCMVAvailable SinceJan. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
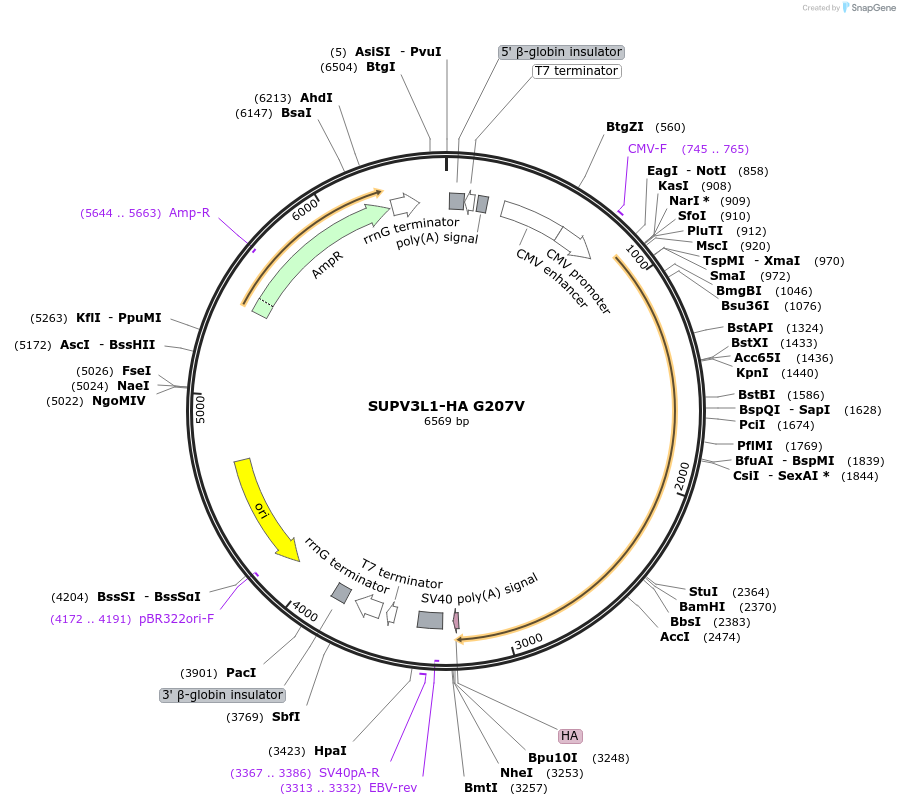
SUPV3L1-HA G207V
Plasmid#232349PurposeExpresses human SUPV3L1/SUV3 (G207V) in mammalian cells with HA tagDepositorAvailable SinceFeb. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
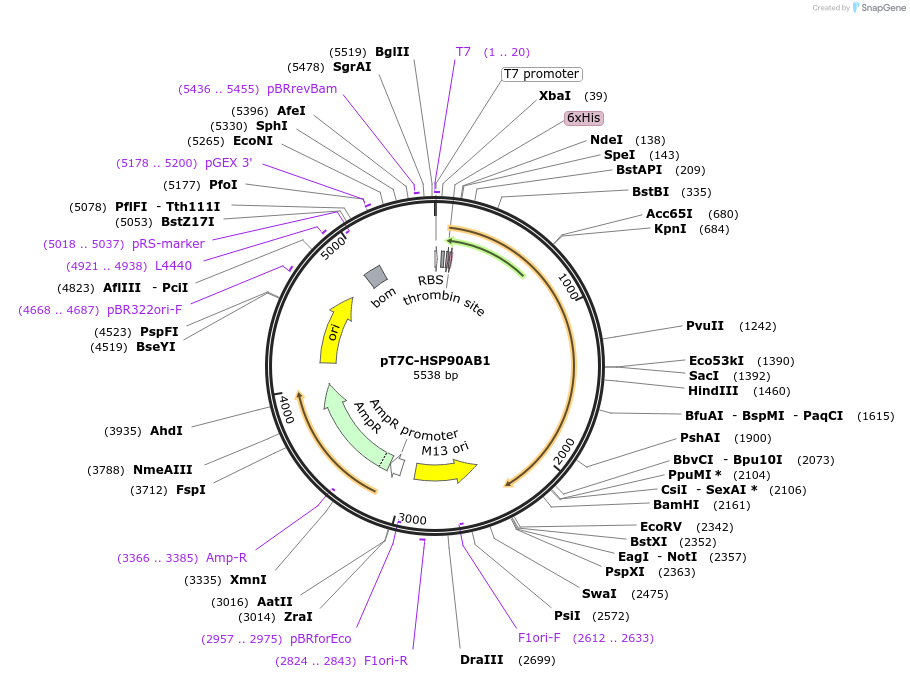
pT7C-HSP90AB1
Plasmid#177659PurposeRecombinant protein synthesis of human HSP90AB1DepositorAvailable SinceMarch 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
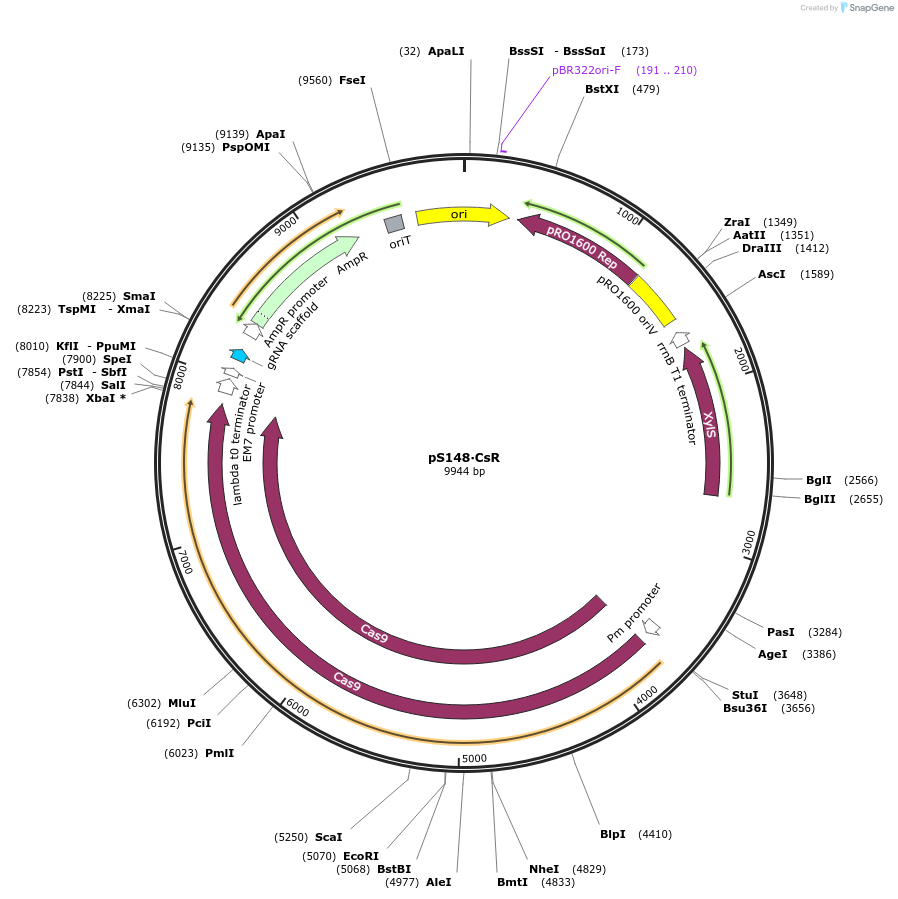
pS148∙CsR
Plasmid#197858PurposeExpresses Streptococcus pyogenes' cas9 and a tailored sgRNA for counterselection in gram-negative bacteriaDepositorInsertgRNA scaffold - Ap/Cb resistance cassette - incP oriT
UseCRISPR and Synthetic BiologyPromoterPEM7Available SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
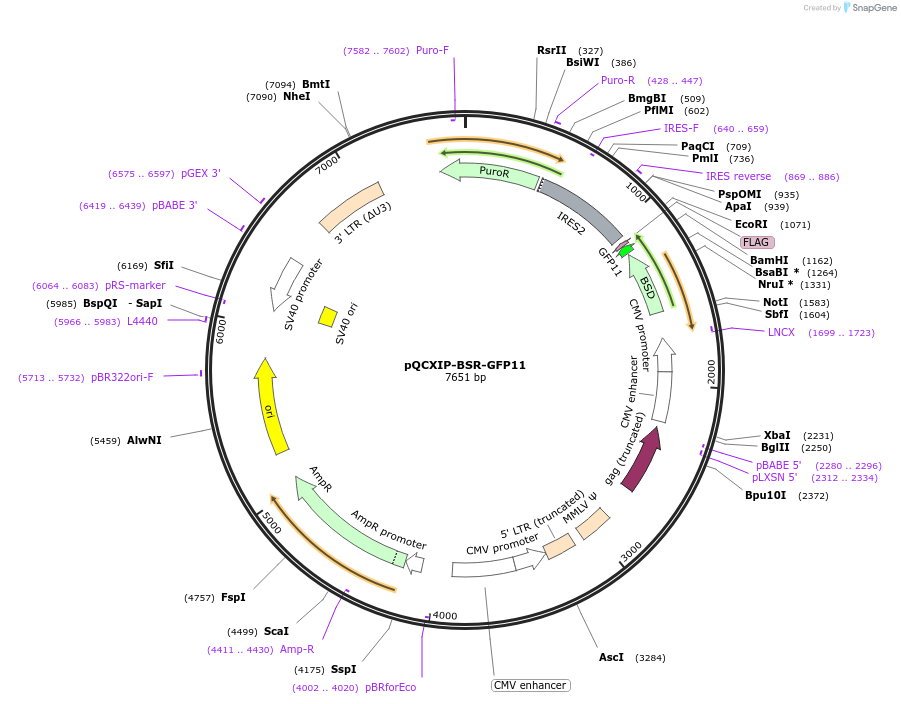
pQCXIP-BSR-GFP11
Plasmid#68716PurposeSplit GFP assayDepositorInsertbsr
UseRetroviralTagsFLAG and GFP11ExpressionMammalianAvailable SinceJan. 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
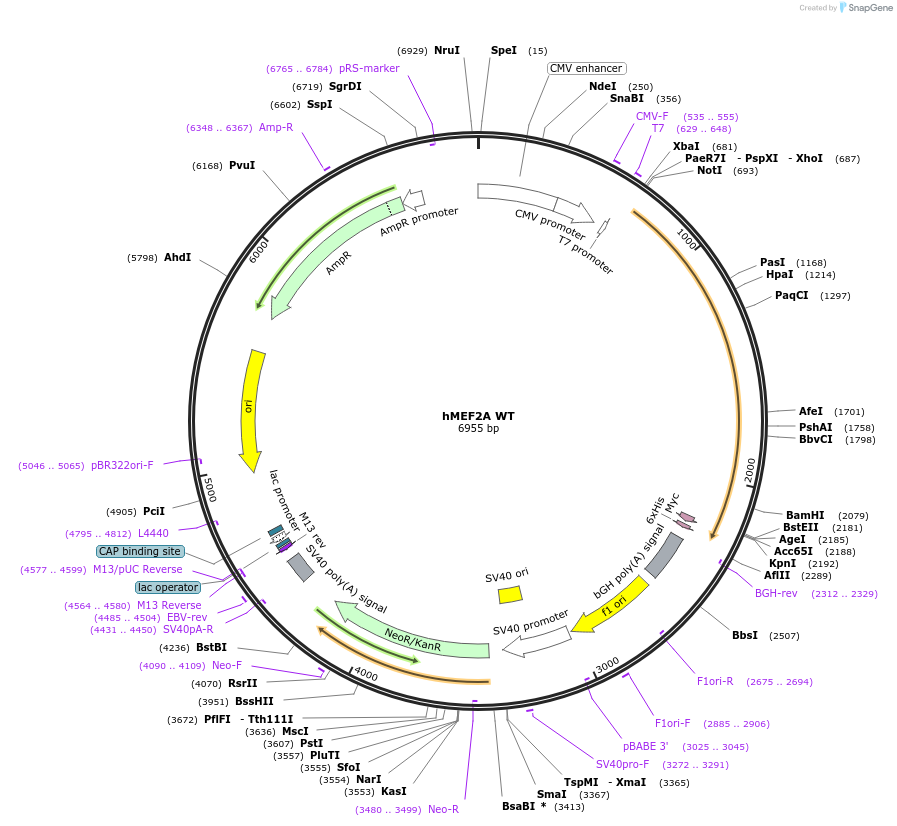
hMEF2A WT
Plasmid#118354PurposeThis plasmid expresses human MEF2A-1 WT.DepositorAvailable SinceDec. 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
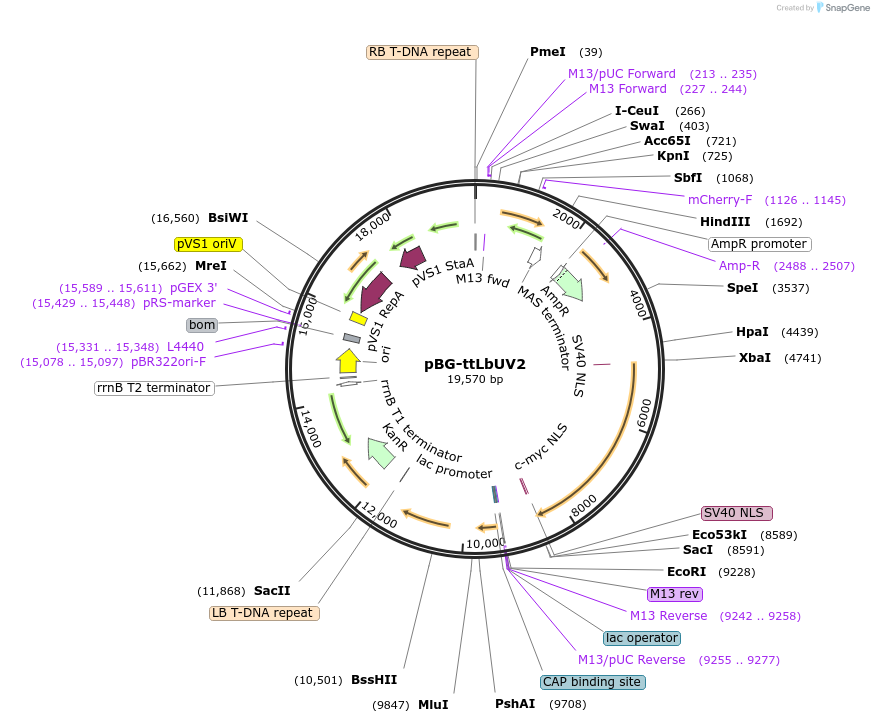
pBG-ttLbUV2
Plasmid#231386PurposeLbCas12a variant with D156R and E795L mutations and harboring same NLS sequence as PEmax but codon-optimized for dicot species; for Agrobacterium-mediated Arabidopsis transformation.DepositorInsertttLbCas12a Ultra V2
UseCRISPRExpressionPlantMutationD156R and E795LAvailable SinceFeb. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
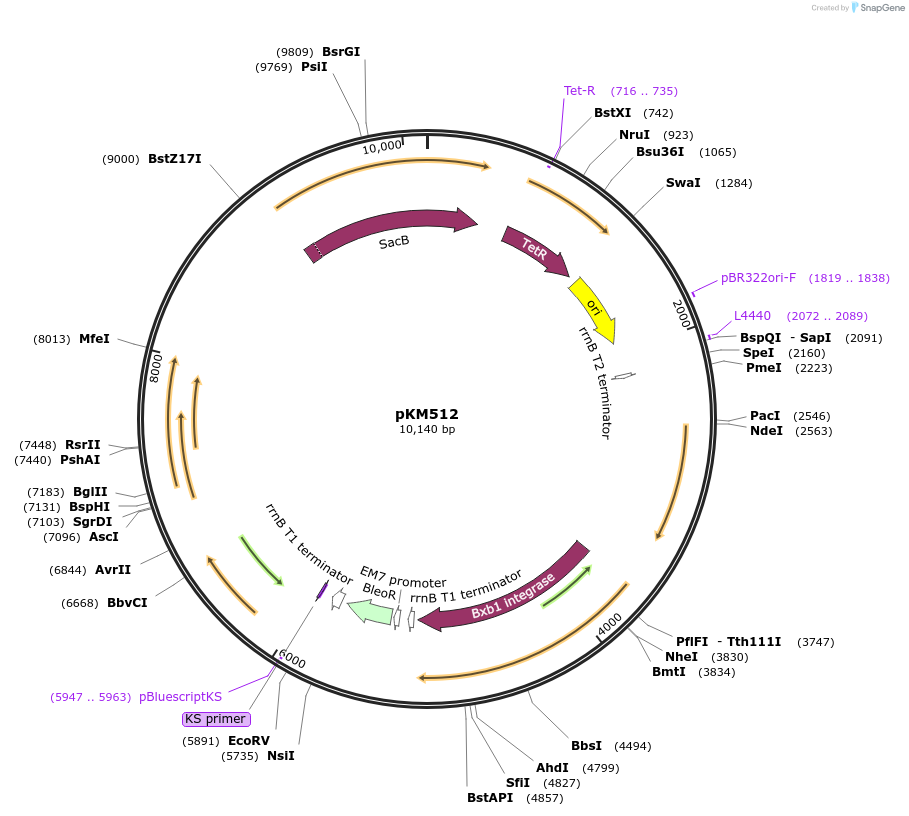
pKM512
Plasmid#140192PurposeFor curing of Orbit-integrated plasmids and associated drug markerDepositorInsertszeocin resistance gene
Bxb1gp47 (directionality factor)
ExpressionBacterialMutationCloned in gp47 directinality factor in place of R…PromoterEM7 and PtetAvailable SinceJan. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
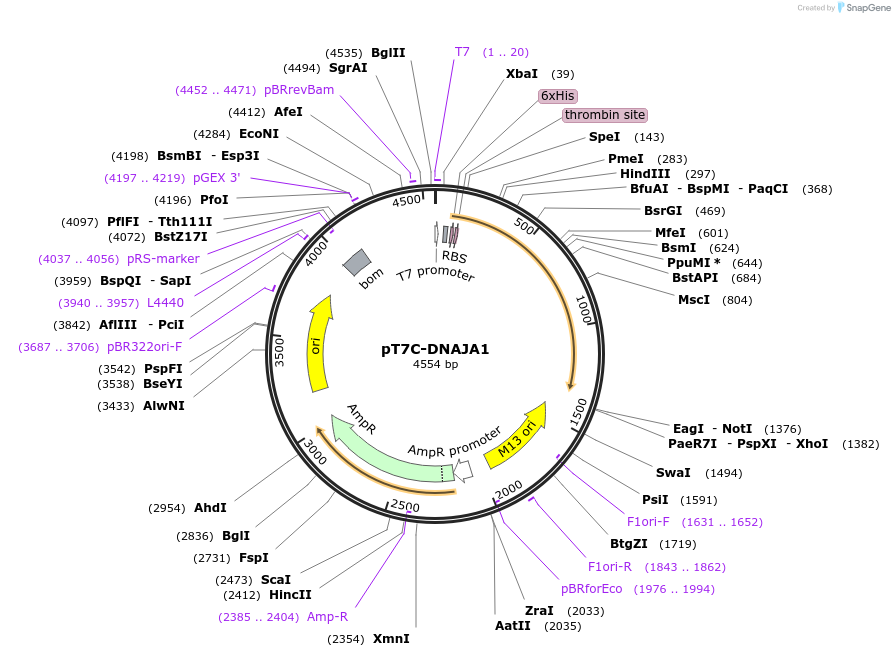
pT7C-DNAJA1
Plasmid#177655PurposeRecombinant protein synthesis of human DNAJA1DepositorAvailable SinceMarch 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
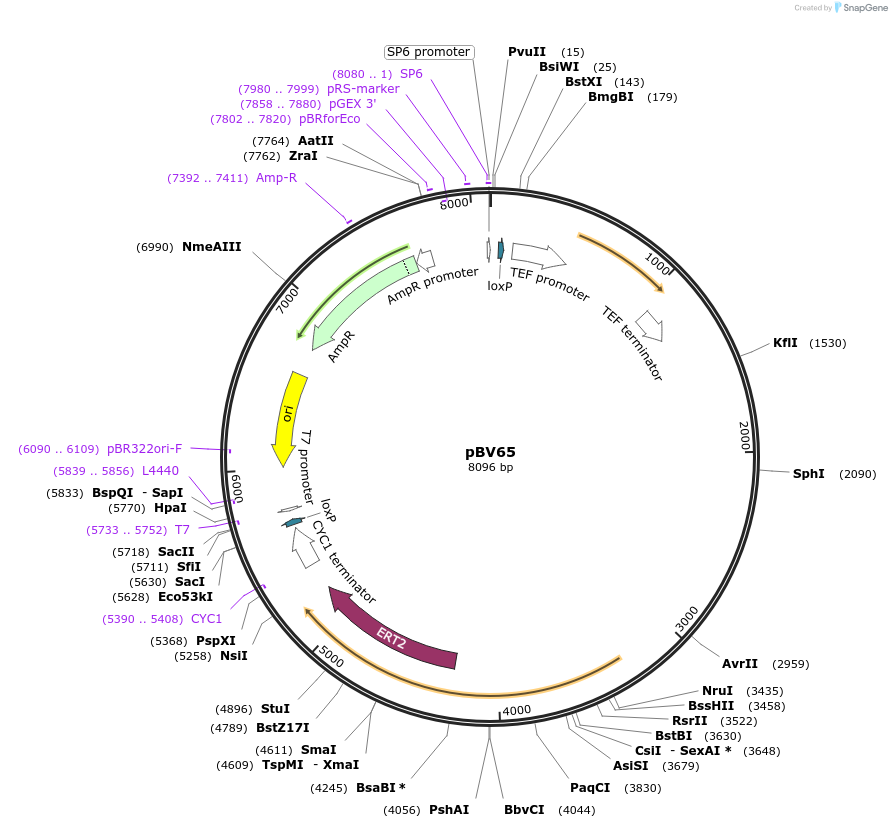
pBV65
Plasmid#108363PurposeA nourseothricin resistance marker used to construct gene disruptions in Candida glabrata. The natMX6 marker can be evicted by growth on media lacking methionine and containing estradiol.DepositorInsertloxP-natMX6-CgMET3prom-cre-loxP
UseGene disruption marker for candida glabrataPromoterC. glabrata MET3Available SinceMay 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
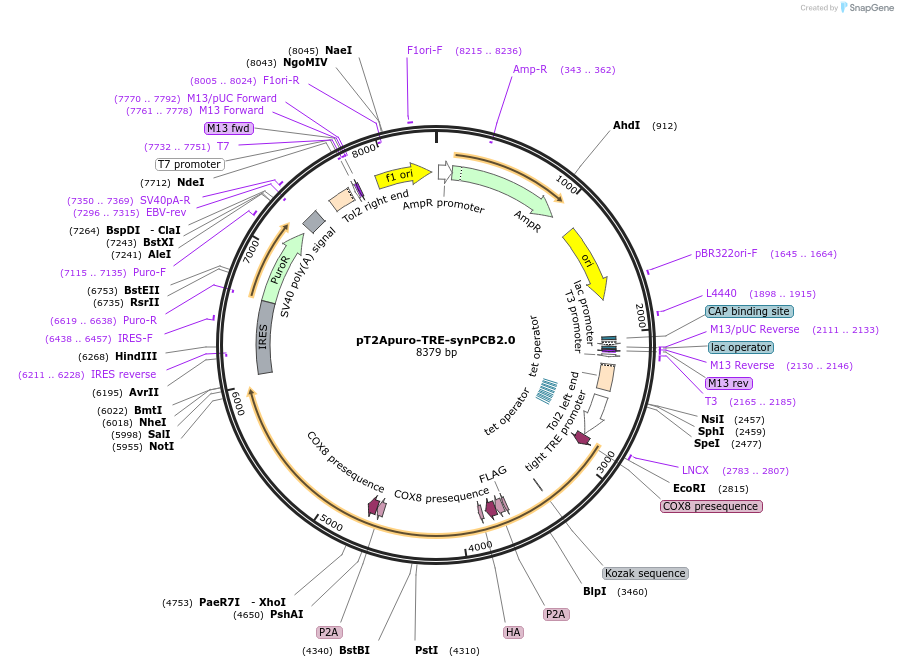
pT2Apuro-TRE-synPCB2.0
Plasmid#139481PurposeExpression vector for phycocyanobilin (PCB) synthesis in mammalian cells.DepositorInsertMTS-PcyA-FLAG-P2A-MTS-HA-HO1-P2A-MTS-Fnr-Fd-T7
UseTol2 transposon donor vectorExpressionMammalianPromoterTet responsive elementAvailable SinceMarch 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
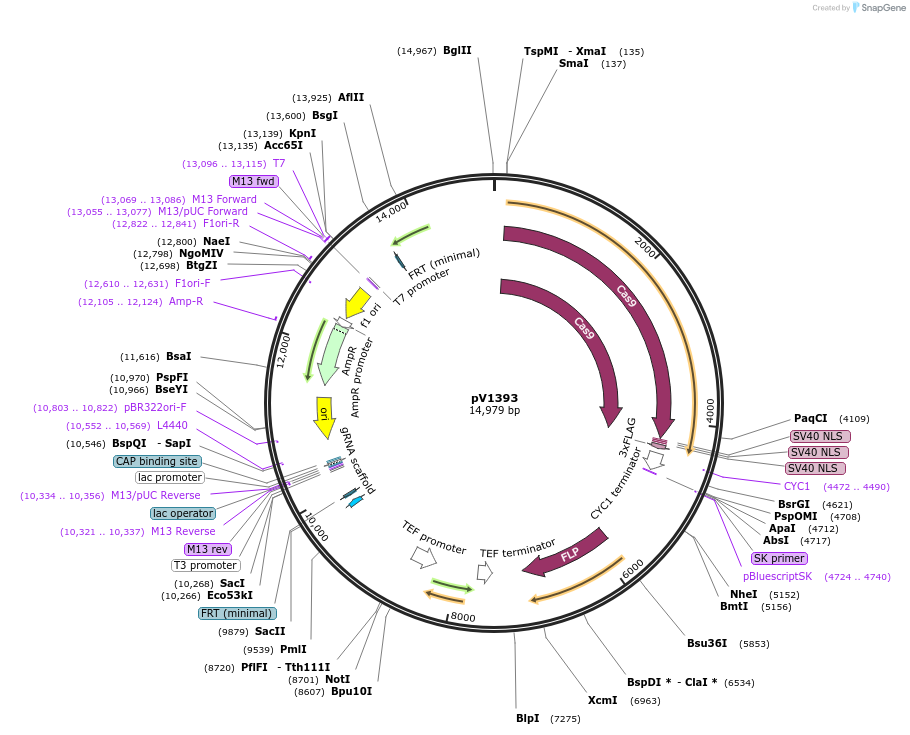
pV1393
Plasmid#111430PurposeCaCas9/gRNA Solo entry plasmid for cloning guides - contains stuffer with BsmBI sites - inserts at Neut5L - Recyclable for serial mutagenesis (Sap2-FLP)DepositorInsertCaCas9/sgRNA-BsmBI stuffer
UseCRISPRExpressionYeastAvailable SinceAug. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
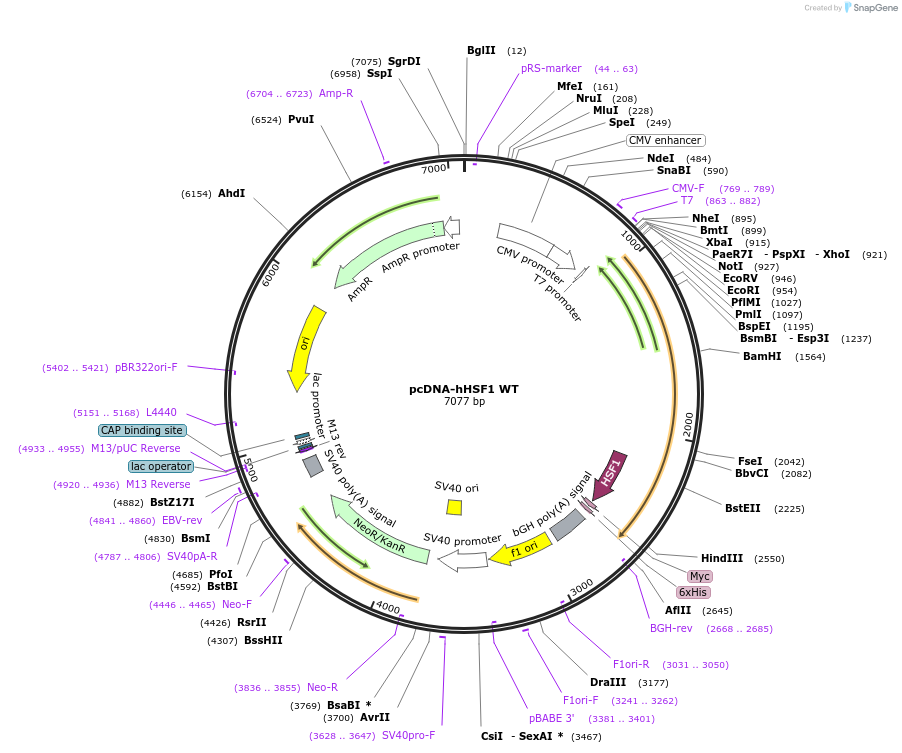
pcDNA-hHSF1 WT
Plasmid#71724Purposeplasmid expressing myc-his tag hHSF1 WTDepositorAvailable SinceMay 25, 2016AvailabilityAcademic Institutions and Nonprofits only -
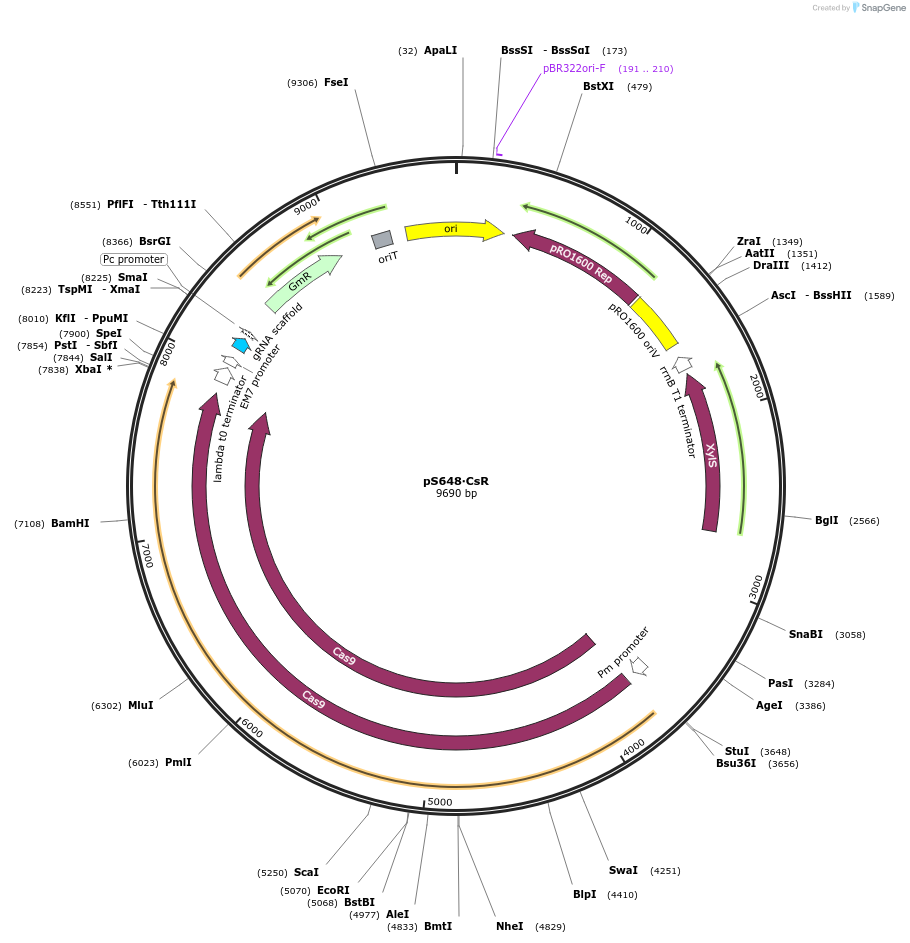
pS648∙CsR
Plasmid#197860PurposeExpresses Streptococcus pyogenes' cas9 and a tailored sgRNA for counterselection in gram-negative bacteriaDepositorInsertgRNA scaffold - Gm resistance cassette - incP oriT
UseCRISPR and Synthetic BiologyPromoterPEM7Available SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
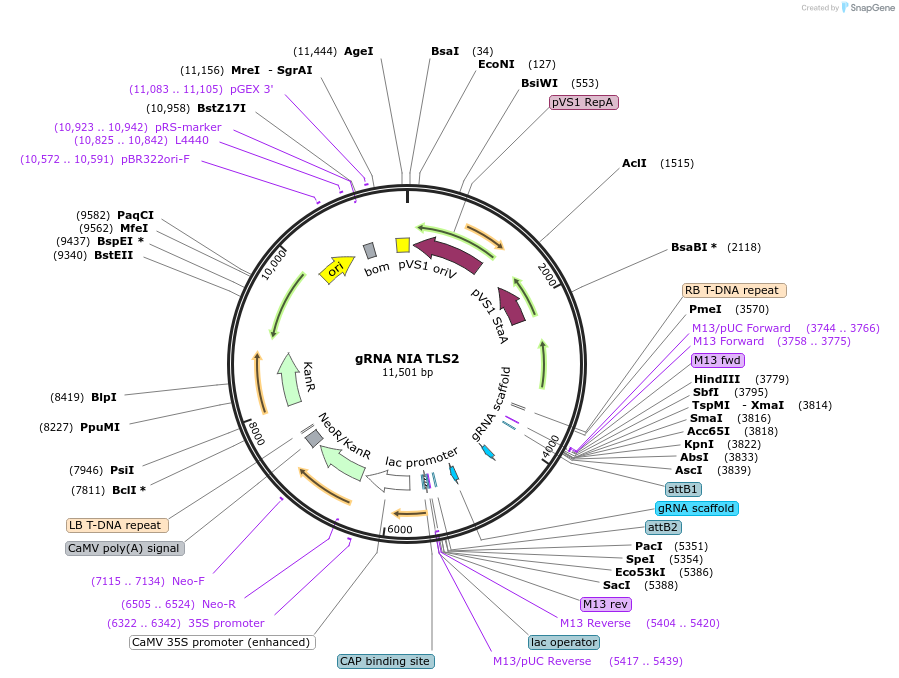
gRNA NIA TLS2
Plasmid#196981PurposeExpresses two gRNAs targeting Arabidopsis NIA1 gene for knockout. Each gRNA is fused to TLS2 mobility signal that confers graft mobility.DepositorInsertAtNIA1 gRNAs fused to TLS2 tags
ExpressionPlantPromoterpU6-26, pU6-29Available SinceMay 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
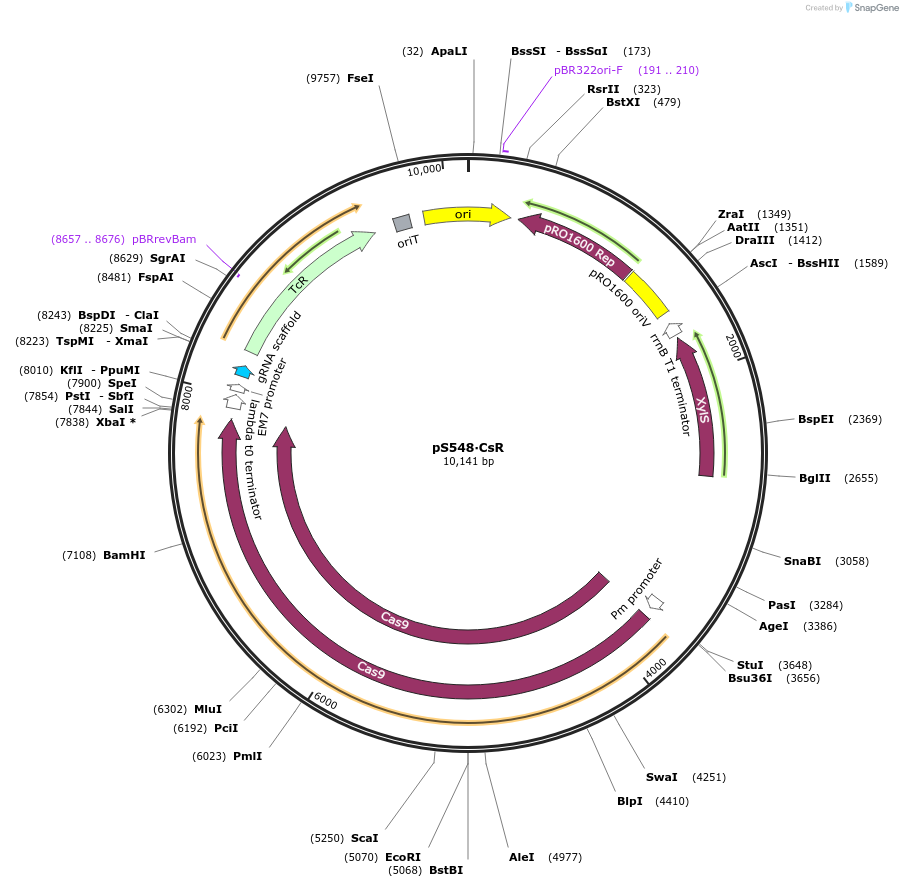
pS548∙CsR
Plasmid#197859PurposeExpresses Streptococcus pyogenes' cas9 and a tailored sgRNA for counterselection in gram-negative bacteriaDepositorInsertgRNA scaffold - Tc resistance cassette - incP oriT
UseCRISPR and Synthetic BiologyPromoterPEM7Available SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
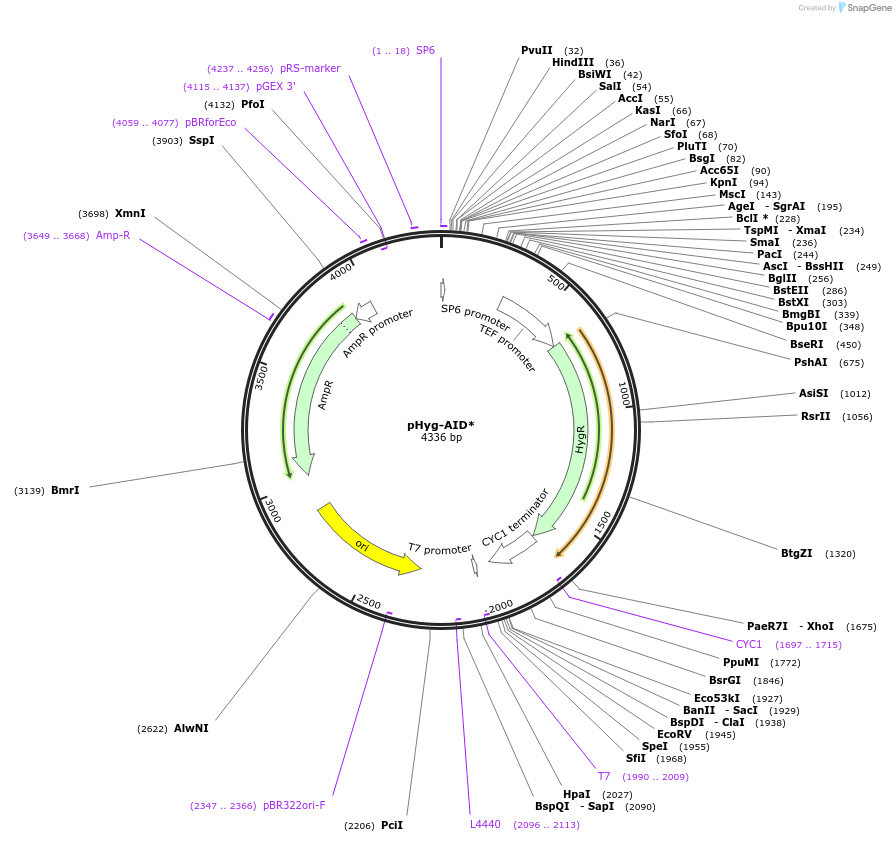
pHyg-AID*
Plasmid#99515PurposeC-terminal AID* degron cassette with HygR marker for S. cerevisiaeDepositorInsertIAA17(71-114) (AXR3 Mustard Weed)
UsePcr template for yeast tagging cassetteMutationinternal inversion of nucleotides 265-312 of the …Available SinceSept. 14, 2017AvailabilityAcademic Institutions and Nonprofits only -
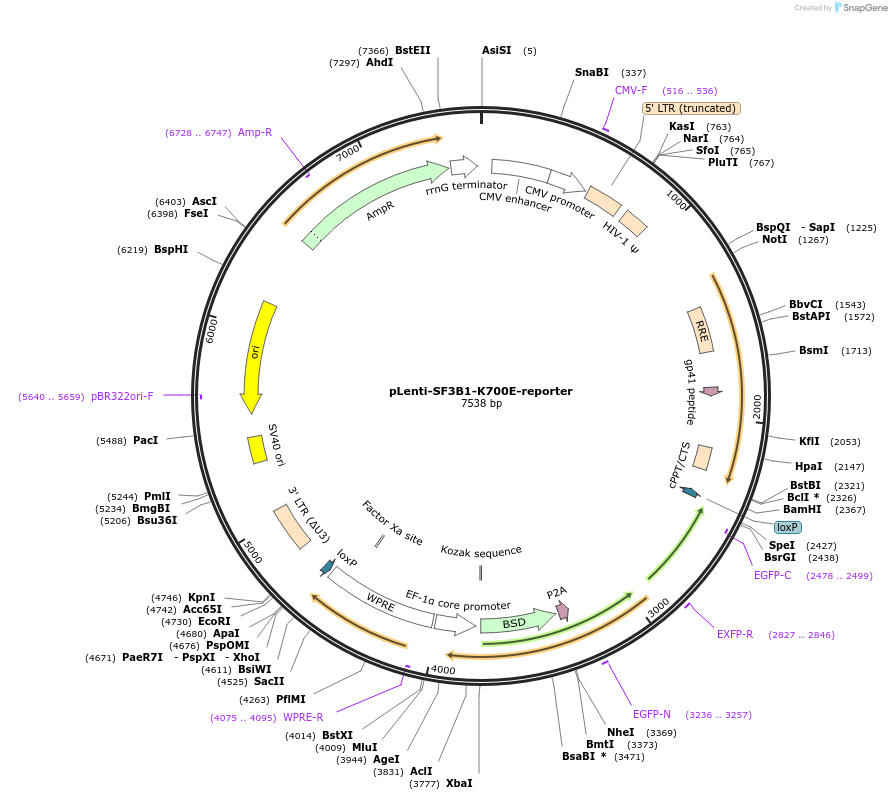
pLenti-SF3B1-K700E-reporter
Plasmid#200318PurposeA fluorescent reporter for SF3B1 K700E mutation. Co-expresses a blasticidin resistance gene for selection. Includes LoxP sites for Cre-mediated deletion of reporter cassette.DepositorInsertBSD-2A-EGFPi
UseLentiviralExpressionMammalianPromoterEF1-aAvailable SinceAug. 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
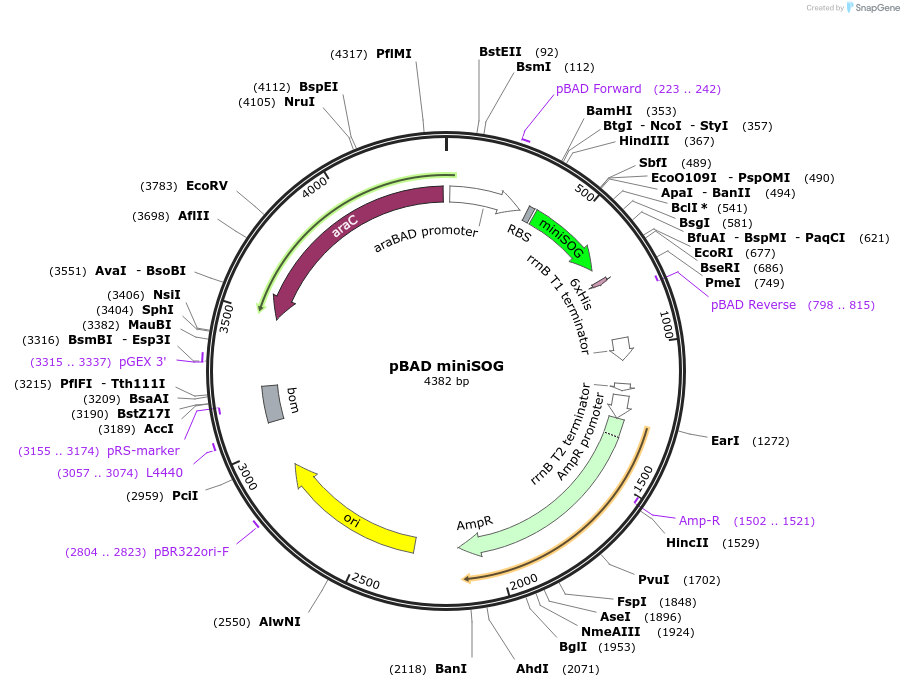
pBAD miniSOG
Plasmid#91774PurposeBacterial expression of a fluorescent protein, mini Singlet Oxygen Generator (miniSOG)DepositorInsertmini Singlet Oxygen Generator
TagsHexa-Histidine tagExpressionBacterialPromoteraraBADAvailable SinceJune 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
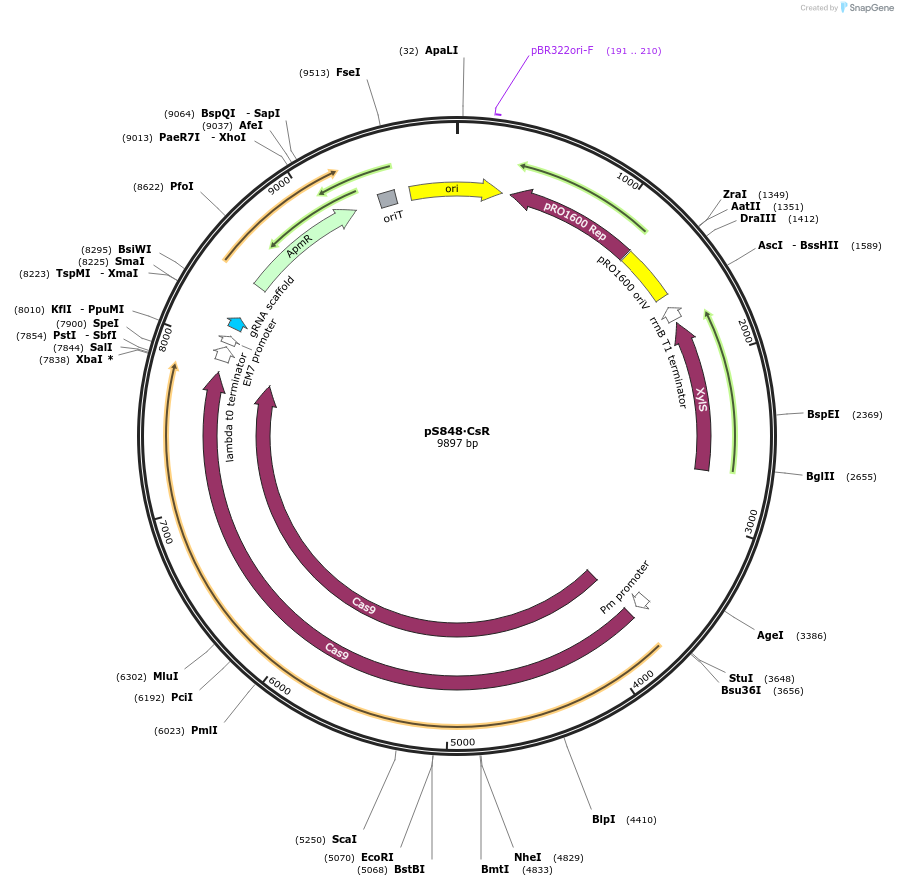
pS848∙CsR
Plasmid#199240PurposeExpresses Streptococcus pyogenes' cas9 and a tailored sgRNA for counterselection in gram-negative bacteriaDepositorInsertgRNA scaffold - Gm resistance cassette - incP oriT
UseCRISPR and Synthetic BiologyPromoterPEM7Available SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
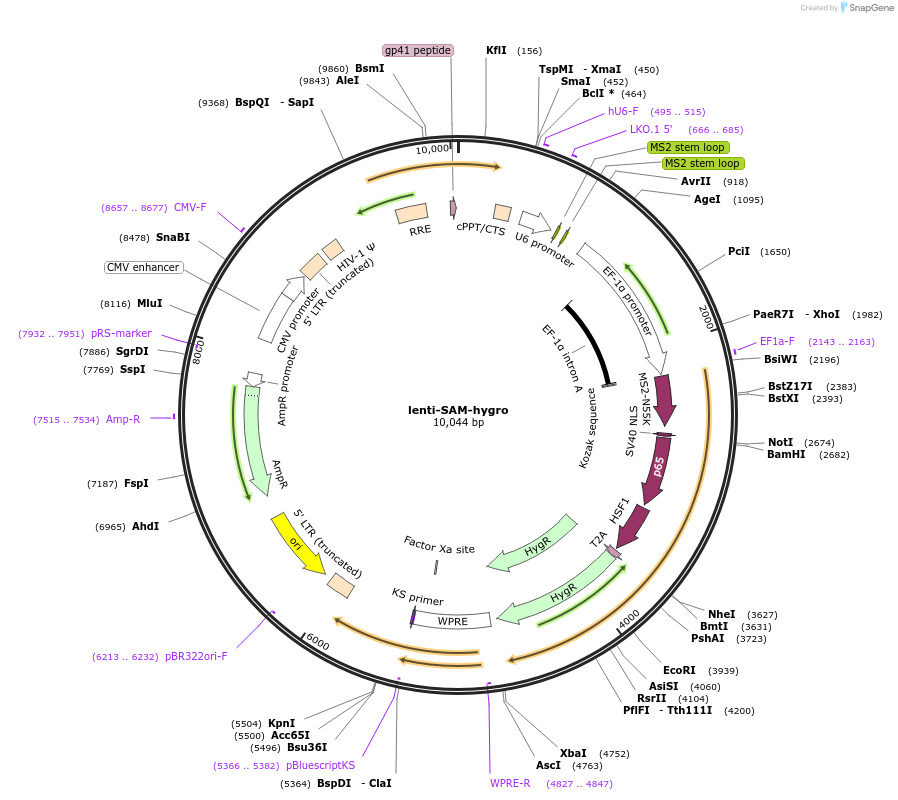
lenti-SAM-hygro
Plasmid#138955PurposeLentiviral vector for co-expressing spCas9, dRNA-MS2, p65 and HSF1 fused to MS2 binding protein, and hygromycin resistance geneDepositorTypeEmpty backboneUseLentiviralExpressionMammalianPromoterU6 AND EF1aAvailable SinceAug. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
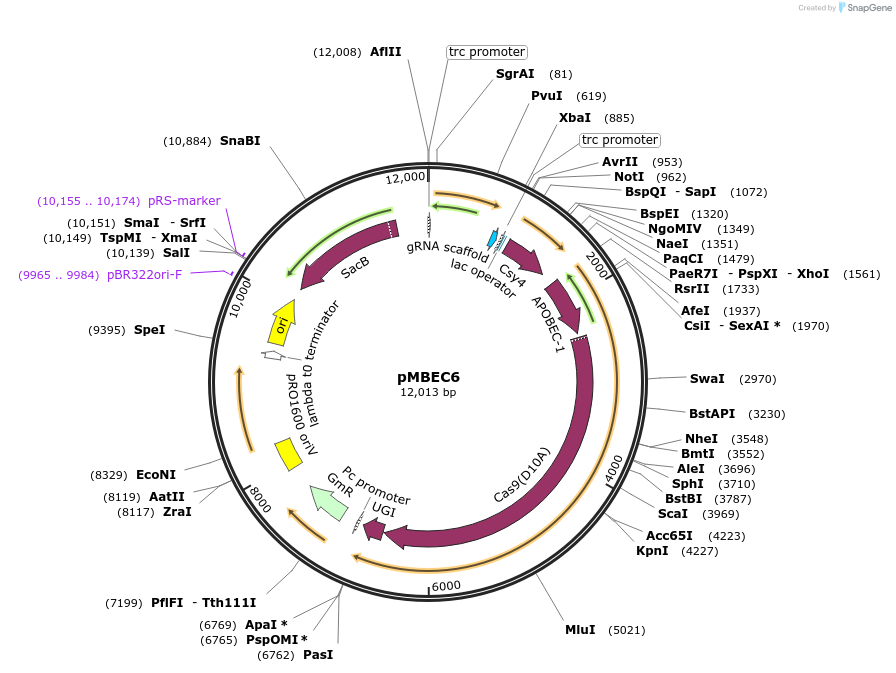
pMBEC6
Plasmid#187598PurposeBase editing plasmid, constitutively expressing cytosine base editor and cas6. Contains GFP, flanked by BsaI restriction sites to introduce spacer and gRNAs. Gentamycin resistanceDepositorInsertnCas9-BEC-UGI; cas6f, gfp
ExpressionBacterialPromotertrcAvailable SinceJuly 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
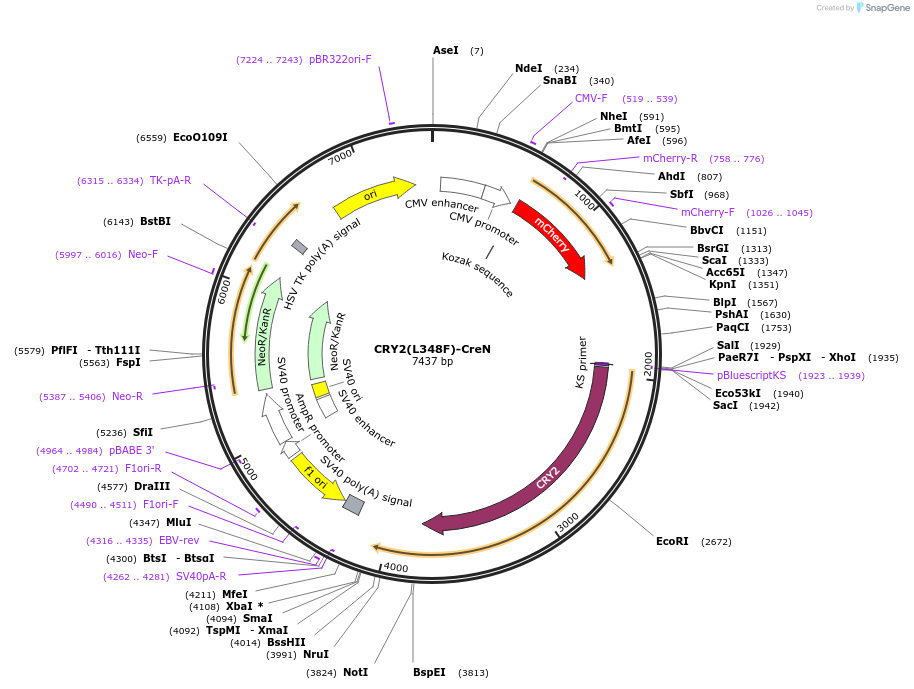
CRY2(L348F)-CreN
Plasmid#75368PurposeExpresses mCherry tag, then IRES, then Arabidopsis CRY2 (with L348F mutation) fused to N-terminal fragment of Cre DNA recombinaseDepositorInsertCRY2 (CRY2 Mustard Weed)
TagsCre recombinase, residues 19-104ExpressionMammalianMutationL348FPromoterCMVAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
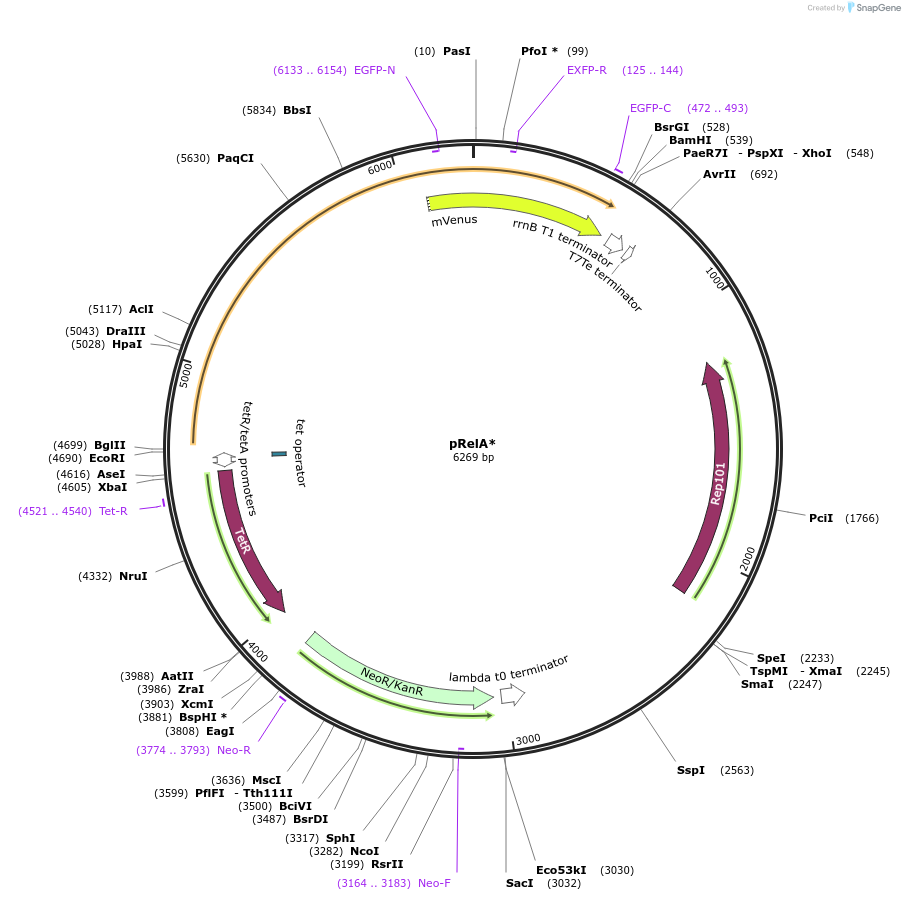
pRelA*
Plasmid#175590PurposeTet inducible, KAN Resistant, fluor labelled (YFP) synthesis domain of the E. coli relA gene on a low copy backbone. Used to induce controllable synthesis of ppGpp in E. coli.DepositorInsertCatalytic domain of E.coli relA gene.
TagsFused with a flexible GS linker to mVenusExpressionBacterialMutationOnly the catalytic domain of the enzyme is used (…Available SinceNov. 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
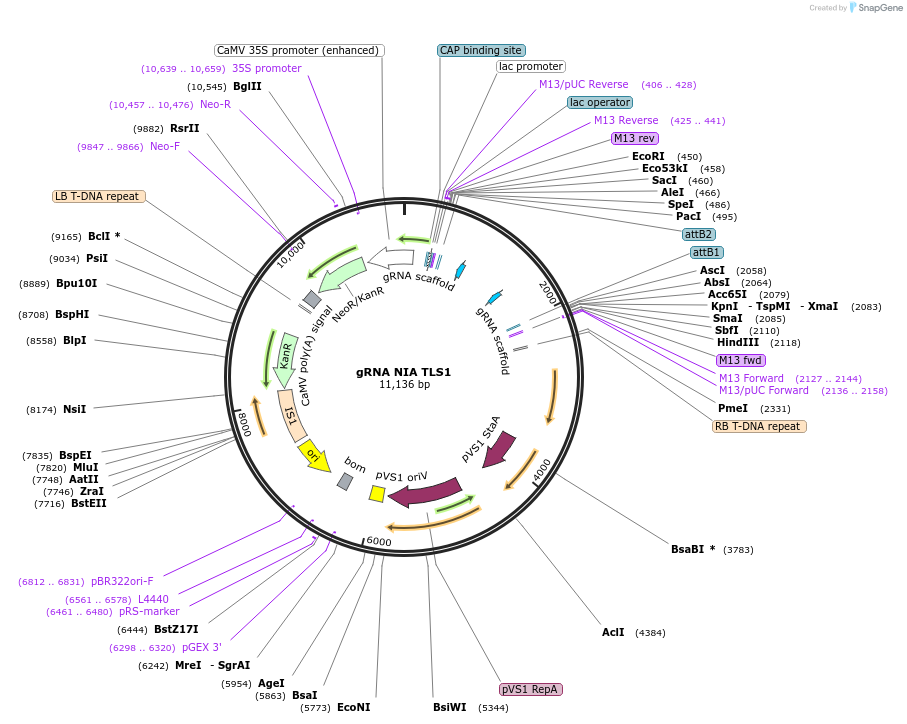
gRNA NIA TLS1
Plasmid#196980PurposeExpresses two gRNAs targeting Arabidopsis NIA1 gene for knockout. Each gRNA is fused to TLS1 mobility signal that confers graft mobility.DepositorInsertAtNIA1 gRNAs fused to TLS1 tags
ExpressionPlantPromoterpU6-26, pU6-29Available SinceMay 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
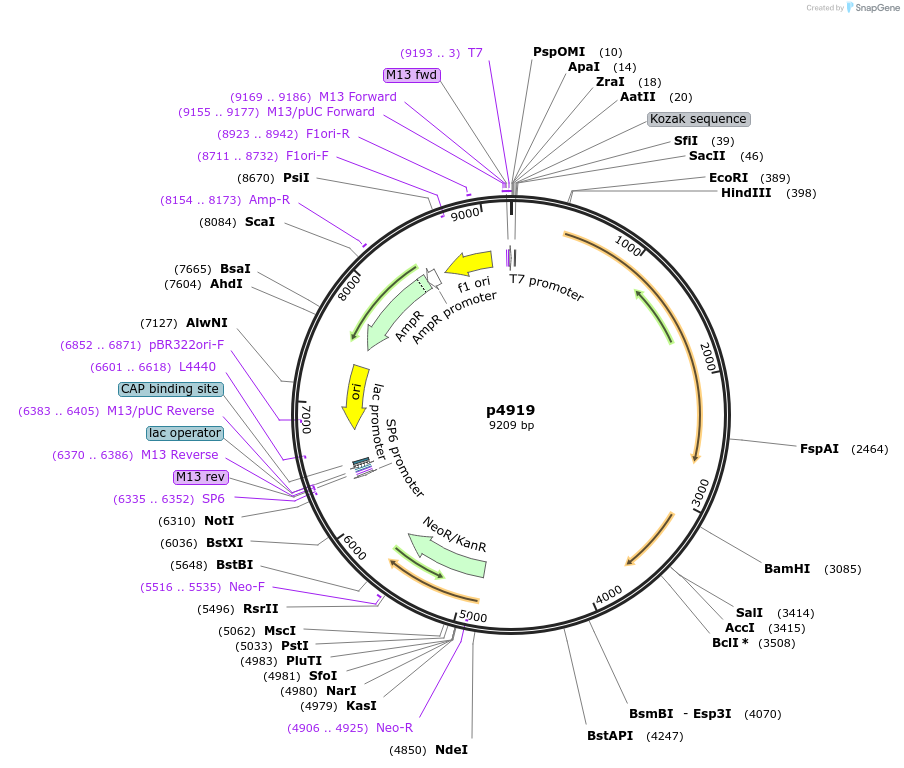
p4919
Plasmid#239357PurposeExpression of Rice Cullin, Rice SKP1, Rice Rbx1, and G418 resistance by read through transcription after integration into the tubulin locusDepositorInsertRice CUL1, Rice SKP1, Rice RBX1 and G418 resistance
TagsCUL1, SKP1 and RBX1 are all Ty epitope taggedPromoternoneAvailable SinceJuly 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
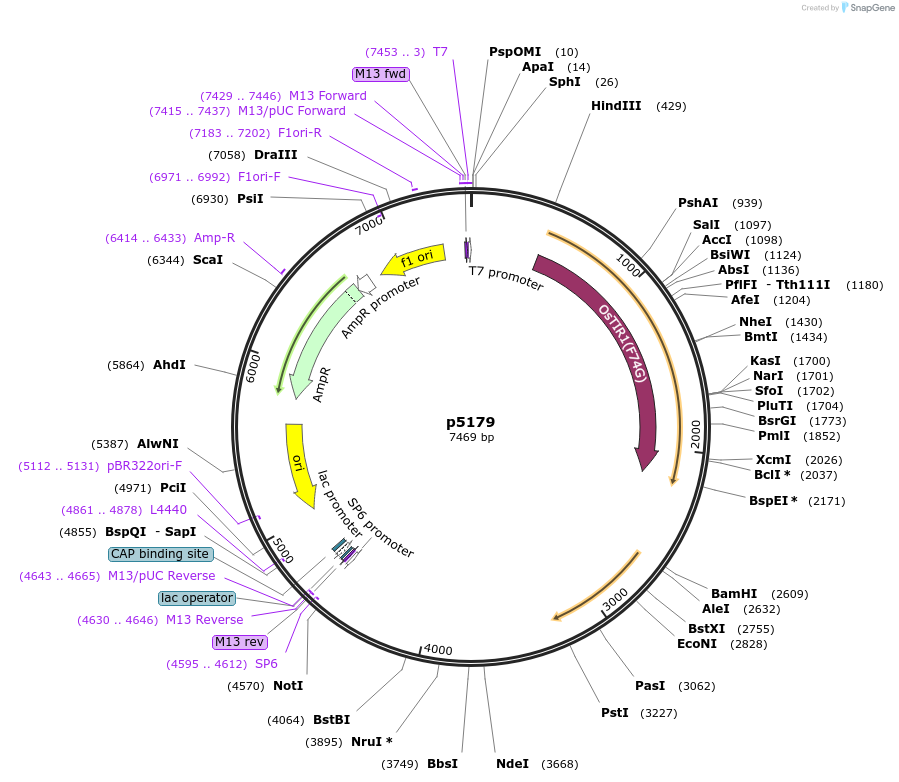
p5179
Plasmid#239358PurposeExrpession of Rice TIR1 F74->G, Rice ARF fragment and blaticidin resistance by read through transaction after integration int the PFR1 locusDepositorInsertRice TIR1, Rice ARF16 and blasticidin resistance
UseBacterialTagsTIR1 and ARF are both Ty-epitope taggedMutationTIR1 F74->G. ARF16 is residues 784 to 959 onlyPromoternoneAvailable SinceJuly 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
pKan-PCUP1-AID*(N)-9myc
Plasmid#99526PurposeN-terminal AID*-9myc degron cassette with CUP1 promoter and KanR marker for S. cerevisiaeDepositorInsertIAA17(71-116) (AXR3 Mustard Weed)
UsePcr template for yeast tagging cassetteTags9xMycPromoterCUP1Available SinceSept. 14, 2017AvailabilityAcademic Institutions and Nonprofits only -
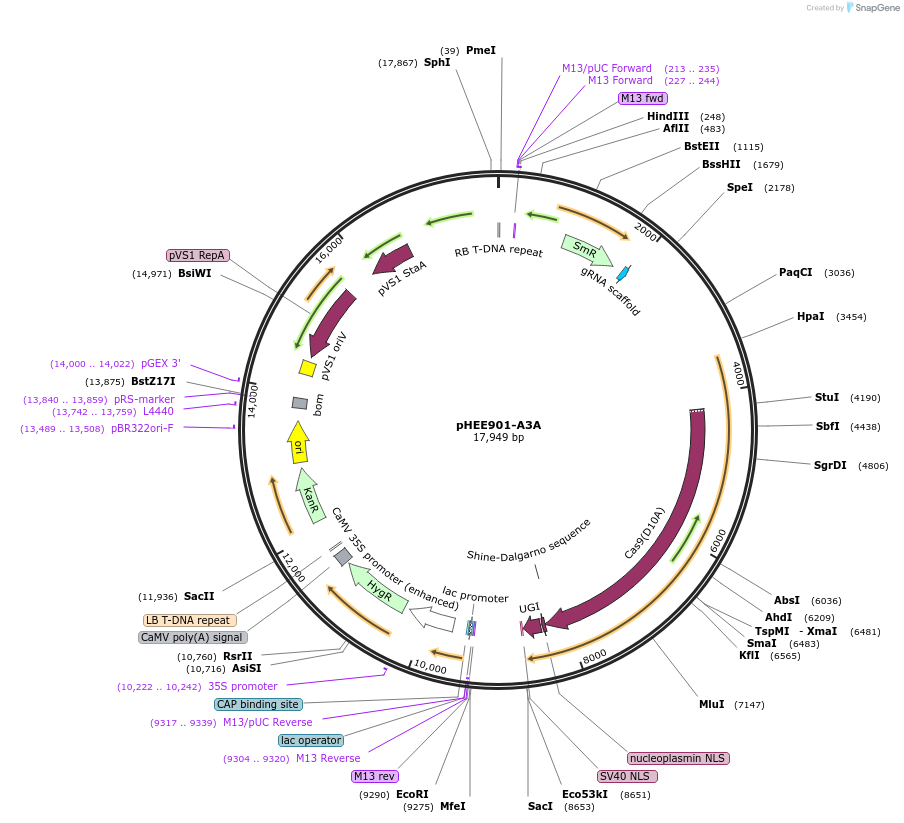
pHEE901-A3A
Plasmid#229044PurposenCas9 mediated-base editing with NGG PAM recognition motif in Arabidopsis. A3A-nCas9-UGI includes D10A in Cas9.DepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
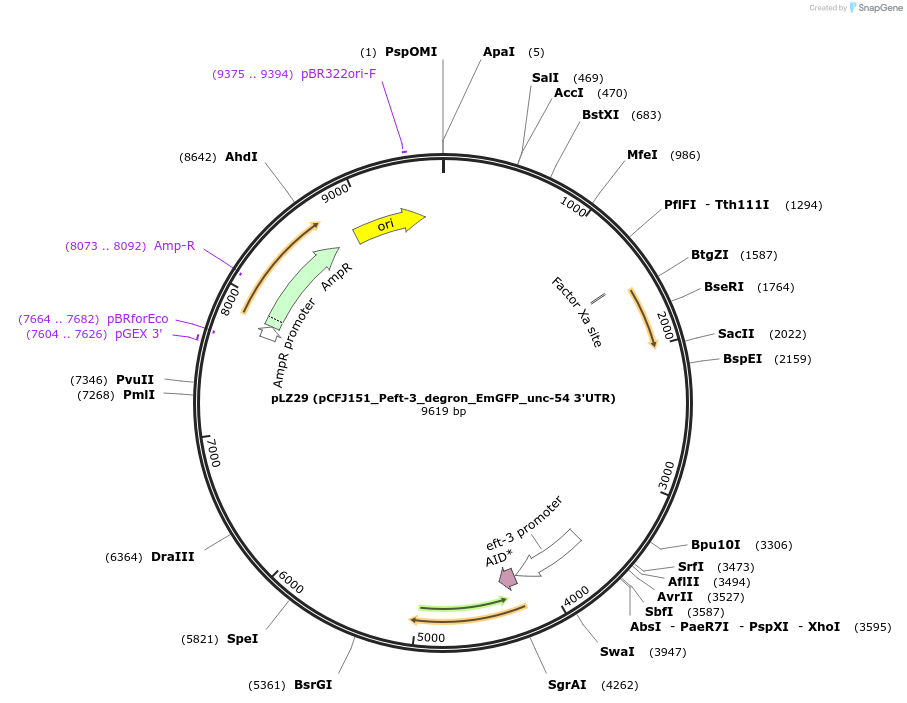
pLZ29 (pCFJ151_Peft-3_degron_EmGFP_unc-54 3'UTR)
Plasmid#71719PurposeExpressing a GFP-tagged degron in the soma of C. elegansDepositorInsertauxin-responsive protein IAA17 (AXR3 Mustard Weed)
TagsEmGFPExpressionWormMutationminimal degron sequence (71-114aa) with start cod…Promotereef-1A.1 (eft-3)Available SinceDec. 17, 2015AvailabilityAcademic Institutions and Nonprofits only