We narrowed to 5,933 results for: chia
-
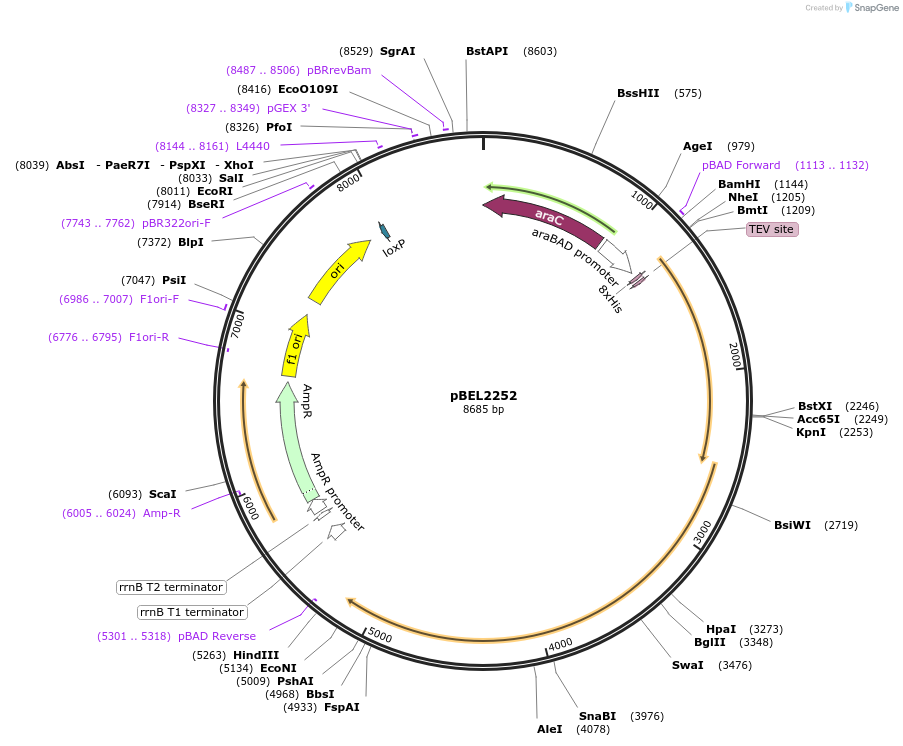
Plasmid#170404PurposeFor expresson of LetA-[LetB PLL6->4] operon from E. coli. N-terminal his tag on LetA, araBAD promoterDepositorTags8xHis2xQH and TEV cleavage site on LetAExpressionBacterialMutationLetA-LetB(Δ700-716, replace with 455-479)Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only
-
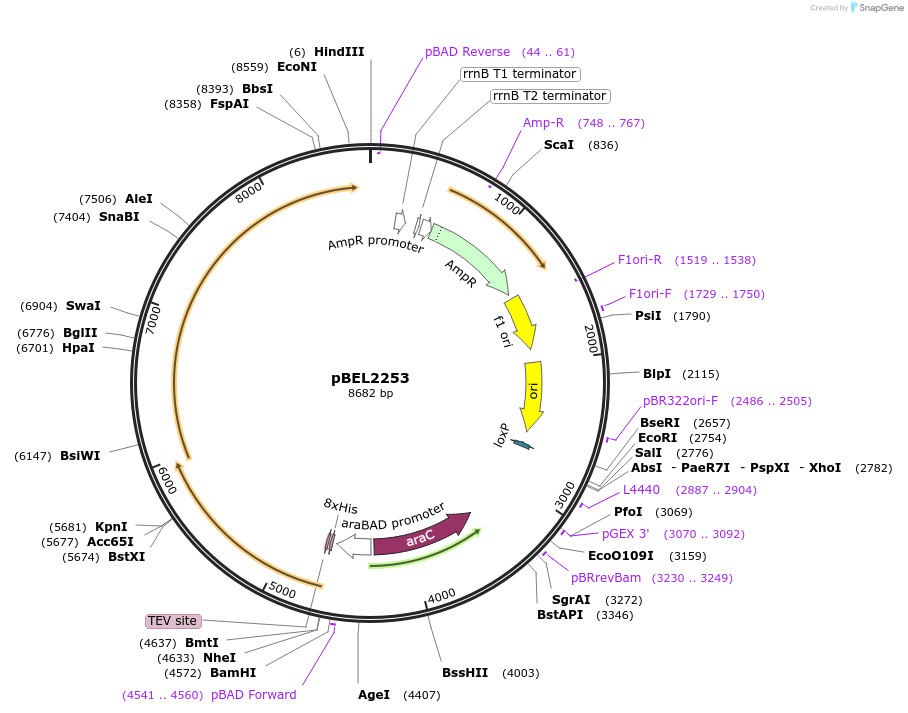
pBEL2253
Plasmid#170405PurposeFor expresson of LetA-[LetB PLL6->7] operon from E. coli. N-terminal his tag on LetA, araBAD promoterDepositorTags8xHis2xQH and TEV cleavage site on LetAExpressionBacterialMutationLetA-LetB(Δ700-717, replace with 812-836)Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
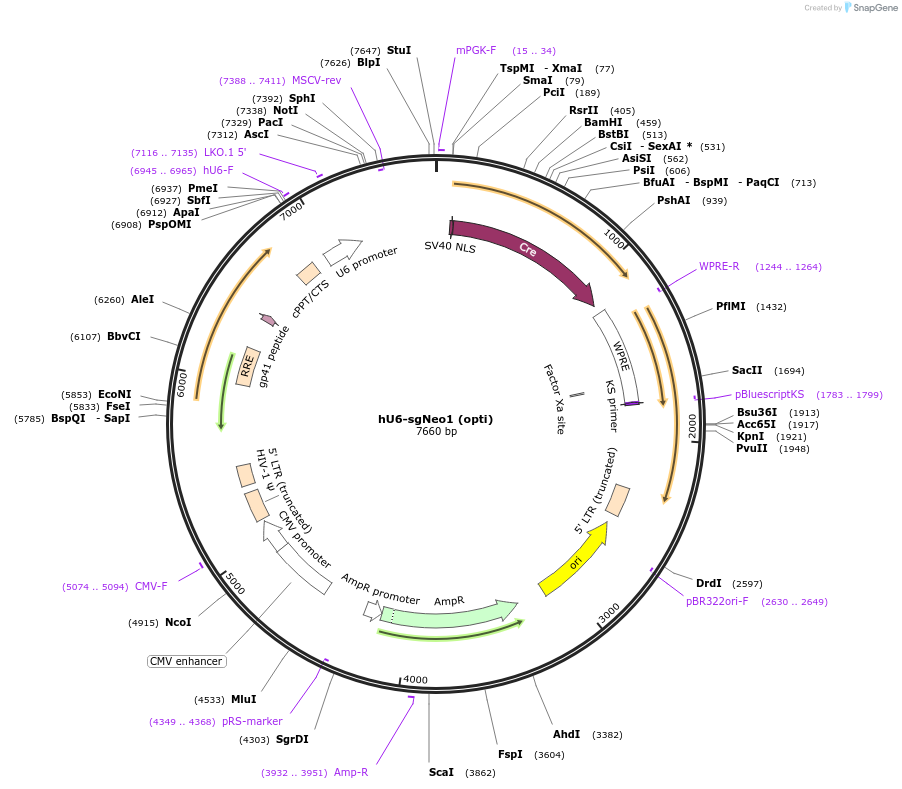
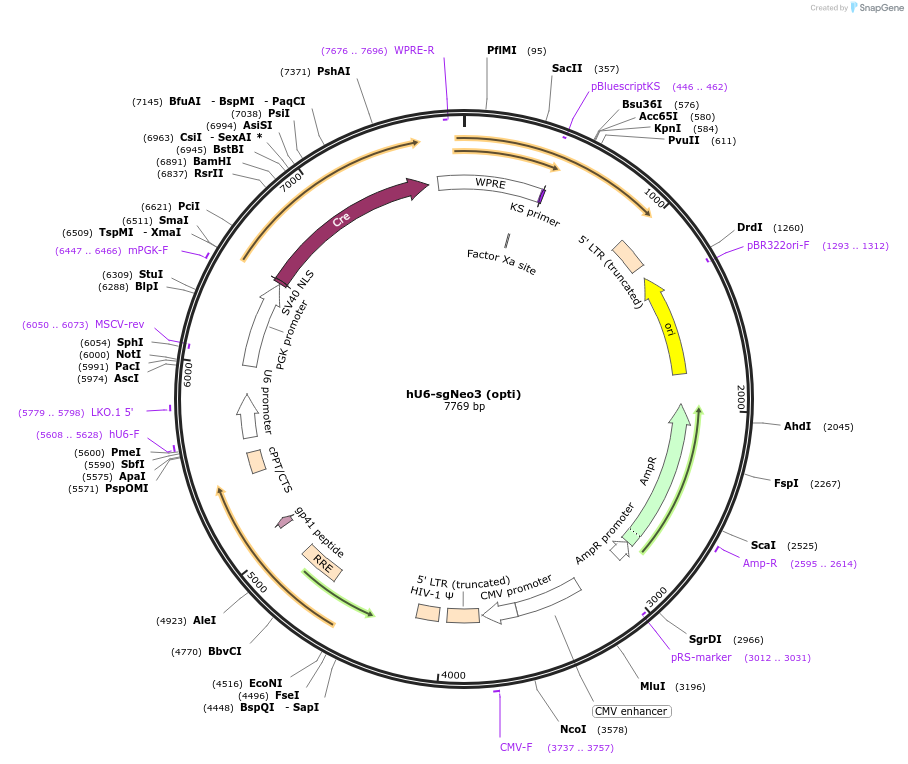
hU6-sgNeo1 (opti)
Plasmid#177240PurposeSame as pLL3.3;U6(BstXI)-XhoI-chimeric RNA;PGK-Cre but XhoI stuffer removed, expresses neomycin gRNADepositorInsertsgNeo1
UseLentiviralPromoterhU6Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
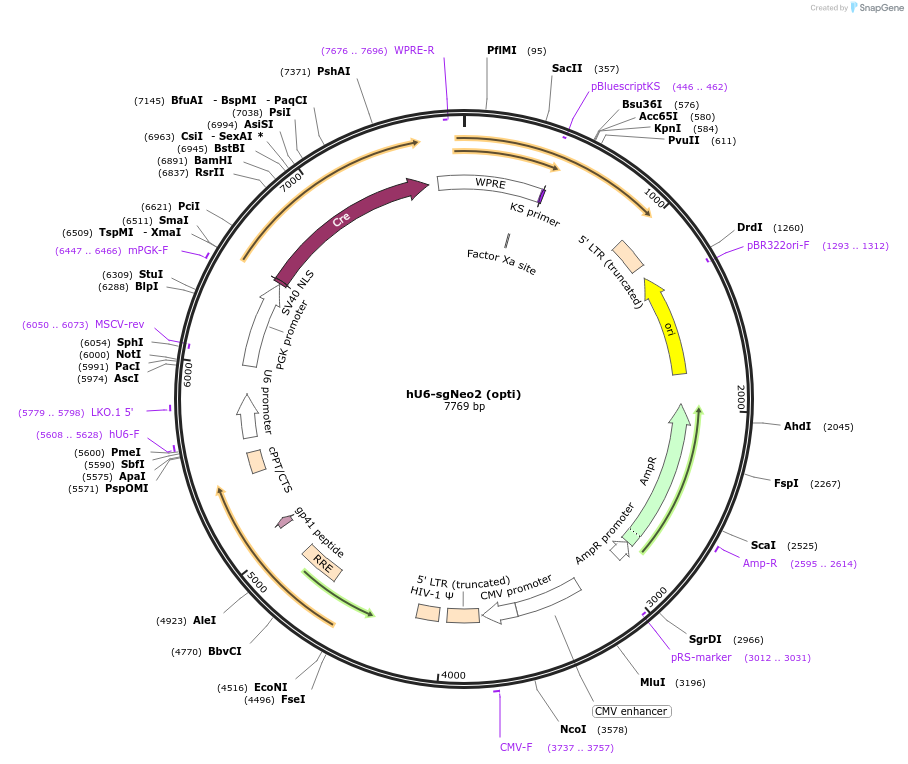
hU6-sgNeo2 (opti)
Plasmid#177241PurposeSame as pLL3.3;U6(BstXI)-XhoI-chimeric RNA;PGK-Cre but XhoI stuffer removed, expresses neomycin gRNADepositorInsertsgNeo2
UseLentiviralPromoterhU6Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
hU6-sgNeo3 (opti)
Plasmid#177242PurposeSame as pLL3.3;U6(BstXI)-XhoI-chimeric RNA;PGK-Cre but XhoI stuffer removed, expresses neomycin gRNADepositorInsertsgNeo3
UseLentiviralPromoterhU6Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
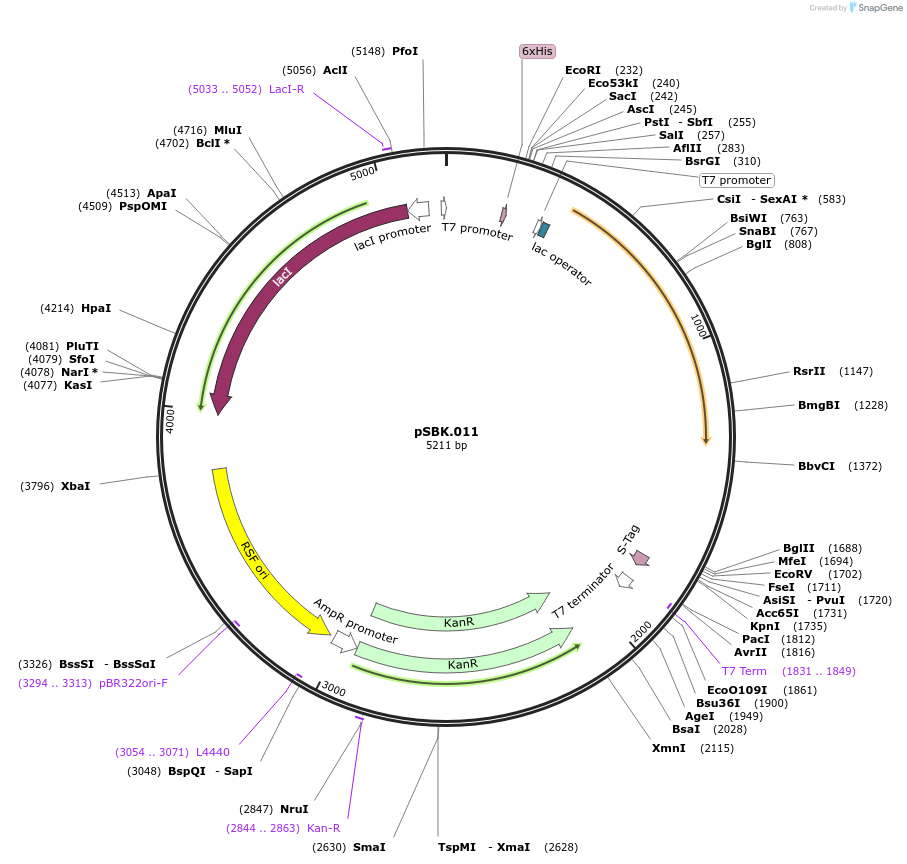
pSBK.011
Plasmid#187211PurposeExpresses retron Eco1 ncRNA v35 (long a1/a2, barcode 3 [CTG-CAG]), and Cas1 + Cas2DepositorInsertsRetron Eco1 ncRNA v35 (long a1/a2, barcode 3 [CTG-CAG])
Cas1 + Cas2
UseSynthetic BiologyExpressionBacterialPromoterT7/LacAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
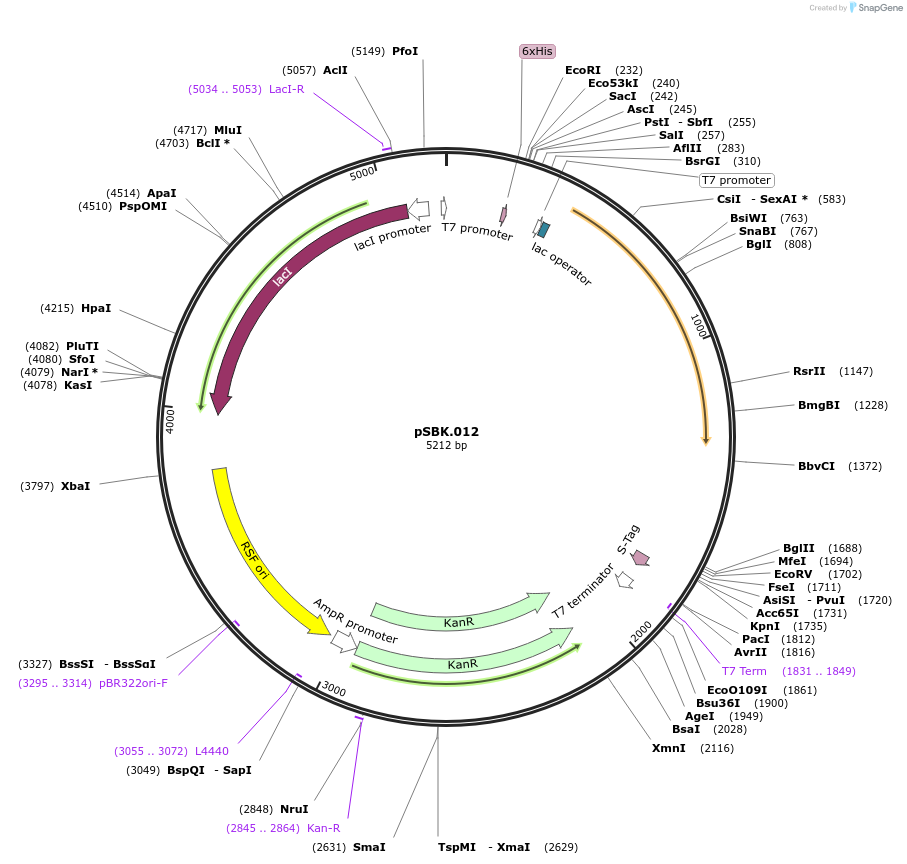
pSBK.012
Plasmid#187212PurposeExpresses retron Eco1 ncRNA v35 (long a1/a2, barcode 4 [GTG-CAC]), and Cas1 + Cas2DepositorInsertsRetron Eco1 ncRNA v35 (long a1/a2, barcode 4 [GTG-CAC])
Cas1 + Cas2
UseSynthetic BiologyExpressionBacterialPromoterT7/LacAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
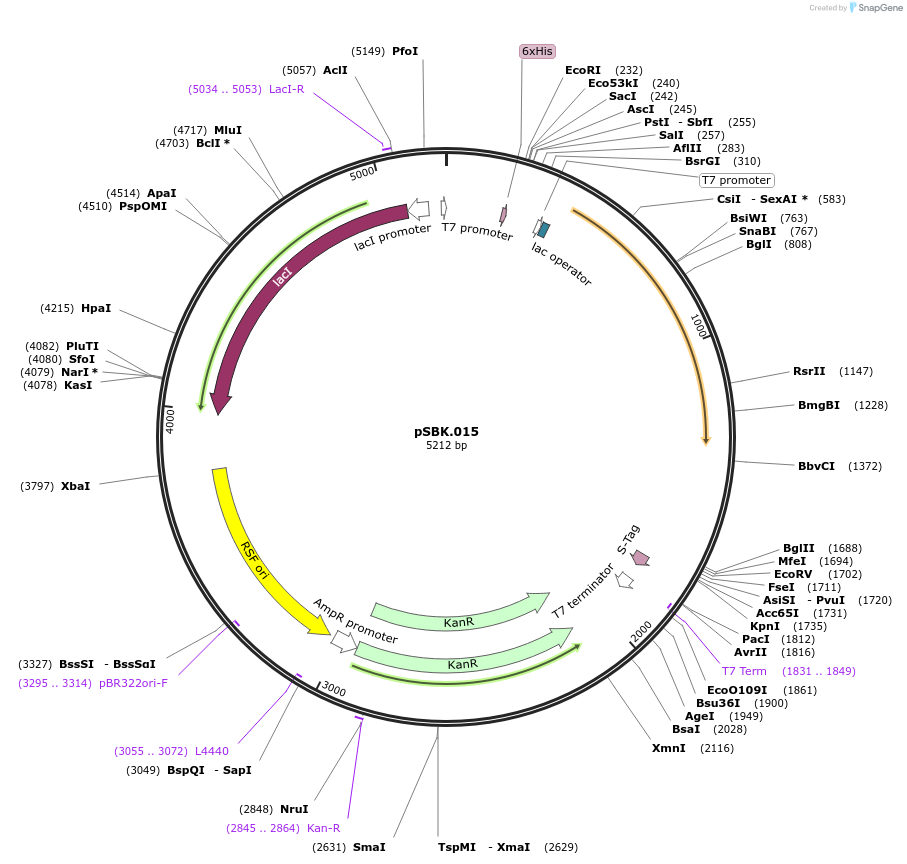
pSBK.015
Plasmid#187216PurposeExpresses retron Eco1 ncRNA v35 (long a1/a2, barcode 7 [GAG-CTC]), and Cas1 + Cas2DepositorInsertsRetron Eco1 ncRNA v35 (long a1/a2, barcode 7 [GAG-CTC])
Cas1 + Cas2
UseSynthetic BiologyExpressionBacterialPromoterT7/LacAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
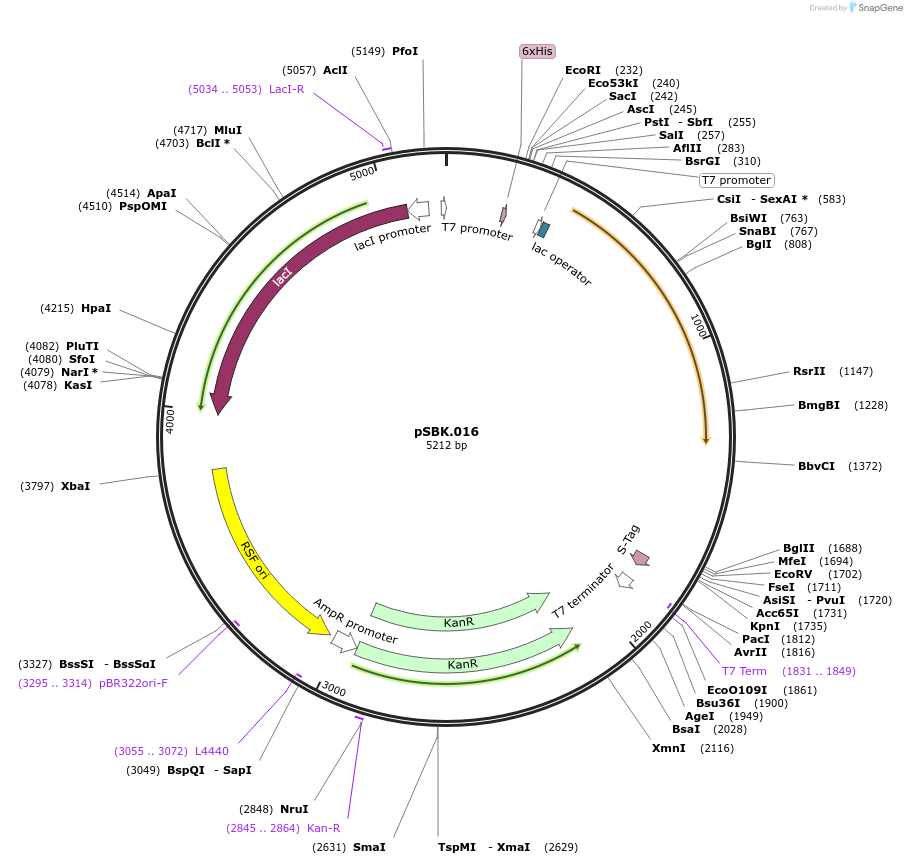
pSBK.016
Plasmid#187217PurposeExpresses retron Eco1 ncRNA v35 (long a1/a2, barcode 8 [GCT-TGC]), and Cas1 + Cas2DepositorInsertsRetron Eco1 ncRNA v35 (long a1/a2, barcode 8 [GCA-TGC])
Cas1 + Cas2
UseSynthetic BiologyExpressionBacterialPromoterT7/LacAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
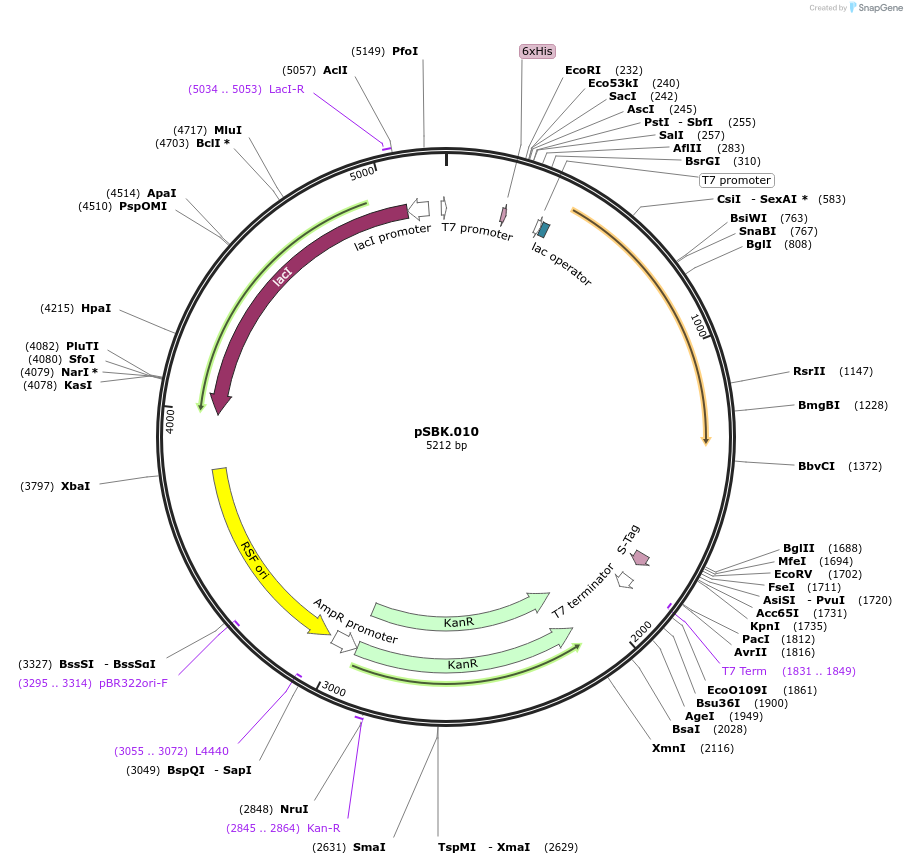
pSBK.010
Plasmid#187210PurposeExpresses retron Eco1 ncRNA v35 (long a1/a2, barcode 2 [GCT-AGC]), and Cas1 + Cas2DepositorInsertsRetron Eco1 ncRNA v35 (long a1/a2, barcode 2 [GCT-AGC])
Cas1 + Cas2
UseSynthetic BiologyExpressionBacterialPromoterT7/LacAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
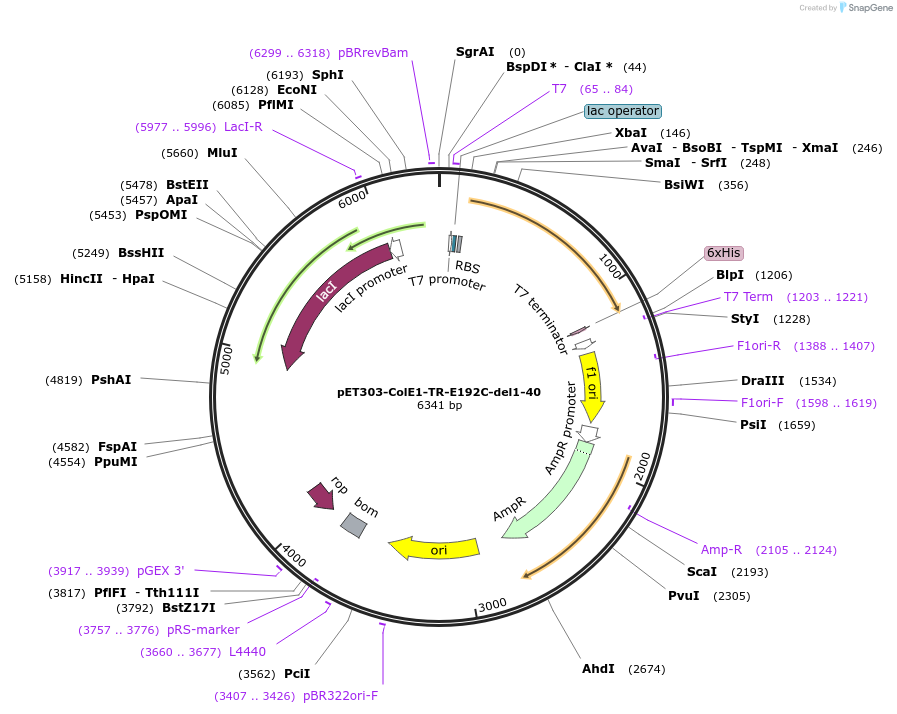
pET303-ColE1-TR-E192C-del1-40
Plasmid#180235PurposeExpresses T and R domains of Colicin E1 with N-terminal 40 residue deletion (residues 41-364) with C terminal cysteine for maleimide chemistryDepositorInsertColicin E1
Tags6x His-tagExpressionBacterialMutationTruncation of ColE1 containing T and R domains (r…PromoterT7Available SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
pDEST-pSP172BSSPE-R3-ccdB/CmR-R5::B1-NLSLacZ-B2
Plasmid#186413PurposeMultisite destination vector for ascidian electroporation of an AttB1/B2 recombined NLSLacZ under control of an AttB3/B5 recombined reg. sequence.DepositorAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
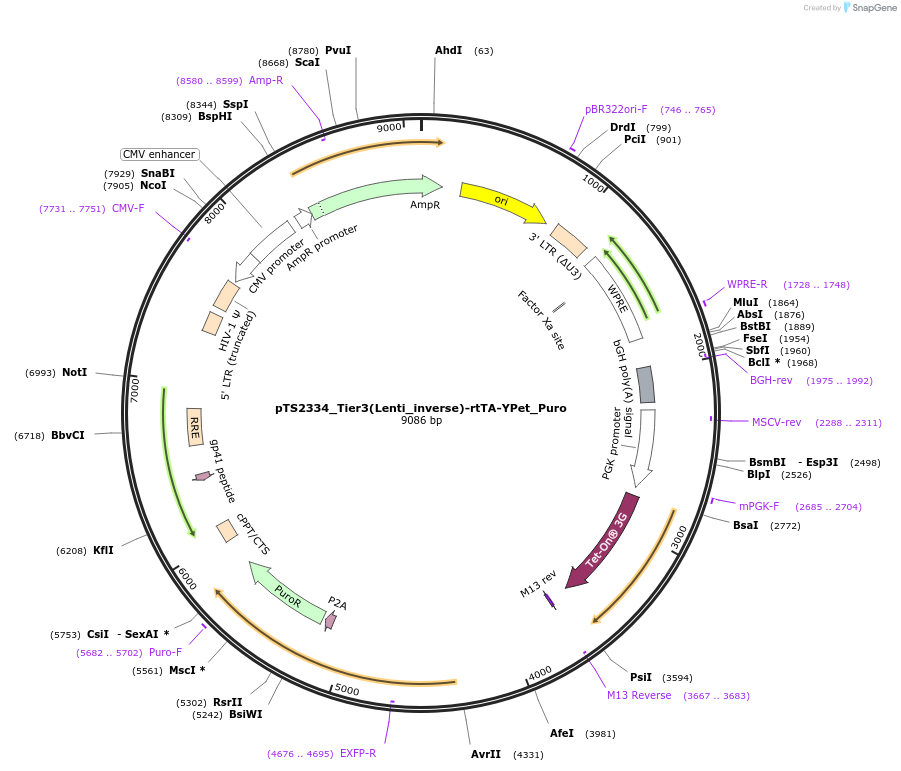
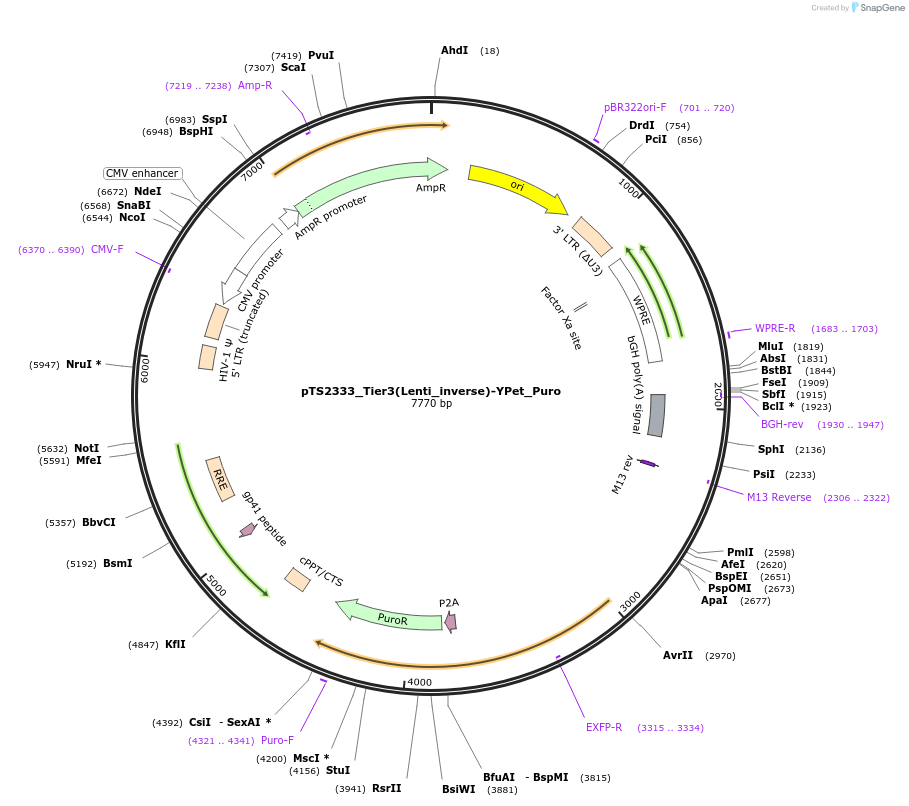
pTS2334_Tier3(Lenti_inverse)-rtTA-YPet_Puro
Plasmid#169665PurposeTier-3 vector for lentiviral stable integration of the rtTA and puromyine - Ypet selection cassette in inverted direction with polyA (PRPBSA-YPet-p2a-PuroR::PPGK-rtTA-pA::A3-pA)DepositorInsertPPGK-driven rtTA expression and PRPBSA-driven YPet and PuroR expression
ExpressionMammalianPromoterPhCMV / PPGK / RPBSAAvailable SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pTS2333_Tier3(Lenti_inverse)-YPet_Puro
Plasmid#169664PurposeTier-3 vector for stable integration via lentiviral transduction in inverted direction with polyA carrying a constitutive expression cassette for puromyine and Ypet (PCMV-5'LTR-Psi-gag-env-PRPBSA-YPet-p2A-PuroR-pA::A2-pA::A3-pA-WPRE-3'LTR)DepositorInsertPRPBSA-driven YPet and PuroR expression
ExpressionMammalianPromoterPhCMV / RPBSAAvailable SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
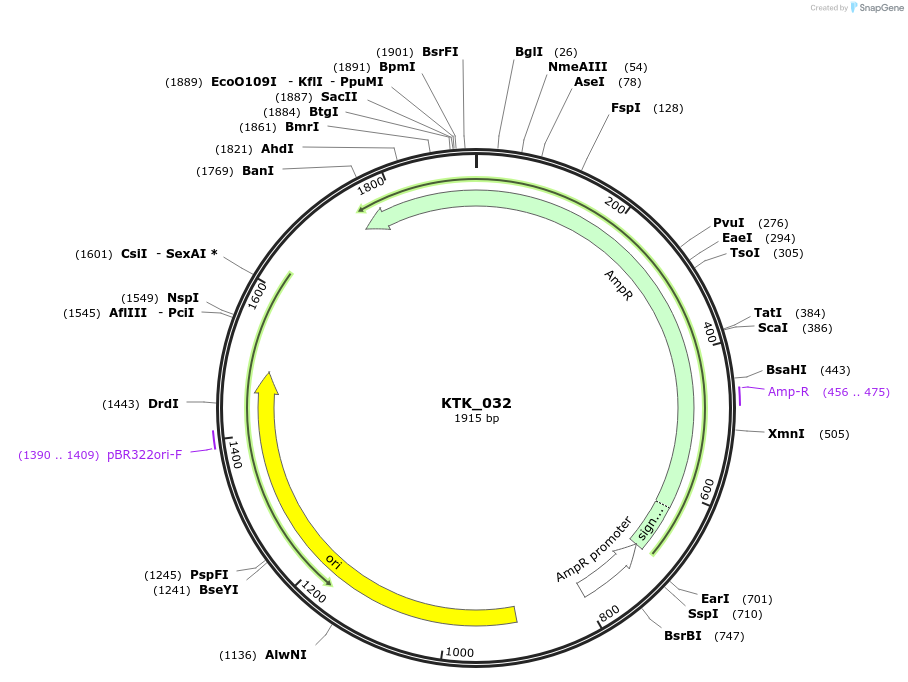
KTK_032
Plasmid#180528PurposeEntry vector containing pLac promoterDepositorInsertpLac promoter
ExpressionBacterialAvailable SinceApril 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
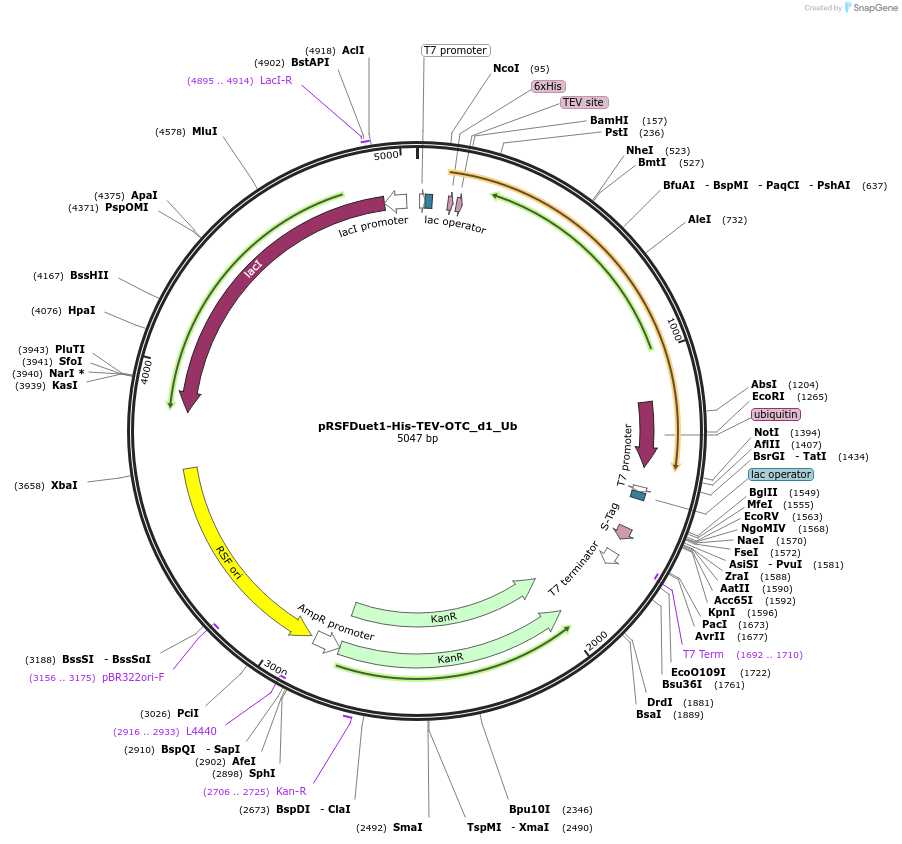
pRSFDuet1-His-TEV-OTC_d1_Ub
Plasmid#170729PurposeScaffold design for studying ubiquitin interacting proteins by cryo-EM.DepositorInsertE. coli ornithine transcarbamylase (OTC), fused to human ubiquitin
Tags6xHis-TEVExpressionBacterialMutationThe C-terminus of OTC was truncated by removing o…PromoterT7Available SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
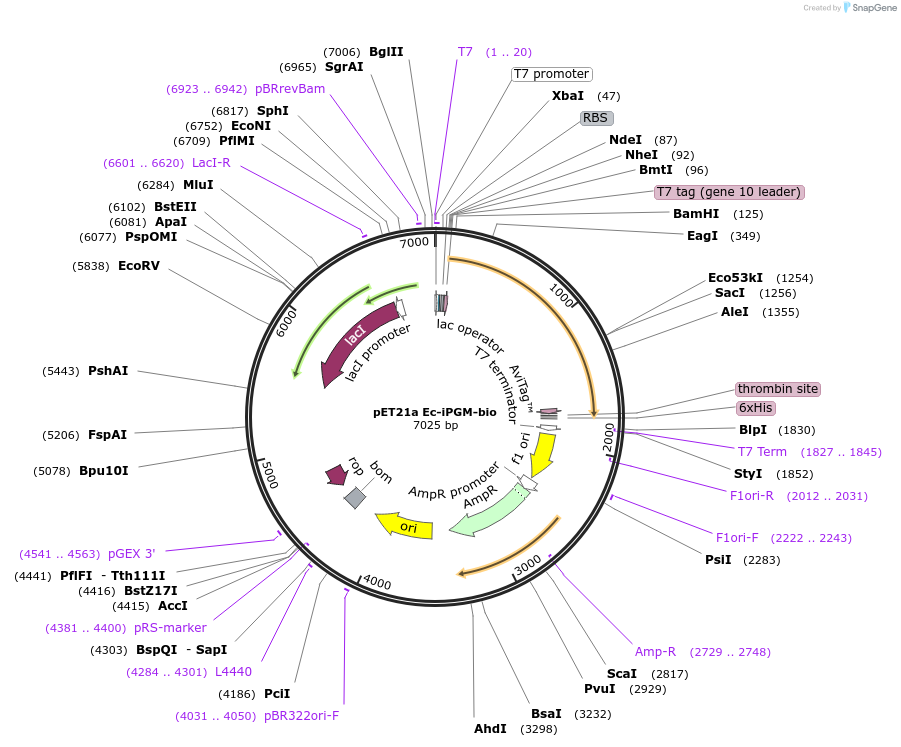
pET21a Ec-iPGM-bio
Plasmid#162575PurposeExpresses E. coli iPGM containing a C-terminal biotinylation site and His tagDepositorInsertE. coli iPGM bio-6His
Tags6His tag, T7 tag, biotinylation sequence, and thr…ExpressionBacterialPromoterT7Available SinceMay 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
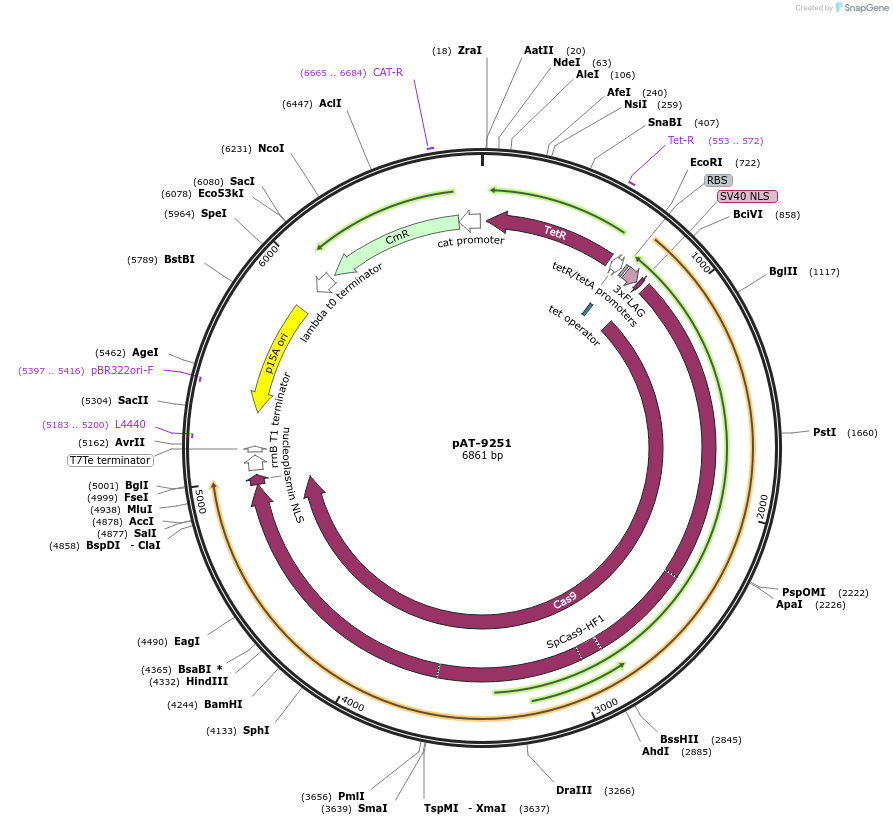
pAT-9251
Plasmid#124226PurposeBacterial SpCas9-HF1 expressionDepositorInsertSpCas9-HF1
UseCRISPRExpressionBacterialPromoterTetR/TetAAvailable SinceFeb. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
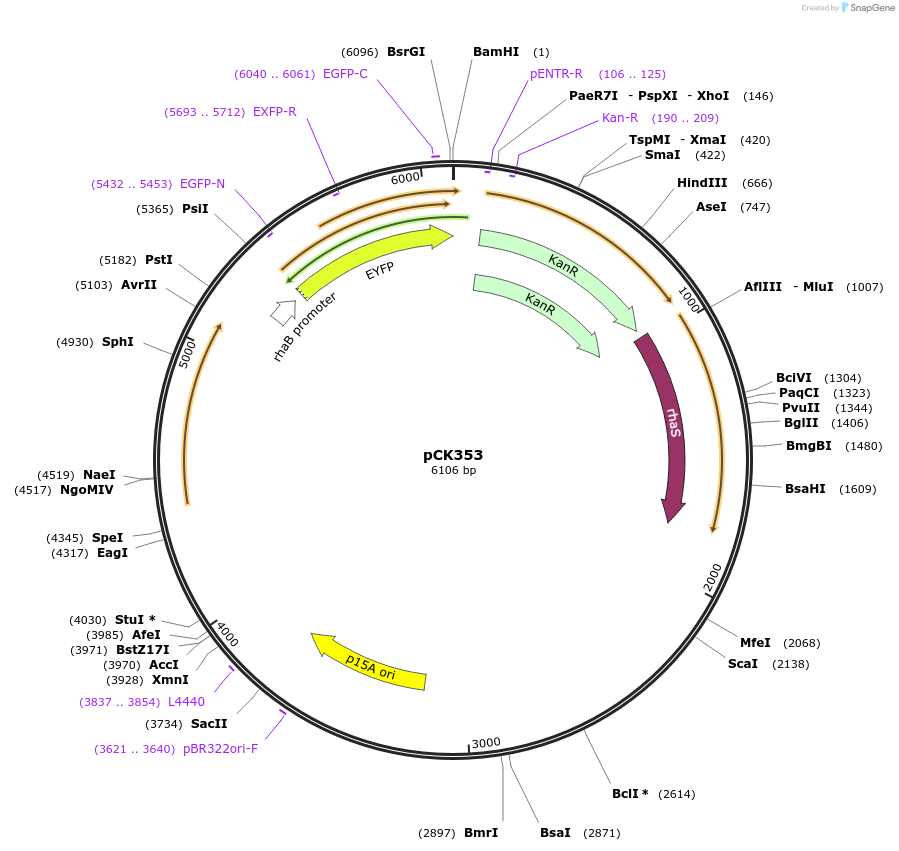
pCK353
Plasmid#129697PurposeFor insulated expression in Synechocystis 6803. As pCK306 (rhaBAD rhamnose-inducible promoter, YFP, an E. coli-Synechocystis shuttle vector, chromosomal-integration sites) + terminator ECK120015170DepositorInsertsPrhaBAD
yfp
rhaS
Terminator ECK120015170
ExpressionBacterialAvailable SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
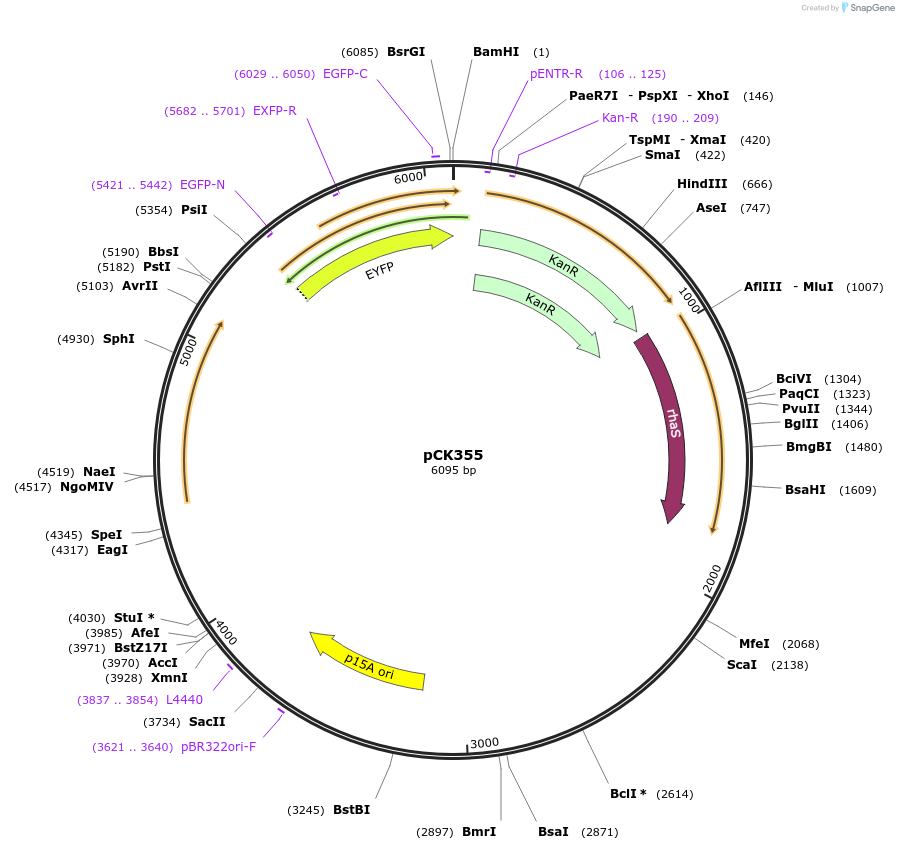
pCK355
Plasmid#129699PurposeFor insulated expression in Synechocystis 6803. As pCK306 (rhaBAD rhamnose-inducible promoter, YFP, an E. coli-Synechocystis shuttle vector, chromosomal-integration sites) + ilvBN terminatorDepositorInsertsPrhaBAD
yfp
rhaS
ilvBN terminator
ExpressionBacterialAvailable SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
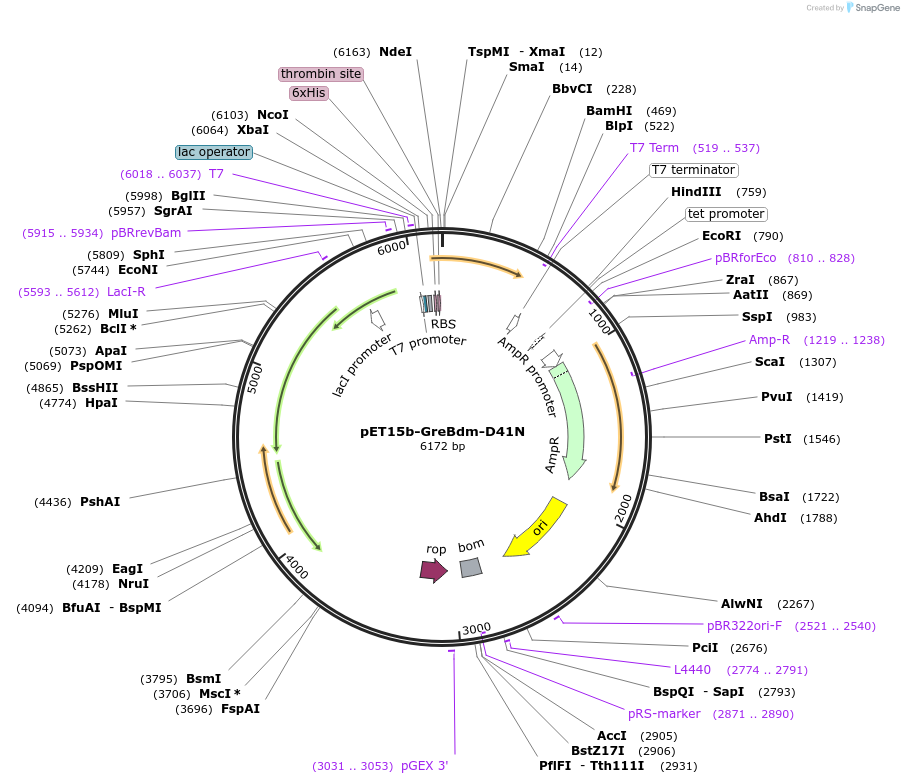
pET15b-GreBdm-D41N
Plasmid#129691Purposeexpress E. coli GreB E82C/C68S/D41NDepositorAvailable SinceSept. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
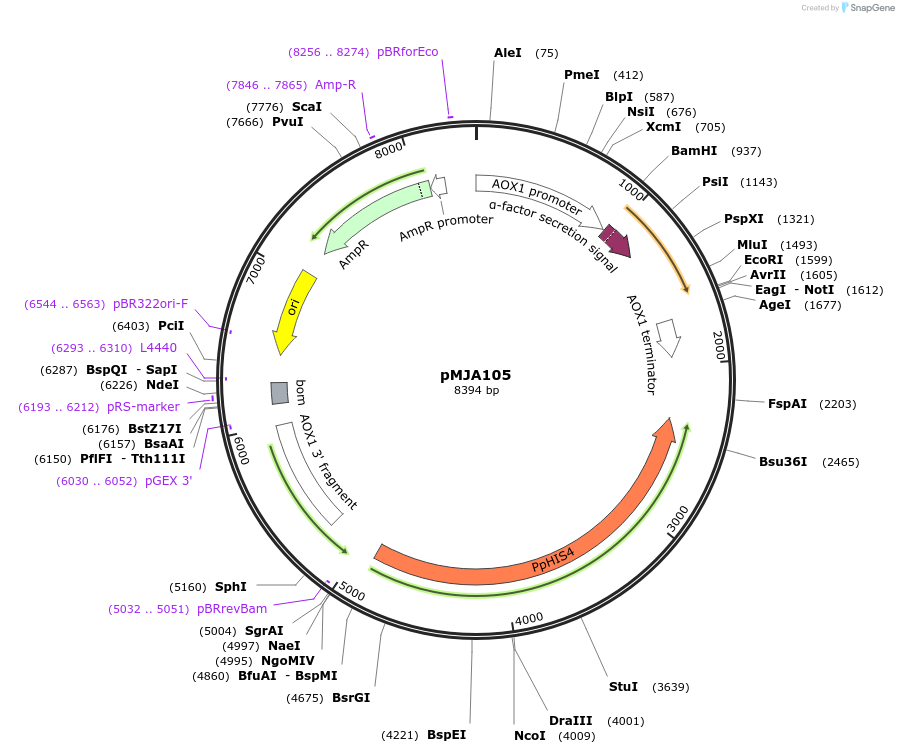
pMJA105
Plasmid#128521PurposeExpresses amyloid human Lysozyme in Pichia pastorisDepositorAvailable SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
pVR2
Plasmid#84719PurposeE. coli RecA promoter followed by stretch with no A's followed by riboswitch for single-round transcriptionDepositorInsertsE. coli RecA promoter
stretch lacking A's followed by 5' UTR of queC gene
ExpressionBacterialMutation12 nt stretch lacking A's inserted before 5&…Available SinceMarch 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
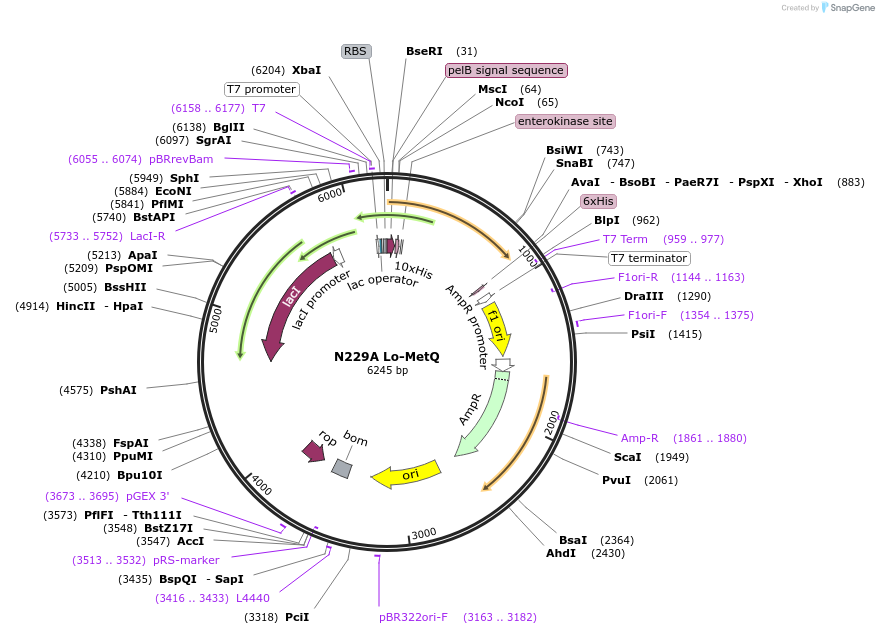
N229A Lo-MetQ
Plasmid#118268PurposeN229A Mutation in Methionine Binding Protein (MetQ) used to solve crystal structureDepositorInsertMetQ (N229A) (metQ E. coli)
Tags10x His Tag and Enterokinase Cleavage SiteExpressionBacterialMutationChanged Asparagine (Asn) 229 to Alanine (Ala)PromoterT7 PromoterAvailable SinceJan. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
-
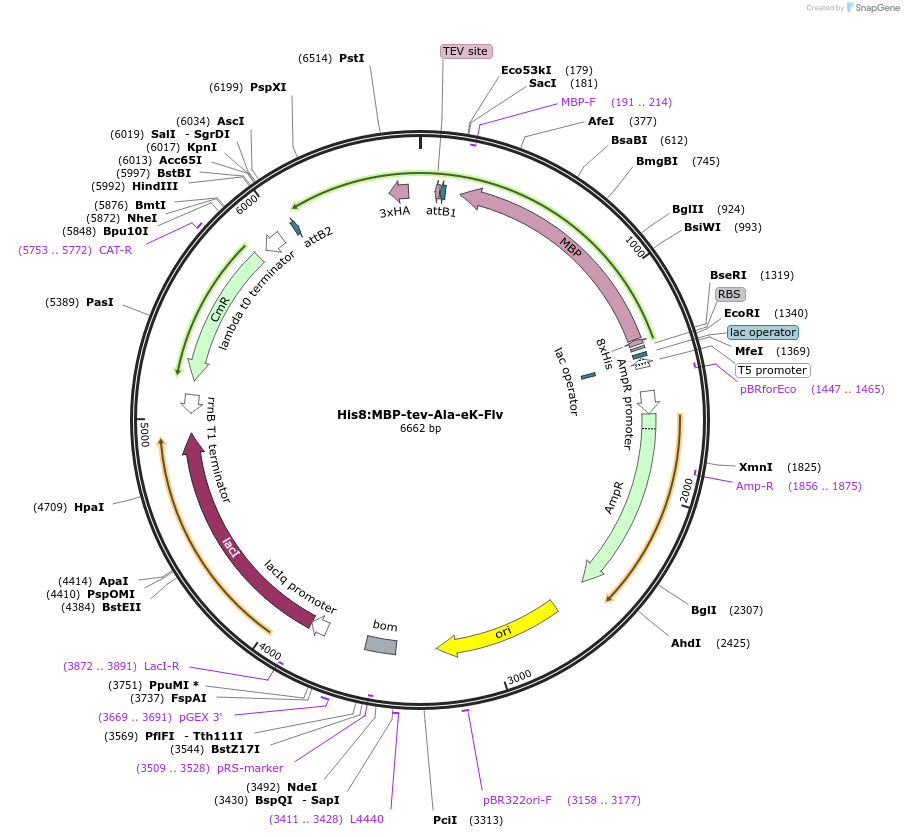
His8:MBP-tev-Ala-eK-Flv
Plasmid#83393Purposebacterial expression vector pVP16 containing a derivative of E. coli lacZ fused with E. coli flavin binding protein MioC to be used in N-end rule in vitro ubiquitination studiesDepositorInsertflavin binding protein MioC
UseSynthetic BiologyTags3xHA, 8xHis, and MBPExpressionBacterialAvailable SinceApril 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
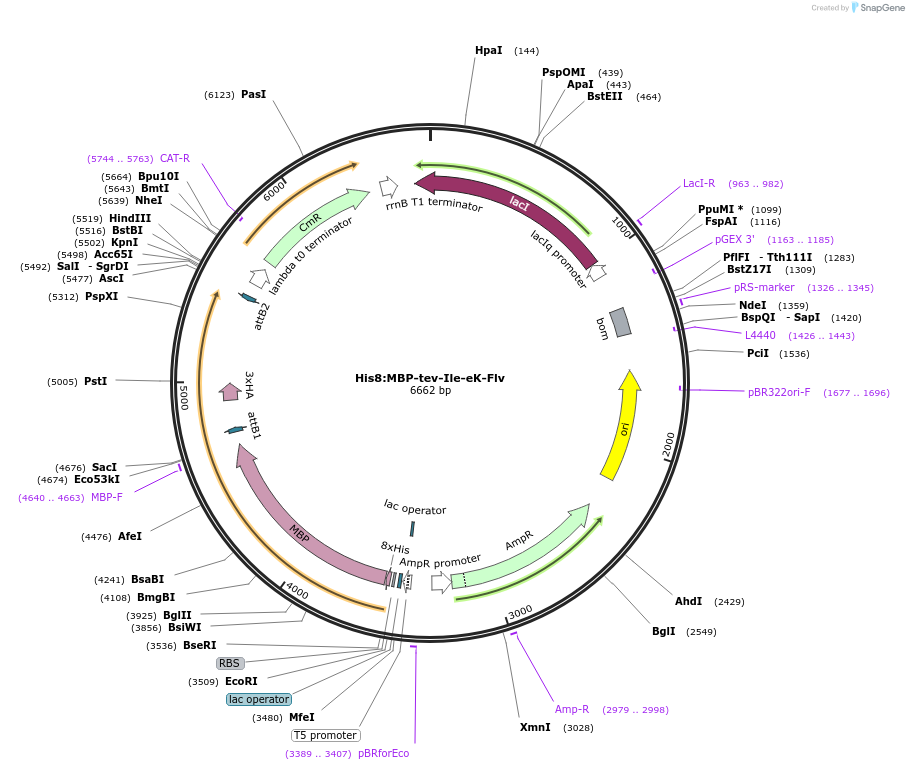
His8:MBP-tev-Ile-eK-Flv
Plasmid#83392Purposebacterial expression vector pVP16 containing a derivative of E. coli lacZ fused with E. coli flavin binding protein MioC to be used in N-end rule in vitro ubiquitination studiesDepositorInsertflavin binding protein MioC
UseSynthetic BiologyTags3xHA, 8xHis, and MBPExpressionBacterialAvailable SinceApril 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
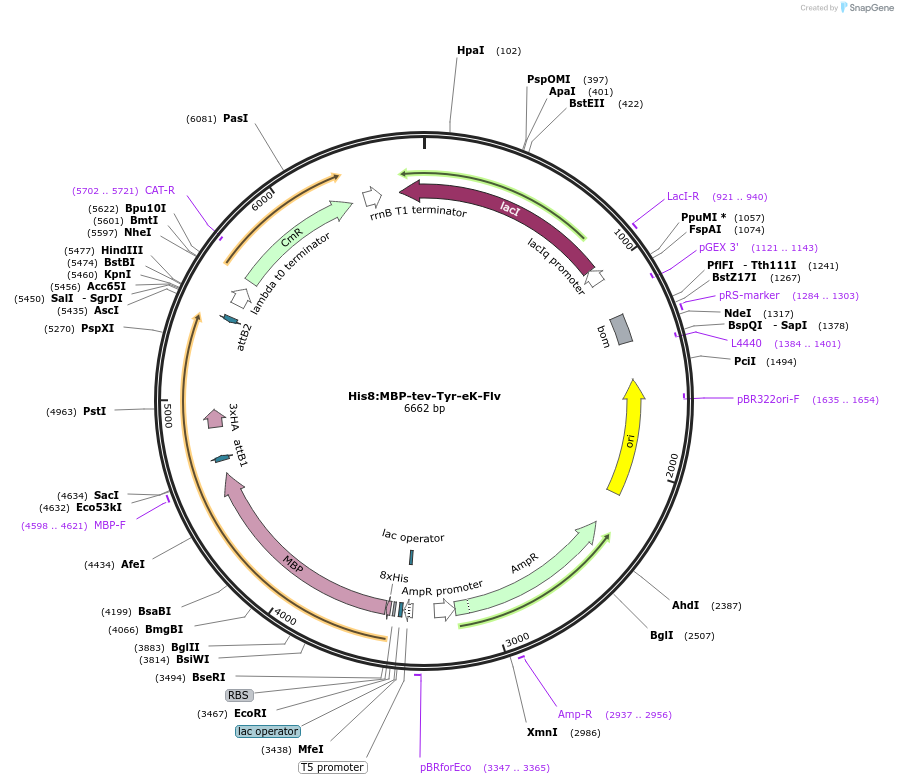
His8:MBP-tev-Tyr-eK-Flv
Plasmid#83391Purposebacterial expression vector pVP16 containing a derivative of E. coli lacZ fused with E. coli flavin binding protein MioC to be used in N-end rule in vitro ubiquitination studiesDepositorInsertflavin binding protein MioC
UseSynthetic BiologyTags3xHA, 8xHis, and MBPExpressionBacterialAvailable SinceApril 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
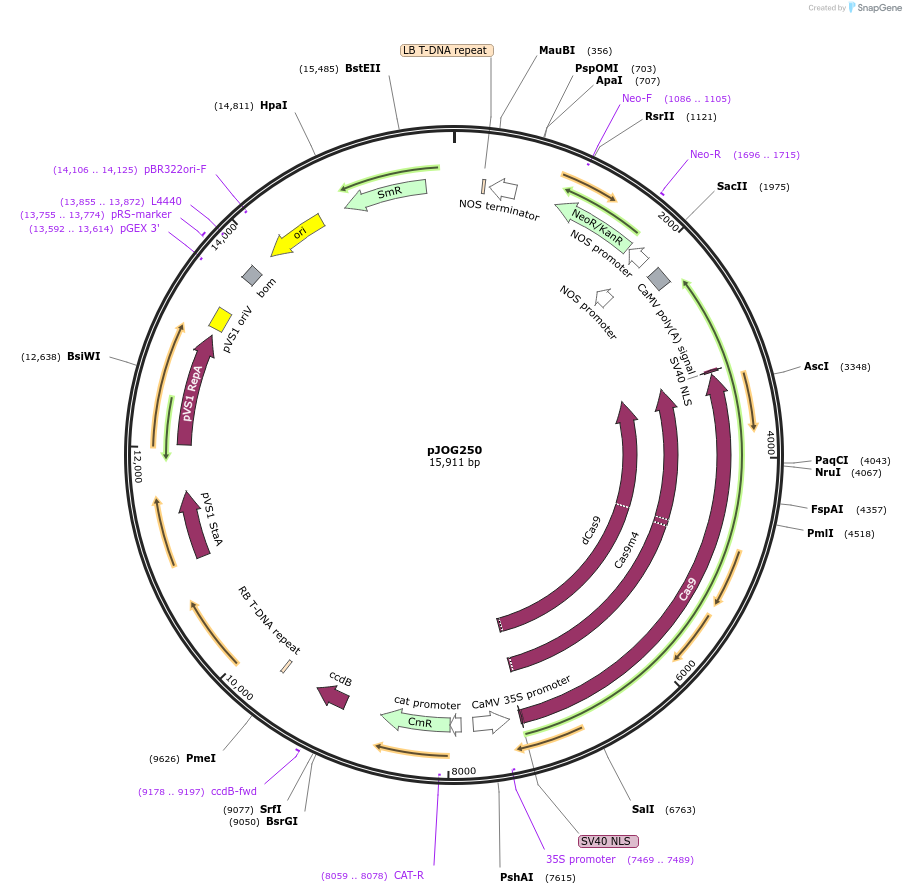
pJOG250
Plasmid#80582Purposerecipient plasmid [multiplexing, activator]; p35S:Cas9 D10A/N863A-Hax3CT, nptIIDepositorInsertspnos:nptII-tnos
p35S:Cas9 D10A/N863A-Hax3(CT)-t35S
BsaI-ccdB_CmR-BsaI
UseCRISPRExpressionPlantMutationD10AAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
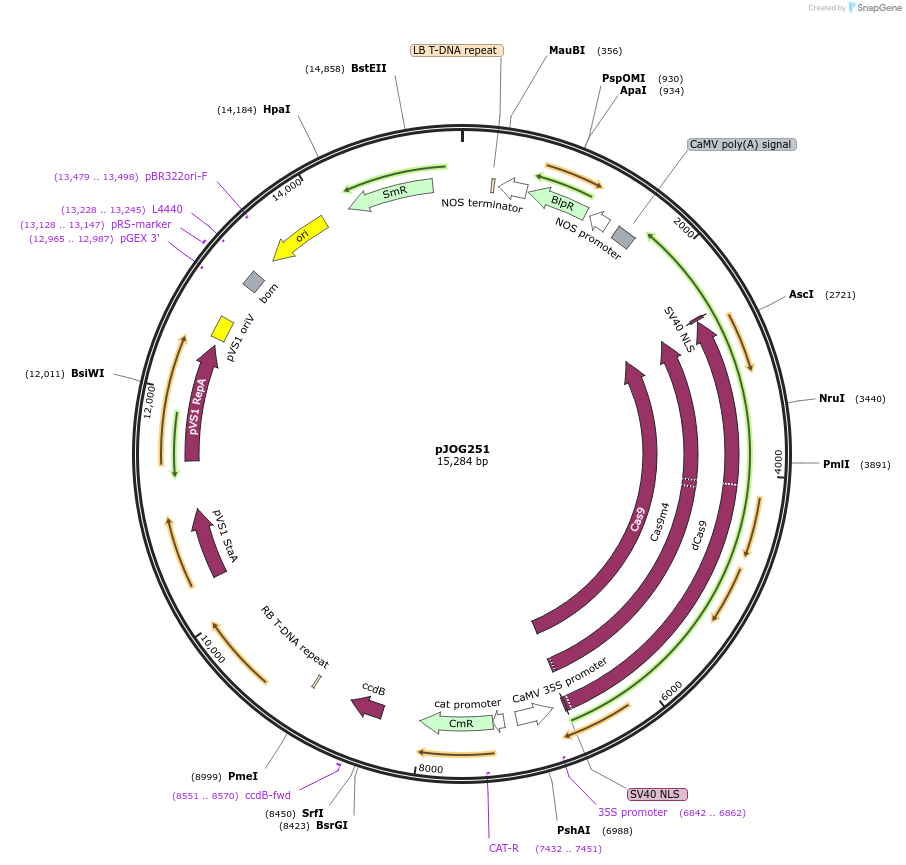
pJOG251
Plasmid#80583Purposerecipient plasmid [multiplexing, activator]; p35S:Cas9 D10A/N863A-Hax3CT, BASTADepositorInsertspnos:PAT-tnos
p35S:Cas9 D10A/N863A-Hax3(CT)-t35S
BsaI-ccdB_CmR-BsaI
UseCRISPRExpressionPlantMutationD10AAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
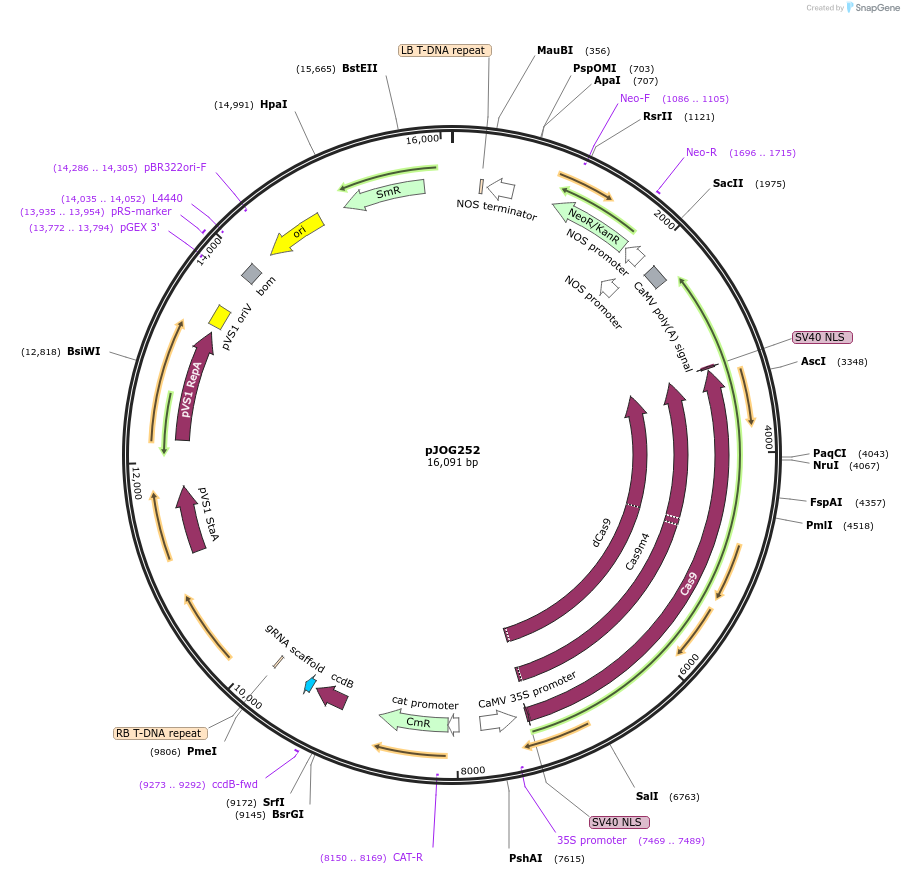
pJOG252
Plasmid#80584Purposerecipient plasmid [single activator]; p35S:Cas9 D10A/N863A-Hax3CT, nptIIDepositorInsertspnos:nptII-tnos
p35S:Cas9 D10A/N863A-Hax3(CT)-t35S
pAtU6-BpiI-ccdB_CmR-BpiI_sgRNA_scaffold
UseCRISPRExpressionPlantMutationD10AAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
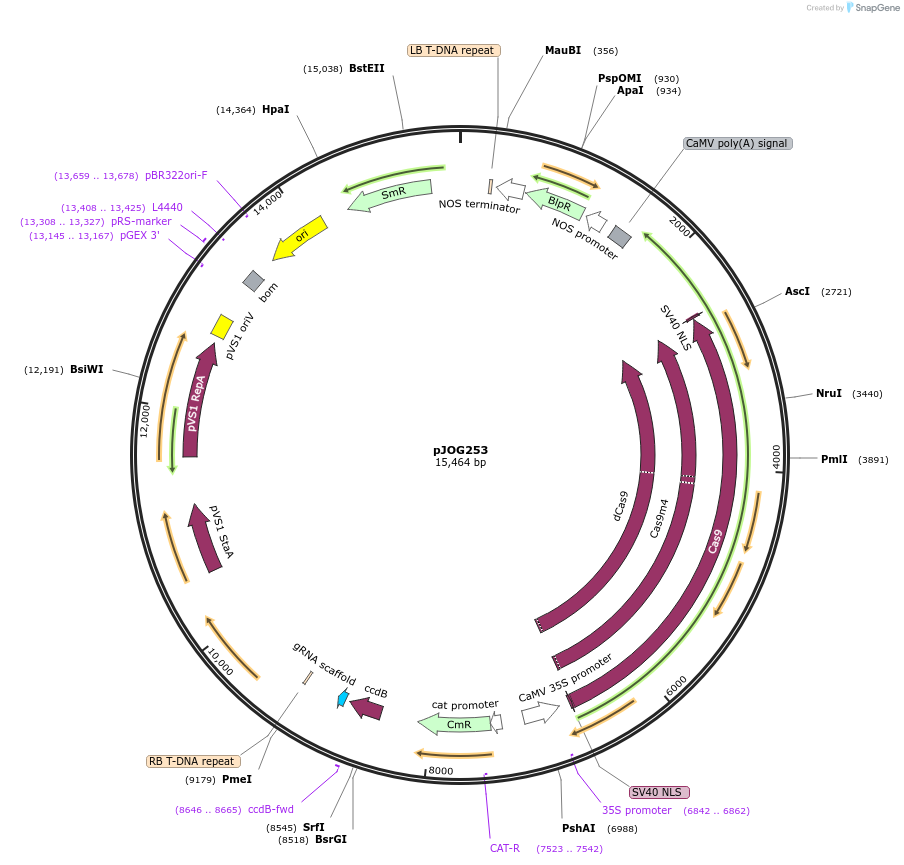
pJOG253
Plasmid#80585Purposerecipient plasmid [single activator]; p35S:Cas9 D10A/N863A-Hax3CT, BASTADepositorInsertspnos:PAT-tnos
p35S:Cas9 D10A/N863A-Hax3(CT)-t35S
pAtU6-BpiI-ccdB_CmR-BpiI_sgRNA_scaffold
UseCRISPRExpressionPlantMutationD10AAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
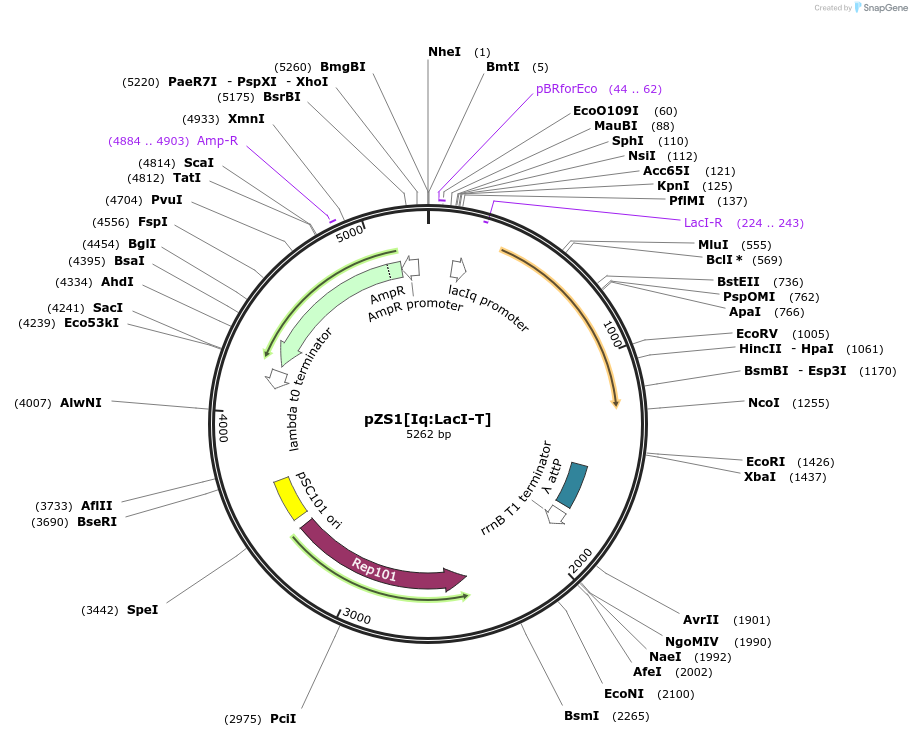
pZS1[Iq:LacI-T]
Plasmid#60756PurposeContains PIq driving expression LacI-T, the Lactose inducible chimera with the TAN DBD.DepositorInsertLacI-T
UseSynthetic BiologyExpressionBacterialMutationLacI without the tetramerization domain and the T…Available SinceJune 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
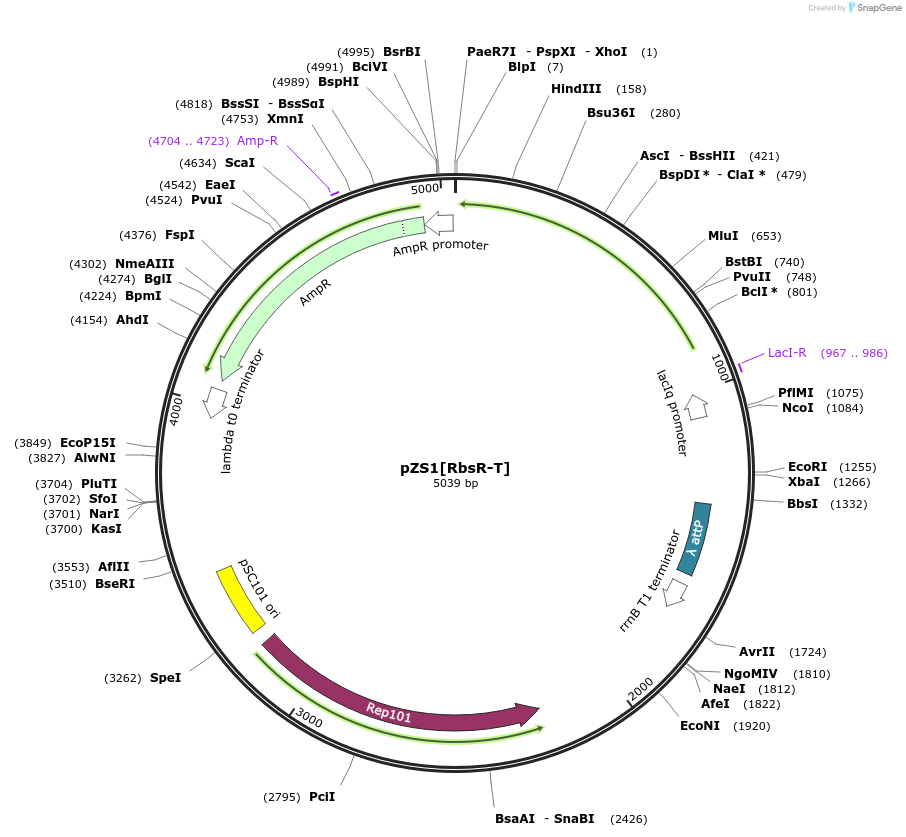
pZS1[RbsR-T]
Plasmid#60751PurposeContains PIq driving expression RbsR-T, the Ribose inducible chimera with the TAN DBD.DepositorInsertRbsR-T
UseSynthetic BiologyExpressionBacterialMutationChimeric LacI/GalR repressor with the TAN DBD and…Available SinceApril 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
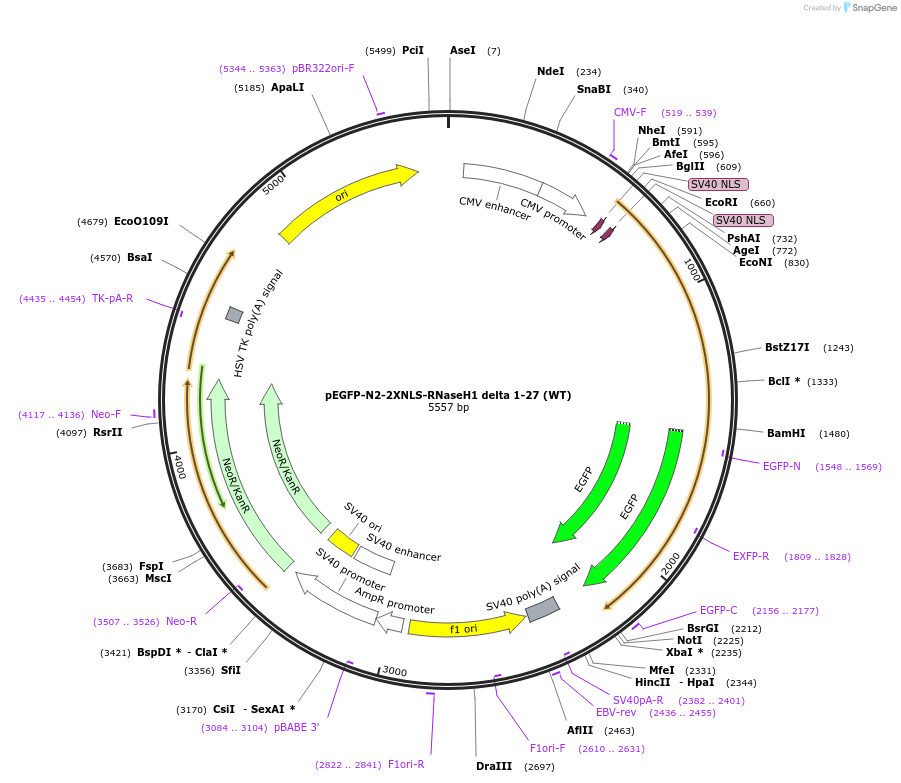
pEGFP-N2-2XNLS-RNaseH1 delta 1-27 (WT)
Plasmid#196702PurposePlasmid for transient mammalian expression of wild type RNase H1 tagged with 2xNLS and EGFP that can be used to specifically degrade the nuclear R-loops.DepositorInserthuman RNaseH1 wild type
ExpressionMammalianAvailable SinceMarch 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
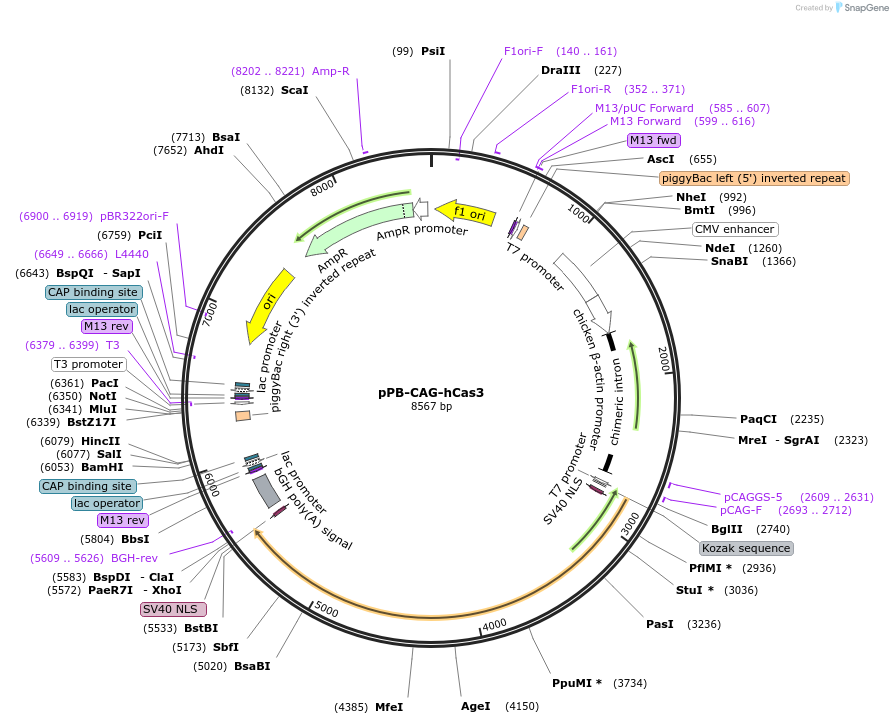
pPB-CAG-hCas3
Plasmid#134920PurposeComponents for genome editing in mammalian cells with pCAG-All-in-one-hCascade and pBS-U6-crRNA-targeted.DepositorInsertCas3 with bpNLS
ExpressionMammalianPromoterCAGAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
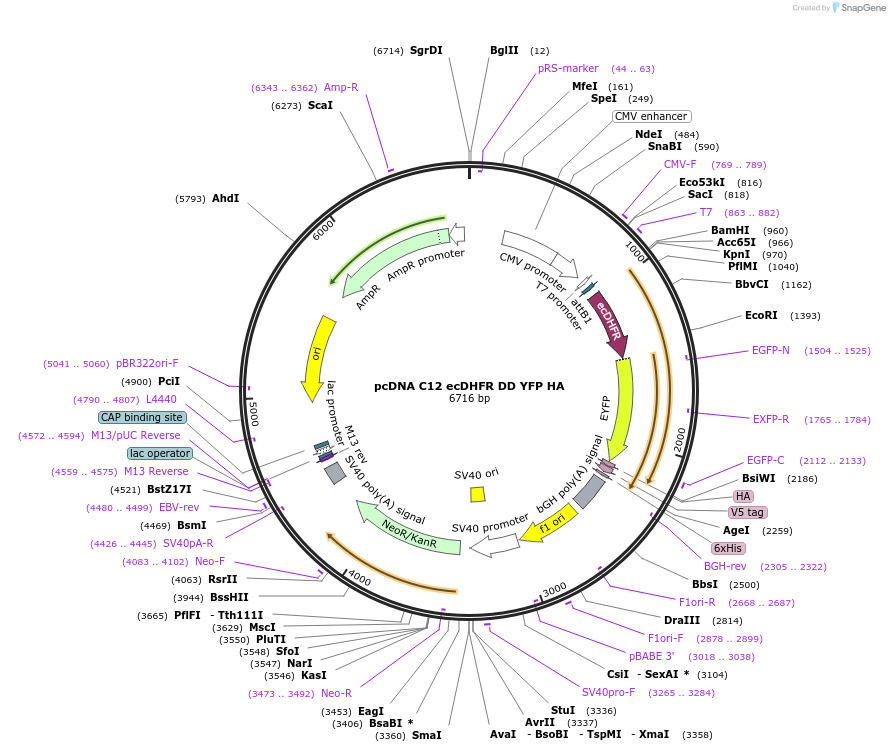
pcDNA C12 ecDHFR DD YFP HA
Plasmid#192815PurposeExpresses an N-terminal enhanced destabilizing domain (DD) version of E. coli DHFR with higher basal turnover in mammalian systems. Contains missense mutations W74R/T113S/E120D/Q146L.DepositorInsertE. coli dihydrofolate reductase
Tagsenhanced yellow fluorescent protein and hemagglut…ExpressionMammalianMutationW74R/T113S/E120D/Q146LPromoterCMVAvailable SinceNov. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
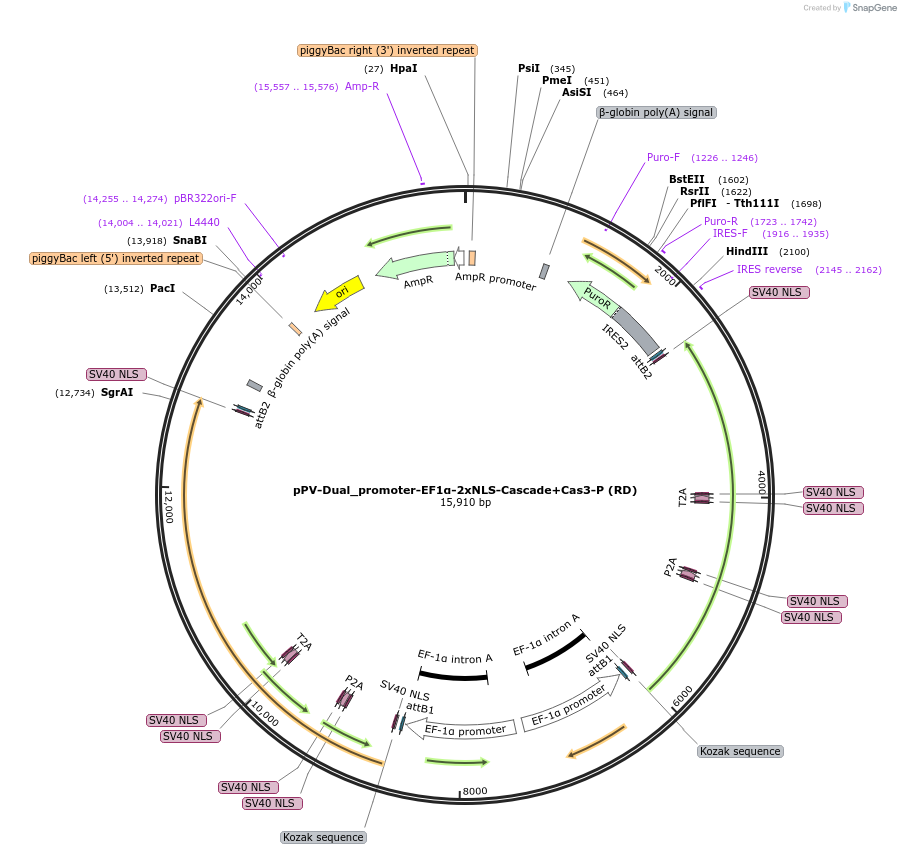
pPV-Dual_promoter-EF1α-2xNLS-Cascade+Cas3-P (RD)
Plasmid#204619PurposeExpression vector of Cascade and Cas3. Puromycin selectable.DepositorInsertCas7, Cas5, Cas8, Cas11, Cas6, and Cas3
UseCRISPRTagsEach protein has C- and N-terminal SV40 NLSPromoterEF1aAvailable SinceAug. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
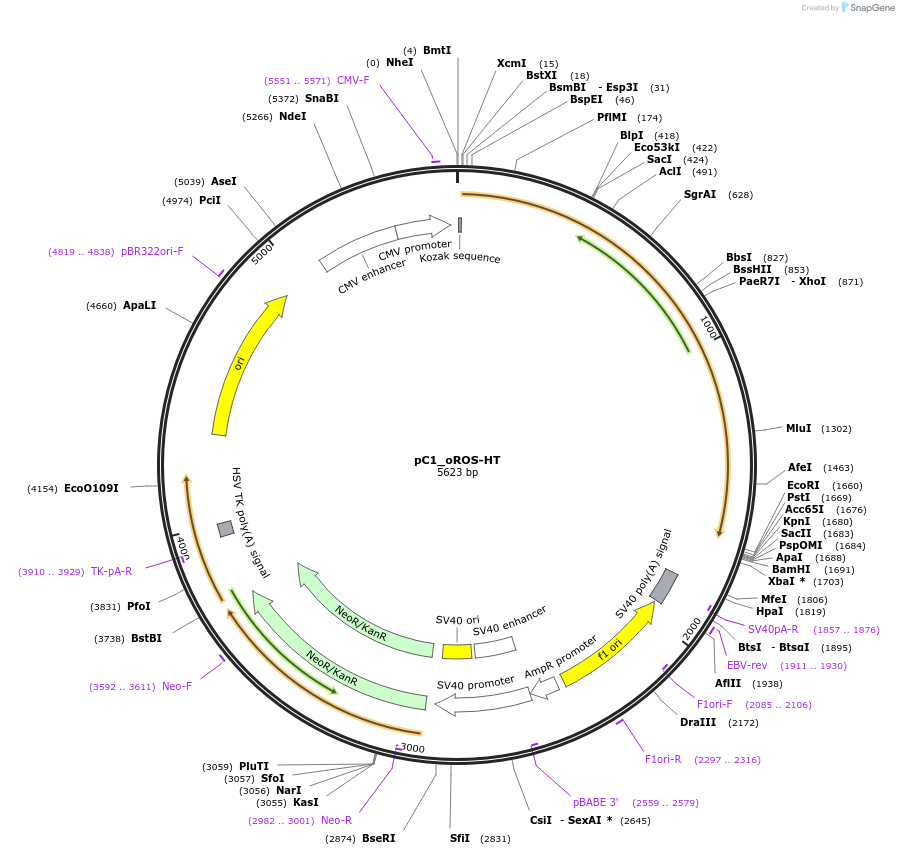
pC1_oROS-HT
Plasmid#216413PurposeExpresses the genetically encoded, chemigenetic hydrogen peroxide sensor oROS-HT in mamalian cells.DepositorInsertoROS-HT
ExpressionMammalianPromoterCMVAvailable SinceJune 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
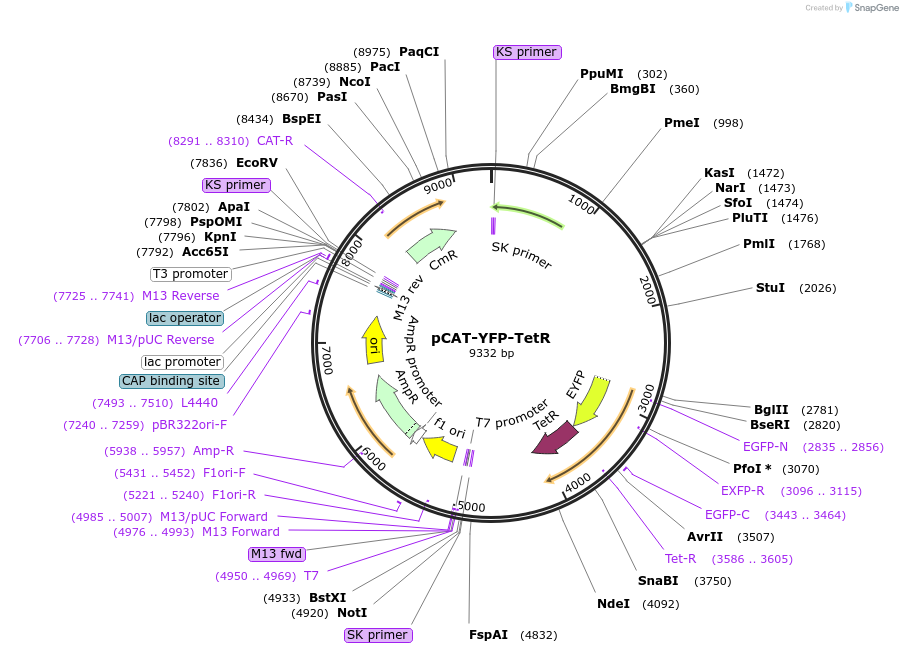
pCAT-YFP-TetR
Plasmid#59018PurposeExpresses a fusion of yellow fluorescent protein (YFP) to the N-terminus of the Escherichia coli Tn10 tet repressor (TetR) driven by the alpha tubulin promoter for use in T. gondiiDepositorInsertYFP-TetR
UseToxoplasma expressionPromoteralpha-tubulinAvailable SinceSept. 29, 2014AvailabilityAcademic Institutions and Nonprofits only -
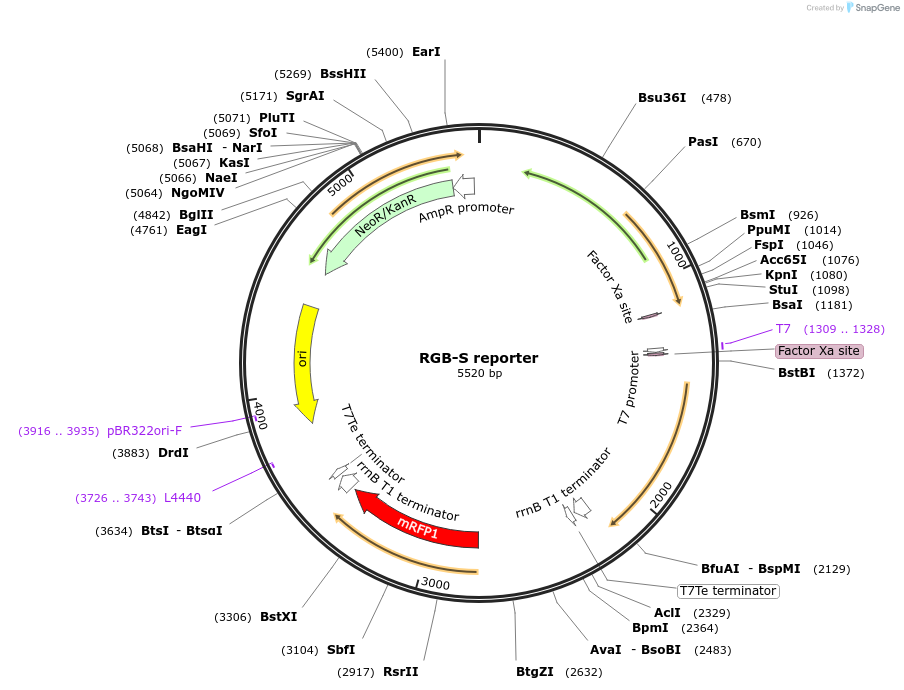
RGB-S reporter
Plasmid#207841PurposeA three-colour stress biosensor for real time analysis of physiological stress, genotoxicity, and cytotoxicity of Escherichia coliDepositorInsertsRpoH sensing construct
SOS sensing construct
RpoS sensing construct
ExpressionBacterialPromoterPosmY, PsulA, and grpEAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
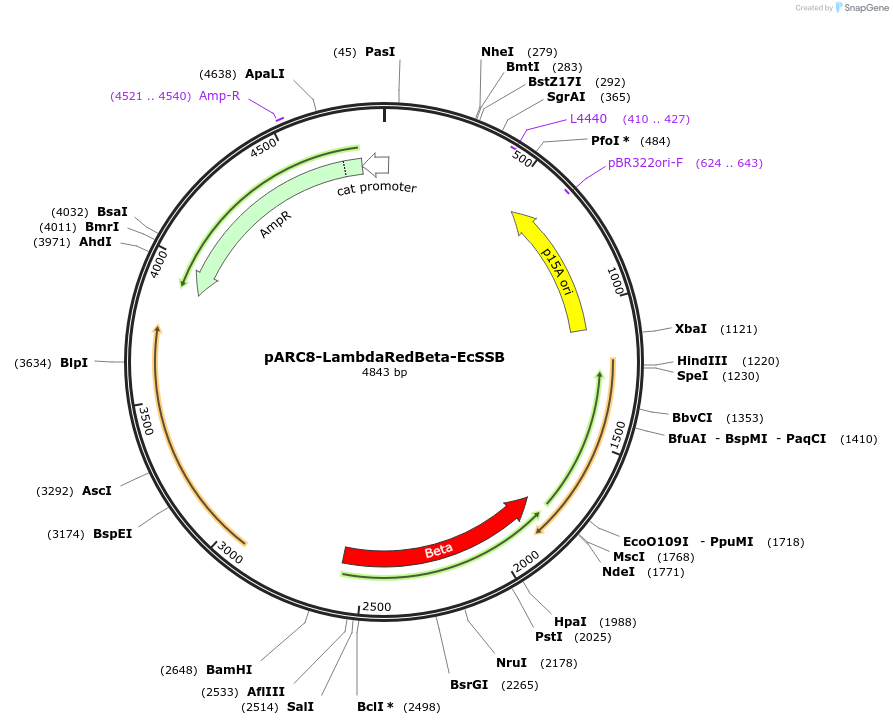
pARC8-LambdaRedBeta-EcSSB
Plasmid#162572PurposeArabinose inducible recombineering plasmid encoding Lambda-Red Beta with EcSSB for use with dsDNA templates and enhanced recombination efficiency.DepositorInsertsLambda Red-Beta
E. coli SSB
ExpressionBacterialPromoterpBADAvailable SinceMay 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
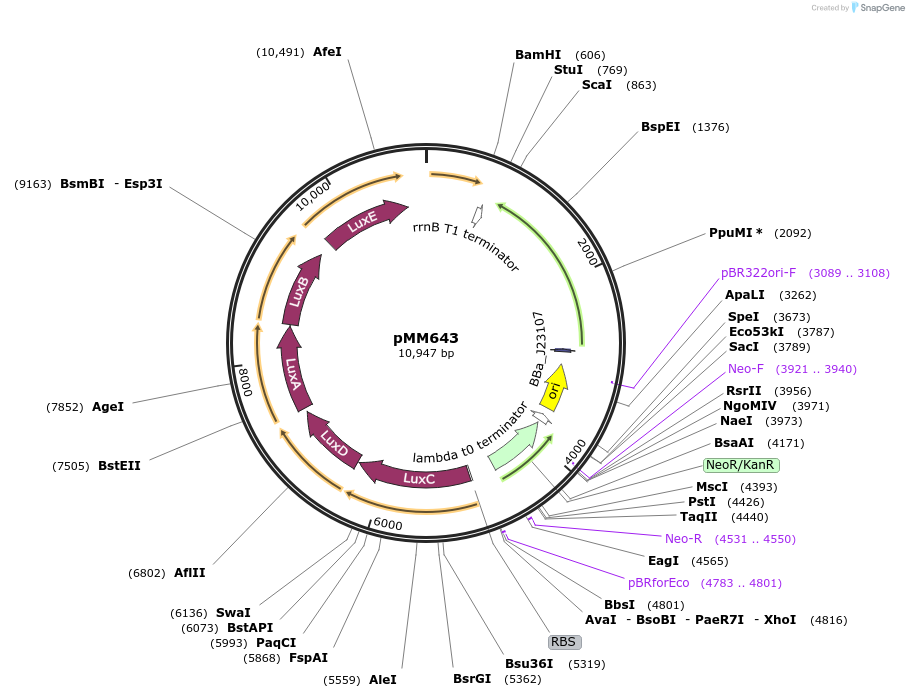
pMM643
Plasmid#112533PurposepZE2-PLhrtO-lux-hrtR-RBS3-chuA - HrtR RBS variant of plasmid pMM627 (Strength 33545.5 AU), ColE1 origin, KanRDepositorInsertsHrtR
ChuA
luxCDABE
ExpressionBacterialMutationA17PPromoterJ23107, PL(HrtO), and ProDAvailable SinceFeb. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
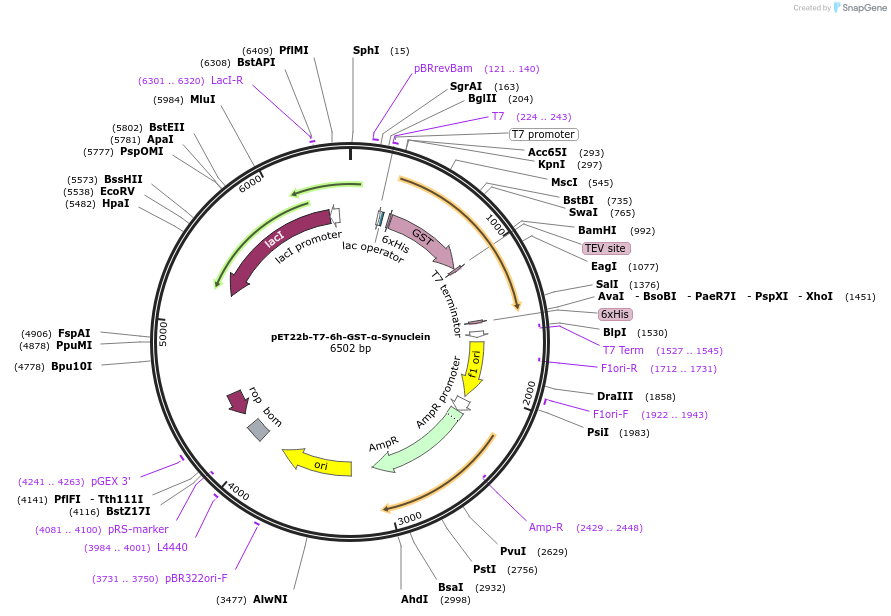
pET22b-T7-6h-GST-α-Synuclein
Plasmid#225224PurposeAlpha synuclein gene fused with gst gene under t7 promoter for bacterial expression of alpha synuclein protein.DepositorInsertsGST
α-Synuclein
Tags6xHis Tag and TEV Cleavage SiteExpressionBacterialPromoterT7Available SinceFeb. 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
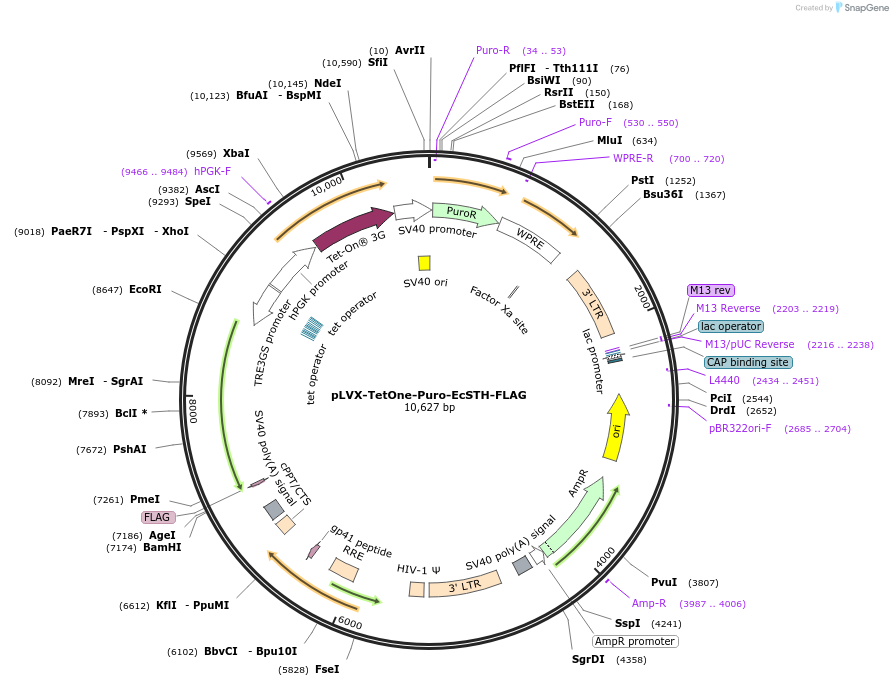
pLVX-TetOne-Puro-EcSTH-FLAG
Plasmid#218628PurposeThis plasmid contains human codon optimized sequence of EcSTH which is a soluble transhydrogenase from E. coli that can be used to increase NADH/NAD+ ratio in mammalian cells.DepositorInsertEcSTH
UseLentiviral and Synthetic BiologyTagsFLAGExpressionMammalianAvailable SinceMay 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
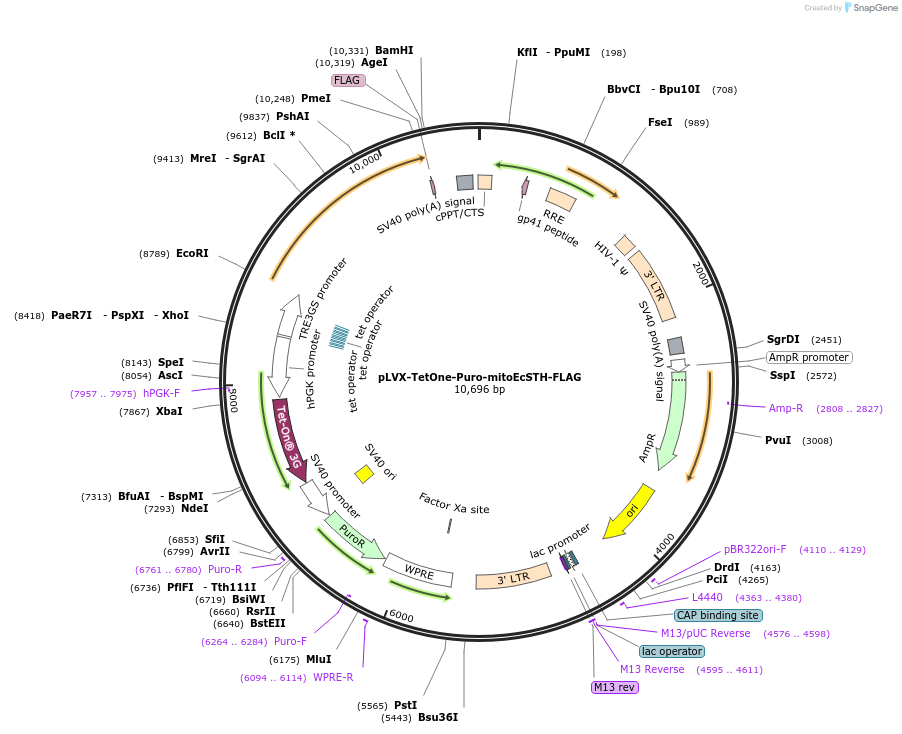
pLVX-TetOne-Puro-mitoEcSTH-FLAG
Plasmid#218629PurposeThis plasmid contains human codon optimized sequence of mitoEcSTH which is a soluble transhydrogenase from E. coli that can be used to increase NADH/NAD+ ratio in mammalian cells.DepositorInsertmitoEcSTH
UseLentiviral and Synthetic BiologyTagsFLAG and MTSExpressionMammalianAvailable SinceMay 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
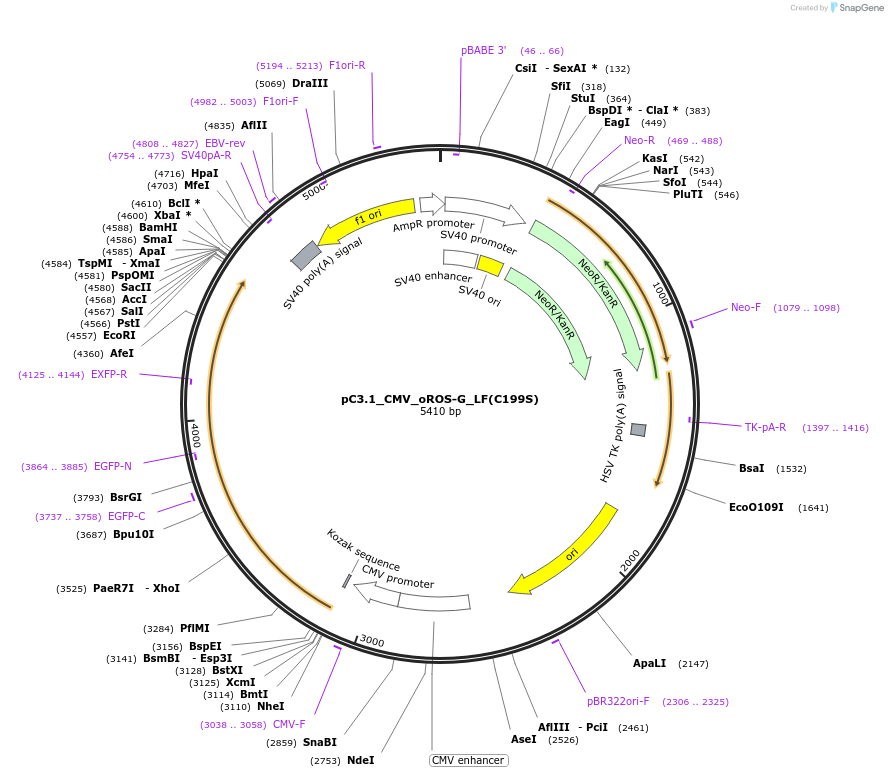
pC3.1_CMV_oROS-G_LF(C199S)
Plasmid#216112PurposeExpresses the loss-of-function mutations C199S of the genetically encoded green fluorescent hydrogen peroxide sensor oROS-G in mamalian cells.DepositorInsertoROS-G_LF(C199S)
ExpressionMammalianPromoterCMVAvailable SinceMay 28, 2024AvailabilityAcademic Institutions and Nonprofits only