We narrowed to 5,715 results for: Feng Zhang
-
Plasmid#48139PurposeNOTE: A new version of this plasmid is now available. See Addgene plasmid 62988.DepositorInserthSpCas9-2A-Puro
UseCRISPRTags2A-Puro and 3XFLAGExpressionMammalianPromoterCbhAvailable SinceOct. 29, 2013AvailabilityAcademic Institutions and Nonprofits only -
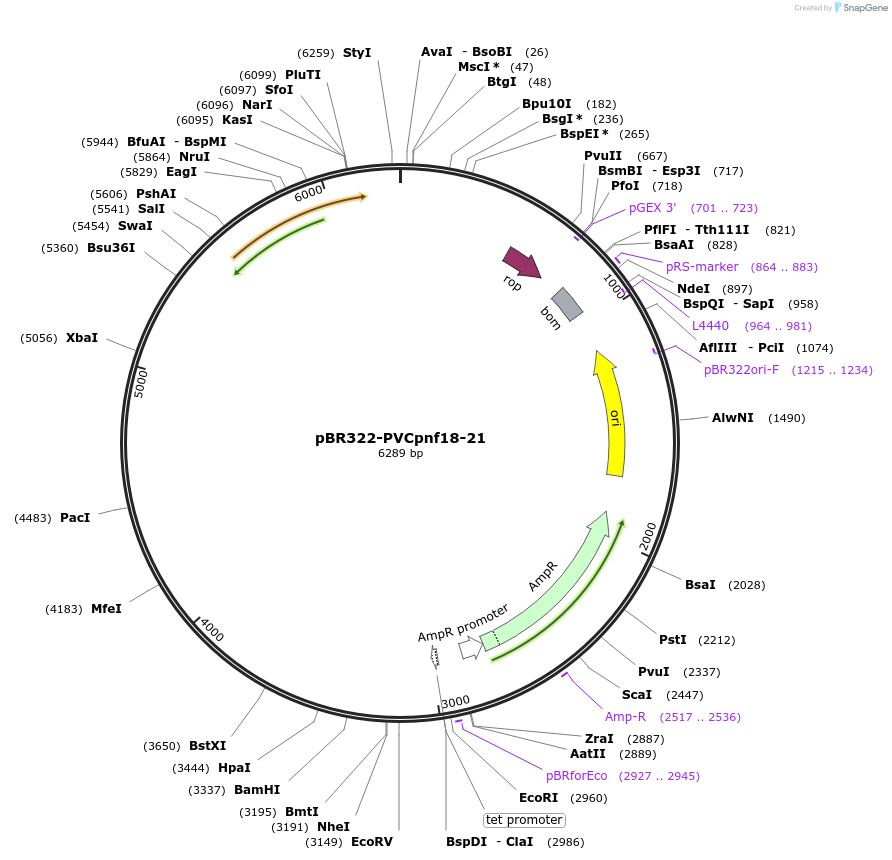
pBR322-PVCpnf18-21
Plasmid#198273PurposeContains only regulatory genes; for generating empty PVCsDepositorInsertPVCpnf18-21
ExpressionBacterialAvailable SinceMarch 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
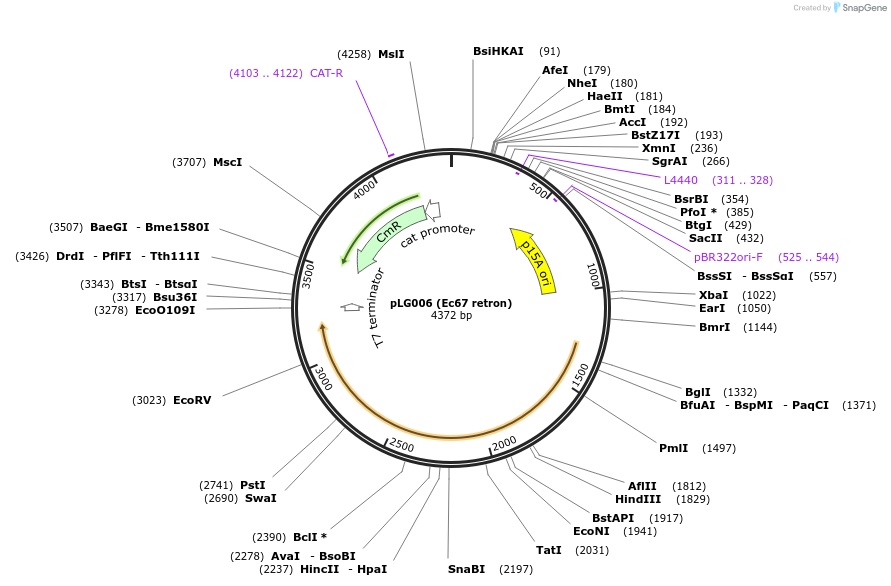
pLG006 (Ec67 retron)
Plasmid#157884PurposeExpresses the Ec67 retron defense systemDepositorInsertEc67 retron
ExpressionBacterialAvailable SinceDec. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
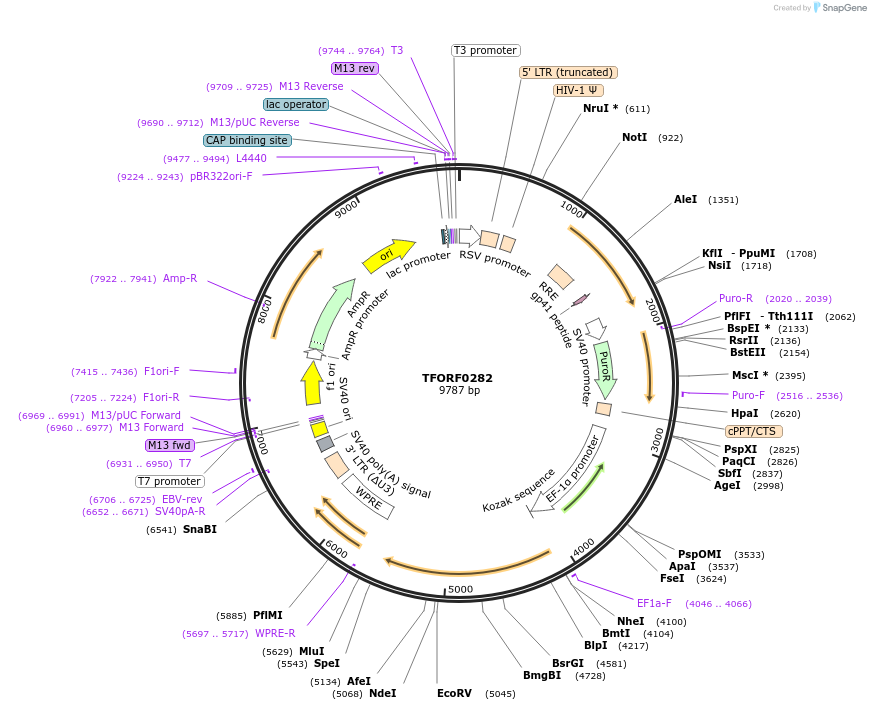
TFORF0282
Plasmid#141561PurposeLentiviral vector for overexpressing the MEF2C transcription factor ORF with a unique 24-bp barcode. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
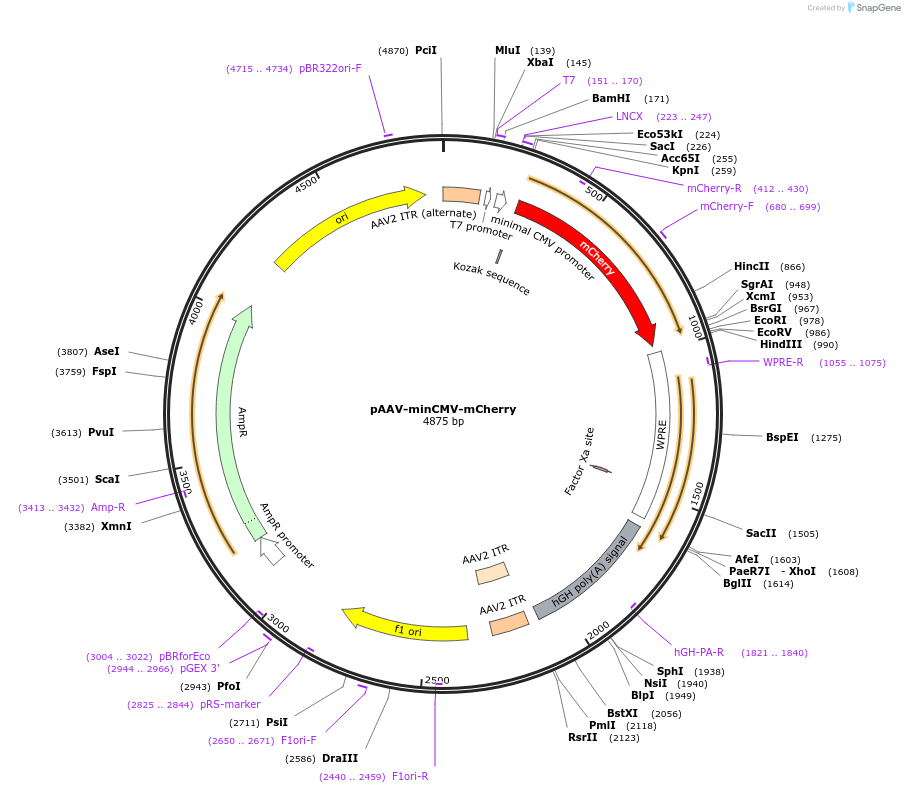
pAAV-minCMV-mCherry
Plasmid#27970DepositorInsertmCherry
UseAAVExpressionMammalianAvailable SinceFeb. 10, 2011AvailabilityAcademic Institutions and Nonprofits only -
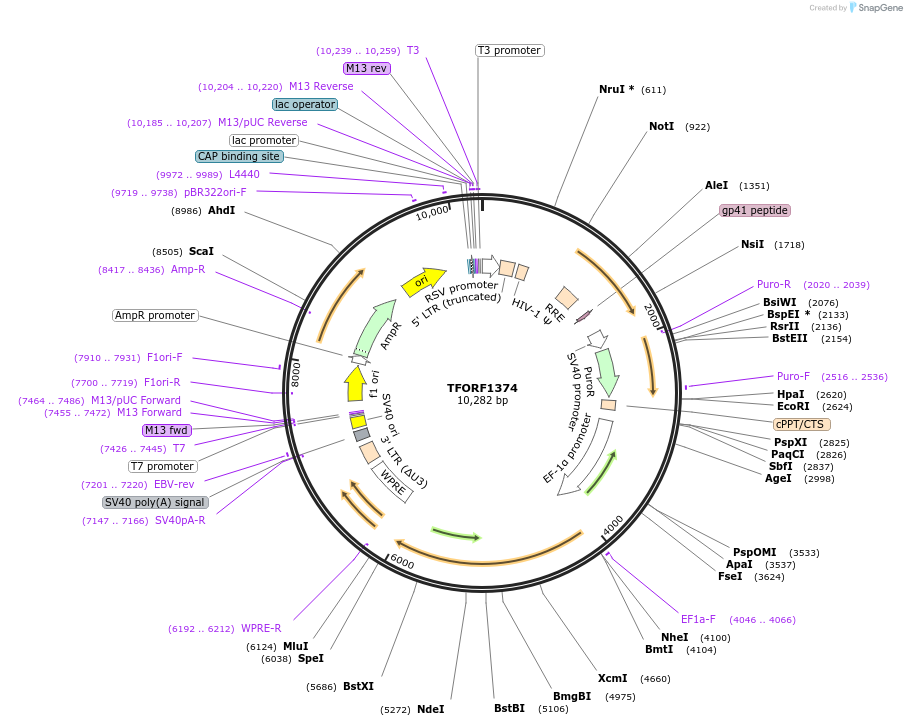
TFORF1374
Plasmid#143955PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceAug. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
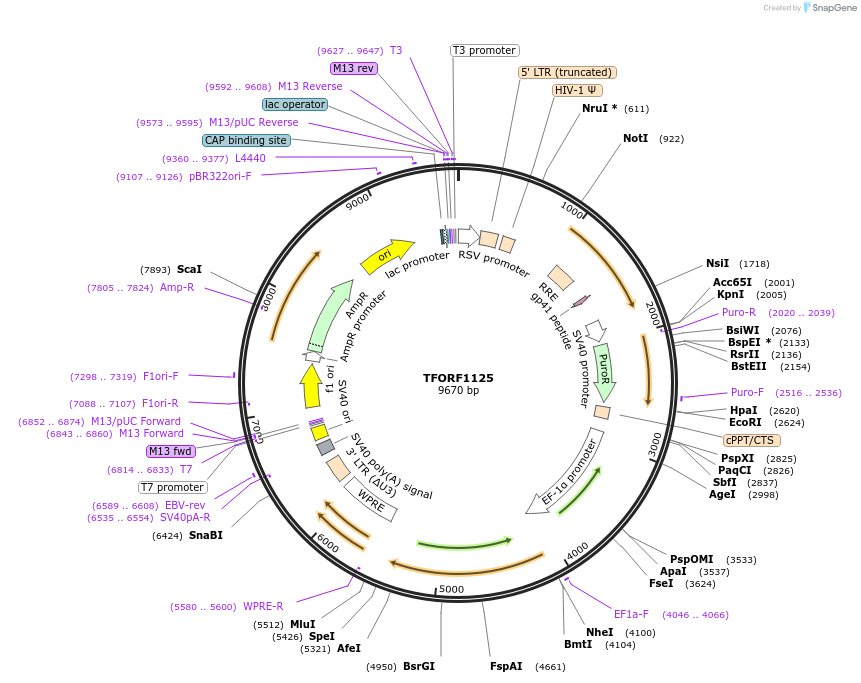
TFORF1125
Plasmid#143093PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
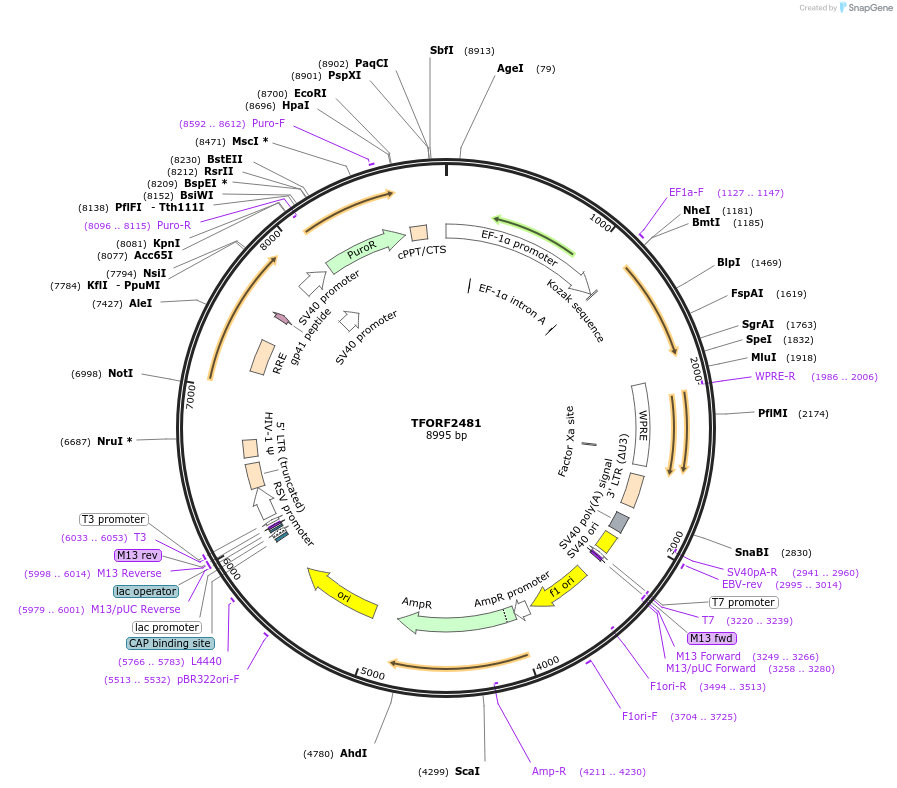
TFORF2481
Plasmid#142948PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceMay 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
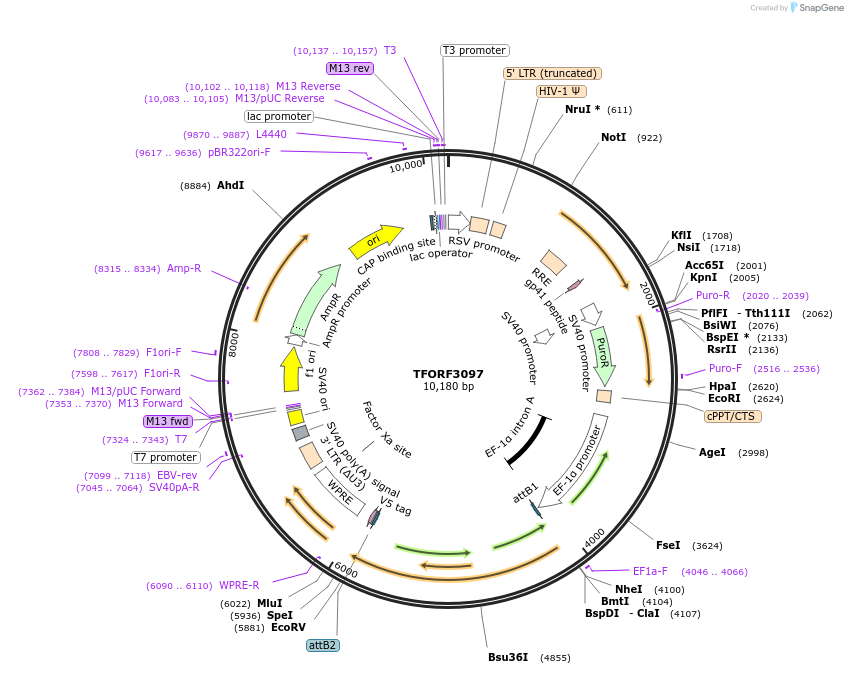
TFORF3097
Plasmid#144573PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceAug. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
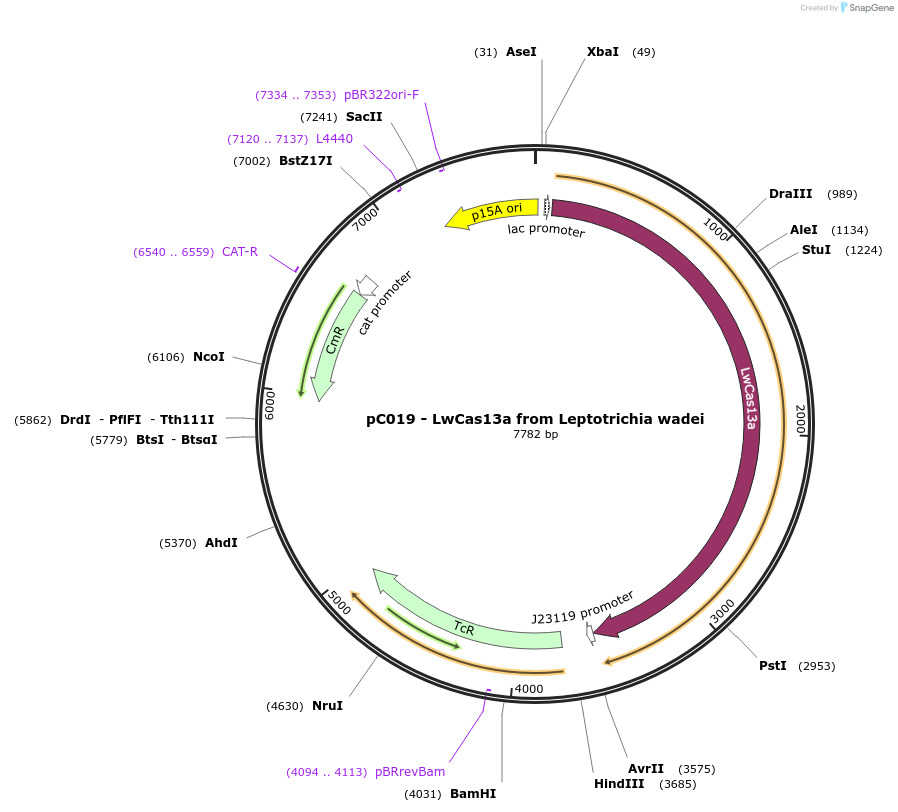
pC019 - LwCas13a from Leptotrichia wadei
Plasmid#91909PurposeExpresses LwCas13a for bacterial expression and contains backbone for spacer cloningDepositorInsertLwCas13a and guide expression cassette
UseCRISPRExpressionBacterialAvailable SinceOct. 4, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits -
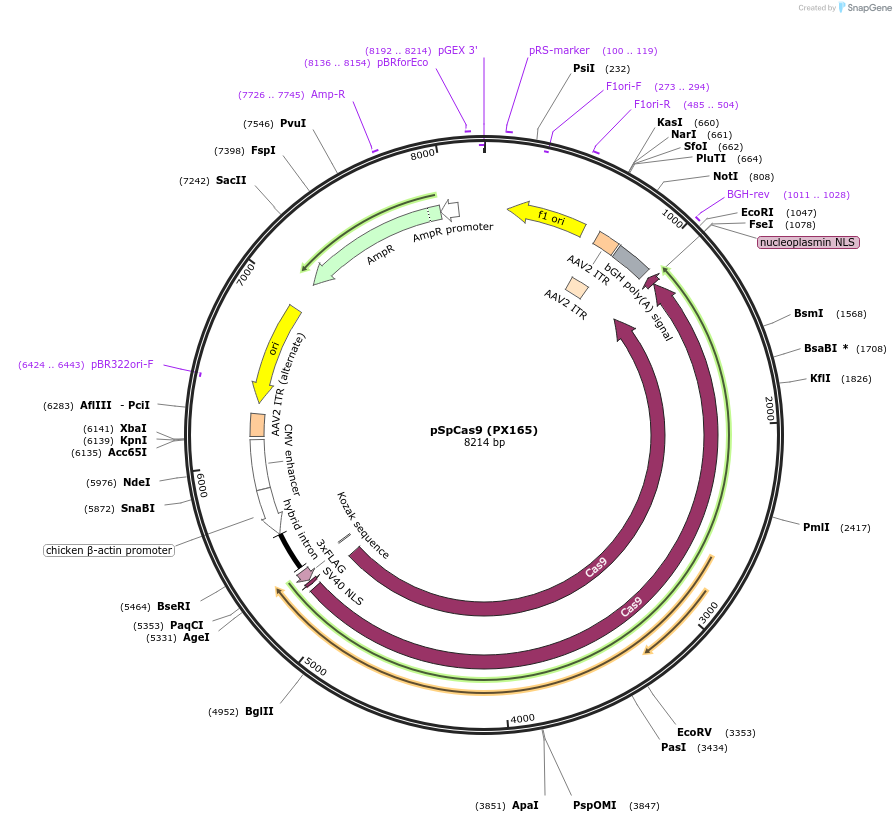
pSpCas9 (PX165)
Plasmid#48137PurposeHuman codon optimized Cas9 nuclease from Streptococcus pyogenes (Cbh-3X-FLAG-NLS-SpCas9-NLS)DepositorInserthSpCas9
UseCRISPRTags3XFLAGExpressionMammalianPromoterCbhAvailable SinceOct. 29, 2013AvailabilityAcademic Institutions and Nonprofits only -
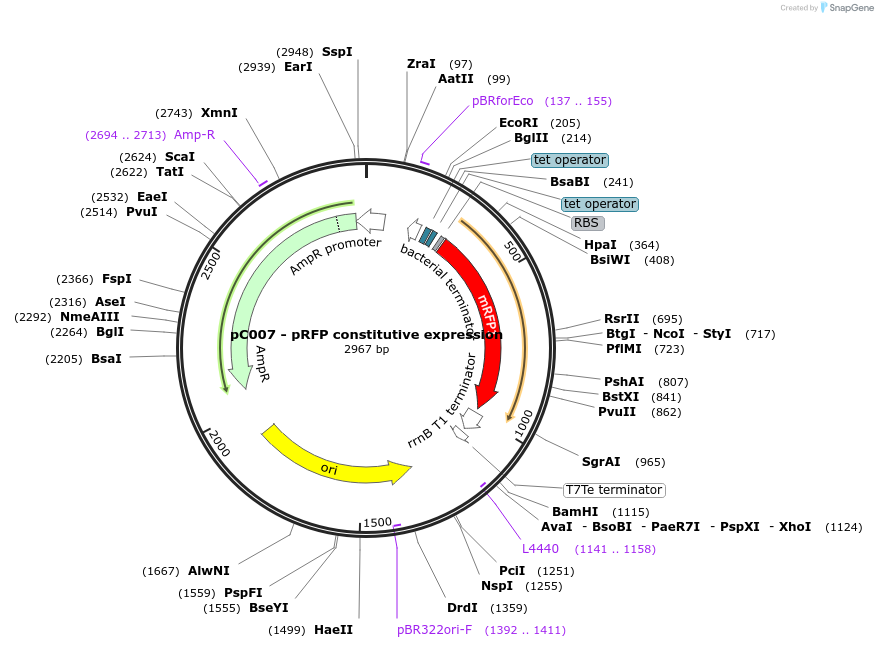
pC007 - pRFP constitutive expression
Plasmid#79156PurposeRFP constitutively expressedDepositorInsertRFP
Available SinceJune 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
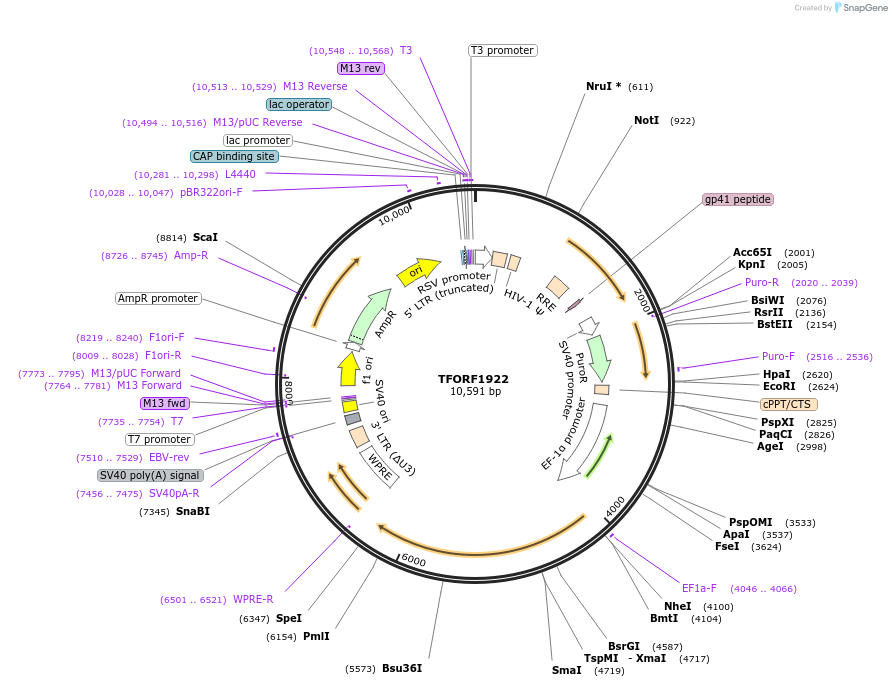
TFORF1922
Plasmid#141888PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
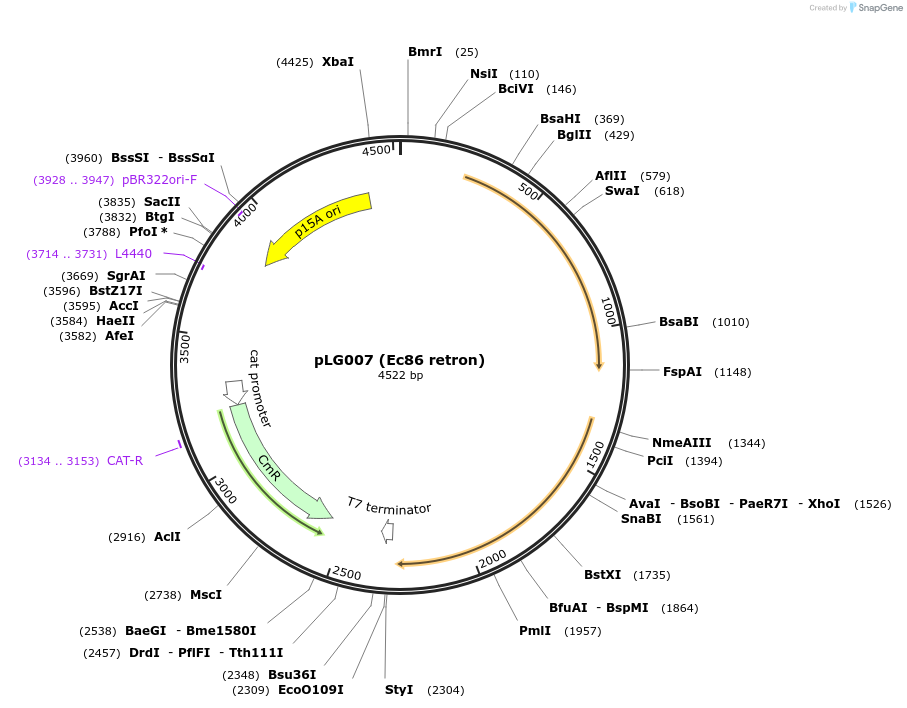
pLG007 (Ec86 retron)
Plasmid#157885PurposeExpresses the Ec86 retron defense systemDepositorInsertEc86 retron
ExpressionBacterialAvailable SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
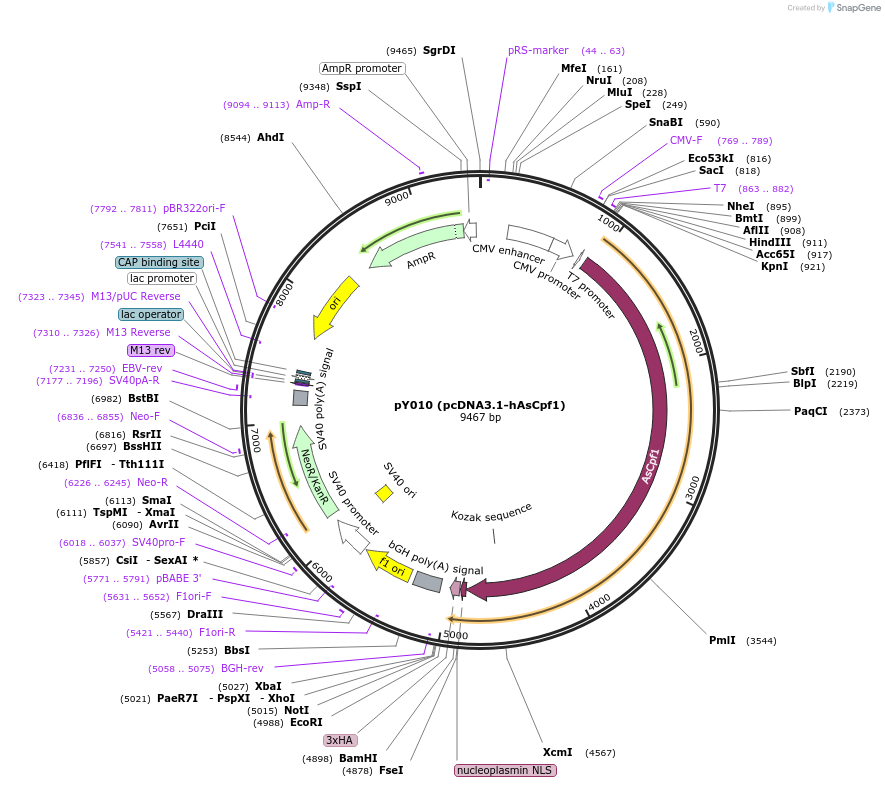
pY010 (pcDNA3.1-hAsCpf1)
Plasmid#69982PurposeExpresses humanized AsCpf1DepositorInserthAsCpf1
UseCRISPRTagsNLS-3xHAExpressionMammalianPromoterCMVAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
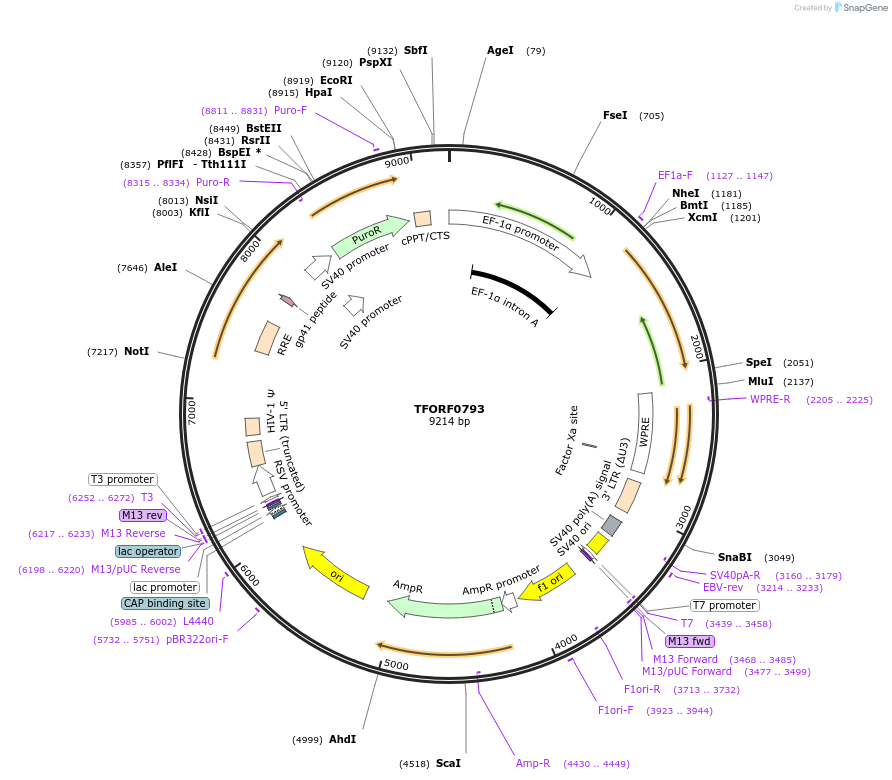
TFORF0793
Plasmid#142692PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceSept. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
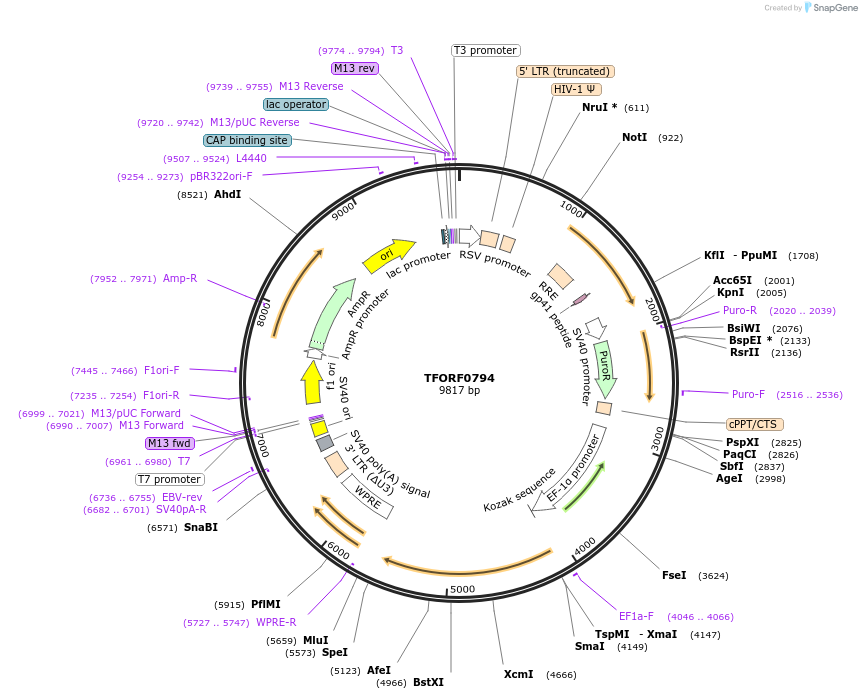
TFORF0794
Plasmid#142239PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
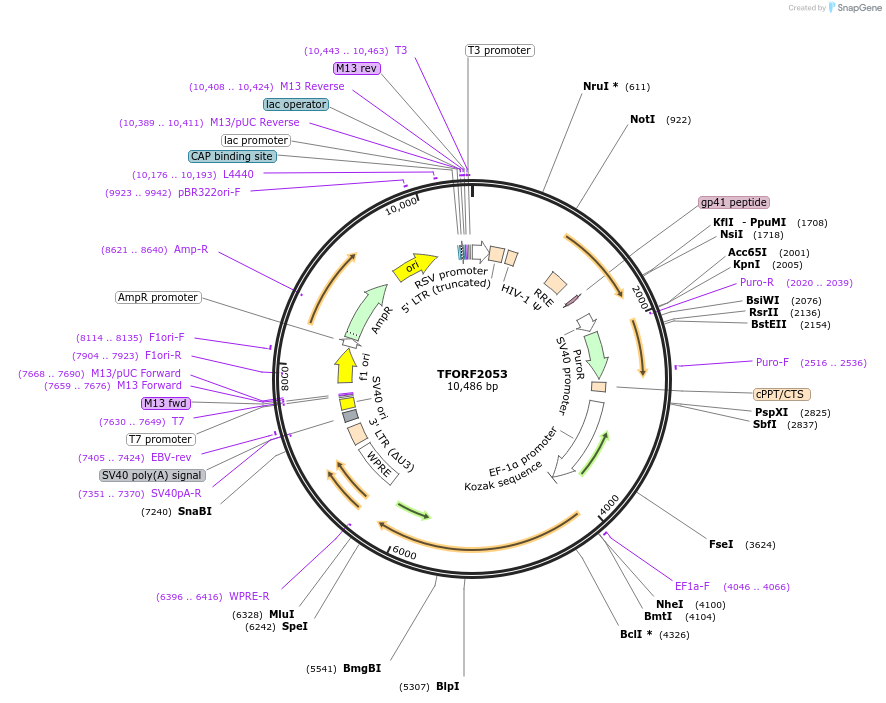
TFORF2053
Plasmid#142912PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceApril 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
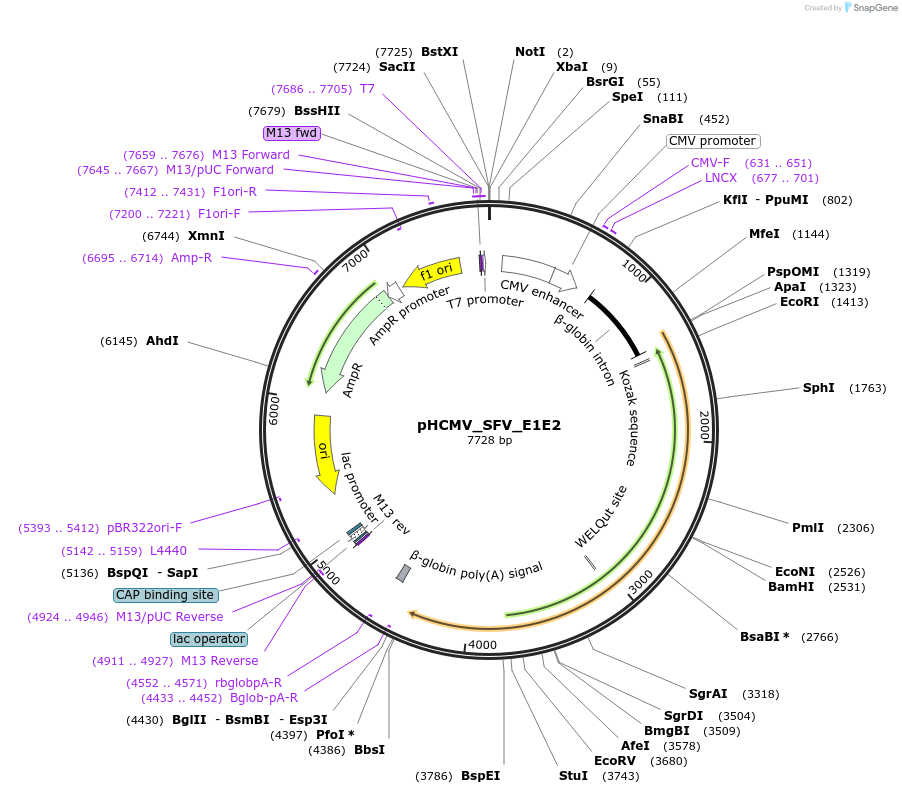
pHCMV_SFV_E1E2
Plasmid#207308PurposeMammalian expression of Semliki Forest virus E1E2 proteinDepositorInsertSFV_E1E2
ExpressionMammalianMutationmammalian codon optimizationPromoterCMV promoterAvailable SinceNov. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
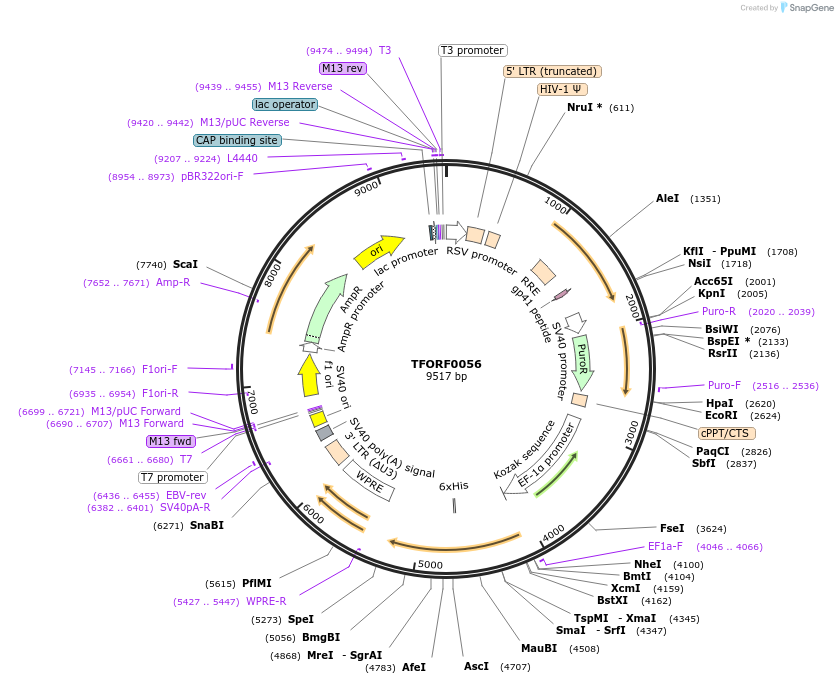
TFORF0056
Plasmid#142529PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 8, 2023AvailabilityAcademic Institutions and Nonprofits only