We narrowed to 19,393 results for: IRE
-
Plasmid#129000PurposepREV1-mRUBY2 cassette plasmid flanked by ConL2 and ConREDepositorInsertConL2-pREV1-mRUBY2-tADH1-ConRE AmpR-ColE1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
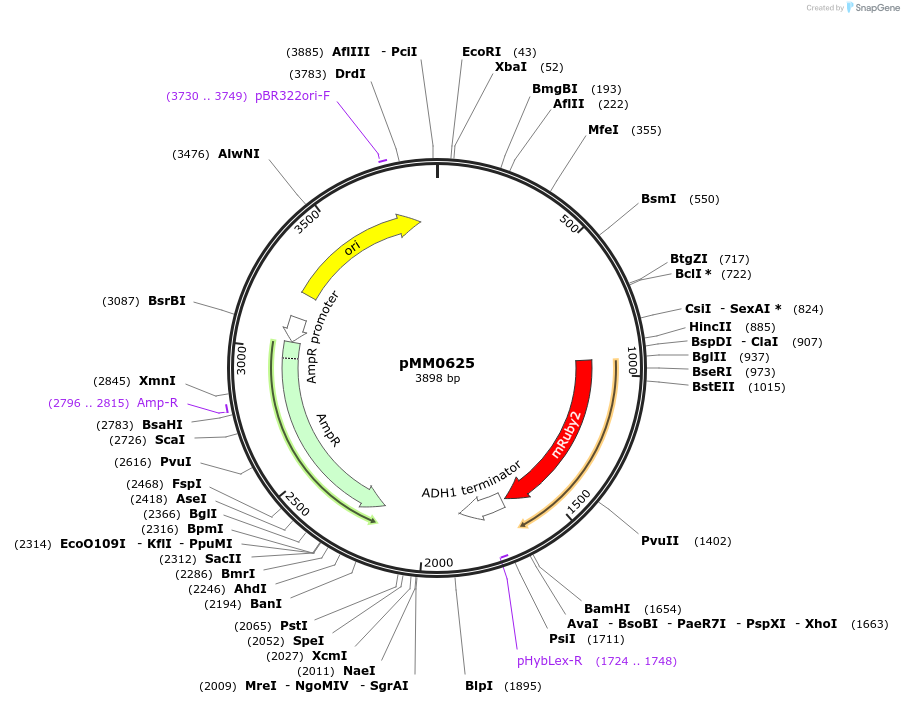
pMM0557
Plasmid#128991PurposeLEU2 integration vector for Zif268DBD-CRY2DepositorInsertUra3 3' homology-pTDH3-SV40NLS-ZiF268-CRY2-tADH1-URA3-Ura3 5' homology
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
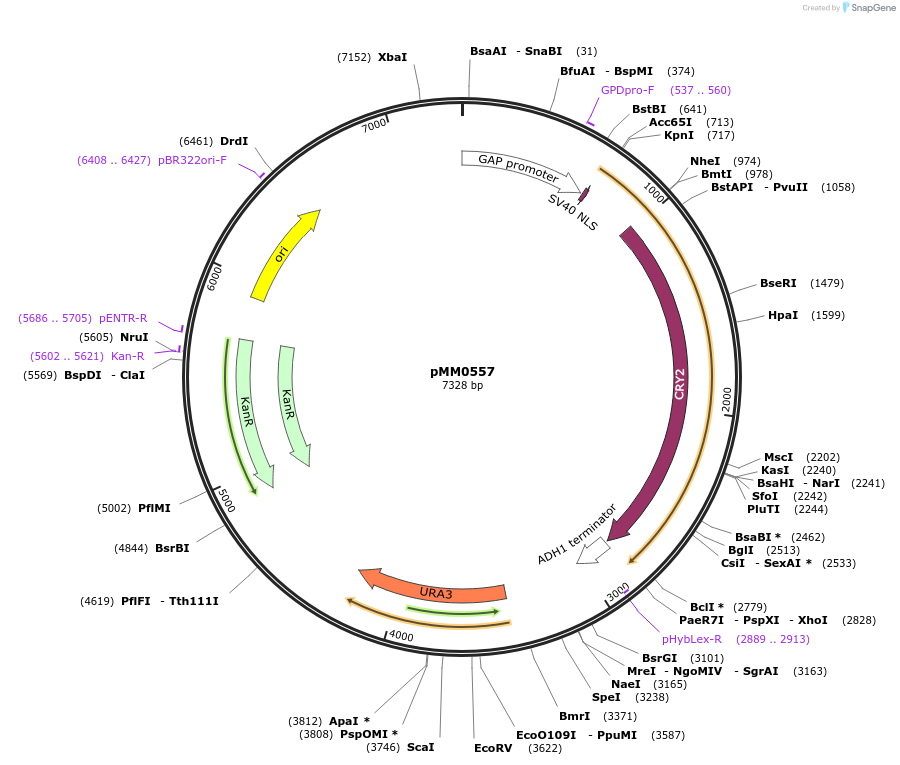
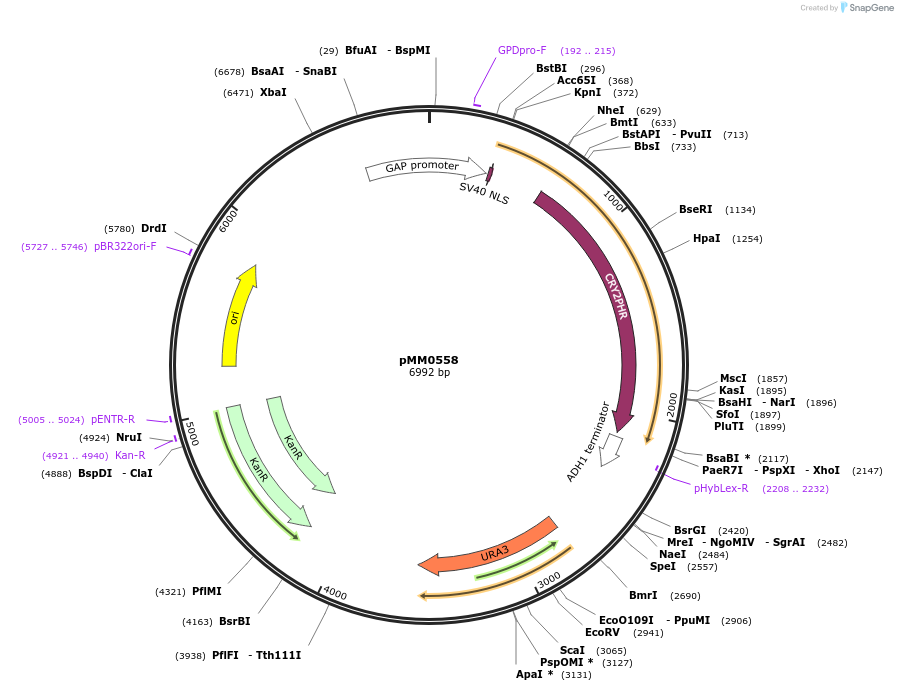
pMM0558
Plasmid#128992PurposeLEU2 integration vector for Zif268DBD-CRY2PHRDepositorInsertUra3 3' homology-pTDH3-SV40NLS-ZiF268-CRY2PHR-tADH1-URA3-Ura3 5' homology
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
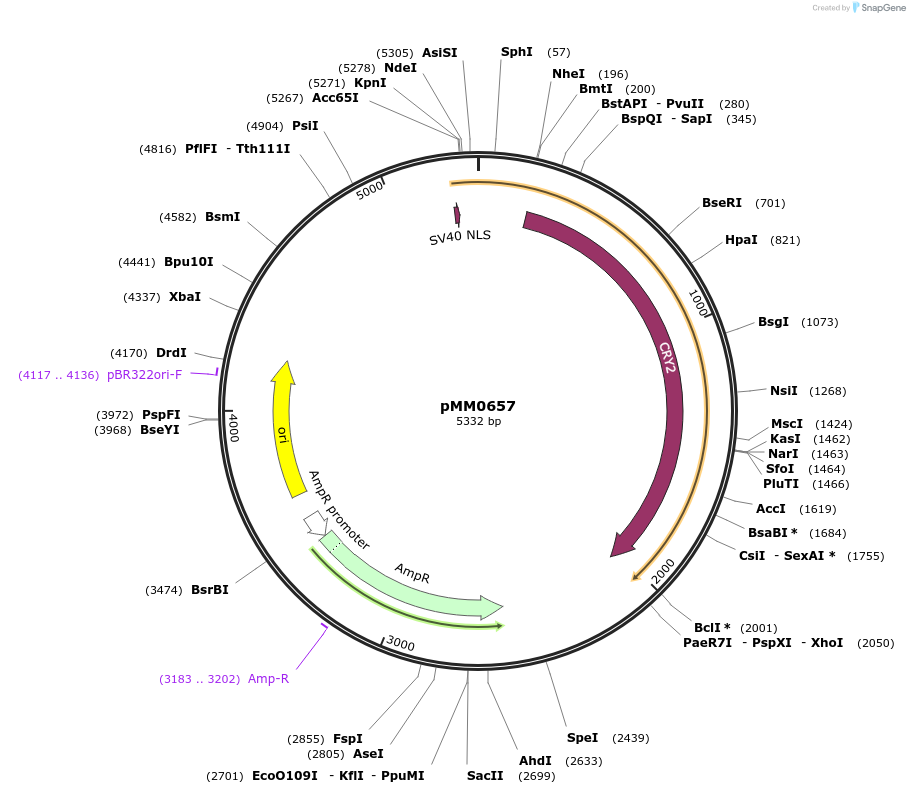
pMM0657
Plasmid#129018PurposepRPL18B-Zif268DBD-CRY2 cassette plasmid flanked by ConL1 and ConR2DepositorInsertConL1-pRPL18B-SV40NLS-ZiF268DBD-CRY2-tSSA1-ConR2-ColE1-AmpR
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
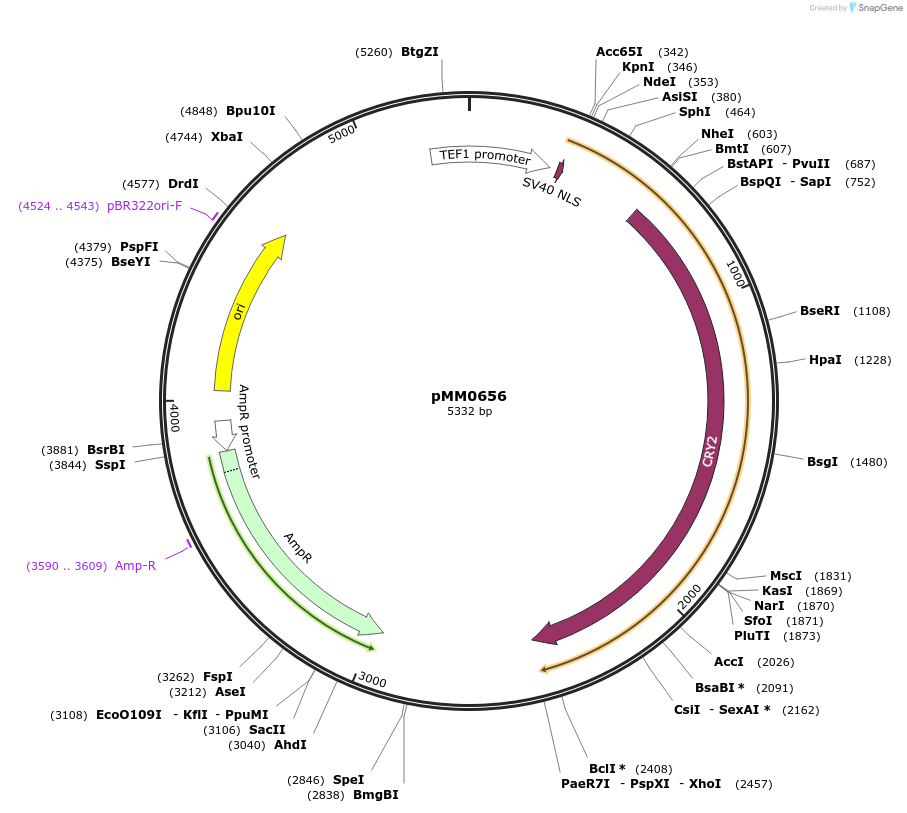
pMM0656
Plasmid#129017PurposepTEF1-Zif268DBD-CRY2 cassette plasmid flanked by ConL1 and ConR2DepositorInsertConL1-pTEF1-SV40NLS-ZiF268DBD-CRY2-tSSA1-ConR2-ColE1-AmpR
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
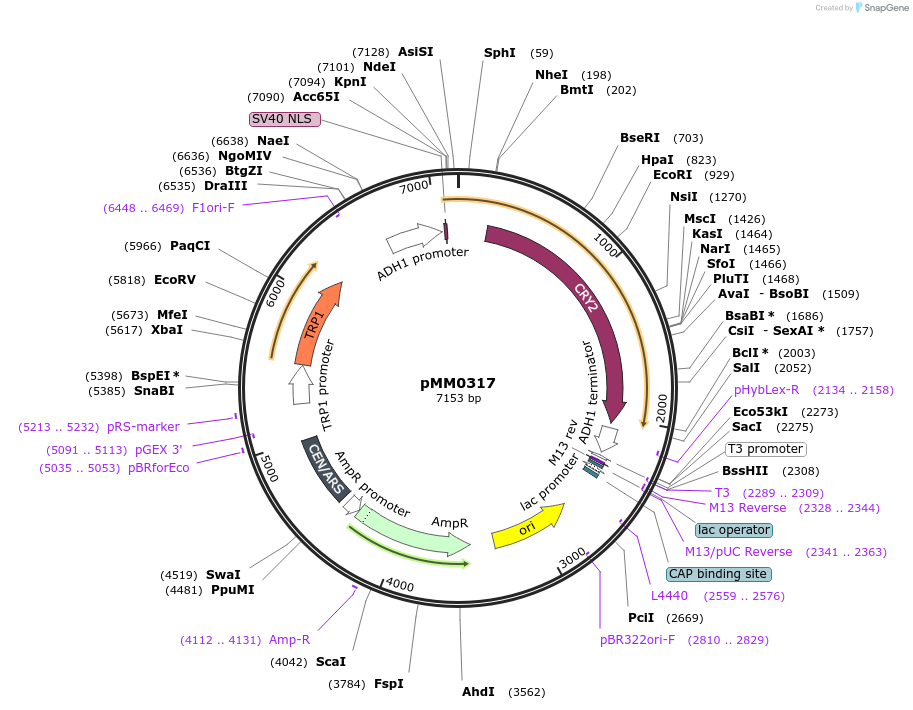
pMM0317
Plasmid#128979PurposeEncodes Zif268DBD-CRY2 under pscADH1DepositorInsertpscADH-SV40NLS-Zif268DBD-CRY2 (L3)-tscADH1 scTRP1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
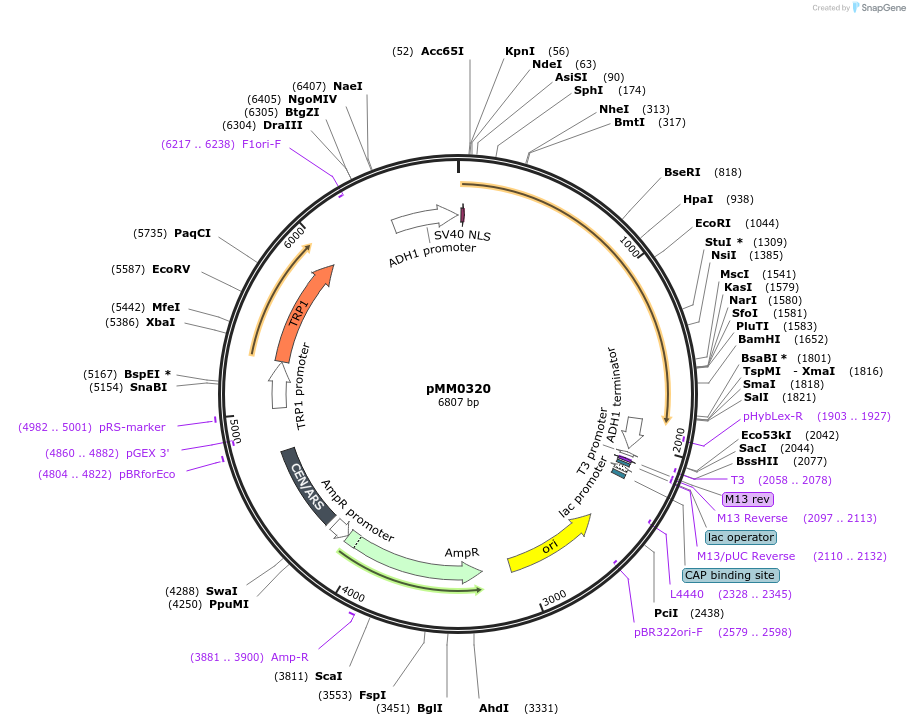
pMM0320
Plasmid#128980PurposeEncodes Zif268DBD-CRY2PHR under pscADH1DepositorInsertpscADH1-SV40NLS-ZIF268DBD-CRY2PHR (L3)-tscADH1 TRP1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
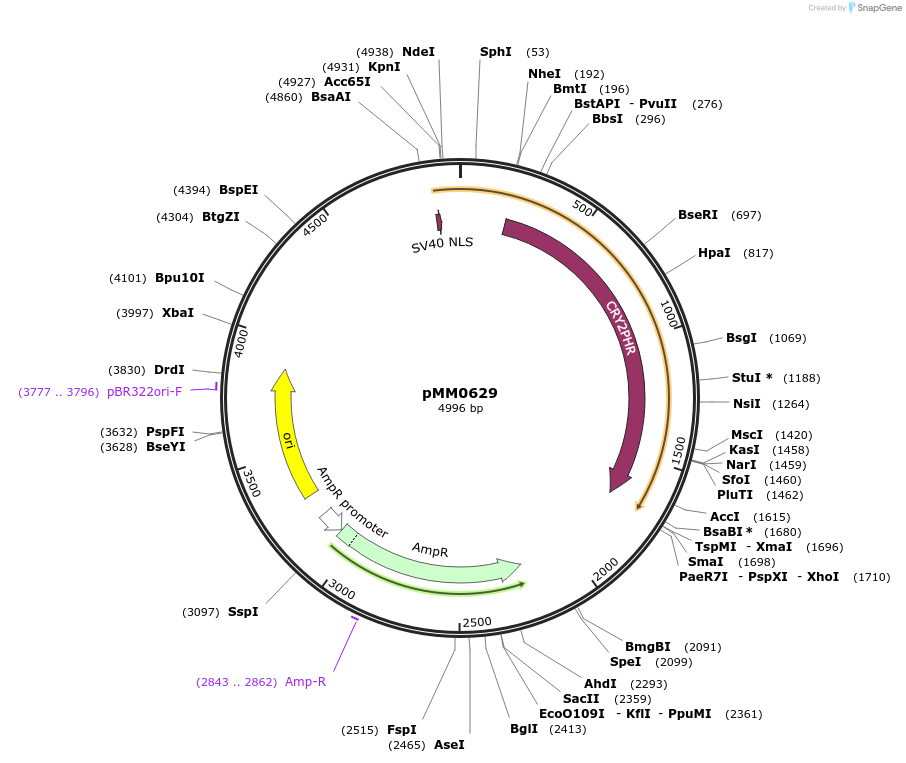
pMM0629
Plasmid#129004PurposepRNR2-Zif268DBD-CRY2PHR cassette plasmid flanked by ConL1 and ConR2DepositorInsertConL1-pRNR2-SV40NLS-ZiF268DBD-CRY2PHR-tSSA1-ConR2 AmpR-ColE1
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
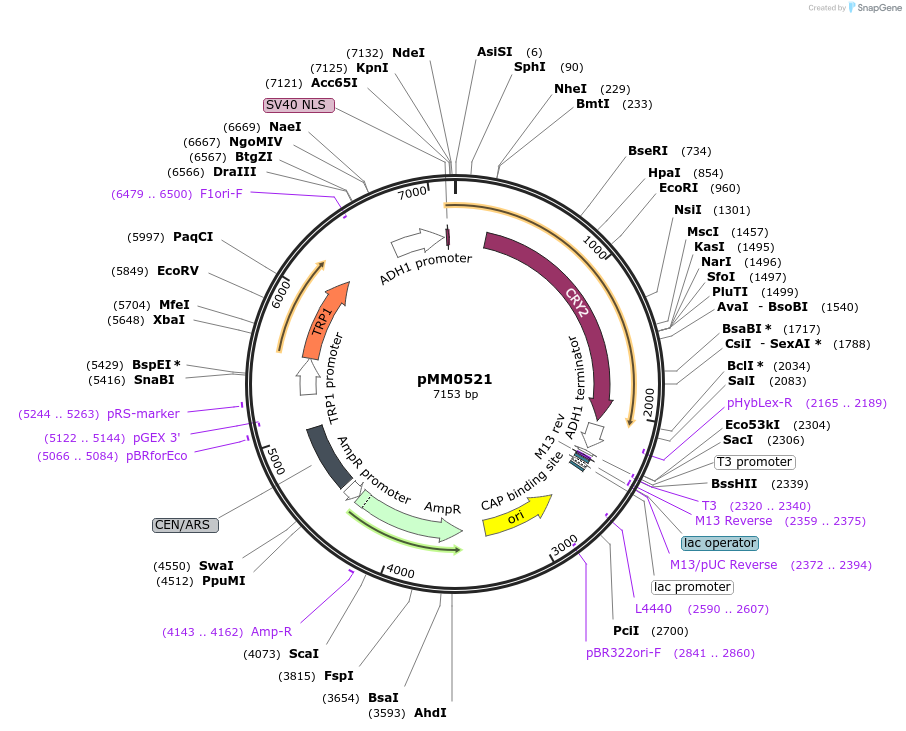
pMM0521
Plasmid#128984PurposeExpression plasmid for ZIF268DBD-L3-CRY2DepositorInsertSV40NLS-Zif268DBD-CRY2 (L3) scTRP1
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
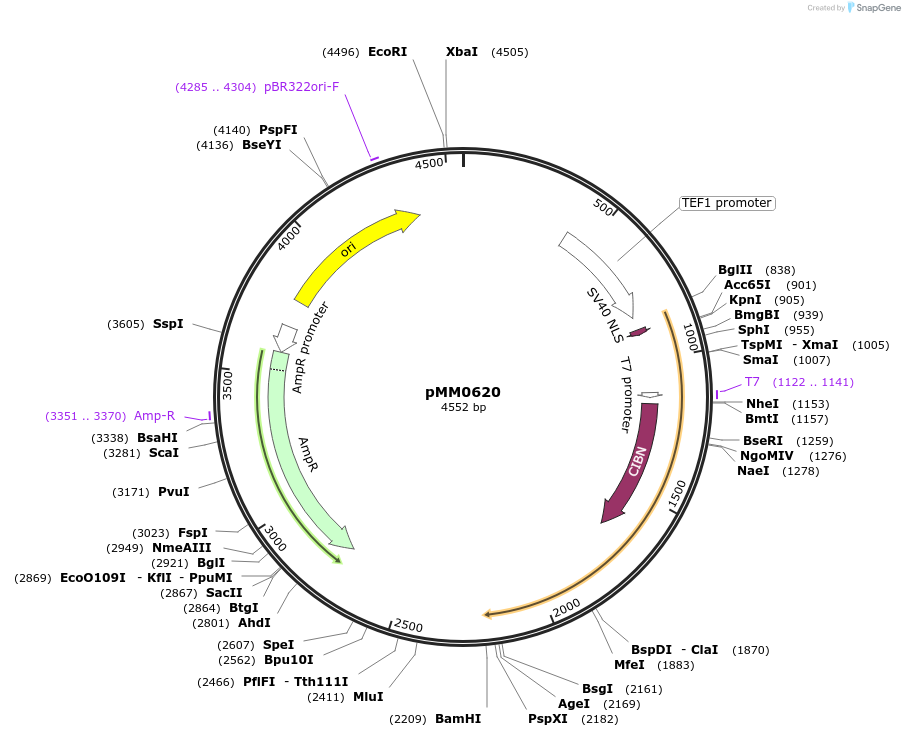
pMM0620
Plasmid#128995PurposepTEF1-VP16-CIB1 cassette plasmid flanked by ConLS and ConR1DepositorInsertConLS_pTEF1-SV40NLS-VP16-CIB1-tENO1-ConR1 AmpR Col-E1
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
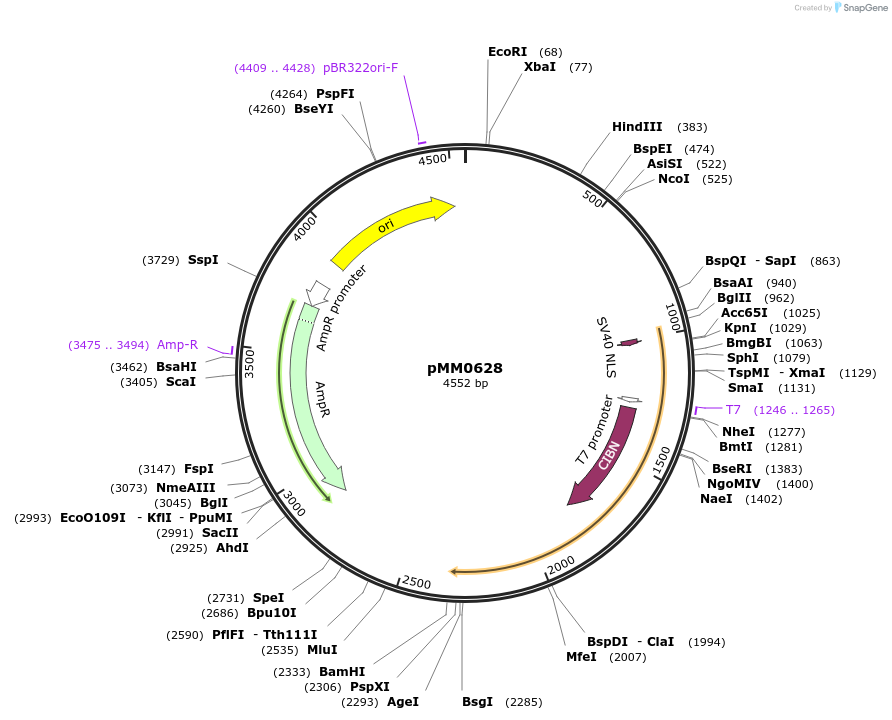
pMM0628
Plasmid#129003PurposepRNR2-VP16-CIB1 cassette plasmid flanked by ConLS and ConR1DepositorInsertConLS_pRNR2-SV40NLS-VP16-CIB1-tENO1-ConR1 AmpR Col-E1
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
pCMV-myc-Rem2 RNAiR2 short
Plasmid#51596Purposeexpresses RNAiR myc-tagged rat Rem2 (short version, missing 69 AA at N terminus)DepositorInsertRem2 (Rem2 Rat)
TagsmycExpressionMammalianMutationsilent mutations at shRNA 1 and 2 recognition sit…PromoterCMVAvailable SinceApril 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
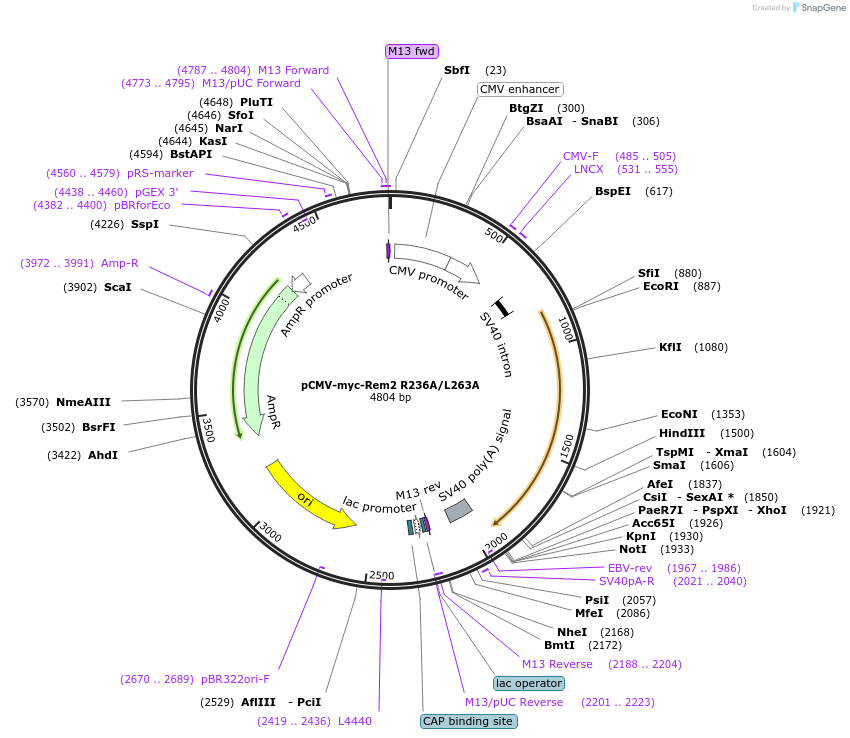
pCMV-myc-Rem2 R236A/L263A
Plasmid#51589Purposeexpresses RNAiR myc-tagged Rem2 non-Ca channel interactingDepositorInsertRem2 (Rem2 Mouse)
TagsmycExpressionMammalianMutationR236A and L263A; silent mutations at shRNA 1 and …PromoterCMVAvailable SinceApril 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
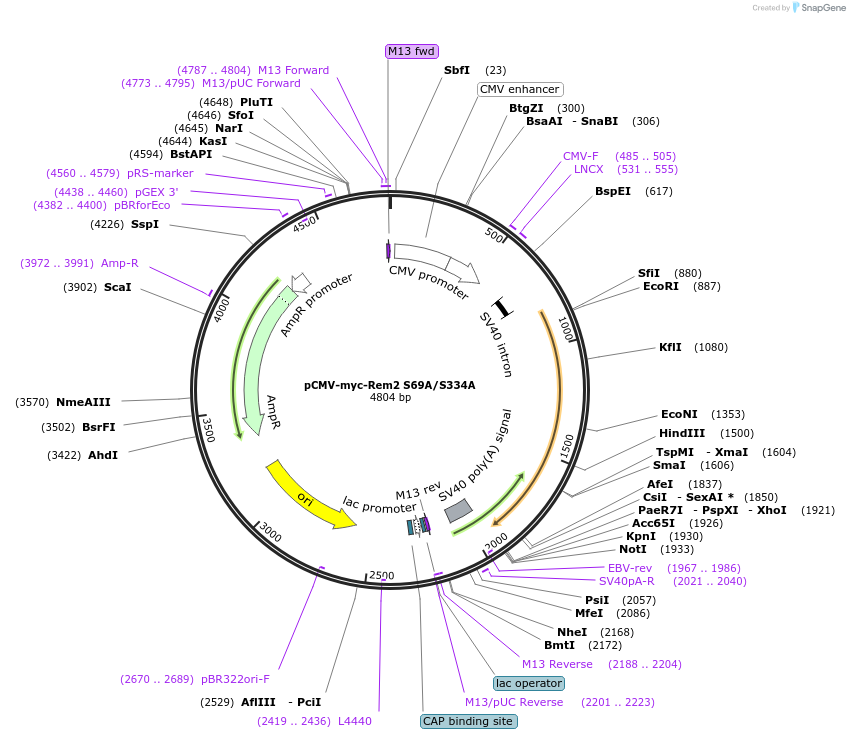
pCMV-myc-Rem2 S69A/S334A
Plasmid#51588Purposeexpresses RNAiR myc-tagged Rem2 non-14-3-3 interactingDepositorInsertRem2 (Rem2 Mouse)
TagsmycExpressionMammalianMutationS69A and S334A; silent mutations at shRNA 1 and 2…PromoterCMVAvailable SinceApril 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
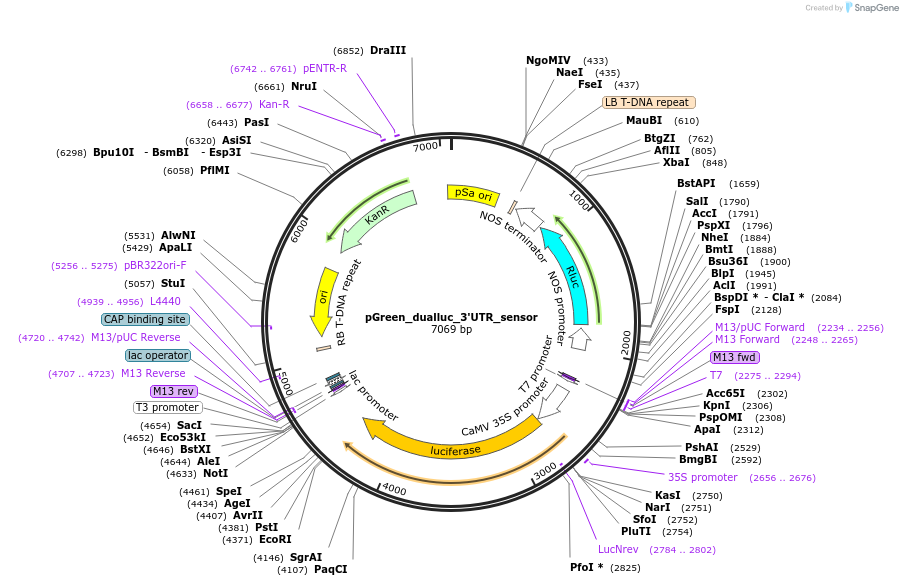
pGreen_dualluc_3'UTR_sensor
Plasmid#55206PurposeDual-luciferase reporter for microRNA targeting in plantsDepositorInsertsRenilla luciferase
Firefly luciferase
UseLuciferase; Plant transient expressionAvailable SinceJuly 15, 2014AvailabilityAcademic Institutions and Nonprofits only -
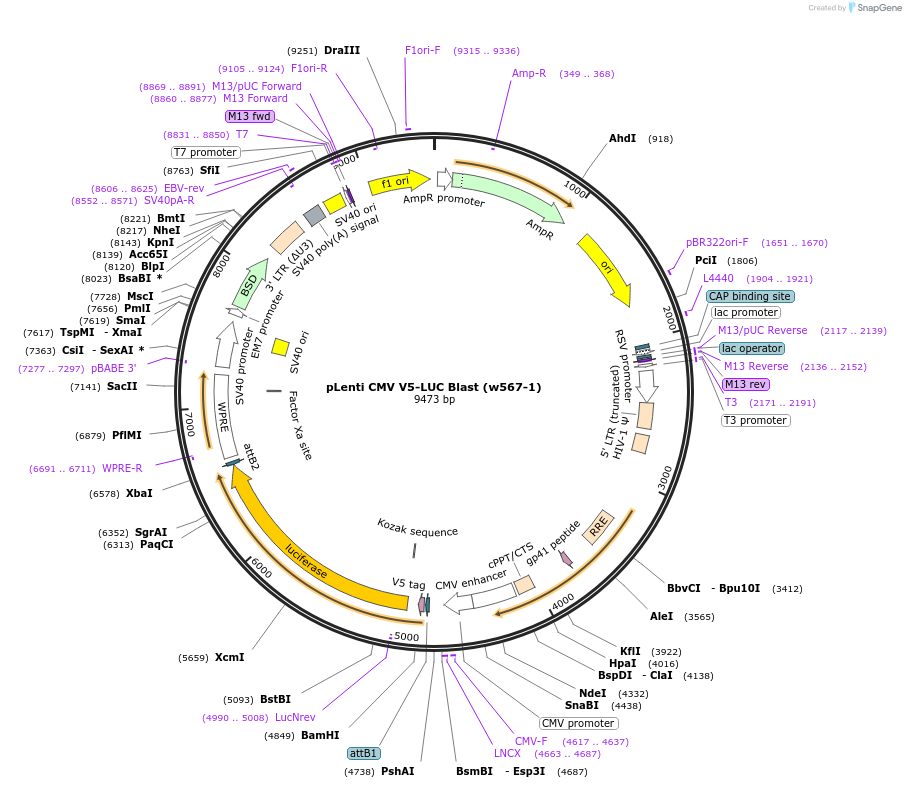
pLenti CMV V5-LUC Blast (w567-1)
Plasmid#21474Purpose3rd gen lentiviral V5-Luciferase expression vector, CMV promoter, BlastDepositorInsertV5-LUC
UseLentiviral and LuciferaseTagsV5ExpressionMammalianAvailable SinceJuly 22, 2009AvailabilityAcademic Institutions and Nonprofits only -
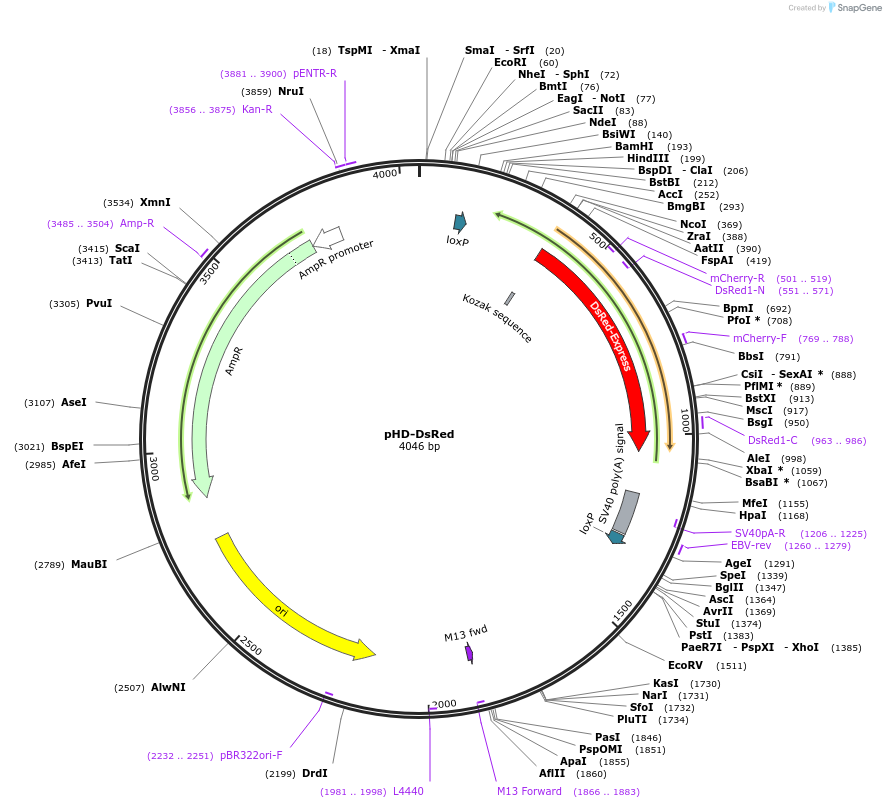
pHD-DsRed
Plasmid#51434PurposeVector for generating dsDNA donors for homology-directed repair. Contains the visible marker 3xP3-DsRed.DepositorInsertLoxP-3xP3-DsRed-LoxP
Promoter3xP3Available SinceFeb. 27, 2014AvailabilityAcademic Institutions and Nonprofits only -
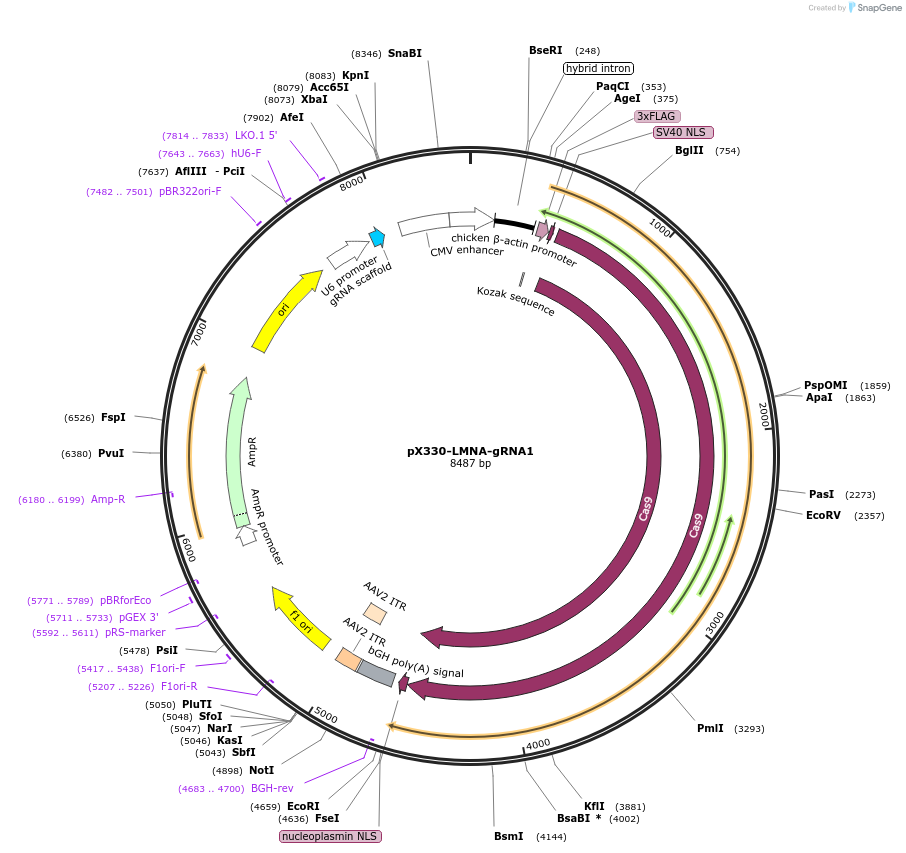
pX330-LMNA-gRNA1
Plasmid#122507Purposeexpresses WT spCas9 and a chimeric gRNA targeting human LMNADepositorInsertLMNA-gRNA1
UseCRISPRExpressionMammalianPromoterU6Available SinceApril 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
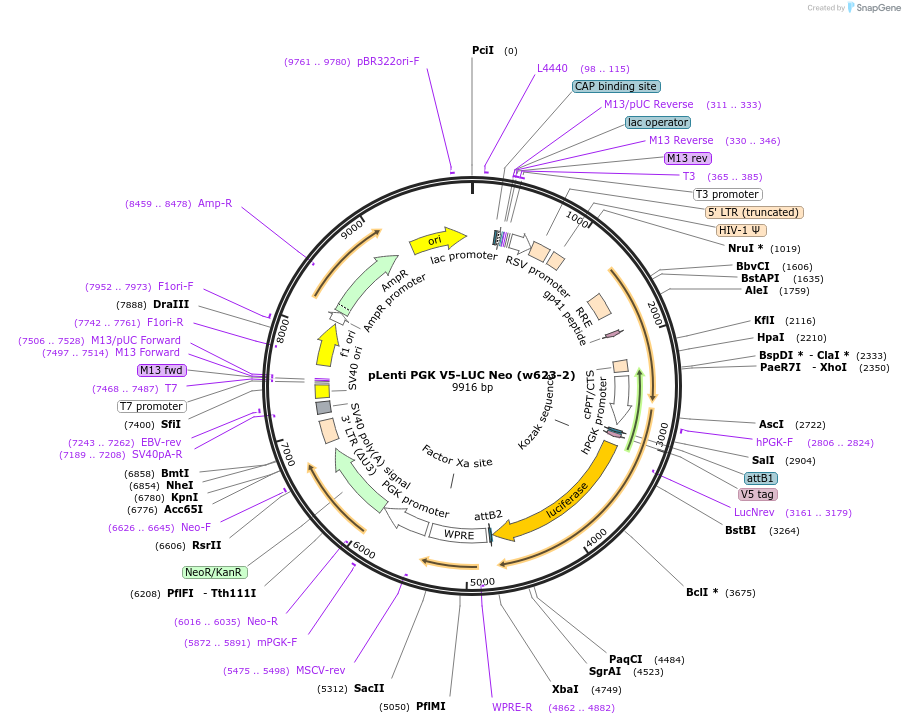
pLenti PGK V5-LUC Neo (w623-2)
Plasmid#21471Purpose3rd gen lentiviral V5-Luciferase expression vector, PGK promoter, NeoDepositorInsertV5-LUC
UseLentiviral and LuciferaseTagsV5ExpressionMammalianAvailable SinceJuly 22, 2009AvailabilityAcademic Institutions and Nonprofits only -
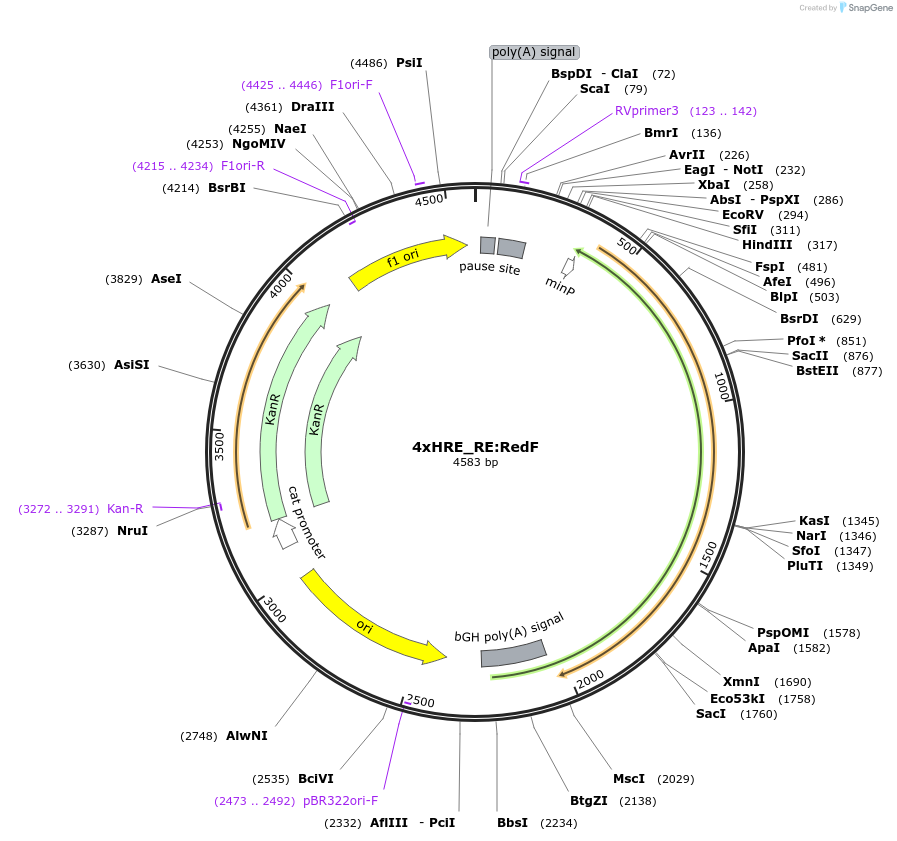
4xHRE_RE:RedF
Plasmid#178314PurposeHRE RedFirefly luciferase reporterDepositorInsertHRE RedFirefly reporter
UseLuciferase and Synthetic BiologyExpressionMammalianAvailable SinceJuly 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
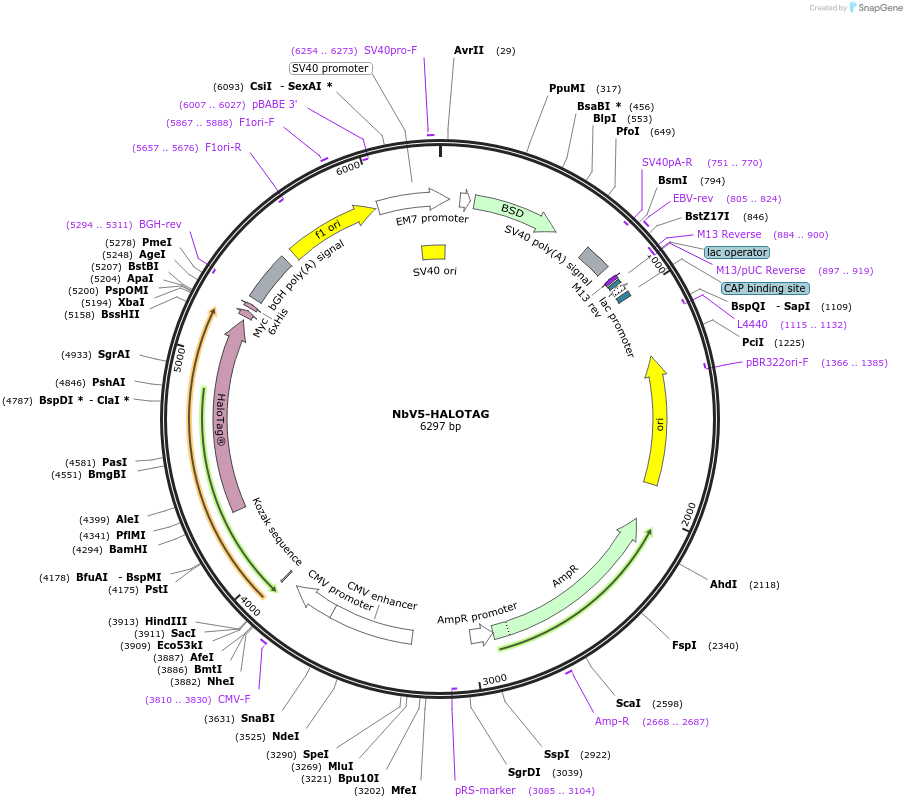
NbV5-HALOTAG
Plasmid#201482PurposeIndirect detection of V5-tagged protein using the HaloTag protein labeling technology.DepositorInsertNbV5-HALOTAG
TagsHaloTagExpressionMammalianPromoterCMVAvailable SinceAug. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
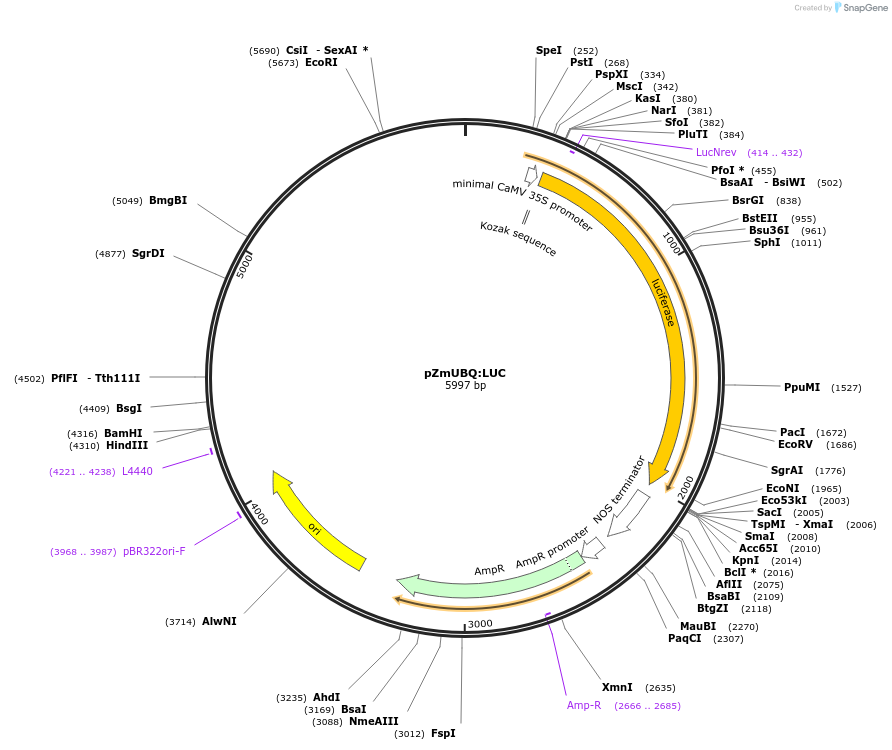
pZmUBQ:LUC
Plasmid#131860PurposeTransient expression of firefly luciferase from the Zea mays ubiquitin promotorDepositorInsertluciferase
ExpressionPlantPromoterZmUBQAvailable SinceOct. 24, 2019AvailabilityAcademic Institutions and Nonprofits only -
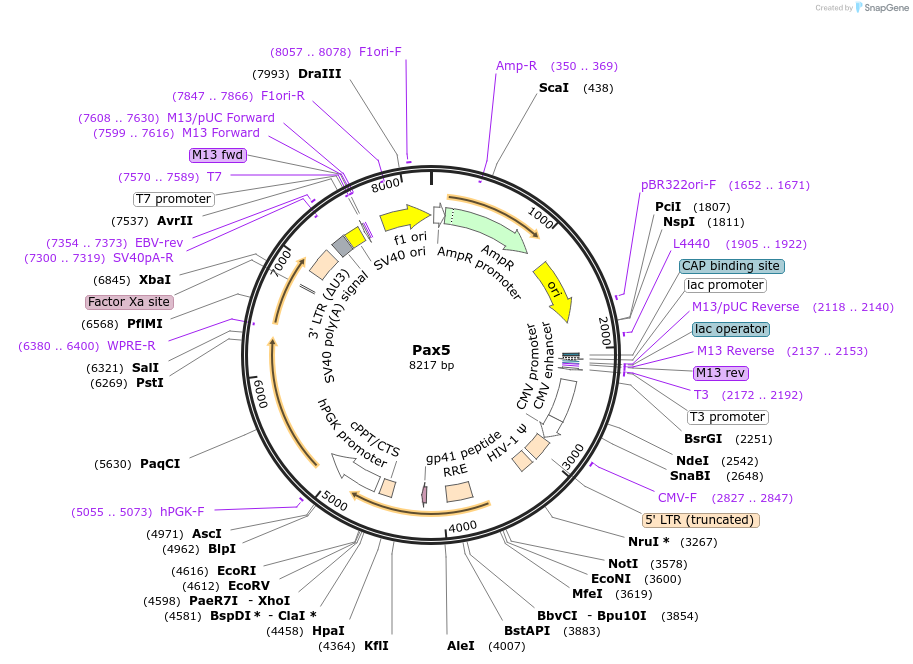
Pax5
Plasmid#35003DepositorAvailable SinceMarch 6, 2012AvailabilityAcademic Institutions and Nonprofits only -
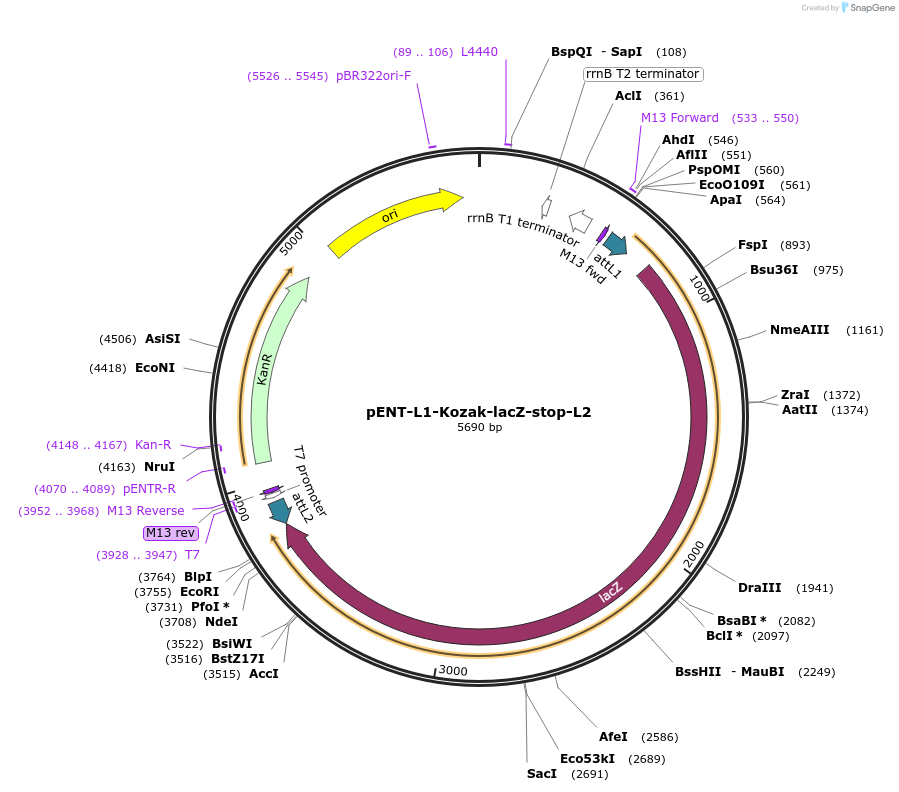
pENT-L1-Kozak-lacZ-stop-L2
Plasmid#186359PurposeEntry clone with ORF encoding lacZ flanked by Gateway recombination sequences.DepositorInsertlacZ (lacZ Synthetic, E. coli)
UseExpression of lacz reporterAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
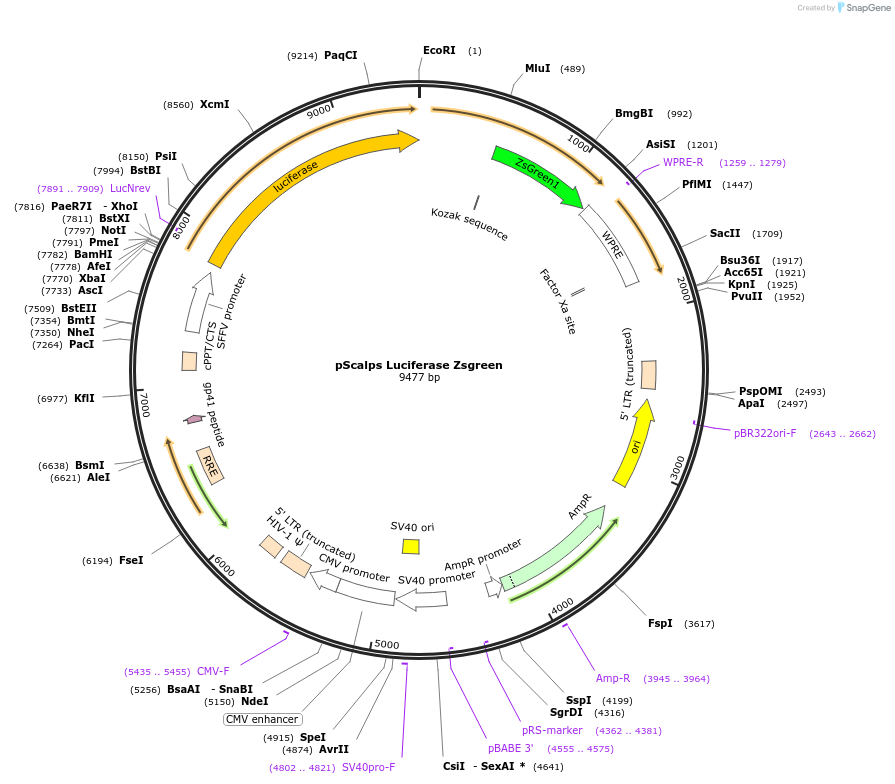
pScalps Luciferase Zsgreen
Plasmid#184784PurposeLentiviral vector with luciferase gene under SFFV promoter and Zsgreen under Cyclophilin promoterDepositorInsertFirefly Luciferase and Zsgreen
UseLentiviral and LuciferasePromoterSFFVAvailable SinceOct. 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
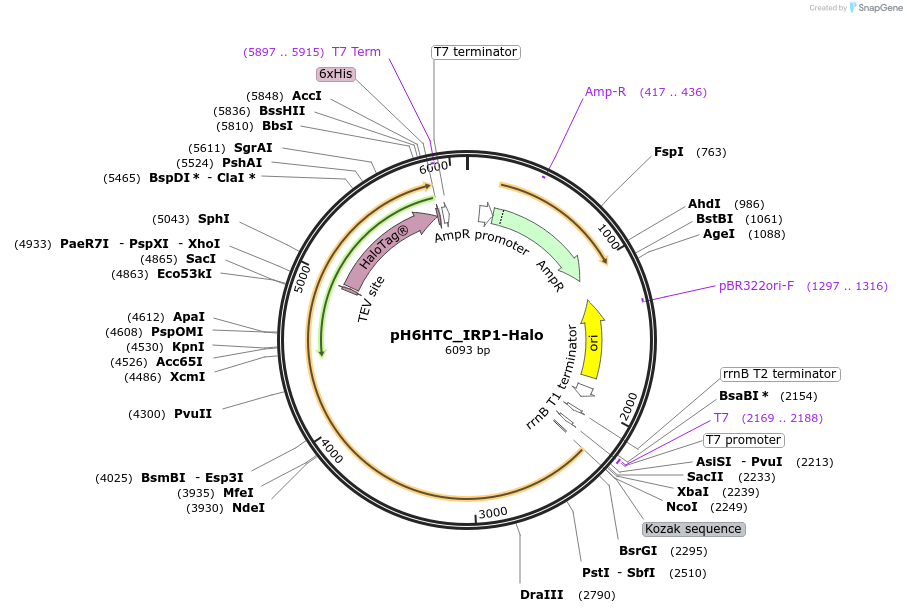
pH6HTC_IRP1-Halo
Plasmid#175305PurposeProduction of HaloTag fusion proteinDepositorAvailable SinceOct. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
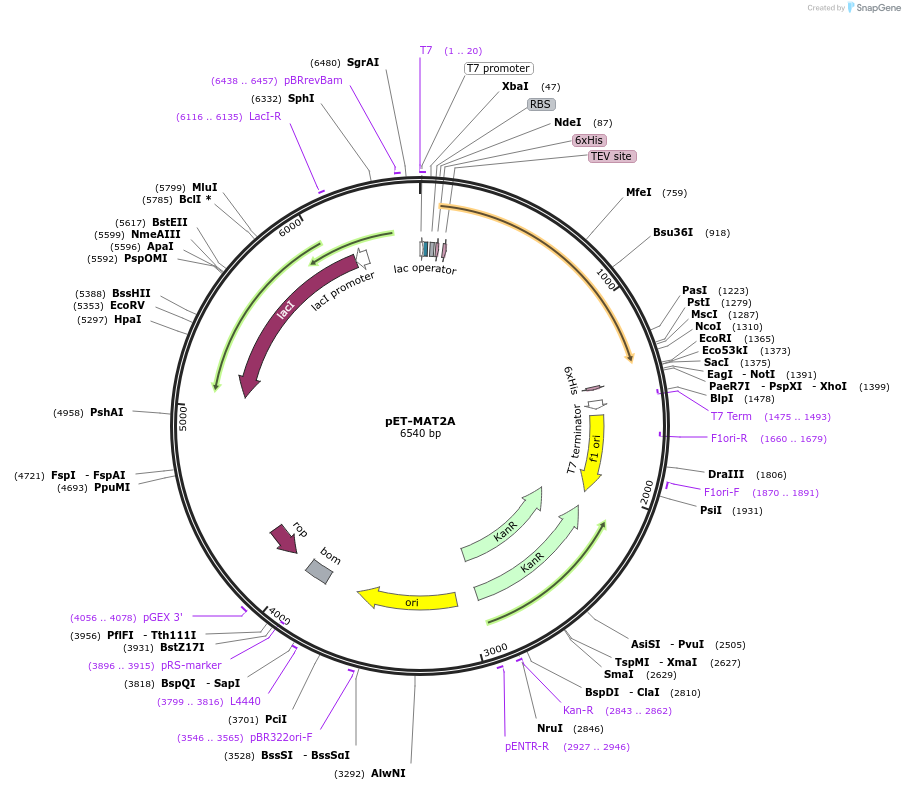
pET-MAT2A
Plasmid#164820PurposeExpresses MAT2A in E. coli BL21(DE3)DepositorAvailable SinceFeb. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
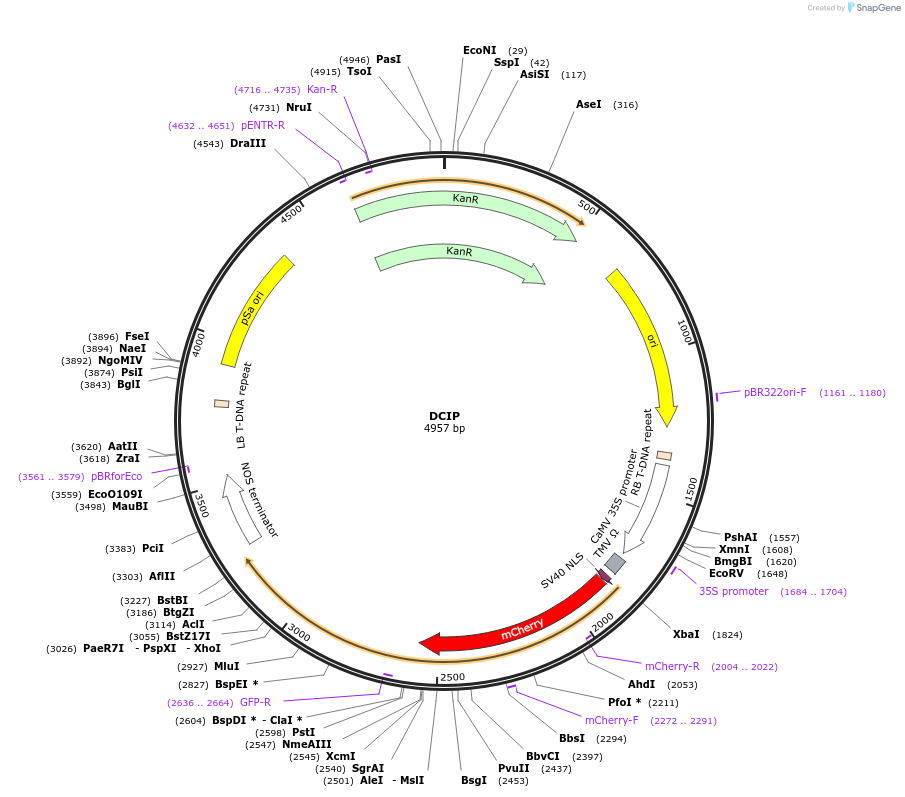
DCIP
Plasmid#193860PurposeEncodes SV40-mCherry-GFP1-10 in pDGB1-aR backbone for plant expression. Requires pSOUP for replication in agrobacterium.DepositorInsertNLS-mCherry-GFP1-10
TagsSV40 NLSExpressionPlantPromoter35SAvailable SinceAug. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
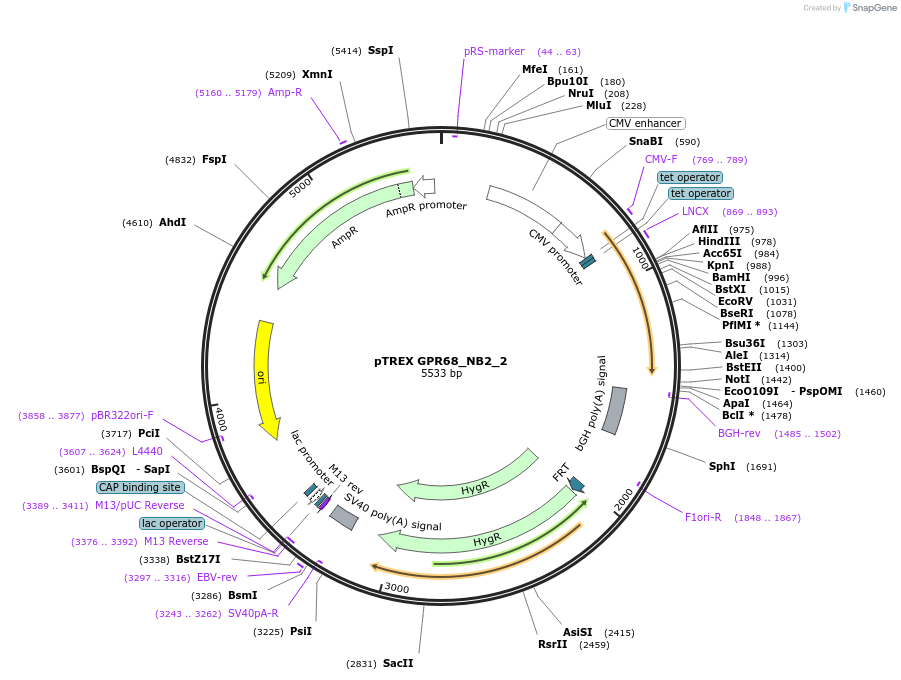
pTREX GPR68_NB2_2
Plasmid#127691PurposePlasmid contains a nanobody that directly interacts with the cytoplasmic surface of GPR68.DepositorInsertGPR68_NB2_2
ExpressionMammalianAvailable SinceJuly 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
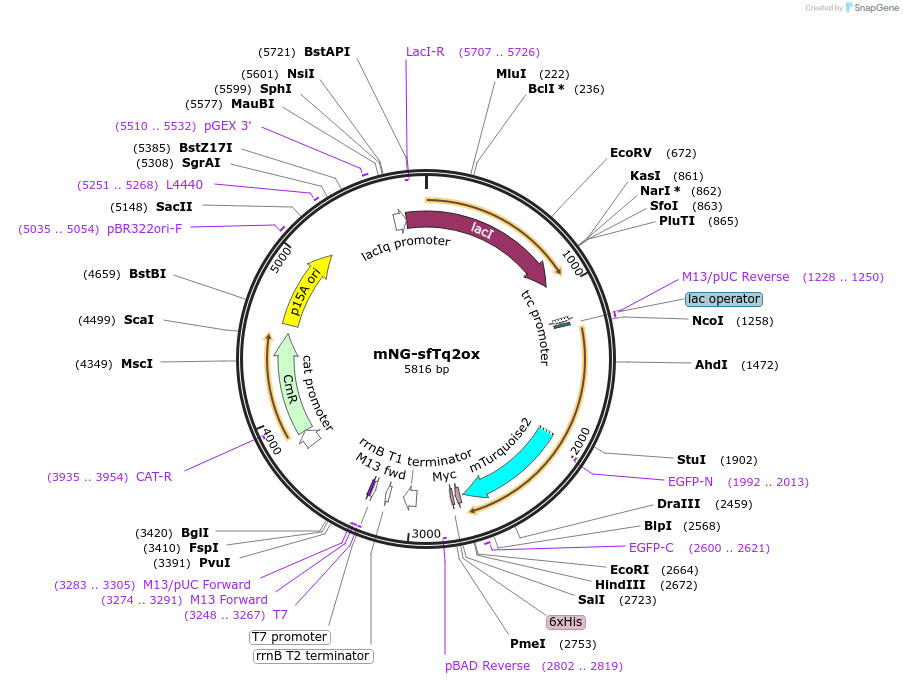
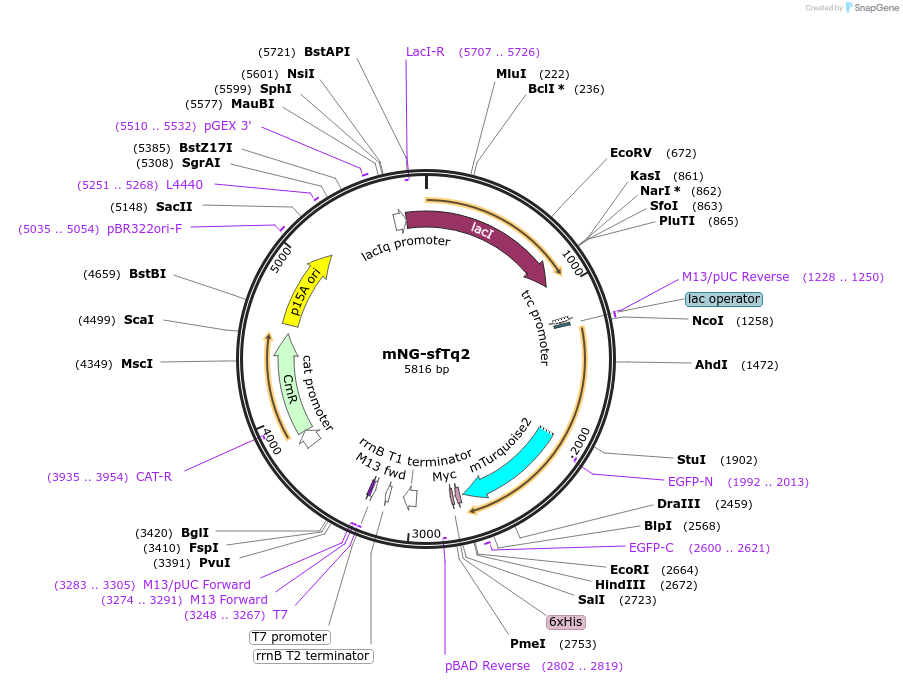
mNG-sfTq2ox
Plasmid#124219PurposeCytoplasmic mNeongreen superfolder mTurquoise2 ox tandem, ~60 % energy transfer. pNM045 - pSAV057-mNG-sfTq2oxDepositorInsertmNG-sfTq2ox
ExpressionMammalianAvailable SinceApril 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
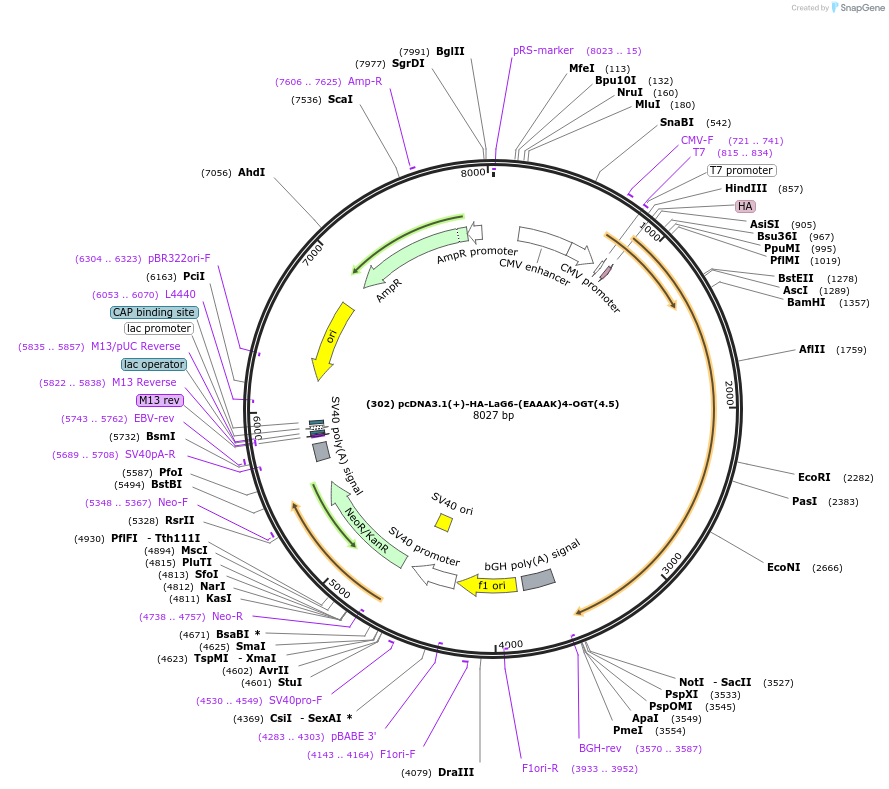
(302) pcDNA3.1(+)-HA-LaG6-(EAAAK)4-OGT(4.5)
Plasmid#168192PurposeHA-tagged GFP nanobody Lag6 (310 nM affinity) fused to a truncated OGT of 4 TPRsDepositorInsertHA-LaG6-(EAAAK)4-OGT(4)
TagsHA-TagExpressionMammalianPromoterT7/CMVAvailable SinceMay 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
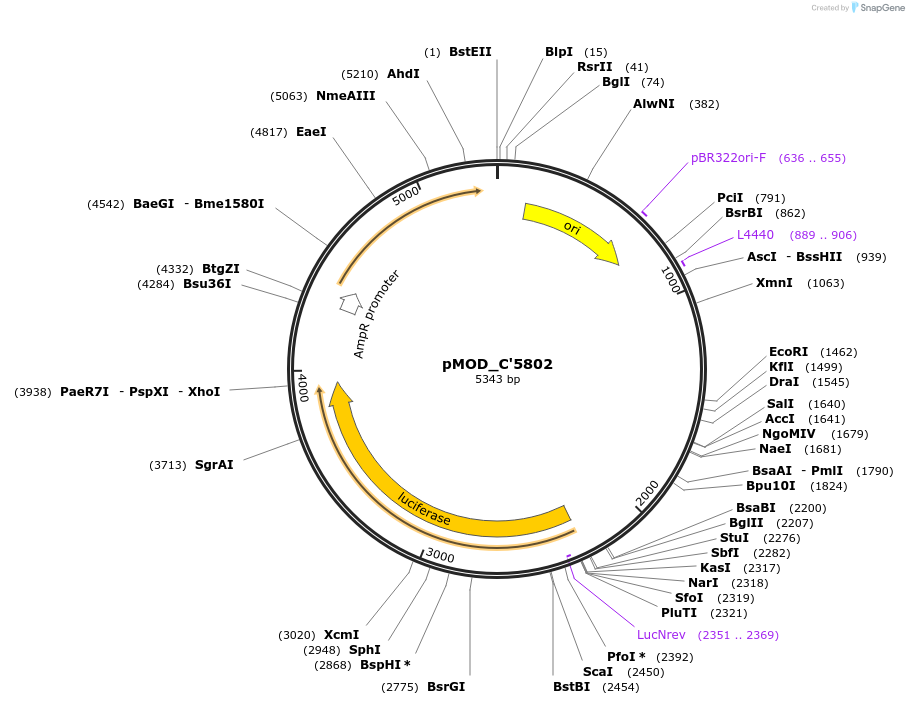
pMOD_C'5802
Plasmid#176879PurposeHigh constitutive expression of Firefly Luciferase in plantsDepositorInsertAtUbi10:Luc+:RbcsE9
UseLuciferaseAvailable SinceMay 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
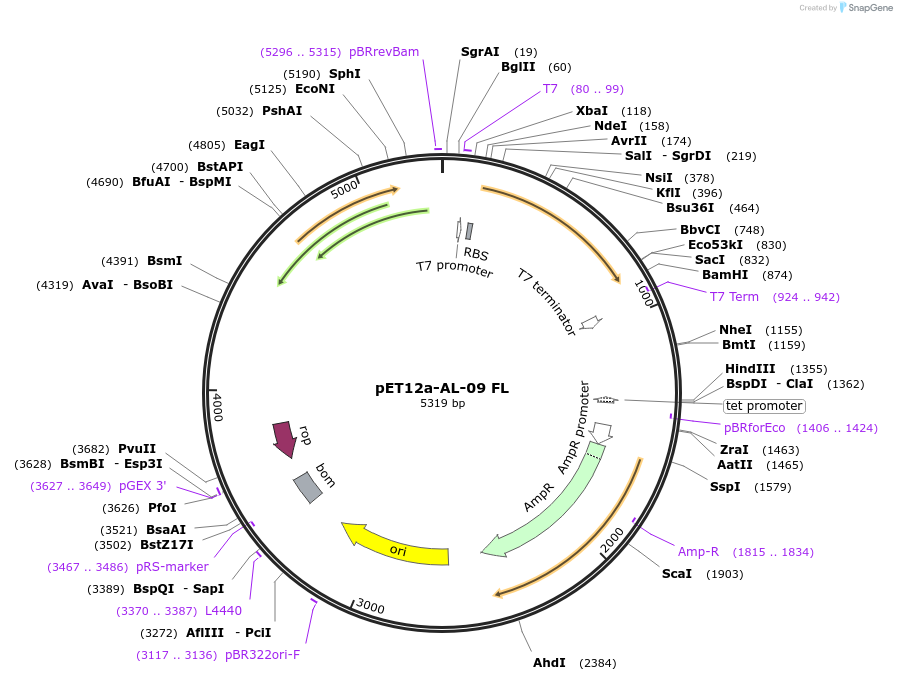
pET12a-AL-09 FL
Plasmid#47081DepositorAvailable SinceAug. 1, 2013AvailabilityAcademic Institutions and Nonprofits only -
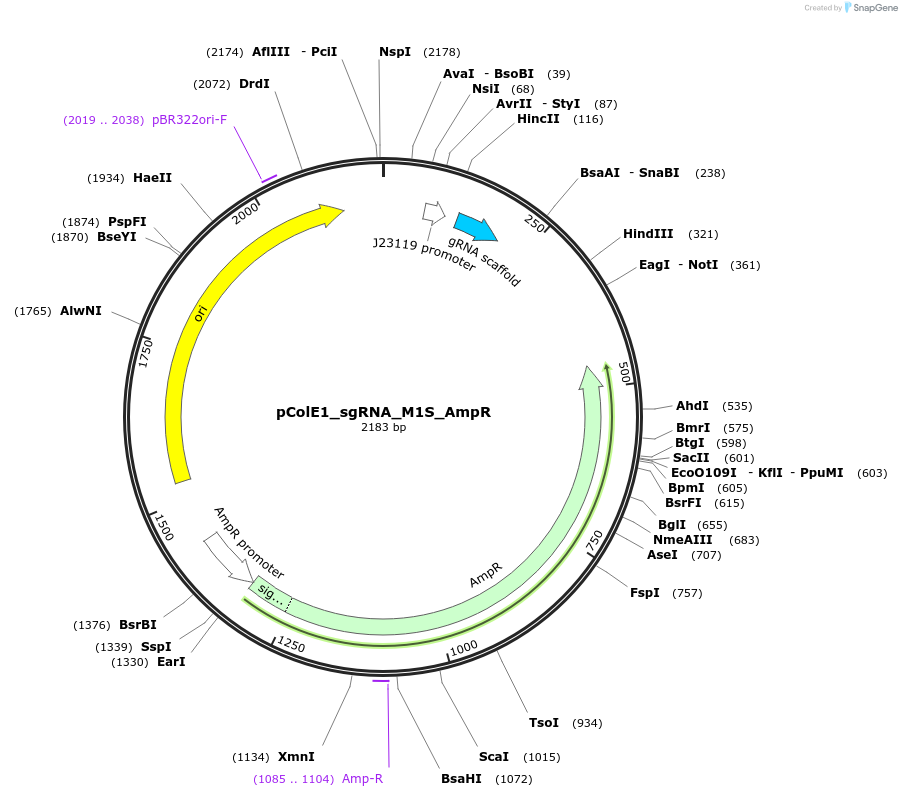
pColE1_sgRNA_M1S_AmpR
Plasmid#119153PurposeEncodes sgRNA targeting position 6 in pColE1_70a_deGFP_KanR with one mismatch in the seed regionDepositorInsertsgRNA targeting position 6 in pColE1_70a_deGFP_KanR, with one mismatch in the seed region
UseCrisprPromoterJ23119Available SinceDec. 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
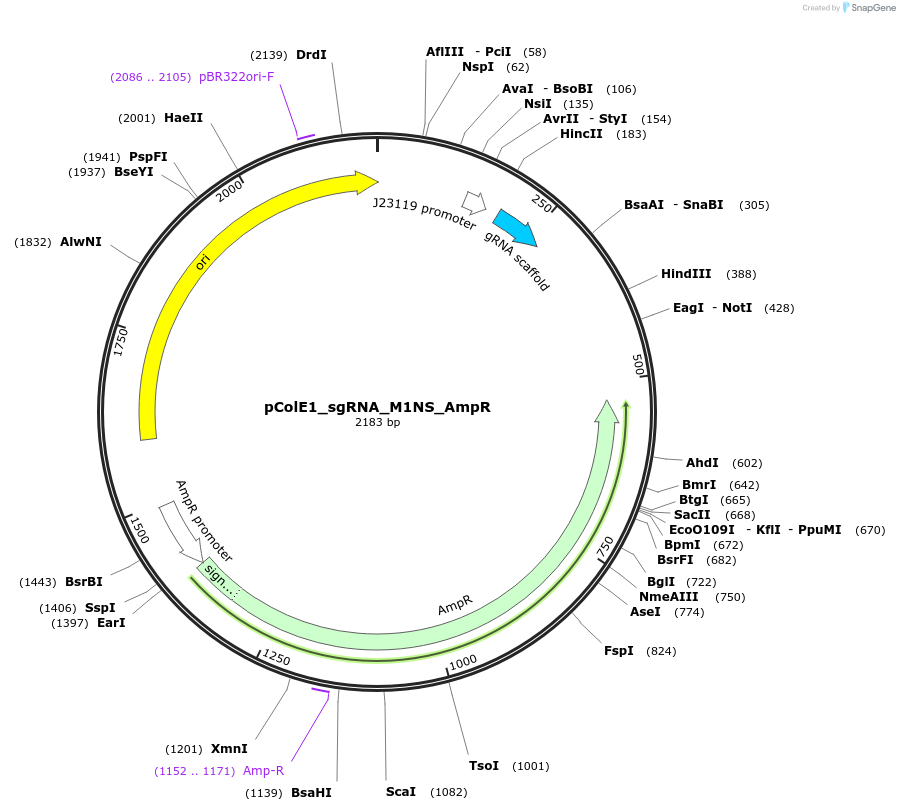
pColE1_sgRNA_M1NS_AmpR
Plasmid#119155PurposeEncodes sgRNA targeting position 6 in pColE1_70a_deGFP_KanR with one mismatch in the non-seed regionDepositorInsertsgRNA targeting position 6 in pColE1_70a_deGFP_KanR, with one mismatch in the non-seed region
UseCrisprPromoterJ23119Available SinceNov. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
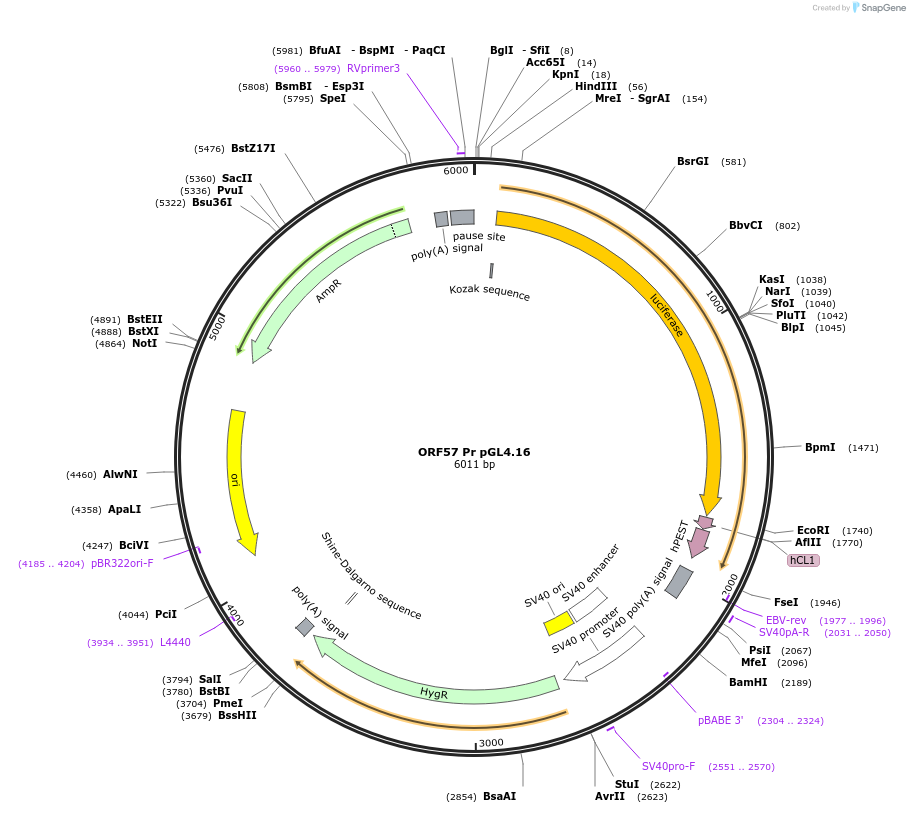
ORF57 Pr pGL4.16
Plasmid#120378PurposeFirefly luciferase reporter with KSHV ORF57 early gene promoterDepositorInsertORF57 Promoter
UseLuciferasePromoterORF57 PromoterAvailable SinceSept. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
mNG-sfTq2
Plasmid#124218PurposeCytoplasmic mNeongreen superfolder mTurquoise2 tandem, ~60 % energy transfer. pNM044 - pSAV057-mNG-sfTq2DepositorInsertmNG-sfTq2
ExpressionBacterialAvailable SinceMay 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
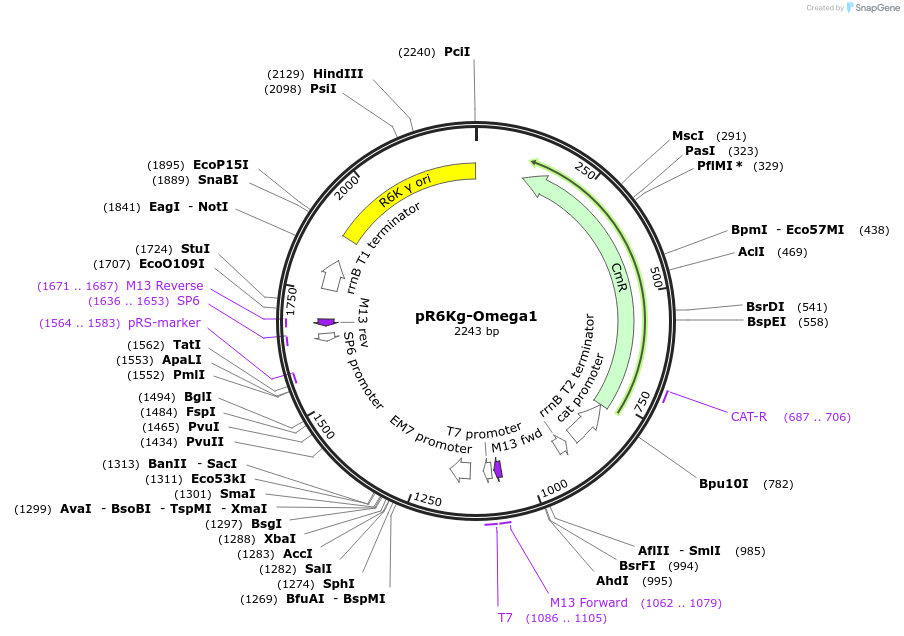
pR6Kg-Omega1
Plasmid#165864PurposeOmega level R6K_ origin GB Cloning vector, Conditionally replicative ori, requires pir gene expression requires pir gene expression requires EC100D or EC100D116 bacterial strain (DH10B derived)DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
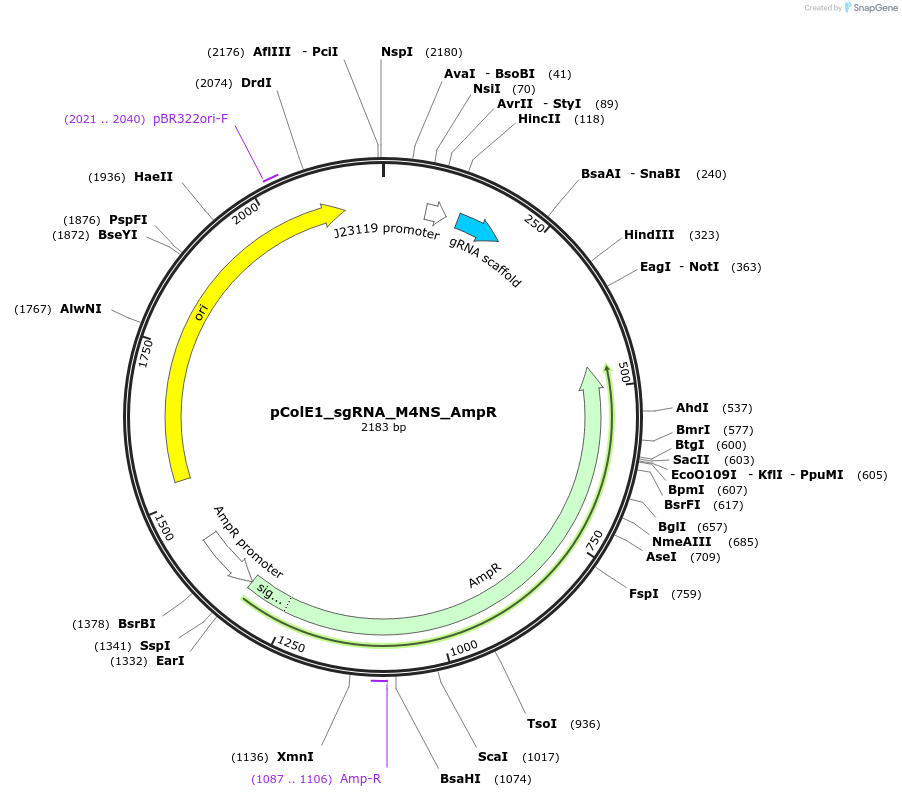
pColE1_sgRNA_M4NS_AmpR
Plasmid#119156PurposeEncodes sgRNA targeting position 6 in pColE1_70a_deGFP_KanR with four mismatches in the non-seed regionDepositorInsertsgRNA targeting position 6 in pColE1_70a_deGFP_KanR, with four mismatches in the non-seed region
UseCrisprPromoterJ23119Available SinceNov. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
pX330-sgRNA_Dgcr8_1
Plasmid#73525PurposeExpresses a human codon-optimized SpCas9 and sgRNA targeting Dgcr8DepositorInsertDGCR8
UseCRISPRExpressionMammalianPromoterhU6Available SinceJan. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
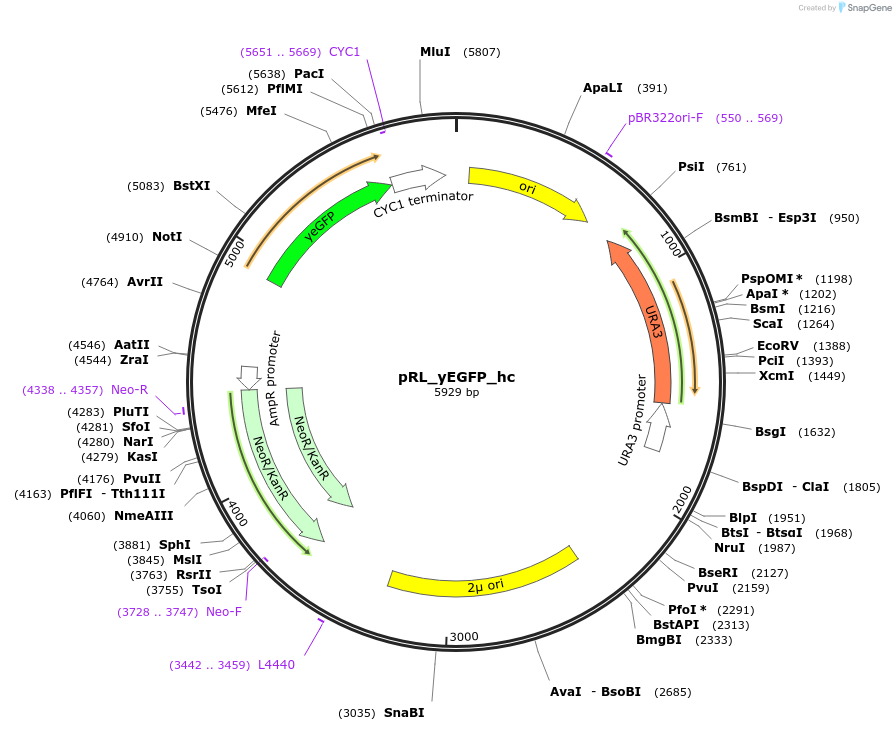
pRL_yEGFP_hc
Plasmid#98746PurposeReporter plasmid for the PhiReX system. Encodes the reporter gene yEGFP under the control of the CYC1 minimal promoter with upstream synTALE-DBS.DepositorInsertYeast-enhanced green fluorescent protein
UseSynthetic BiologyExpressionYeastPromotersyntheticAvailable SinceAug. 14, 2017AvailabilityAcademic Institutions and Nonprofits only -
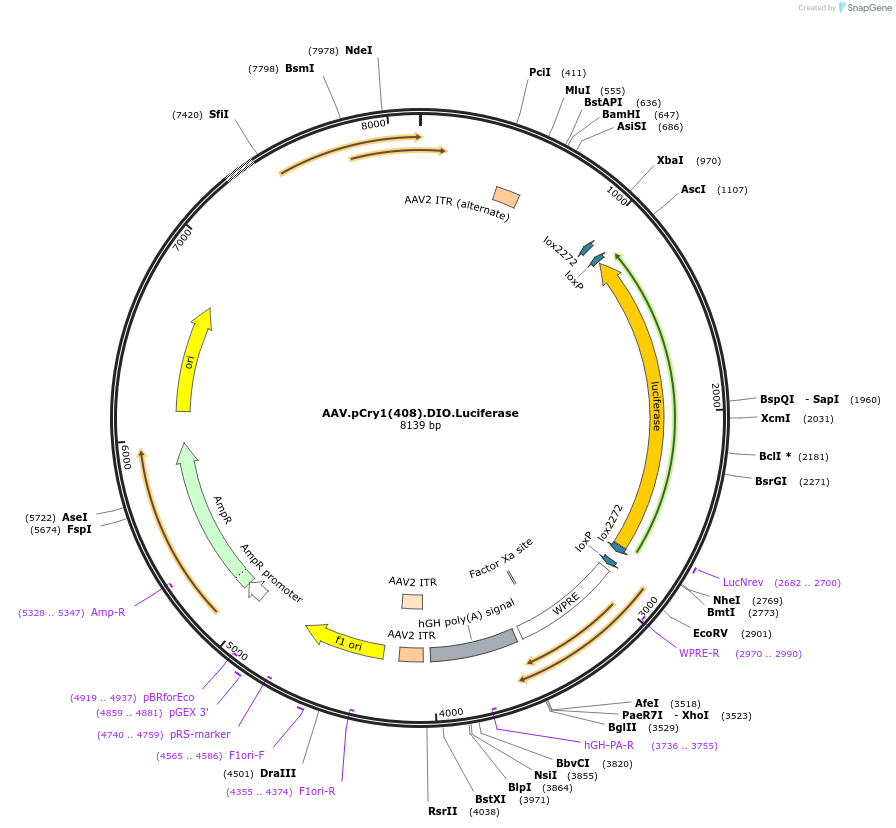
AAV.pCry1(408).DIO.Luciferase
Plasmid#192623PurposeCre-dependent expression of firefly luciferase, driven by a minimal mouse Cry1 promoterDepositorInsertluciferase
UseAAVExpressionMammalianAvailable SinceNov. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
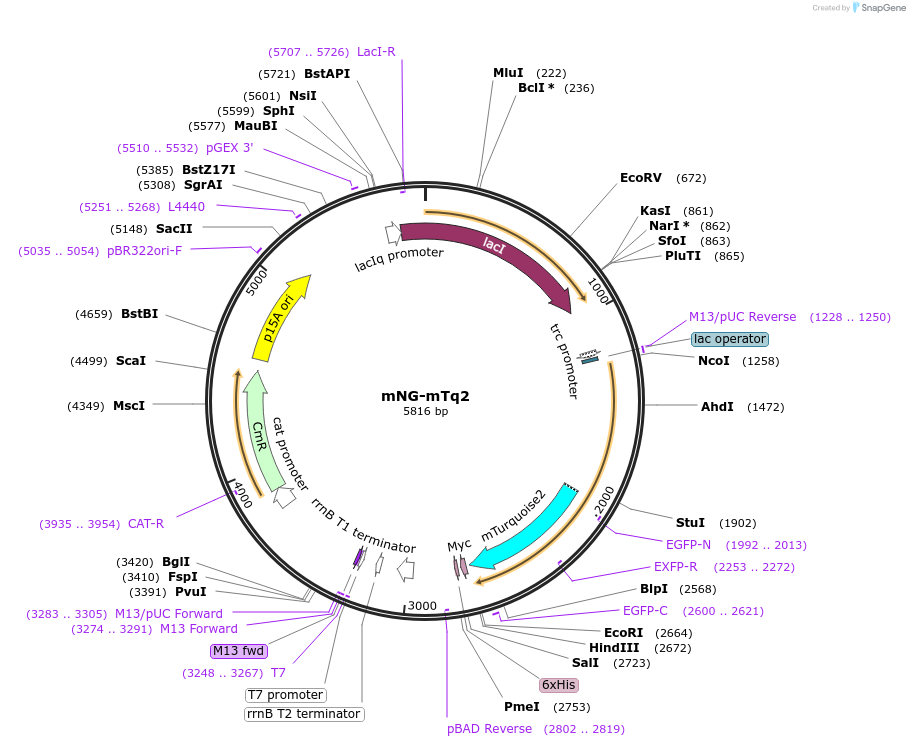
mNG-mTq2
Plasmid#124217PurposeCytoplasmic mNeongreen mTurquoise2 tandem, ~60 % energy transfer. pNM043 - pSAV057-mNG-mTq2DepositorInsertmNG-mTq2
ExpressionBacterialAvailable SinceMay 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
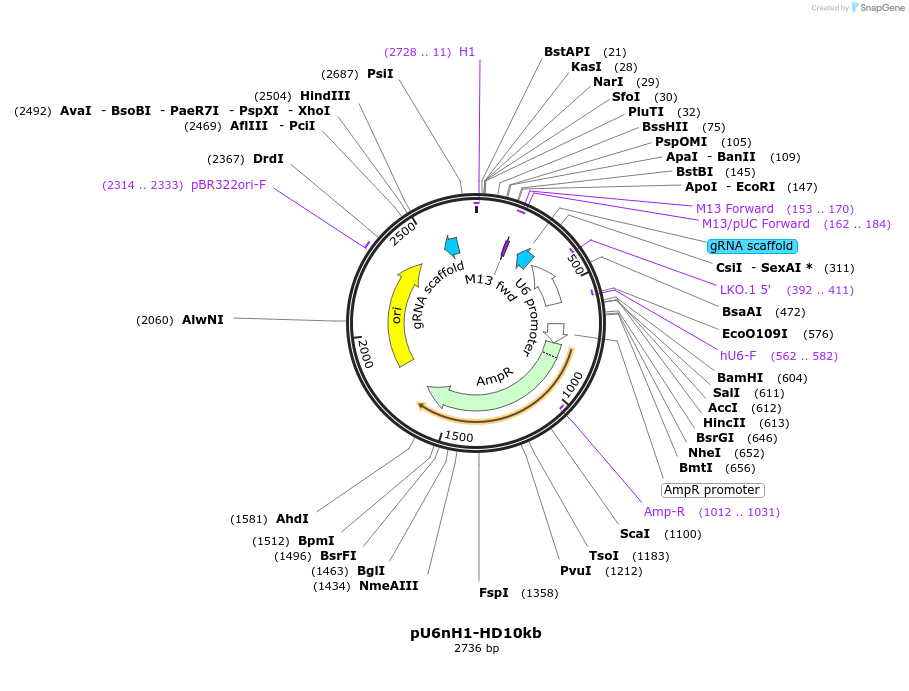
pU6nH1-HD10kb
Plasmid#172657PurposeExpressing paired pegRNAs from human U6 and H1 promoters to make 10204-bp deletion on HPRT1 geneDepositorInsertpegRNA-HD10kbA/pegRNA-HD10kbB
UseCRISPRAvailable SinceOct. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
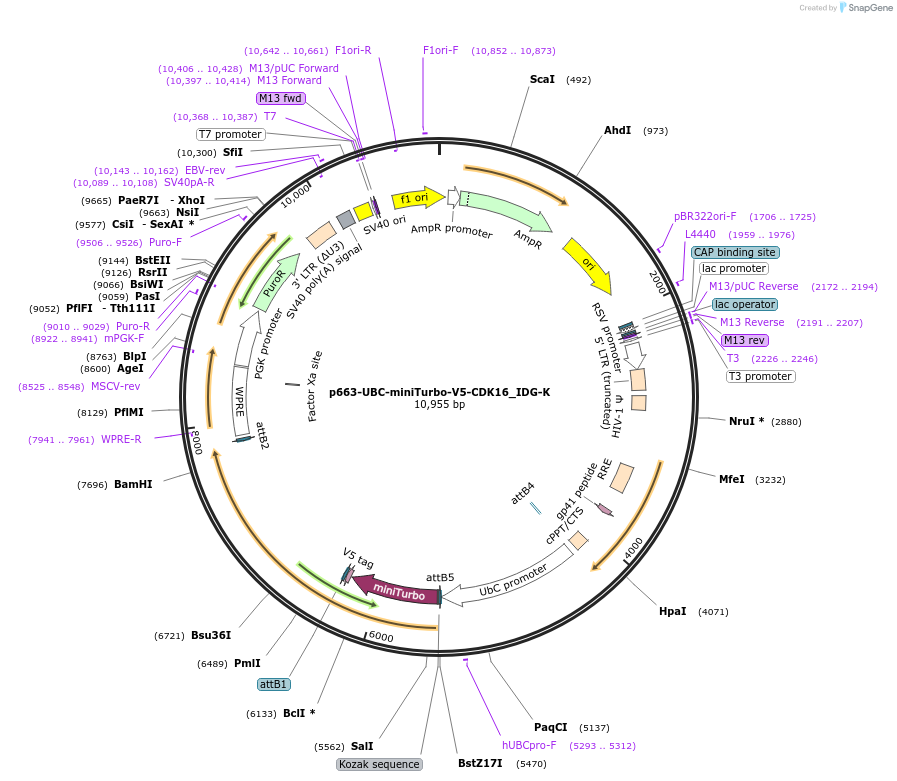
p663-UBC-miniTurbo-V5-CDK16_IDG-K
Plasmid#158173PurposeGateway destination clone of CDK16 (human) tagged with N-terminal miniTurbo-V5 for generating protein-proximity networks using proximity-dependent biotinylation proteomicsDepositorInsertminiTurbo-V5-CDK16 (CDK16 Human)
UseLentiviral; Gateway destinationTagsminiTurbo-V5ExpressionMammalianPromoterUbiquitinAvailable SinceAug. 31, 2020AvailabilityAcademic Institutions and Nonprofits only -
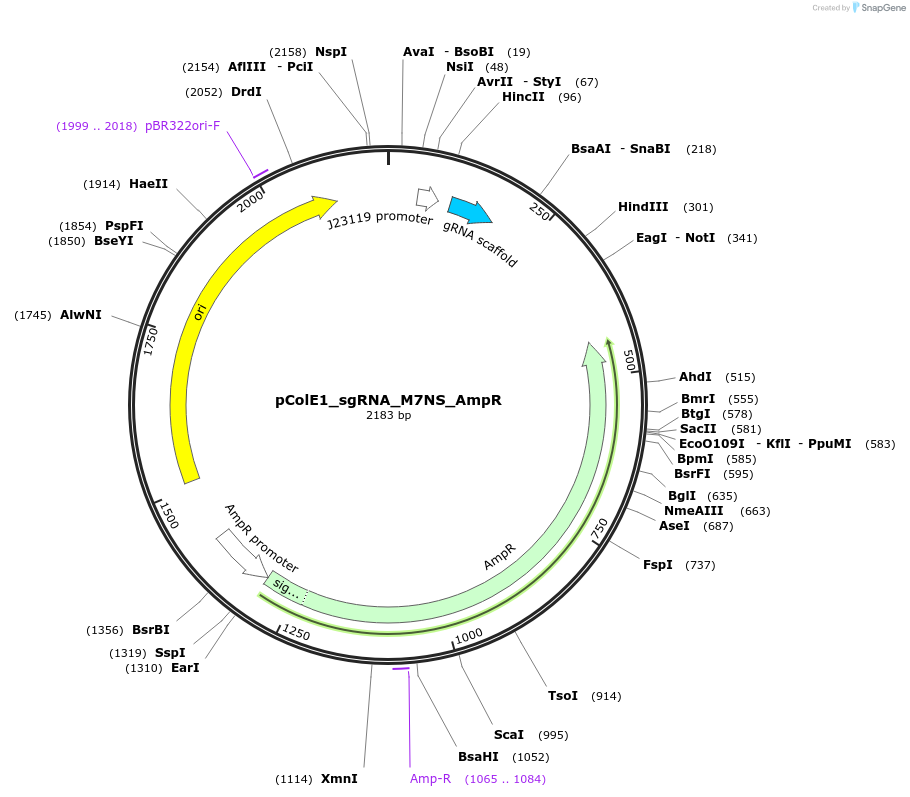
pColE1_sgRNA_M7NS_AmpR
Plasmid#119157PurposeEncodes sgRNA targeting position 6 in pColE1_70a_deGFP_KanR with seven mismatches in the non-seed regionDepositorInsertsgRNA targeting position 6 in pColE1_70a_deGFP_KanR, with seven mismatches in the non-seed region
UseCrisprPromoterJ23119Available SinceNov. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
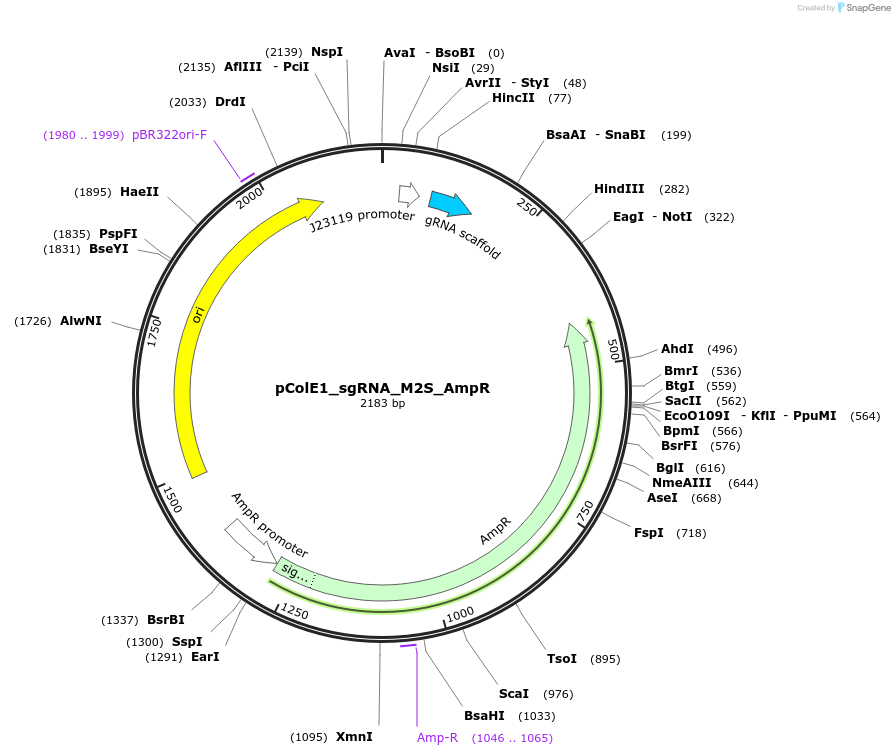
pColE1_sgRNA_M2S_AmpR
Plasmid#119154PurposeEncodes sgRNA targeting position 6 in pColE1_70a_deGFP_KanR with two mismatches in the seed regionDepositorInsertsgRNA targeting position 6 in pColE1_70a_deGFP_KanR, with two mismatches in the seed region
UseCrisprPromoterJ23119Available SinceNov. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
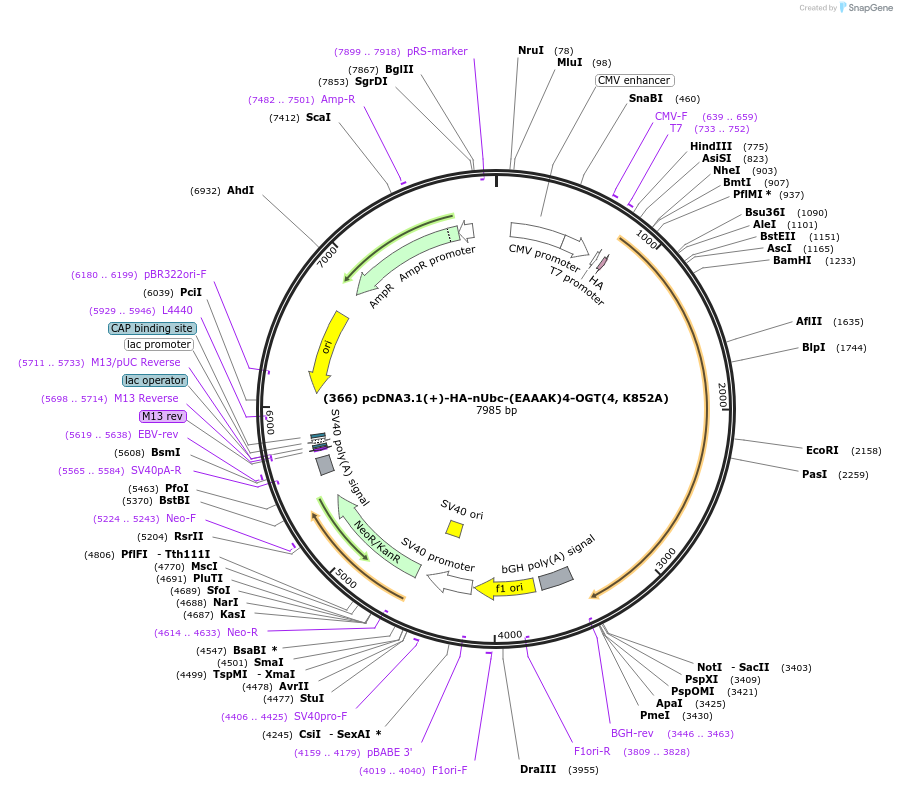
(366) pcDNA3.1(+)-HA-nUbc-(EAAAK)4-OGT(4, K852A)
Plasmid#168189PurposeHA-tagged Ubc6E nanobody fused to an inactive truncated OGT of 4 TPRs with K852A mutationDepositorInsertHA-nUbc6E-(EAAAK)4-OGT(4,K852A)
TagsHA-TagExpressionMammalianPromoterT7/CMVAvailable SinceMay 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
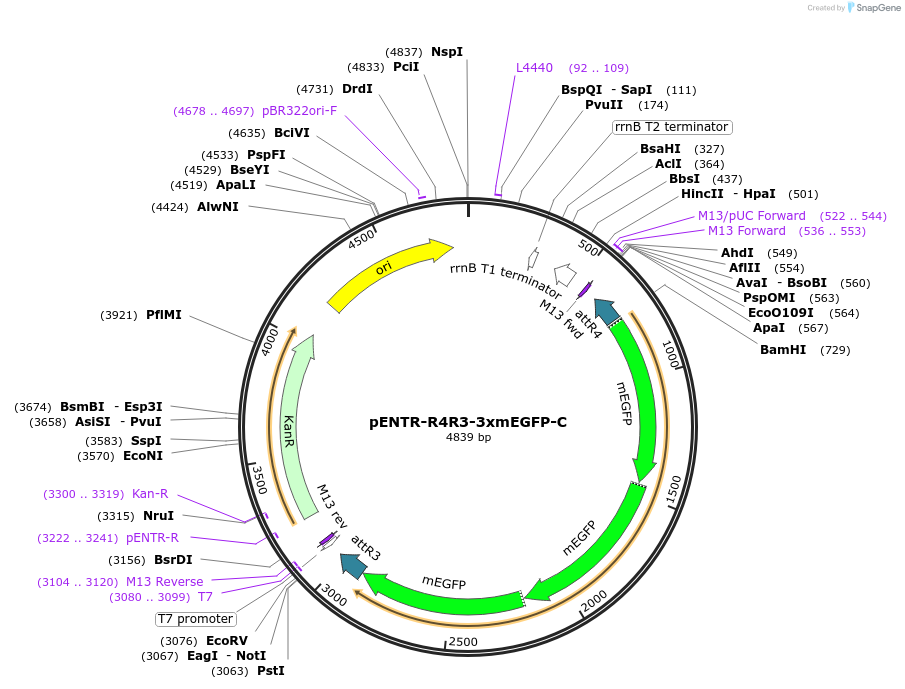
pENTR-R4R3-3xmEGFP-C
Plasmid#113751PurposeGateway multi-site entry clone for second position 3x mEGFP (C-terminal tag). Introduced into the genome via homology-directed repair after an LR reaction with homology sequences.DepositorInsertmEGFP-mEGFP-mEGFP
UseCRISPR; Gateway entry vectorTags3xmEGFPAvailable SinceSept. 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
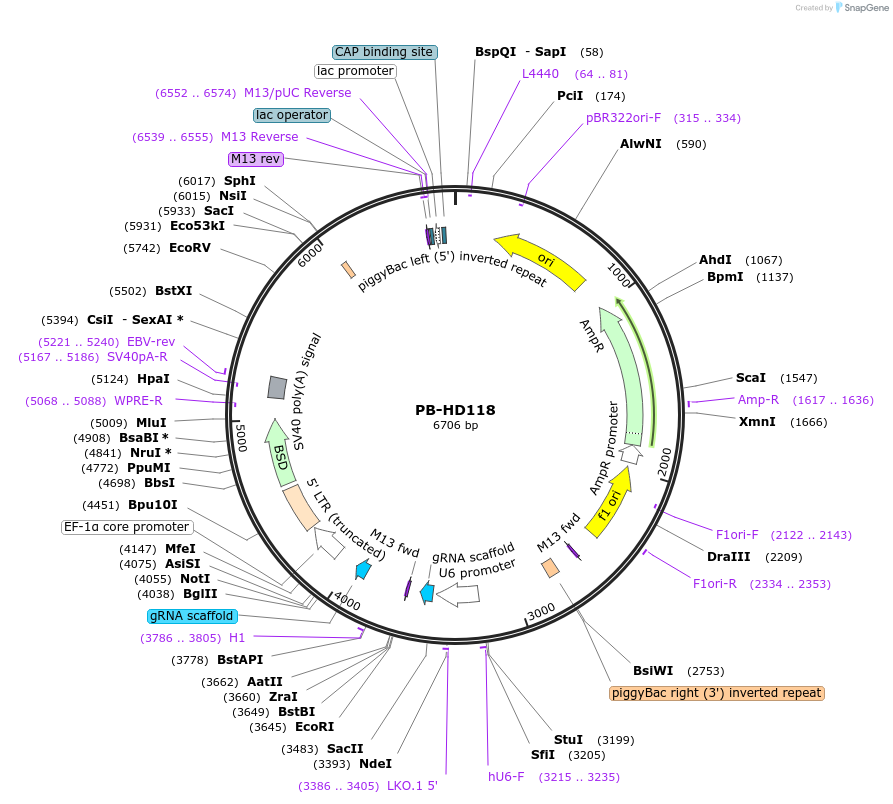
PB-HD118
Plasmid#172655PurposeExpressing paired pegRNAs from human U6 and H1 promoters to make 118-bp deletion on HPRT1 geneDepositorInsertpegRNA-HD118A/pegRNA-HD118B
UseCRISPRAvailable SinceOct. 22, 2021AvailabilityAcademic Institutions and Nonprofits only