8,173 results
-
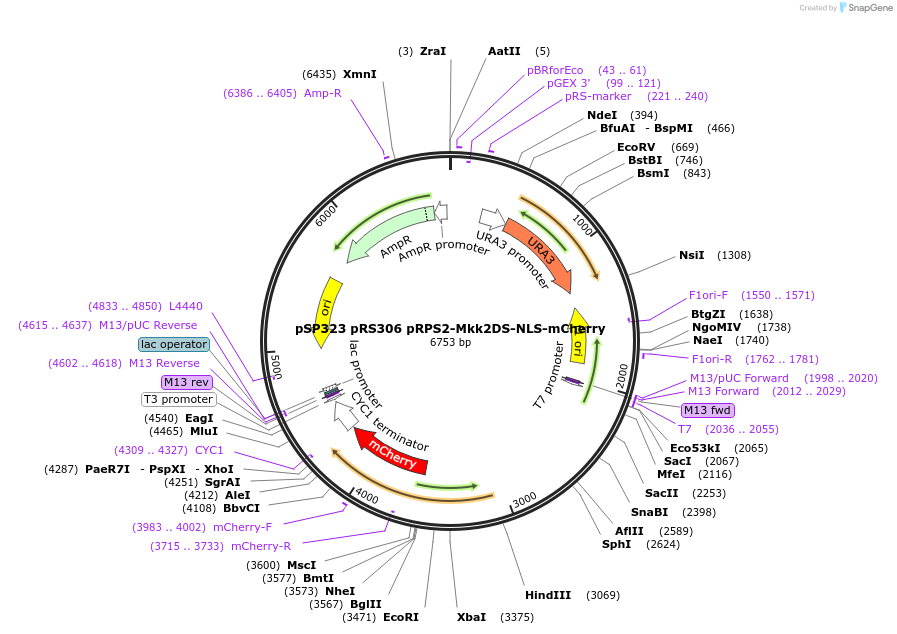
Plasmid#68280PurposeSKARS: Mpk1 MAPK activity sensor: Mkk2DS (1-100) 2xNLS mCherryDepositorAvailable SinceSept. 14, 2015AvailabilityAcademic Institutions and Nonprofits only
-
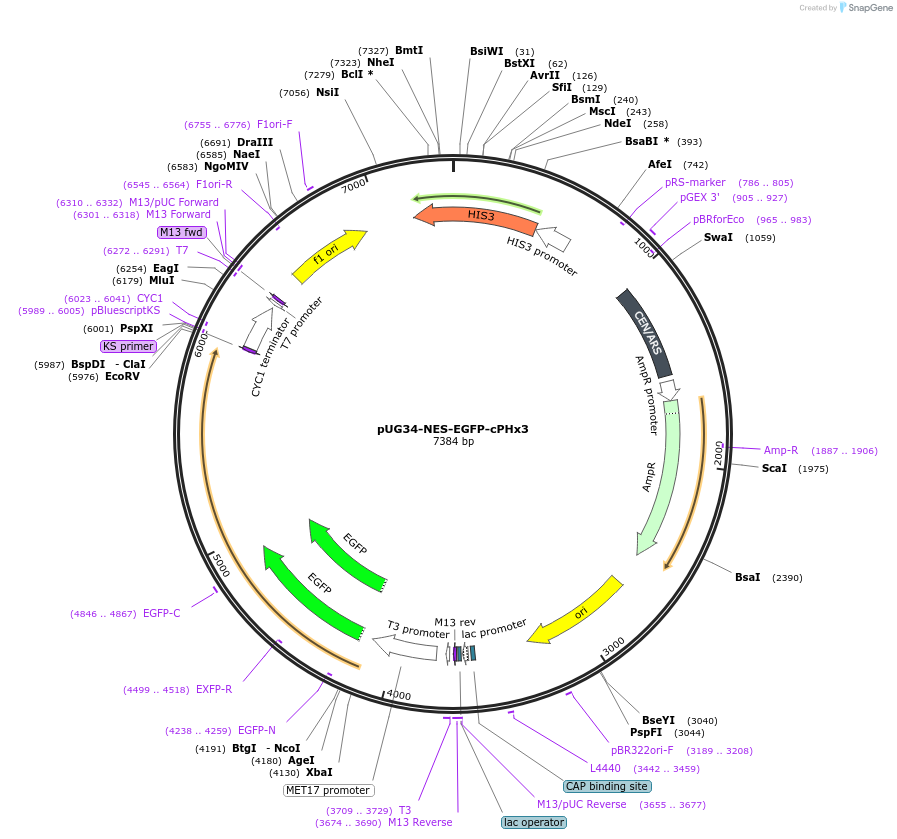
pUG34-NES-EGFP-cPHx3
Plasmid#183669PurposePtdIns(3,4)P2 biosensor for expression in yeastDepositorInsertPLEKHA1 (PLEKHA1 Frog, Human)
TagsX. laevis map2k1.L(32-44)-EGFPExpressionYeastMutationAmino acids 169-329 (tandem trimer)PromoterMET25Available SinceMay 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
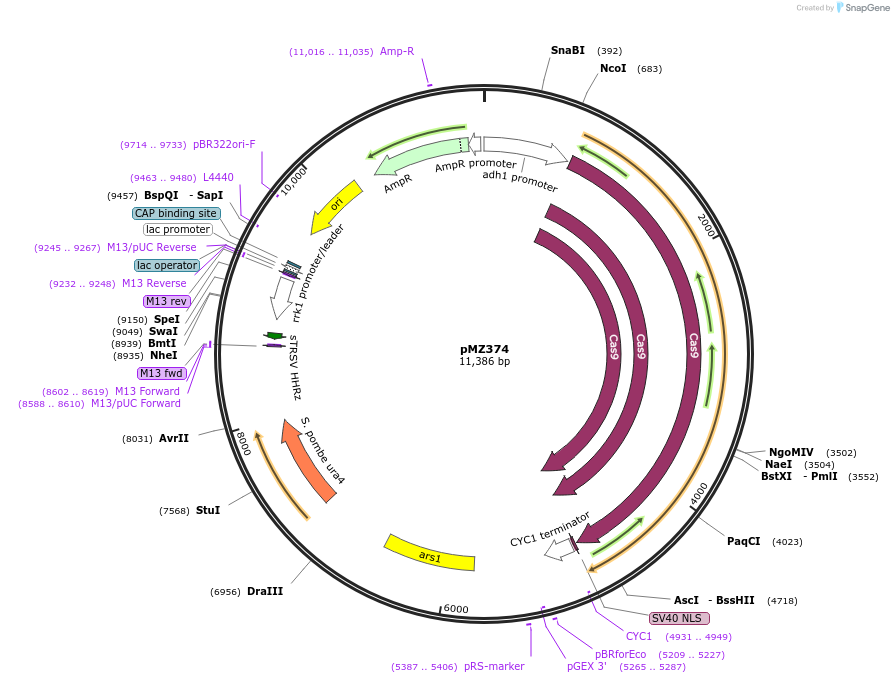
pMZ374
Plasmid#59896PurposeCombination adh1:cas9/rrk1:sgRNA for CRISPR genome editing in fission yeast: Empty sgRNA target (CspCI placeholder) (see comments).DepositorInsertsCas9
rrk1:sgRNA
UseCRISPRExpressionYeastMutationSilent muation of the CspCI sitePromoteradh1 and rrk1Available SinceOct. 29, 2014AvailabilityAcademic Institutions and Nonprofits only -
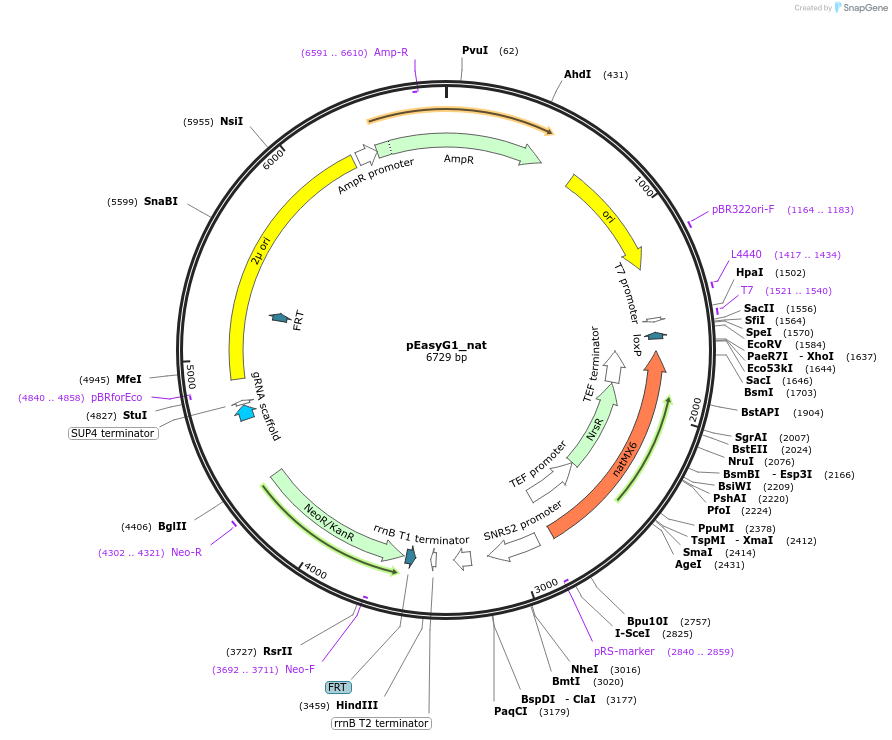
pEasyG1_nat
Plasmid#184911PurposeTemplate to generate via PCR a single gRNA for expression in S. cerevisiaeDepositorInsertgRNA scaffold
UseCRISPRExpressionBacterial and YeastAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
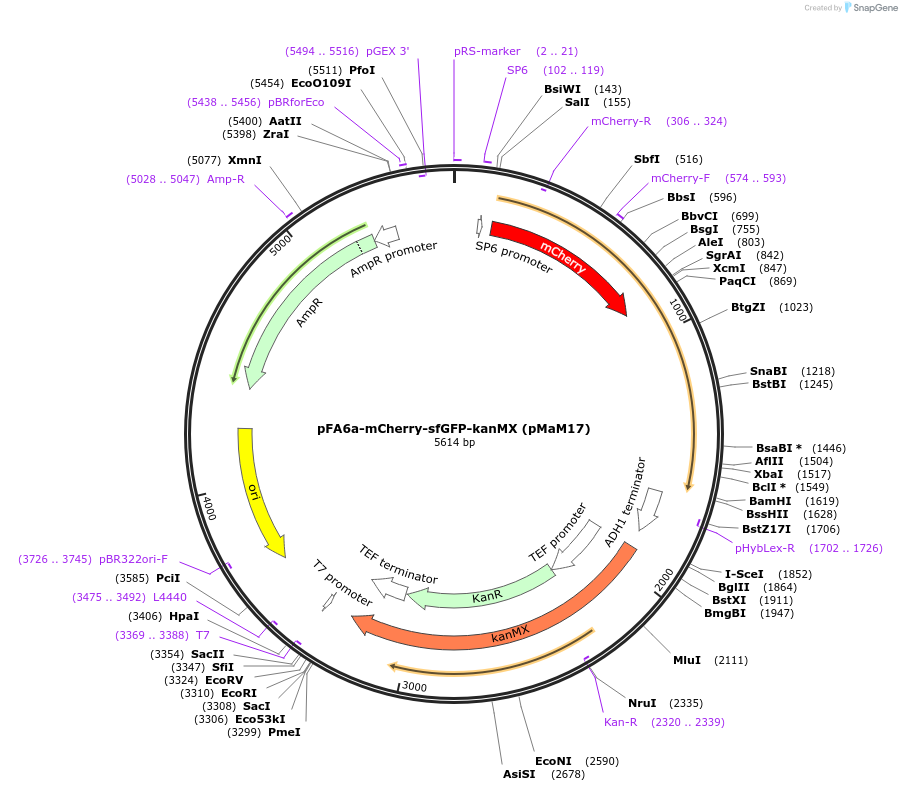
pFA6a-mCherry-sfGFP-kanMX (pMaM17)
Plasmid#173452PurposeContains mCherry-sfGFP tandem fluorescent protein timerDepositorInsertmCherry-sfGFP-ADH1ter
ExpressionYeastPromoterno promoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
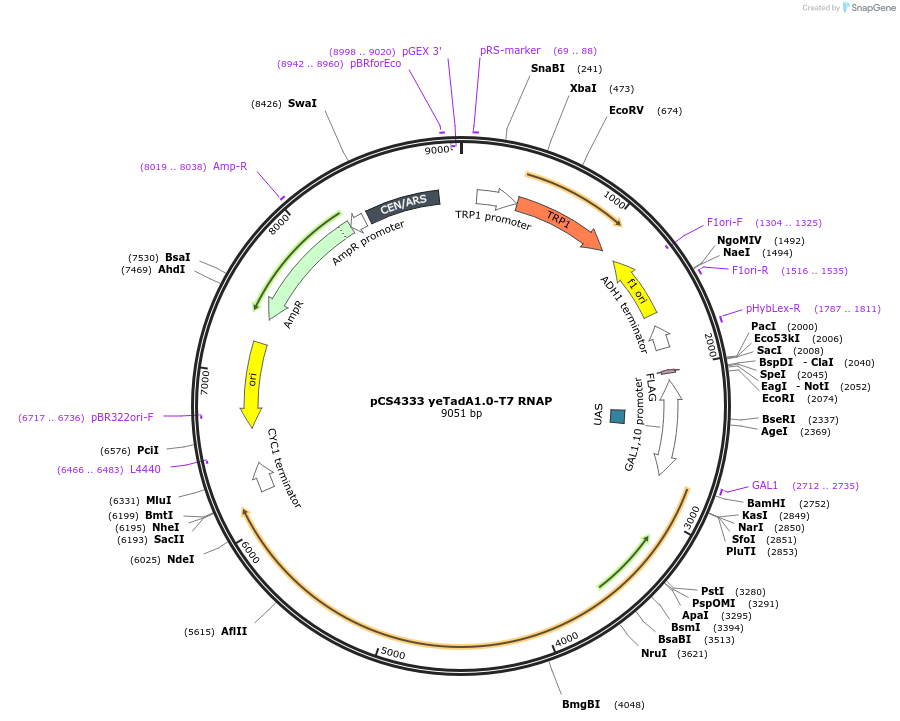
pCS4333 yeTadA1.0-T7 RNAP
Plasmid#137735PurposepCS4333 CEN/ARS vector, PGAL1-yeTadA1.0-100AA linker-T7 RNAP-TCYC1, TRP1 selectable markerDepositorInsertyeTadA1.0-T7 RNAP
ExpressionBacterial and YeastMutationE366G (See depositor comment below)Available SinceJan. 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
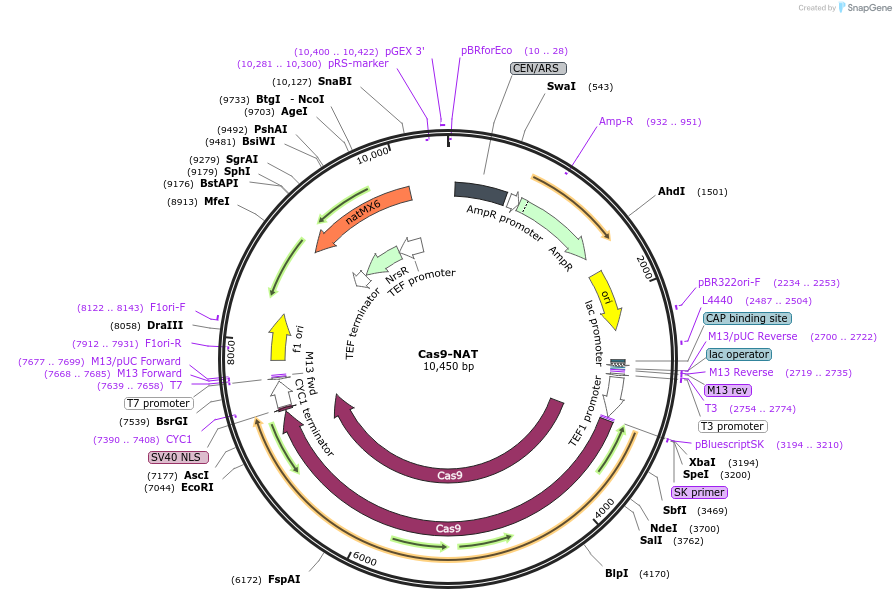
Cas9-NAT
Plasmid#64329PurposeCas9 expression cassette carried by a yeast single-copy episome plasmidDepositorInsertHuman-optimized S. pyogenes Cas9 with Clonnat marker gene expression cassette
UseCRISPRExpressionYeastAvailable SinceApril 29, 2015AvailabilityAcademic Institutions and Nonprofits only -
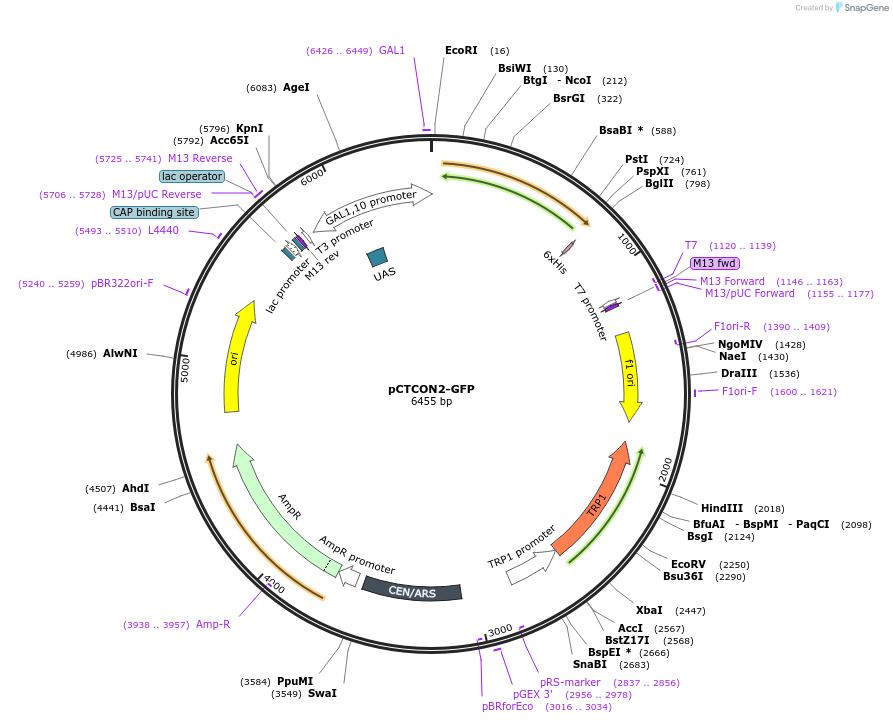
pCTCON2-GFP
Plasmid#158130PurposeSingle fluorescent protein reporter (GFP) for ncAA incorporation (wild-type construct)DepositorInsertsfGFP
ExpressionYeastPromoterGAL1-10Available SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
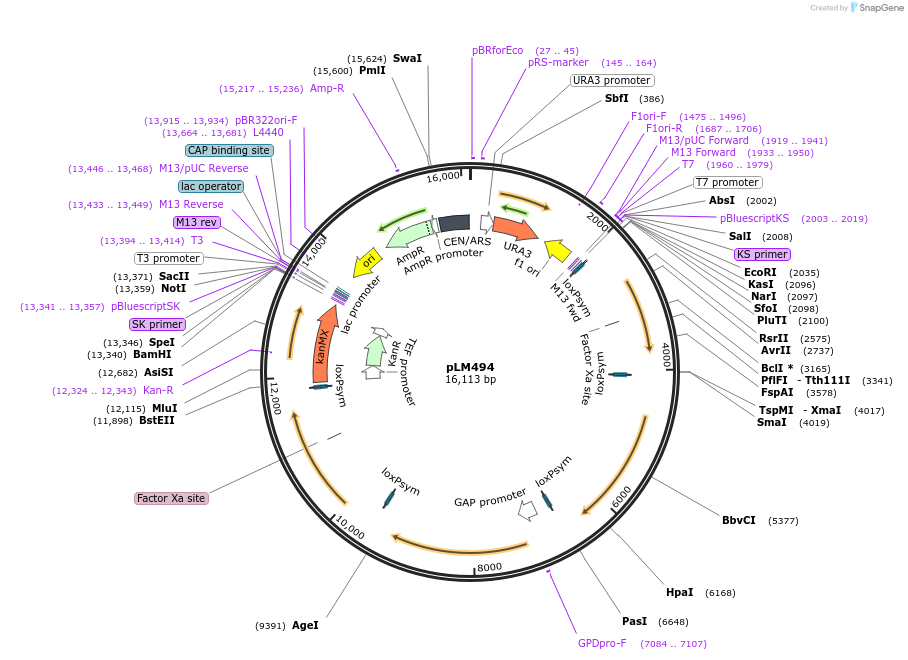
pLM494
Plasmid#100539PurposeCEN/ARS plasmid harboring expression cassettes for crtI, crtYB, crtE and tHMG1 of the β-carotene pathway, each flanked by loxPsym sites.DepositorInsertsGGPP synthase
phytoene desaturase
phytoene-beta carotene synthase
HMG CoA reductase
UseCre/Lox and Synthetic BiologyExpressionYeastPromoterPGK1, TDH3, TIP1, and ZEO1Available SinceJune 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
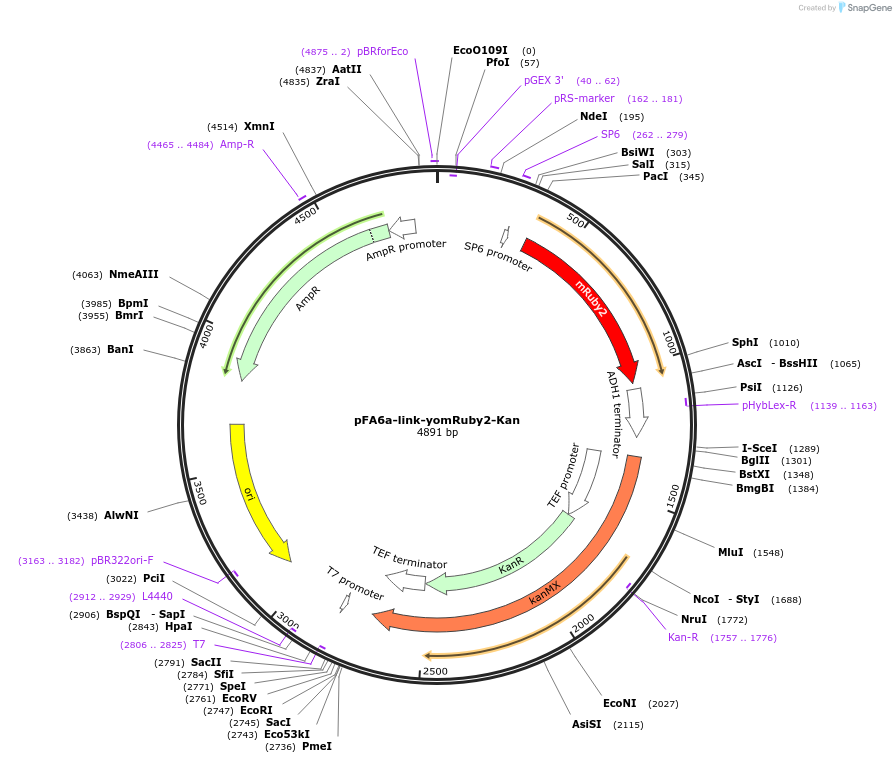
pFA6a-link-yomRuby2-Kan
Plasmid#44953PurposeYeast tagging vector to create gene fusion with yeast optimized mRuby2 with Kan selectionDepositorInsertyomRuby2
ExpressionYeastAvailable SinceMay 9, 2013AvailabilityAcademic Institutions and Nonprofits only -
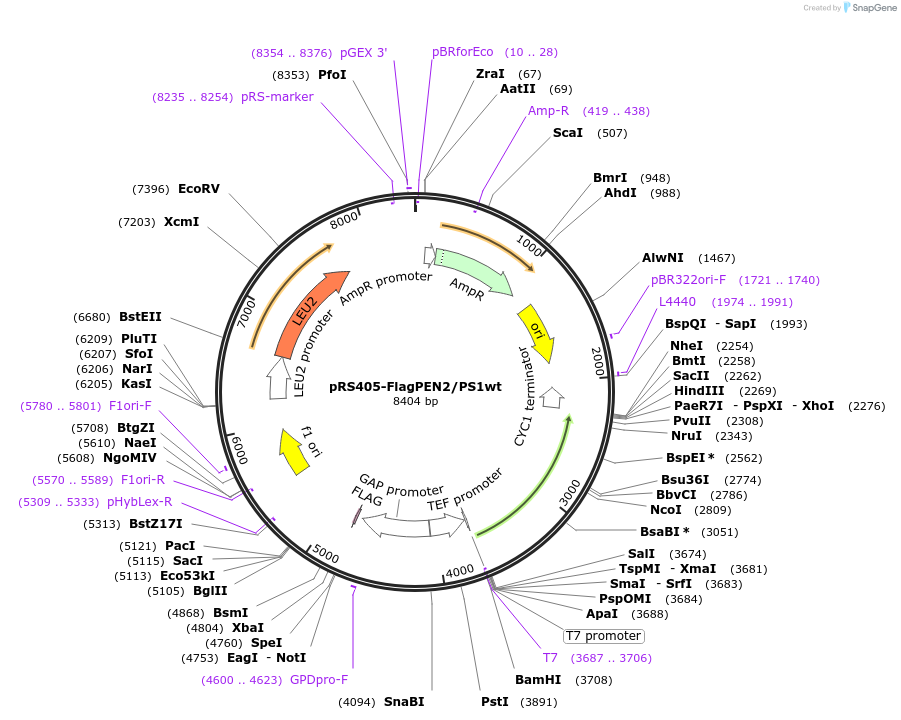
pRS405-FlagPEN2/PS1wt
Plasmid#207803PurposeYeast integrative vector for expression of FlagPEN2 and Presenilin 1DepositorInsertGamma-secretase subunits PEN-2 and presenilin1 wt
ExpressionYeastAvailable SinceJan. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
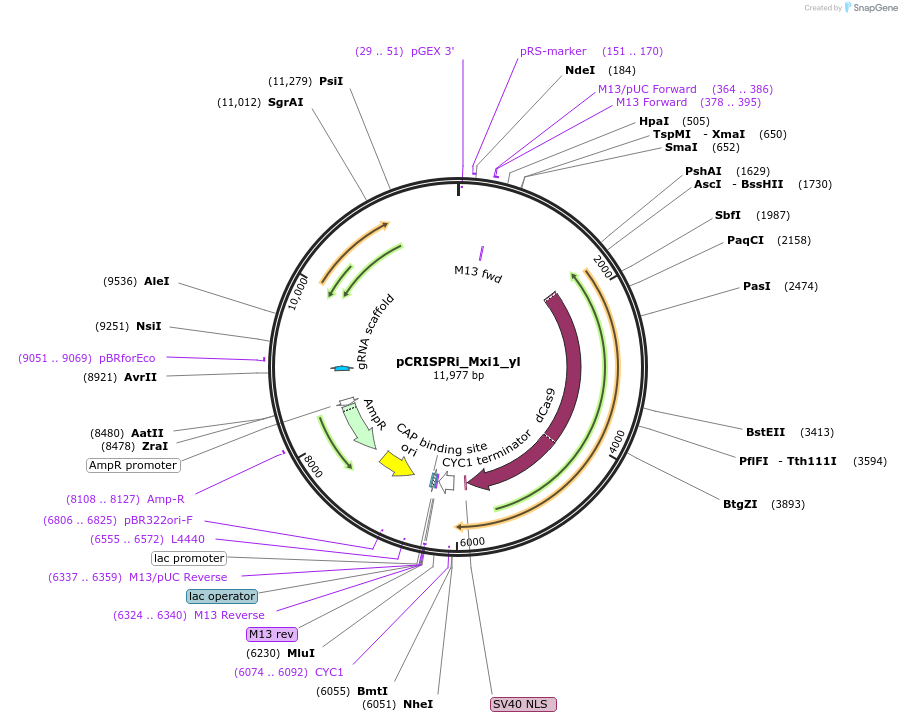
pCRISPRi_Mxi1_yl
Plasmid#91248PurposeCRISPR-dCas9-Mxi1 vector for Yarrowia lipolytica, expressing dCas9-Mxi1 and AvrII site for sgRNA insertionDepositorInsertsCodon optimized dCas9-Mxi1
sgRNA expression cassette
UseCRISPR and Synthetic BiologyExpressionYeastPromoterSCR1'-tRNA and UAS1B8-TEF(136)Available SinceAug. 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
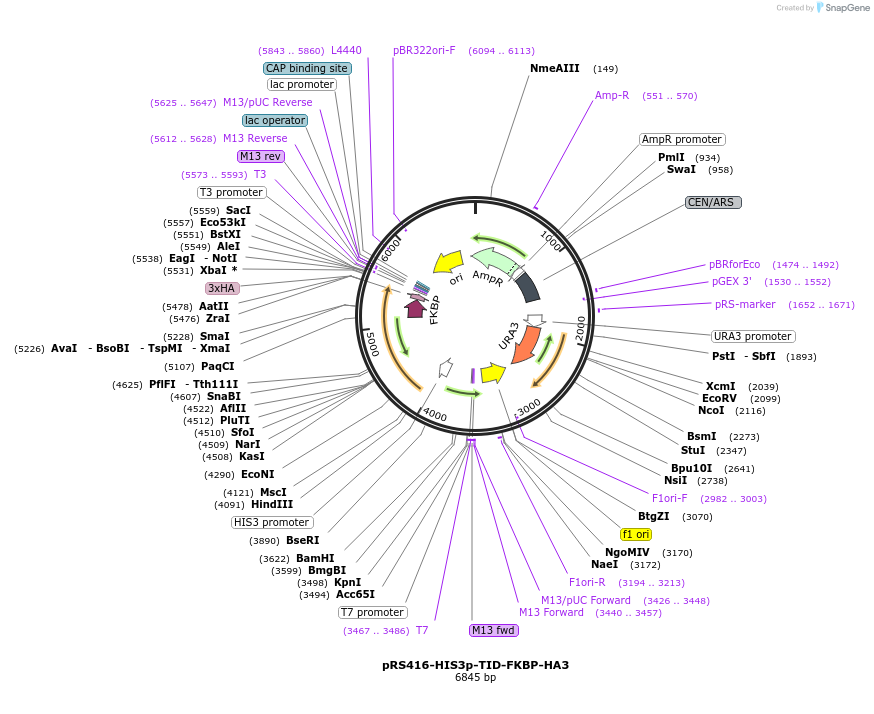
pRS416-HIS3p-TID-FKBP-HA3
Plasmid#228006PurposeExpresses TurboID-FKBP fusion from HIS3 promoter; yeast BioIDDepositorInsertTurboID-FKBP
TagsHA tagExpressionYeastPromoterHIS3Available SinceNov. 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
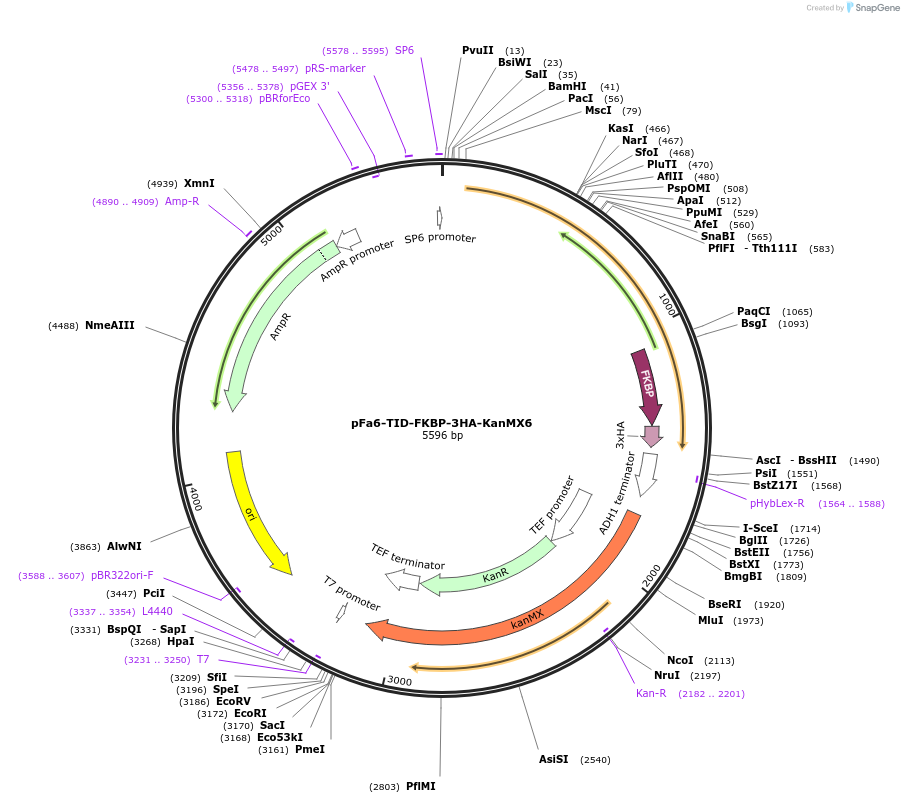
pFa6-TID-FKBP-3HA-KanMX6
Plasmid#228024PurposeTo integrate TID-FKBP fusion at yeast promoter; yeast BioID with KanMX6 markerDepositorInsertTurboID-FKBP
TagsHA tagExpressionYeastAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
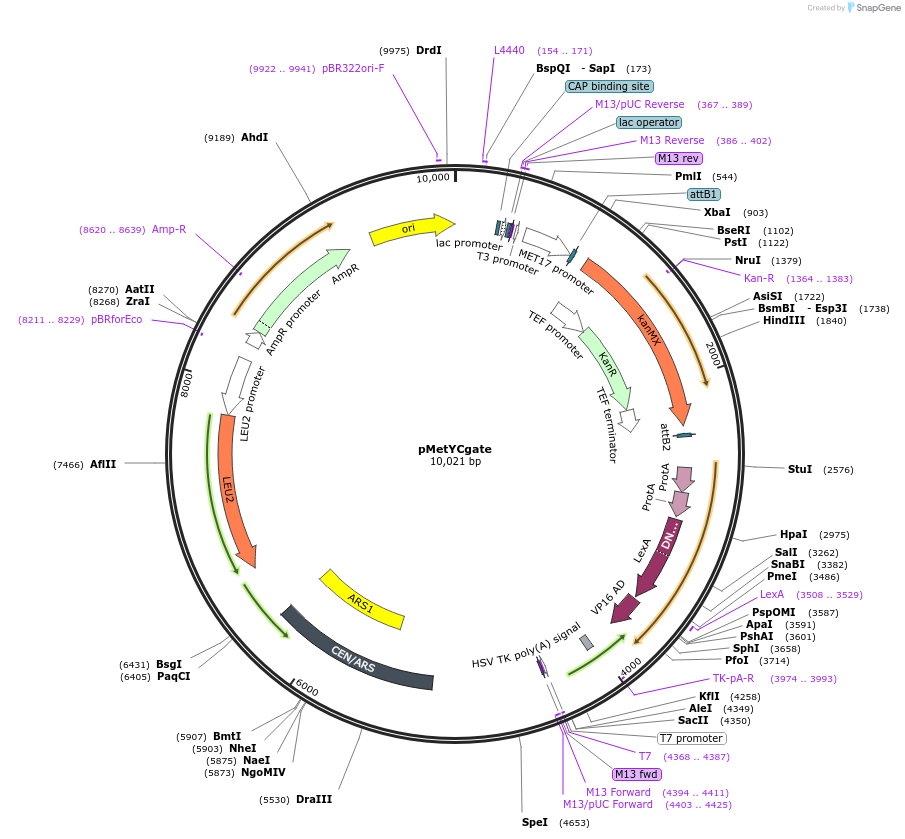
pMetYCgate
Plasmid#105074PurposeYeast mating-based Split Ubiquitin Assay, bait vector for in vivo cloningDepositorTypeEmpty backboneTagsCub-PLVExpressionYeastAvailable SinceFeb. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
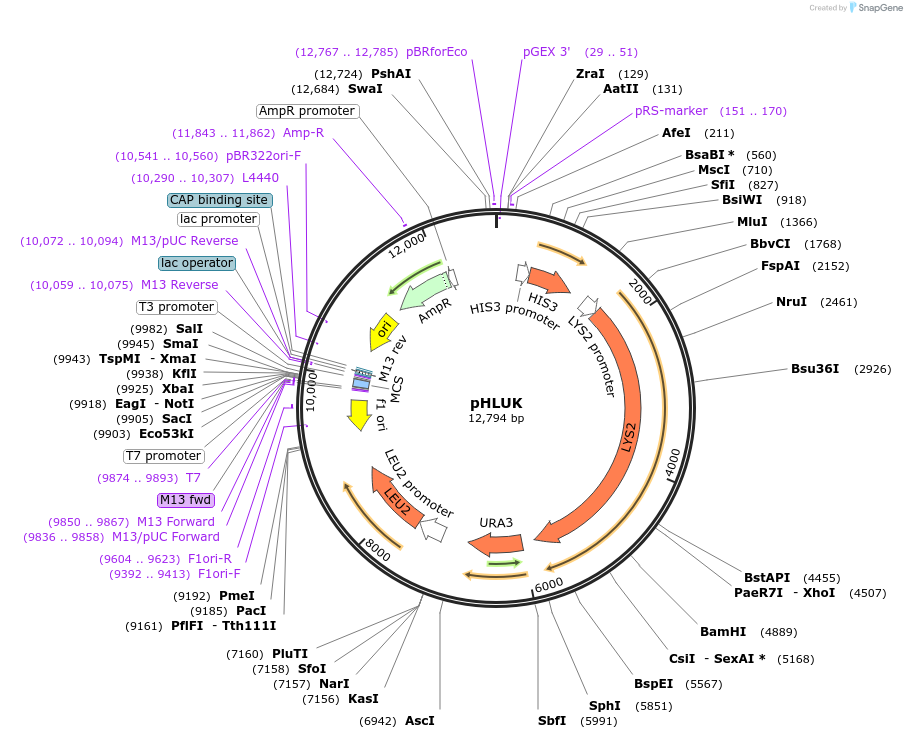
pHLUK
Plasmid#64181Purposeharbours marker genes HIS3, LEU2, URA3 and LYS2DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceSept. 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
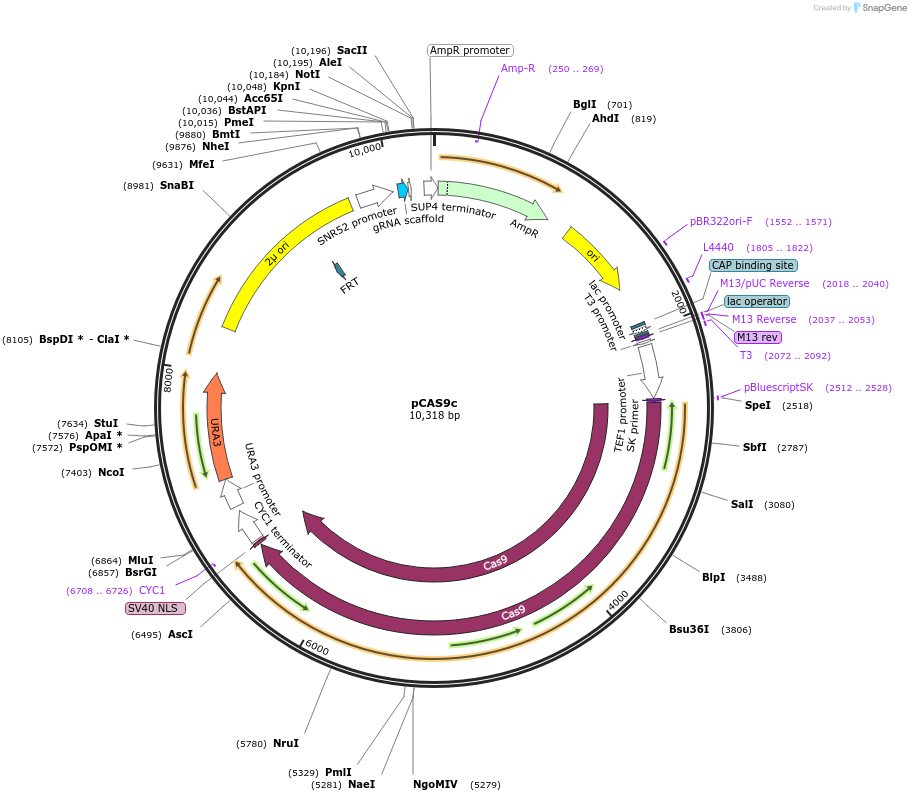
pCAS9c
Plasmid#141250PurposeConstitutive expression of Cas9 for addressing one target; Contains guide RNA expression cassette with stuffer and KpnI-Pme1 restriction sites.DepositorTypeEmpty backboneUseCRISPRExpressionYeastPromoterTEF1 (Cas9), pSNR52 (gRNA)Available SinceAug. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
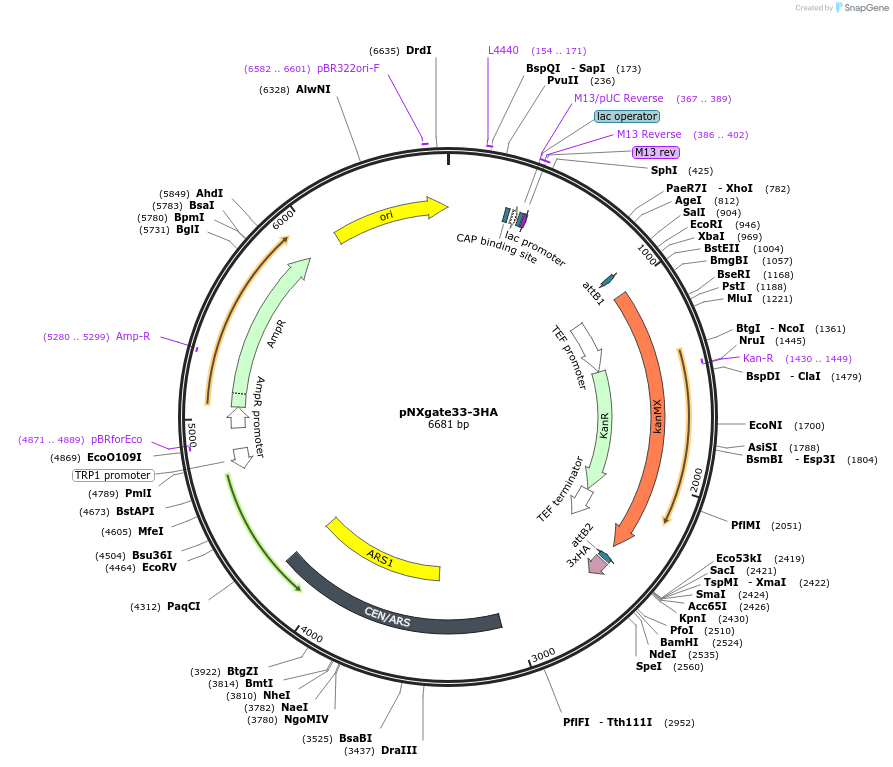
pNXgate33-3HA
Plasmid#105075PurposeYeast mating-based Split Ubiquitin Assay, prey vector for in vivo cloningDepositorTypeEmpty backboneTags3xHA and NubGExpressionYeastAvailable SinceFeb. 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
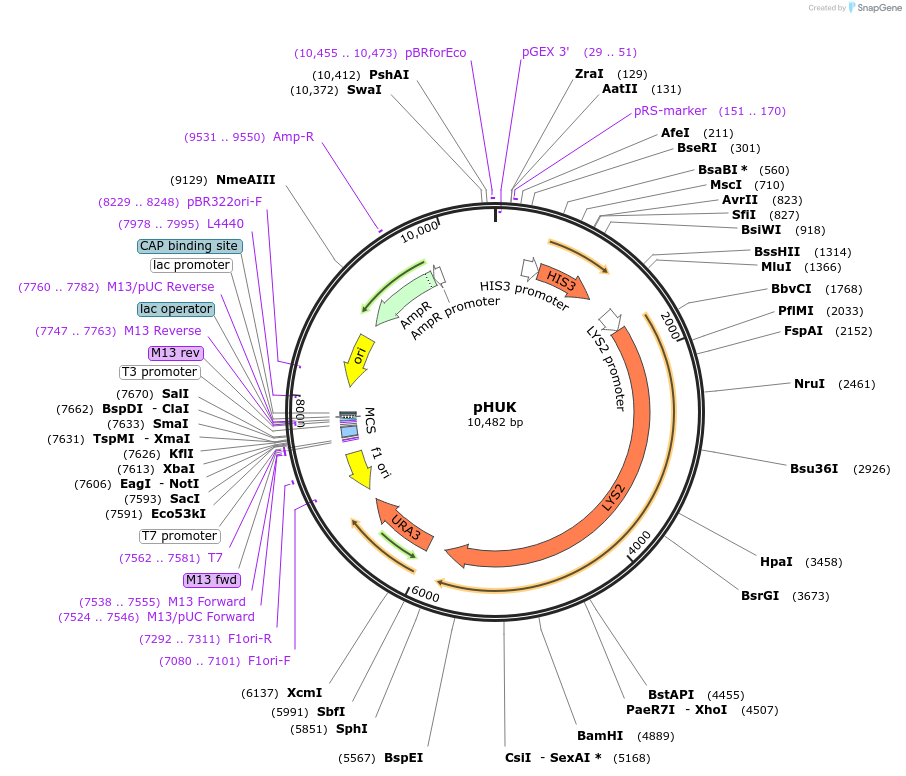
pHUK
Plasmid#64183Purposeharbours marker genes HIS3, URA3 and LYS2DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceSept. 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
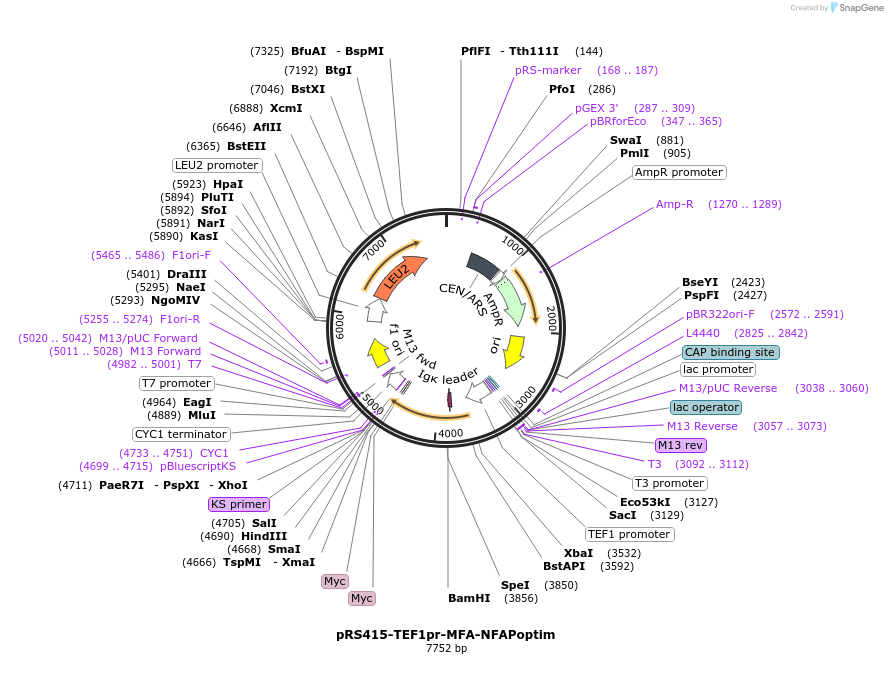
pRS415-TEF1pr-MFA-NFAPoptim
Plasmid#221090PurposeFAP insert for N-terminally tagging genes of interest (optimized FAP for yeast expression + 2xMYC tag) + MFA1 signal sequence to help target constructs to the ERDepositorTypeEmpty backboneExpressionYeastAvailable SinceMarch 11, 2025AvailabilityAcademic Institutions and Nonprofits only