We narrowed to 11,082 results for: CHL
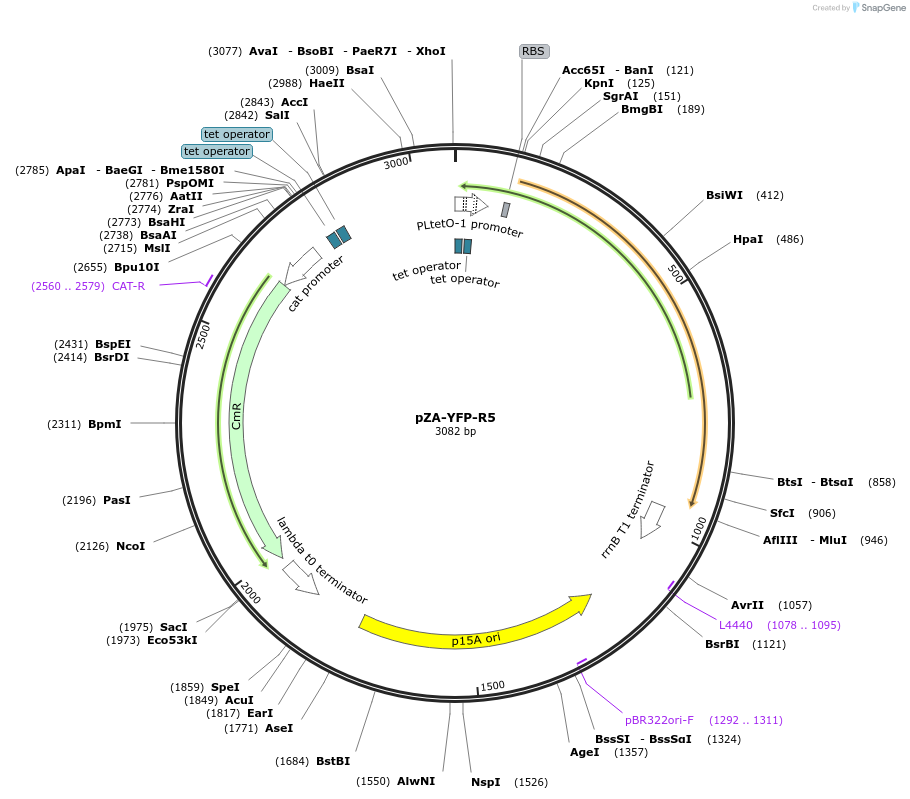
-
Plasmid#106257PurposeExpresses Venus YFP fused with silaffin-R5 silica affinity peptide under the control of TetODepositorInsertYellow fluorescent protein
TagsAdded silaffin R5 peptide to C terminus of YFP af…ExpressionBacterialPromoterpL-TetOAvailable SinceJan. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
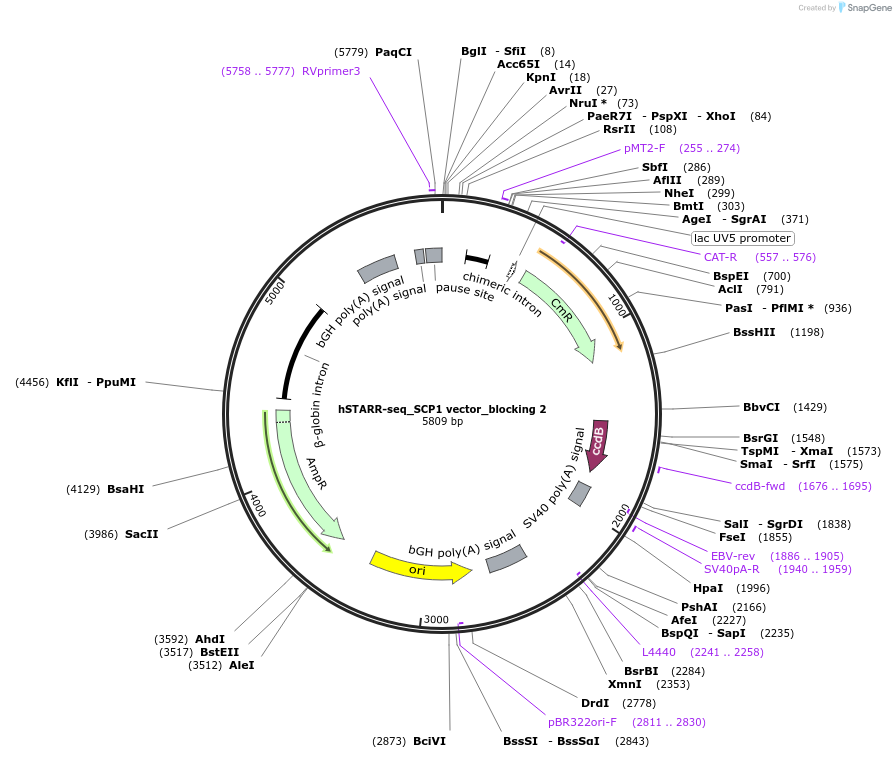
hSTARR-seq_SCP1 vector_blocking 2
Plasmid#99317PurposeVector to measure enhancer activity of candidates from a DNA library by STARR-seq.DepositorTypeEmpty backboneUseHstarr-seq screening vectorExpressionMammalianAvailable SinceSept. 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
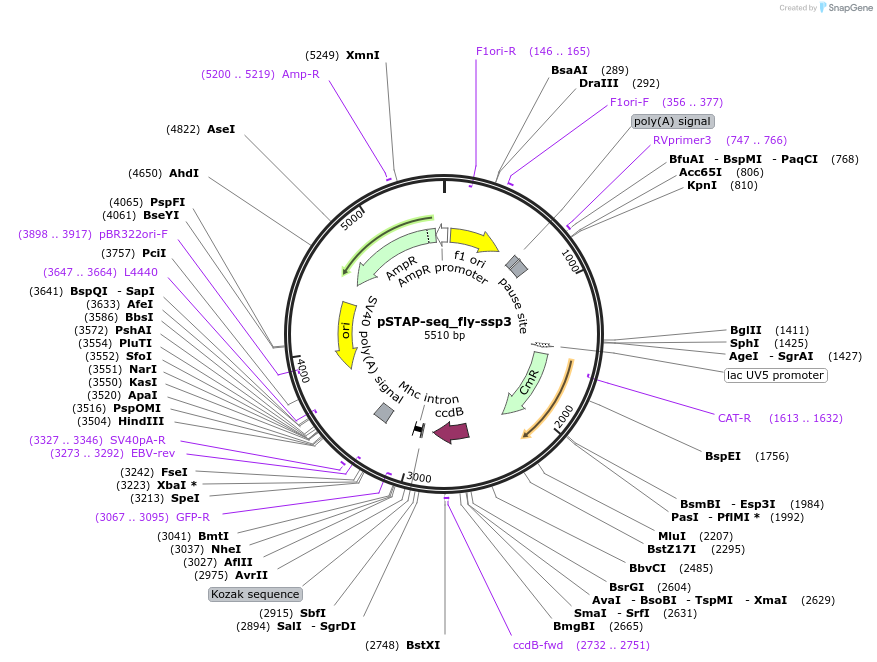
pSTAP-seq_fly-ssp3
Plasmid#86382PurposeVector to measure enhancer responsiveness of candidates from a genomic library (cloned instead of the core promoter) by determining the abundance of transcripts originating from each candidate.DepositorInsertD. melanogaster ssp3 enhancer
UseStap-seq screening vectorAvailable SinceMarch 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
pSTAP-seq_fly-ham
Plasmid#86383PurposeVector to measure enhancer responsiveness of candidates from a genomic library (cloned instead of the core promoter) by determining the abundance of transcripts originating from each candidate.DepositorInsertD. melanogaster ham enhancer
UseStap-seq screening vectorAvailable SinceMarch 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
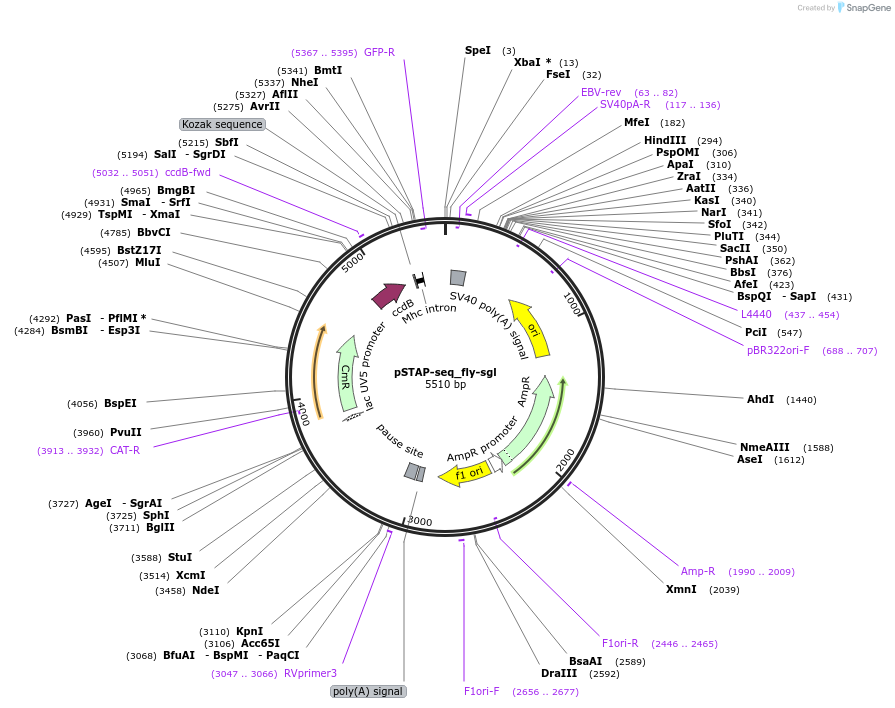
pSTAP-seq_fly-sgl
Plasmid#86384PurposeVector to measure enhancer responsiveness of candidates from a genomic library (cloned instead of the core promoter) by determining the abundance of transcripts originating from each candidate.DepositorInsertD. melanogaster sgl enhancer
UseStap-seq screening vectorAvailable SinceMarch 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
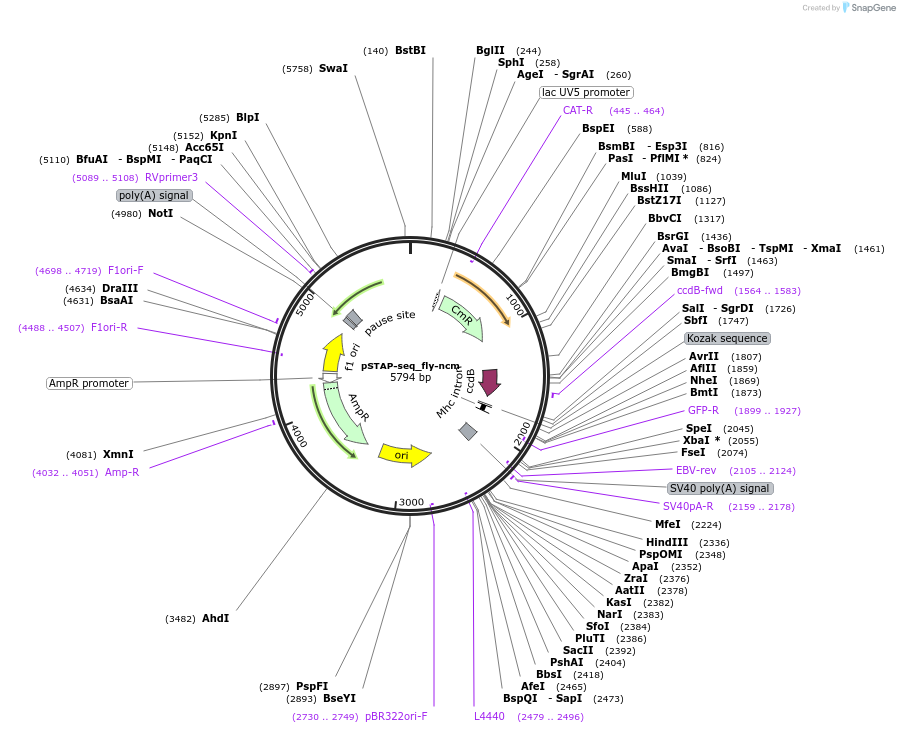
pSTAP-seq_fly-ncm
Plasmid#86385PurposeVector to measure enhancer responsiveness of candidates from a genomic library (cloned instead of the core promoter) by determining the abundance of transcripts originating from each candidate.DepositorInsertD. melanogaster ncm enhancer
UseStap-seq screening vectorAvailable SinceMarch 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
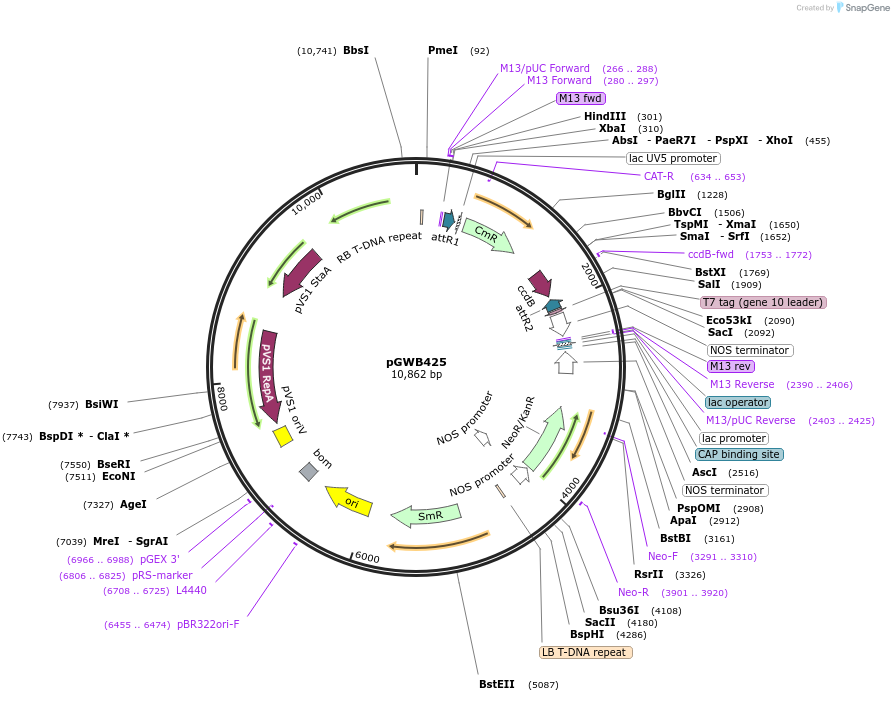
pGWB425
Plasmid#74819PurposeGateway cloning compatible binary vector for C-terminal fusion with T7 (no promoter).DepositorTypeEmpty backboneExpressionPlantAvailable SinceJuly 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
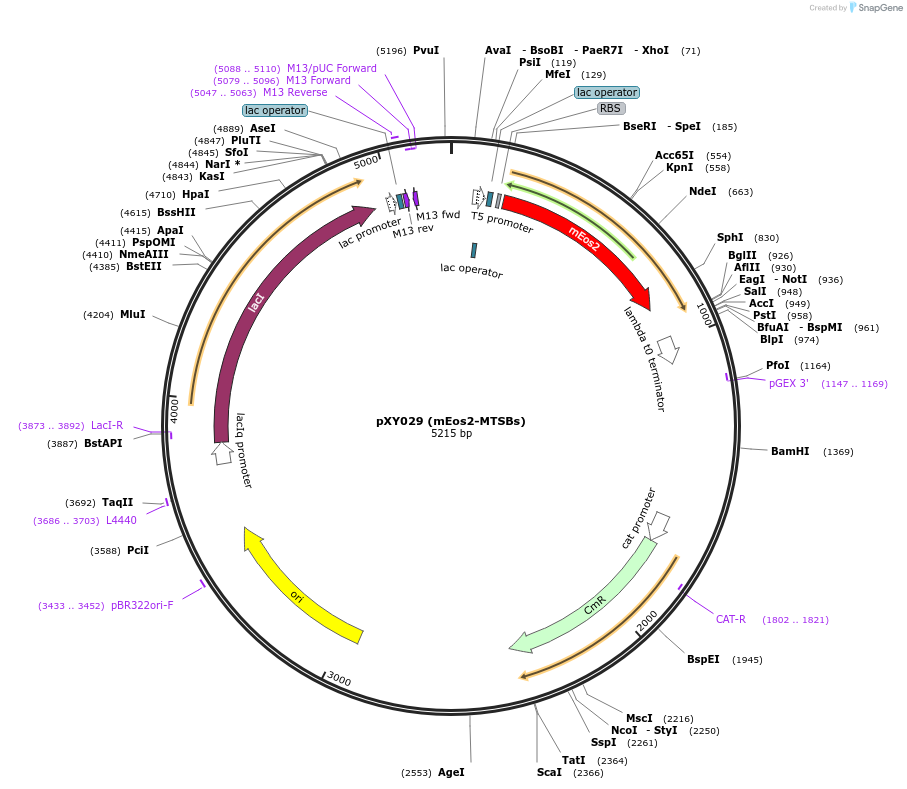
pXY029 (mEos2-MTSBs)
Plasmid#72652PurposeInducible expression of mEos2-MTSBs in bacteriaDepositorInsertmEos2-MTSBs
TagsmEos2ExpressionBacterialPromoterT5-lacAvailable SinceFeb. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
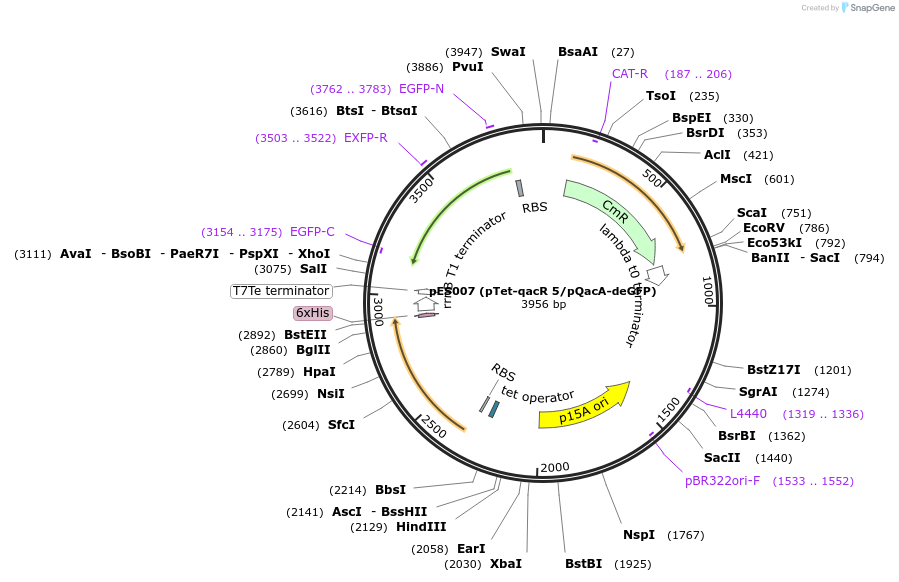
pES007 (pTet-qacR 5/pQacA-deGFP)
Plasmid#69036Purposereporter with qacR 5 on single plasmidDepositorInsertsTagsHis6 tagExpressionBacterialMutationW61Y, E90Q, F102S, M116Q, Y119L, E120QPromoterpQacA and pTetAvailable SinceSept. 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
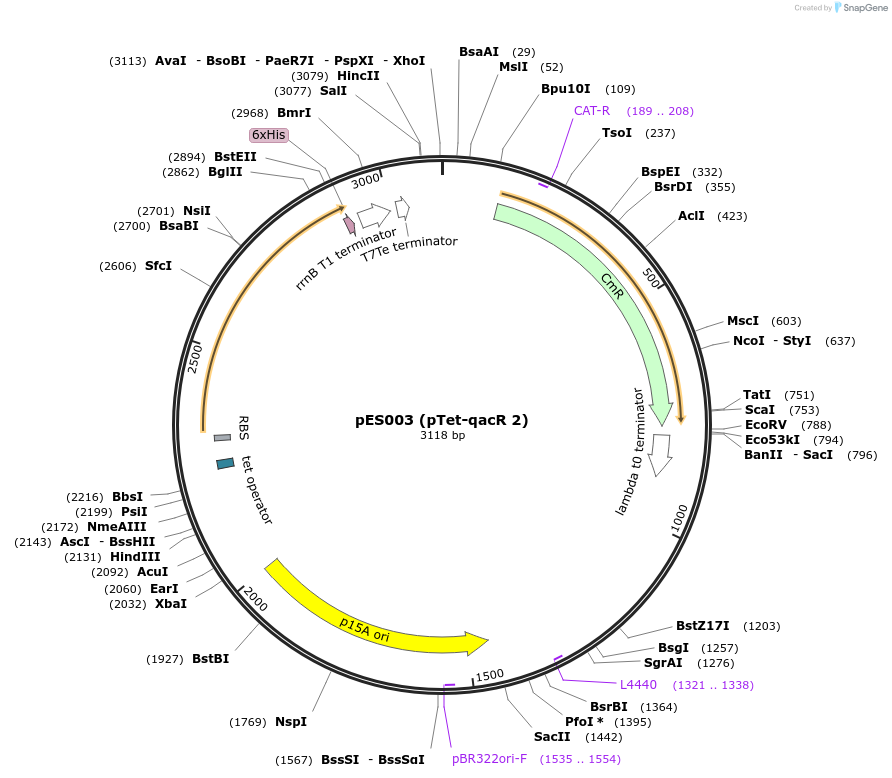
pES003 (pTet-qacR 2)
Plasmid#69032PurposeqacR 2 downstream of a Tet promoterDepositorInsertqacR (qacR Synthetic)
TagsHis6 tagExpressionBacterialMutationE57Q, E58L, W61Y, E90Q, I99Q, M116Q, L119Y, E120Q…PromoterpTetAvailable SinceSept. 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
pCVD419
Plasmid#63754Purpose3.4 kb HindIII fragment from EHEC associated plasmid from E. coli 0157:H7 strain 933 cloned into pBR325. Contains the hlyA gene. Best EHEC probe for detection of this plasmid.DepositorInsertehxA
ExpressionBacterialAvailable SinceApril 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
pREG(R1-R4) #BGV059
Plasmid#48991PurposeRetroviral Destination vector containing AttR1-AttR4 site. Use with 4-way LR reaction with cDNA in entry vector, marker in pBEG R2-L3 plasmids and shRNA in pBEG R3-L4 plasmid (module 2,3,and 4)DepositorTypeEmpty backboneUseRetroviralPromoterCMVAvailable SinceJune 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pVRc33_423
Plasmid#49749DepositorInsertAS33_423
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
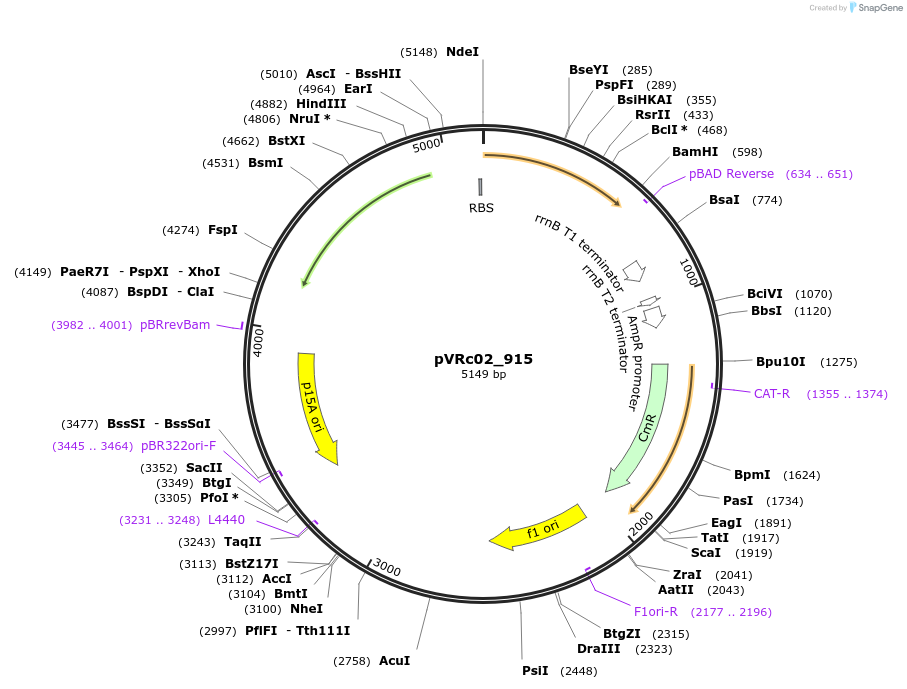
pVRc02_915
Plasmid#49729DepositorInsertAS02_915
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
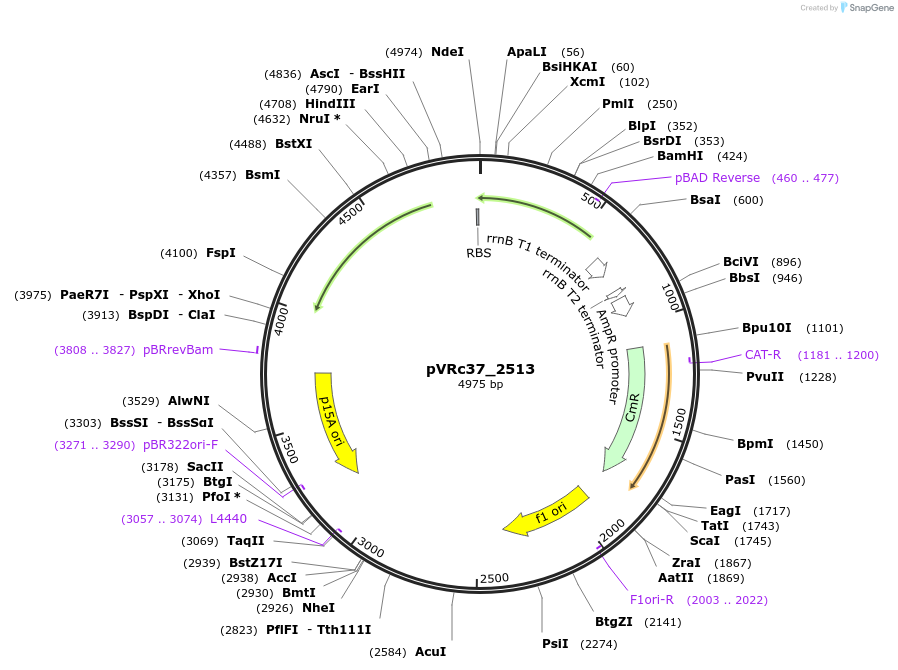
pVRc37_2513
Plasmid#49751DepositorInsertAS37_2513
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
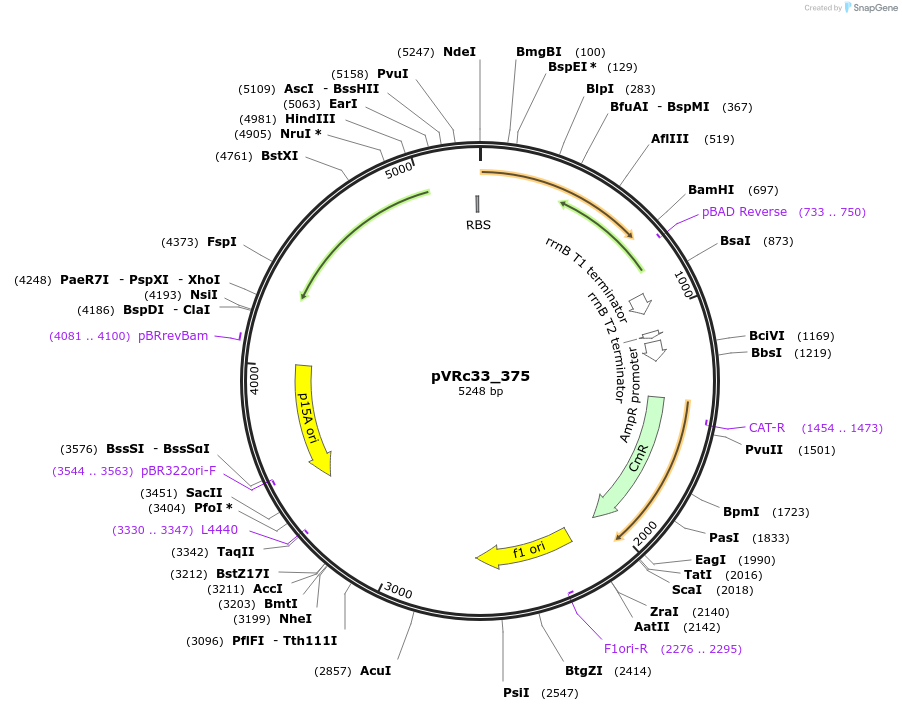
pVRc33_375
Plasmid#49748DepositorInsertAS33_375
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
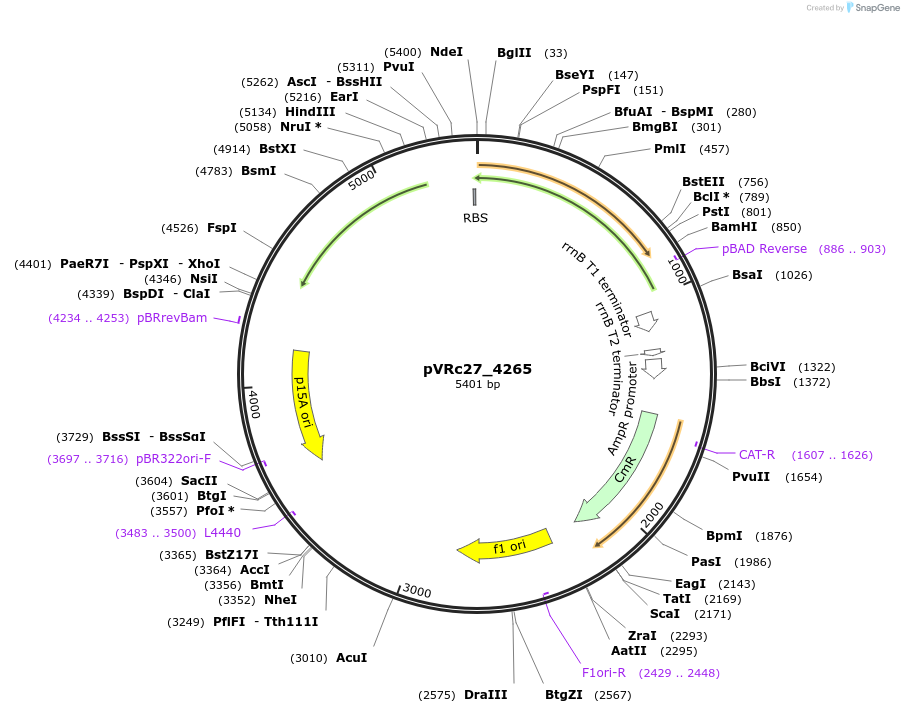
pVRc27_4265
Plasmid#49745DepositorInsertAS27_4265
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
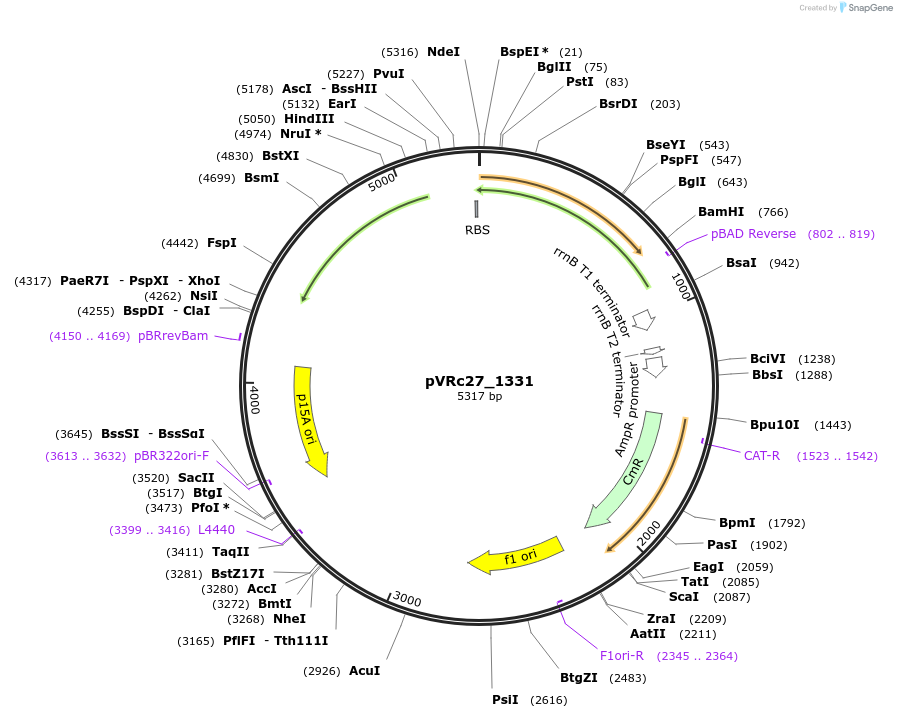
pVRc27_1331
Plasmid#49744DepositorInsertAS27_1331
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
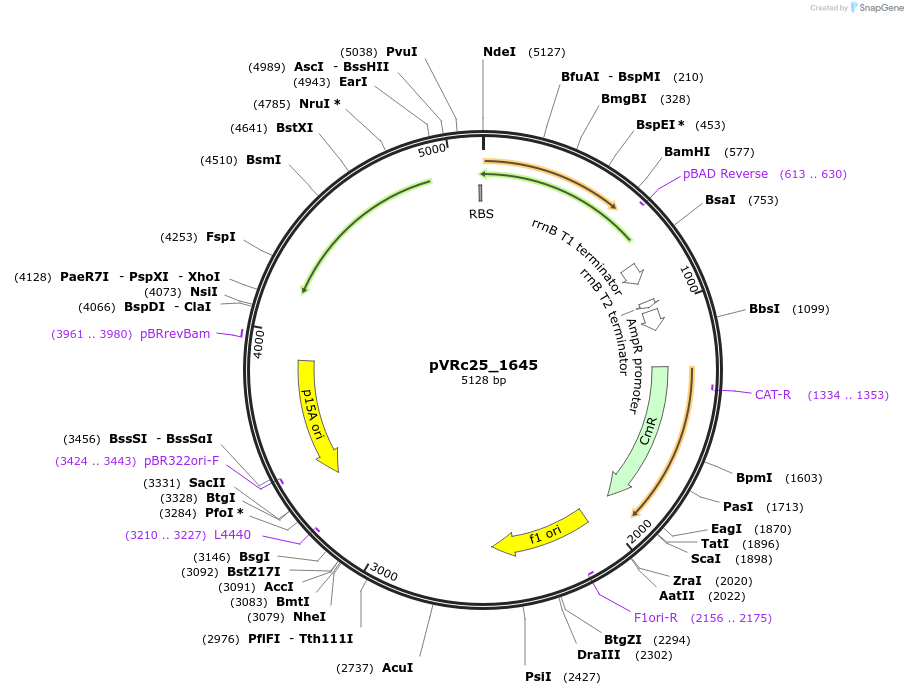
pVRc25_1645
Plasmid#49743DepositorInsertAS25_1645
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
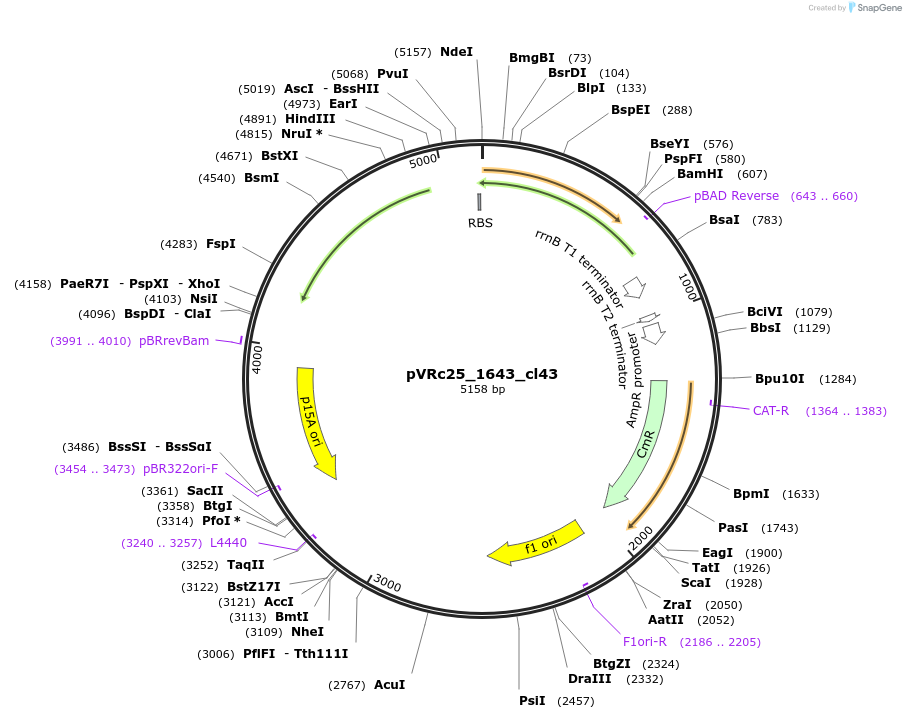
pVRc25_1643_cl43
Plasmid#49742DepositorInsertAS25_1643_cl43
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
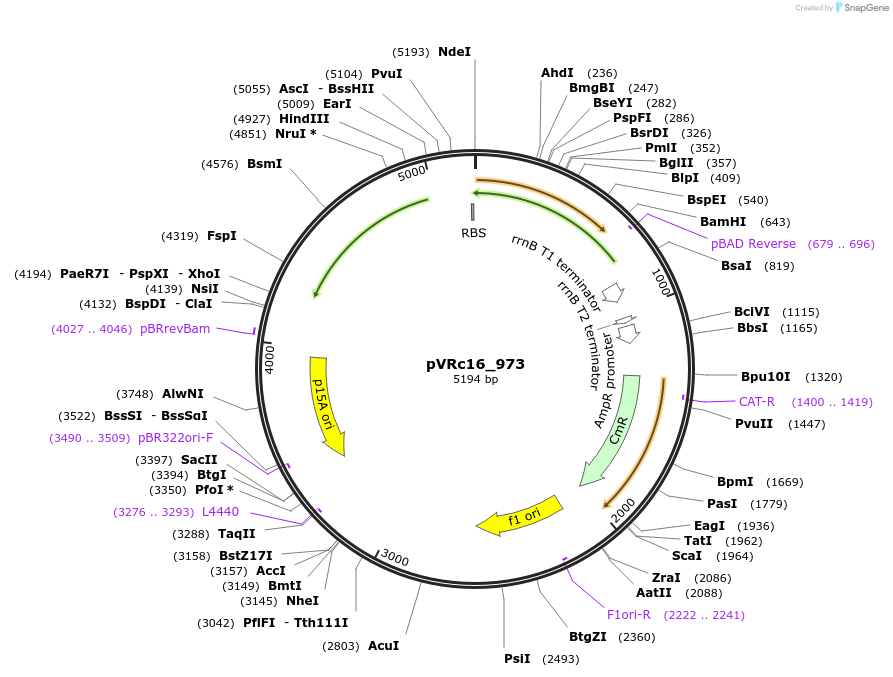
pVRc16_973
Plasmid#49737DepositorInsertAS16_973
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
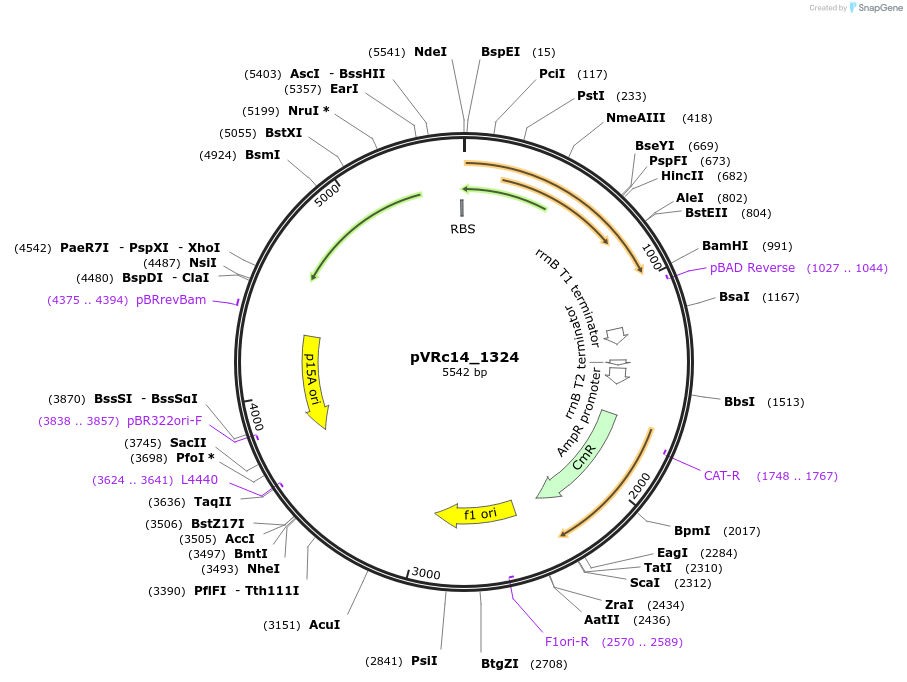
pVRc14_1324
Plasmid#49734DepositorInsertAS14_1324
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
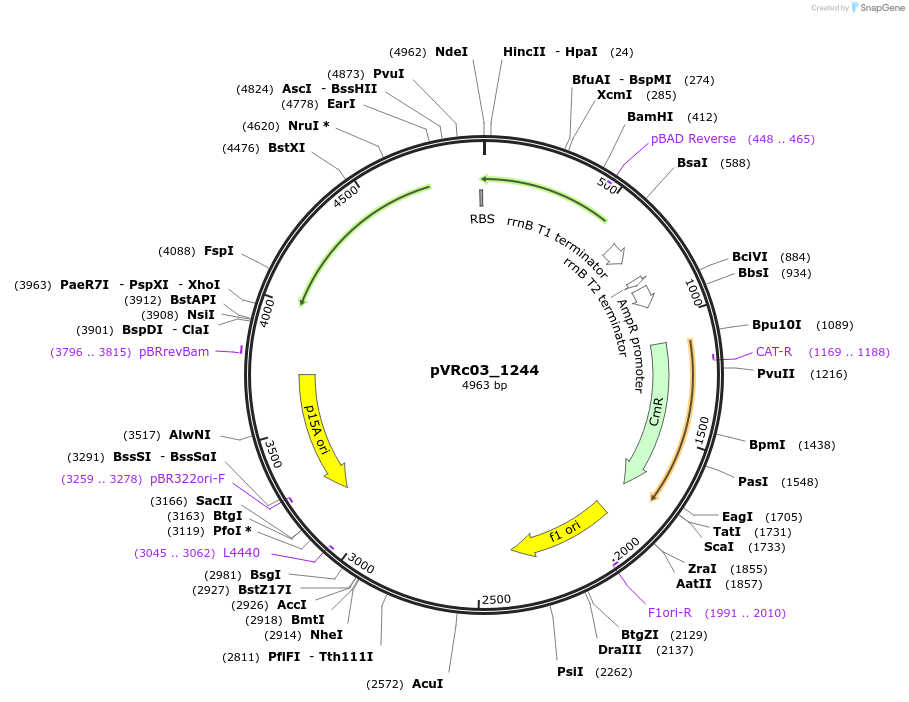
pVRc03_1244
Plasmid#49731DepositorInsertAS03_1244
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
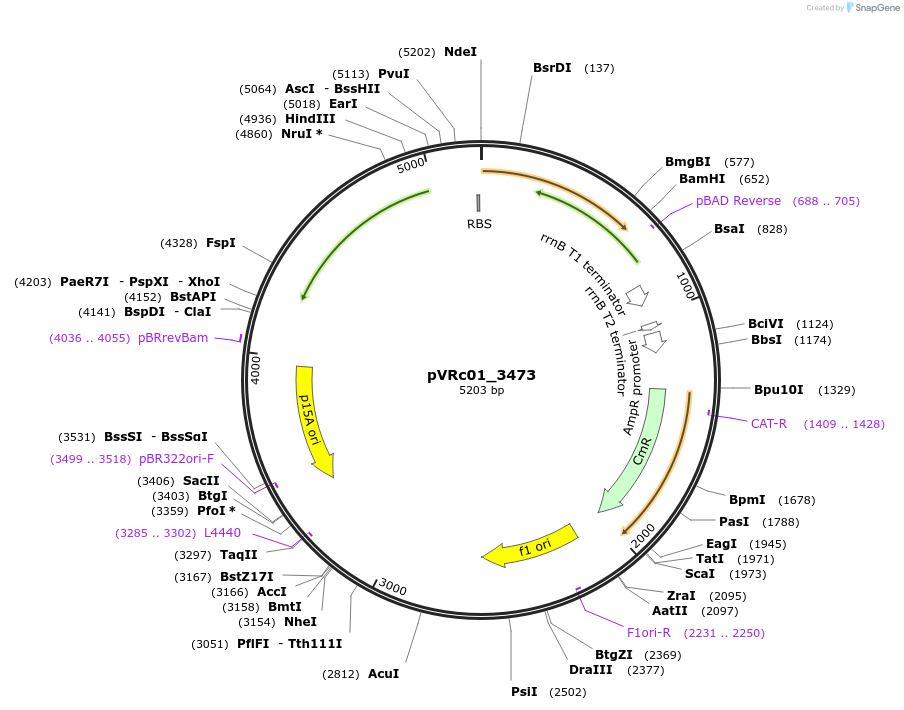
pVRc01_3473
Plasmid#49728DepositorInsertAS01_3473
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
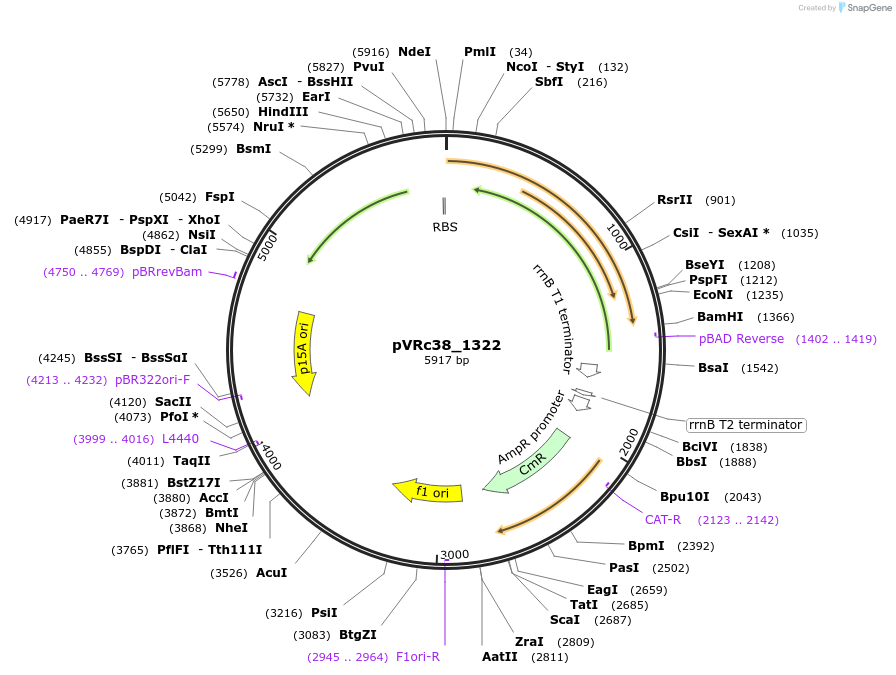
pVRc38_1322
Plasmid#49752DepositorInsertAS38_1322
UseSynthetic BiologyExpressionBacterialMutationCodon optimized for E.coliPromoterpLuxAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
pAC-EsaR-I70V/D91G/V220A
Plasmid#47647PurposepAC-EsaR with a208g, a272g, c659t mutations in esaRDepositorInsertesaR-I70V/D91G/V220A
UseSynthetic BiologyExpressionBacterialMutationChanged Isoleucine 70 to Valine, Aspartic acid 91…PromoterPlacAvailable SinceSept. 6, 2013AvailabilityAcademic Institutions and Nonprofits only -
pET21-MRPL13
Plasmid#31344DepositorInsertmitochondrial ribosomal protein L13 mature form (MRPL13 Human)
TagsHisExpressionBacterialMutationMature form after removal of the predicted mitoch…Available SinceAug. 24, 2011AvailabilityAcademic Institutions and Nonprofits only -
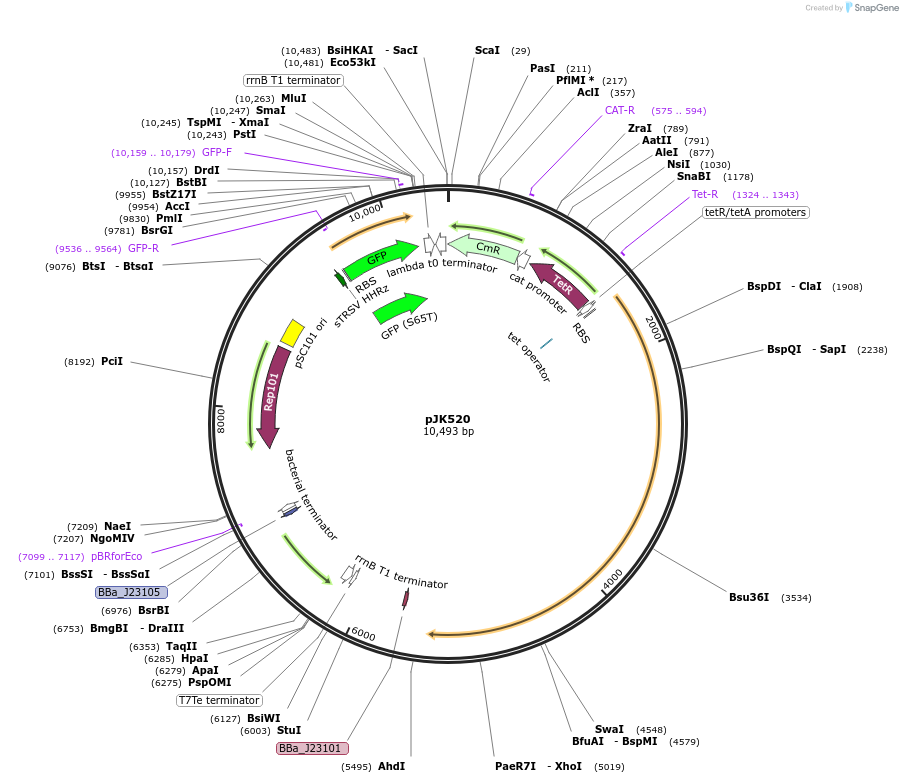
pJK520
Plasmid#155386PurposePphlF-TA1 crRNAs, 2x cascade. dCas12a (D917A) + PA4-mVenusDepositorInsertsdCas12a (F. novicida)
PA4-mVenus
PphlF-TA1 in dCas12a oscillator crRNAs
UseCRISPRExpressionBacterialMutationD917A (nuclease-deactivating)AvailabilityAcademic Institutions and Nonprofits only -
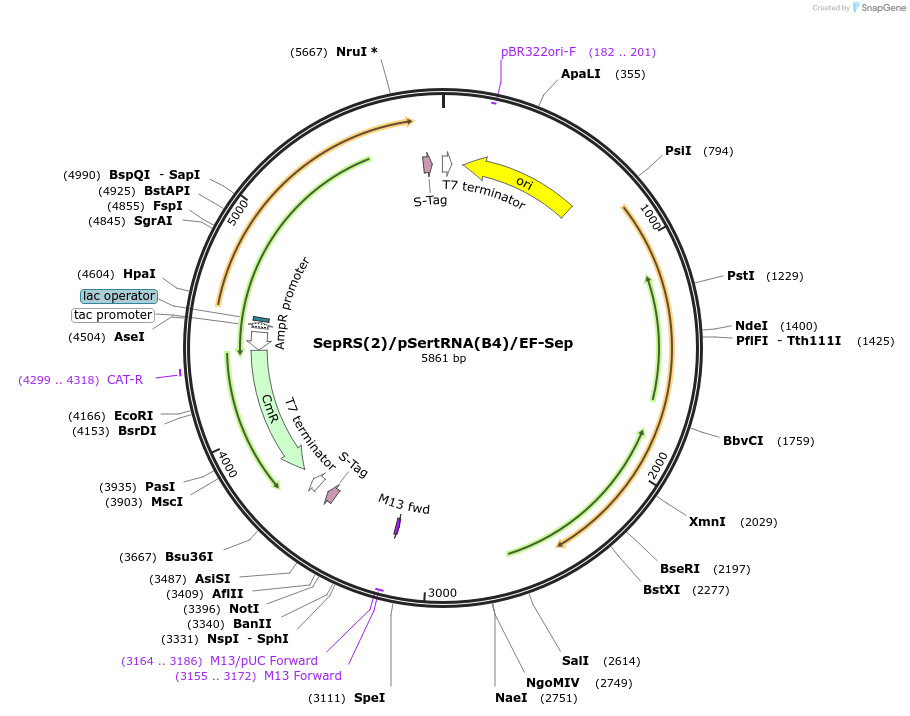
SepRS(2)/pSertRNA(B4)/EF-Sep
Plasmid#173897PurposeThe plasmid contains an orthogonal aminoacyl-tRNA synthetase/tRNACUA pair, which directs the efficient incorporation of phosphoserine (pSer) into recombinant proteins in Escherichia coli.DepositorInsertsSepRS(2)
pSertRNA
EF-Sep
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
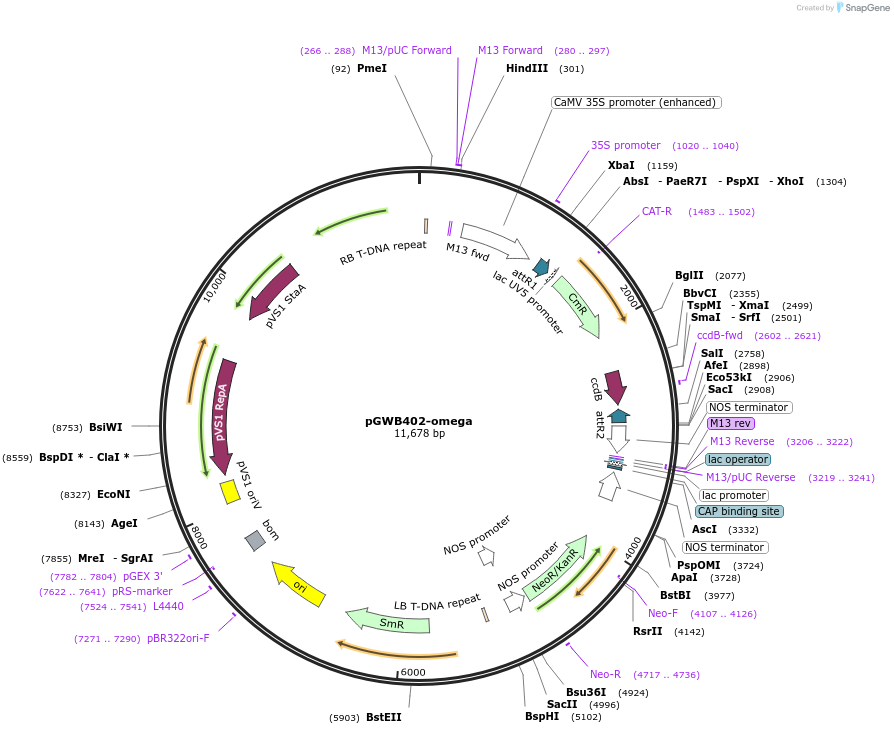
pGWB402-omega
Plasmid#74797PurposeGateway cloning compatible binary vector for expression of gene by 2xCaMV35S enhancer and omega sequence.DepositorTypeEmpty backboneExpressionPlantPromoter2xCaMV35S enhancer and omegaAvailable SinceJuly 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
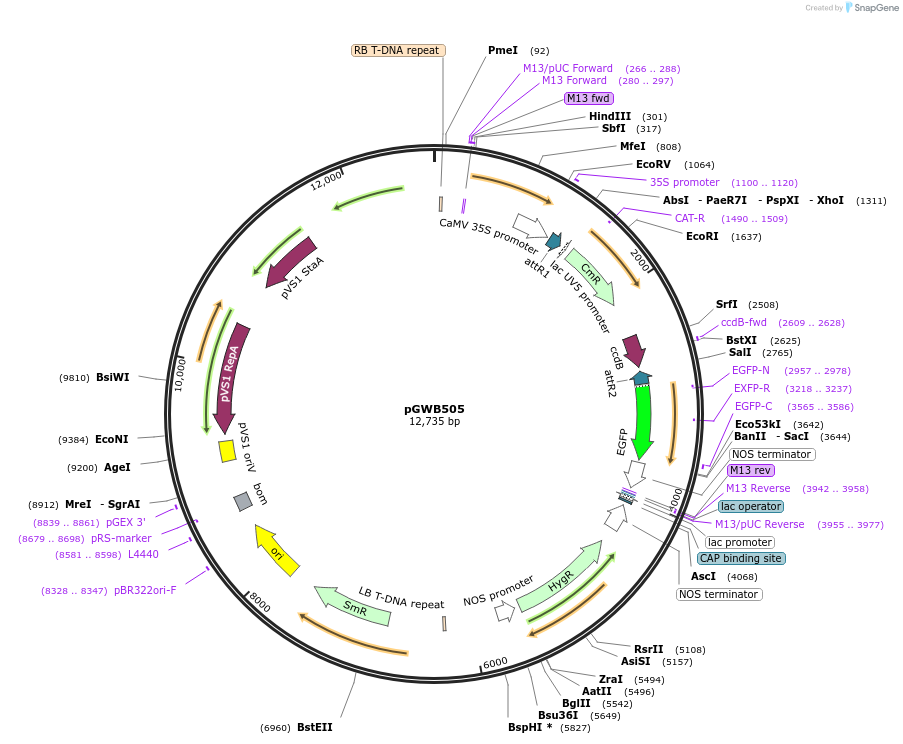
pGWB505
Plasmid#74847PurposeGateway cloning compatible binary vector for C-terminal fusion with sGFP (CaMV35S promoter).DepositorTypeEmpty backboneExpressionPlantPromoterCaMV35SAvailable SinceJuly 18, 2016AvailabilityAcademic Institutions and Nonprofits only -
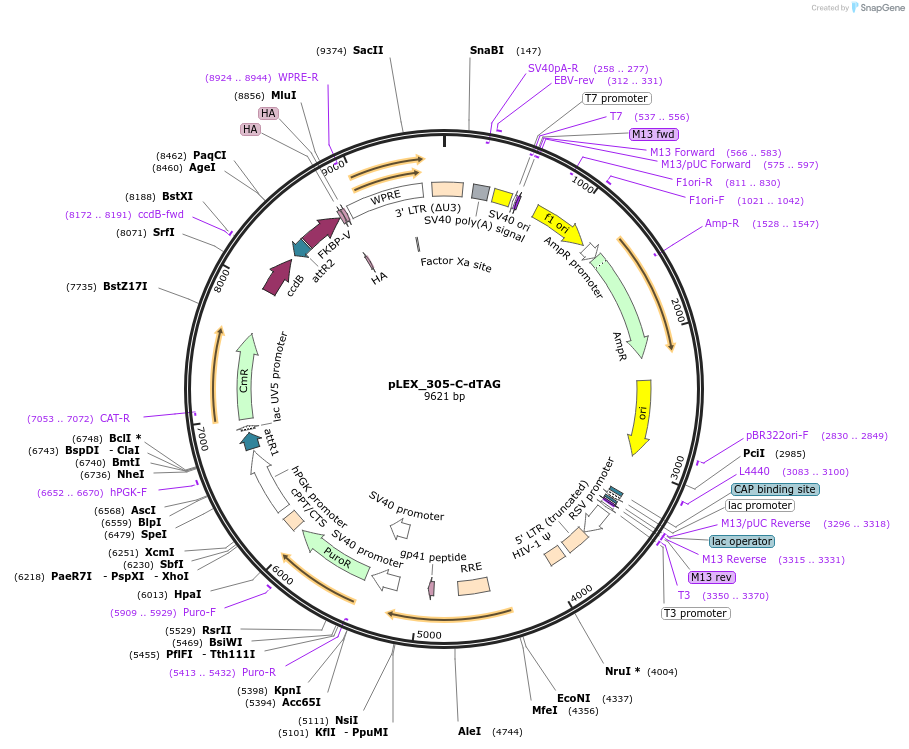
pLEX_305-C-dTAG
Plasmid#91798PurposeLentiviral/gateway cloning vector for C-terminally tagging proteins of interest with the dTAG systemDepositorTypeEmpty backboneUseLentiviralTagsFKBP F36VExpressionMammalianPromoterhPGKAvailable SinceMarch 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
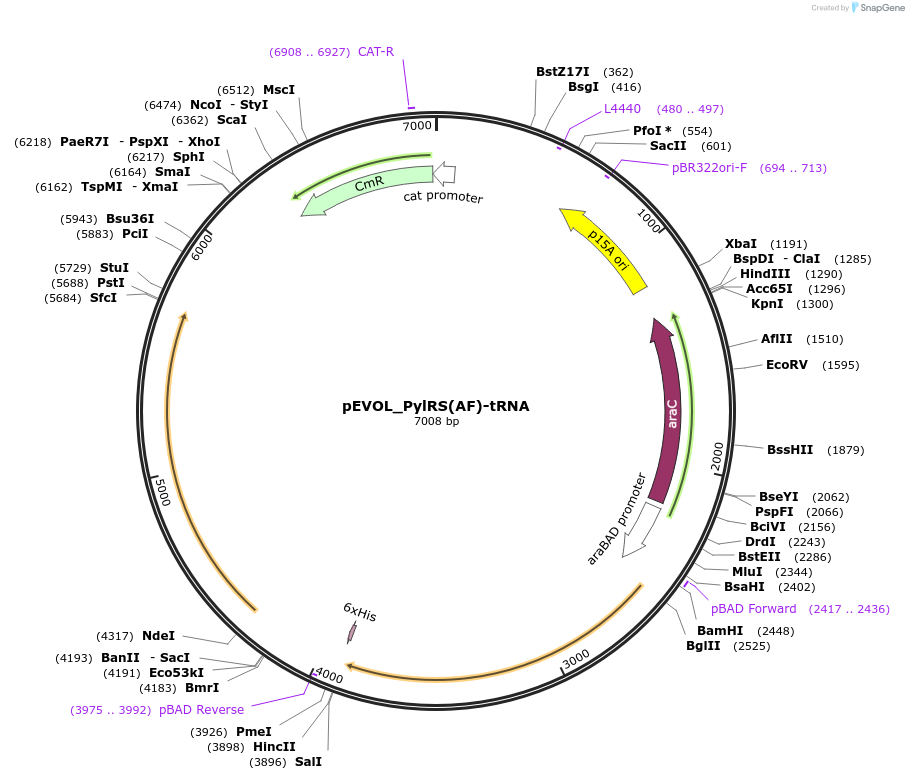
pEVOL_PylRS(AF)-tRNA
Plasmid#223511PurposePylRS (AF)/tRNA Pyl orthogonal pair for genetic code expansion that allows site-specific incorporation of noncanonical amino-acids into a POI using the Amber stop codonDepositorInsertsTags6xHisExpressionBacterialMutationY306A; Y384FPromoteraraBAD, rrnB, and glnS and proKAvailable SinceOct. 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
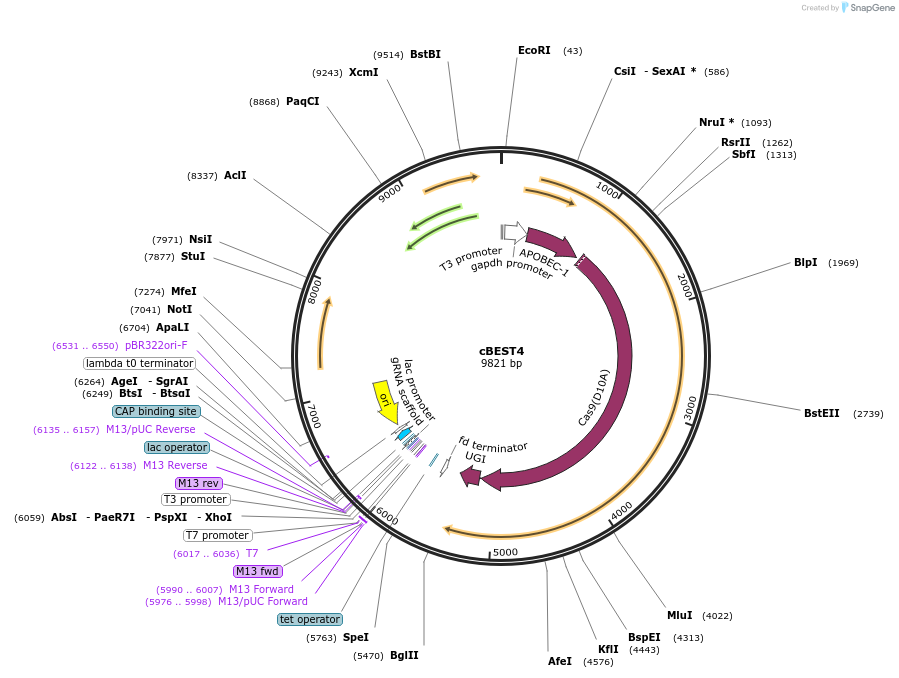
cBEST4
Plasmid#234660PurposeExpresses cytosine base editor - spCas9n (D10A) fused to APOBEC1 and UGI, Include Golden Gate compatible cassette for sgRNA insertionDepositorInsertsspCas9 cytosine base editor
Golden Gate compatible sgRNA insertion cassette
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterP3 and tcp830Available SinceMay 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
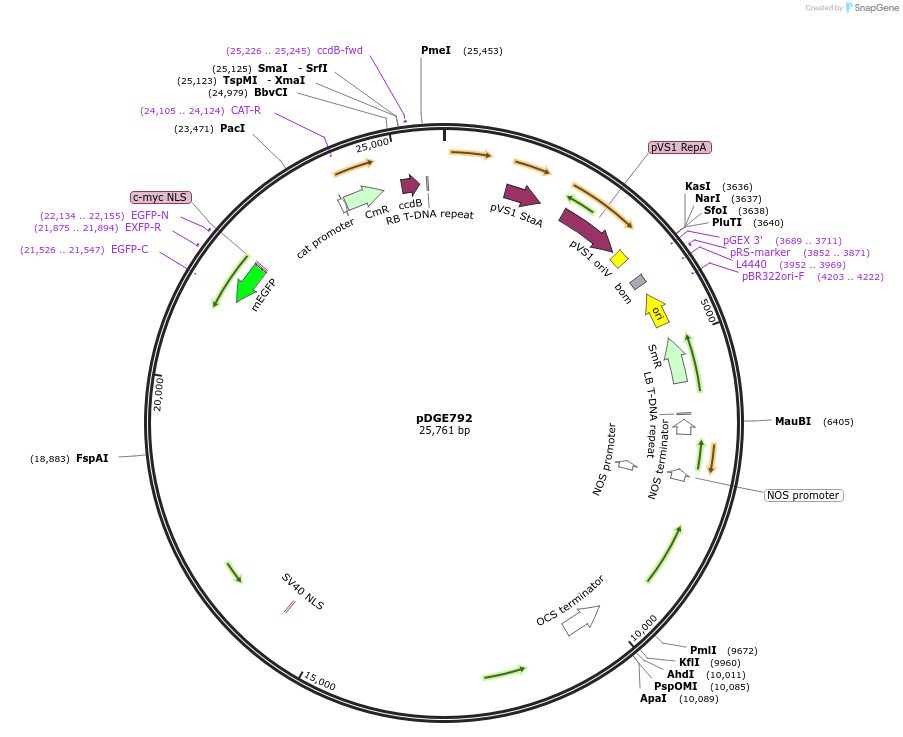
pDGE792
Plasmid#167300PurposeVector for plant genome editing containing pRPS5a:GFP-Cas9 expression cassette, Bar (BASTA/PPT) selection marker, and FCY-UPP and Ca-Bs3 counterselction markers, but not sgRNAs.DepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceMay 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
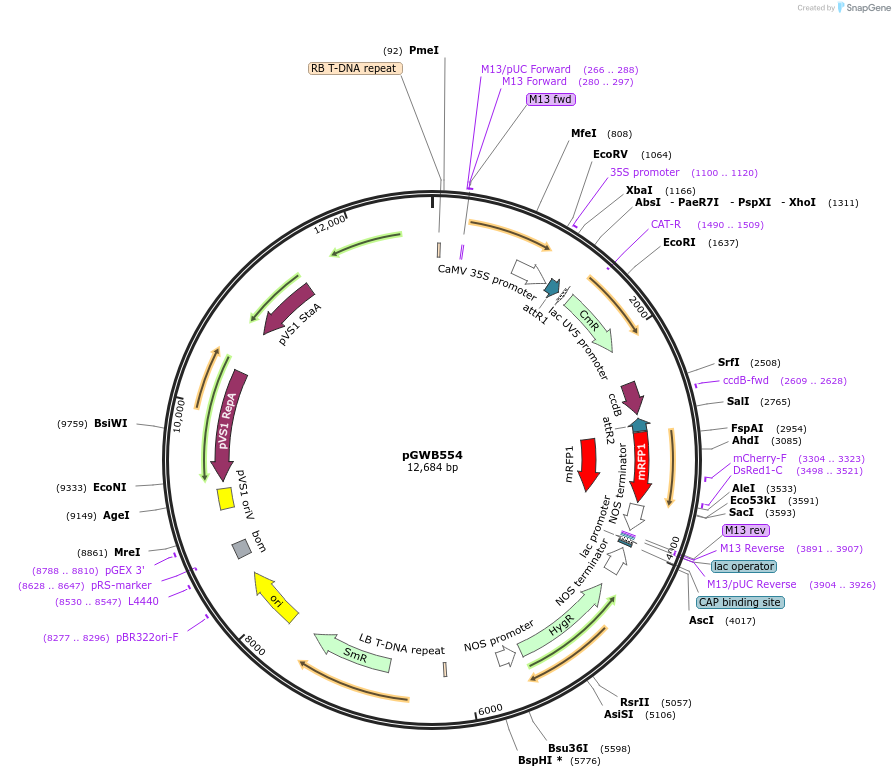
pGWB554
Plasmid#74884PurposeGateway cloning compatible binary vector for C-terminal fusion with mRFP (CaMV35S promoter).DepositorTypeEmpty backboneExpressionPlantPromoterCaMV35SAvailable SinceJuly 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
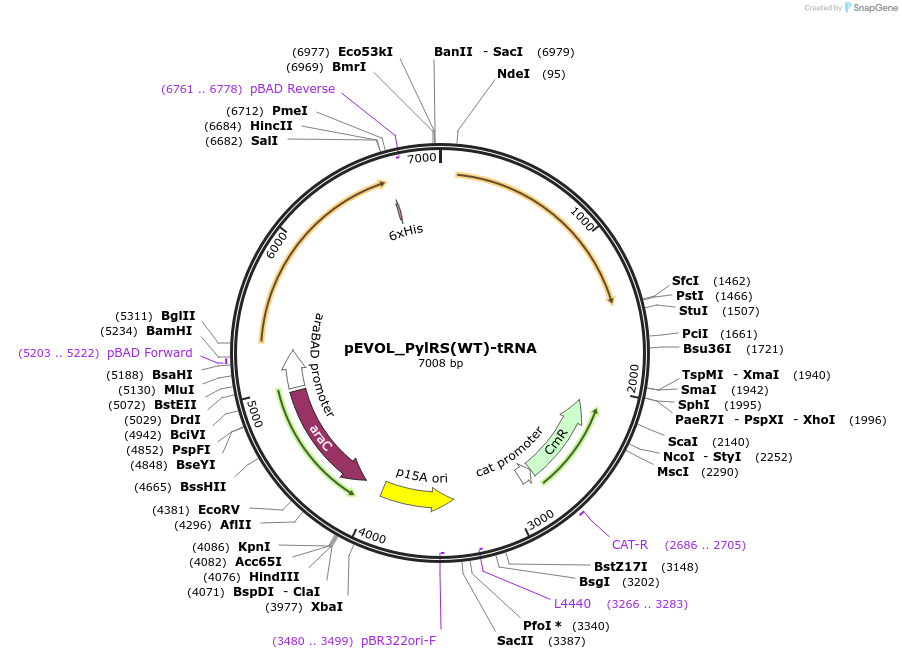
pEVOL_PylRS(WT)-tRNA
Plasmid#223512PurposePylRS (WT)/tRNA Pyl orthogonal pair for genetic code expansion that allows site-specific incorporation of noncanonical amino-acids into a POI using the Amber stop codonDepositorInsertsTags6xHisExpressionBacterialPromoteraraBAD, rrnB, and glnS and proKAvailable SinceJan. 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
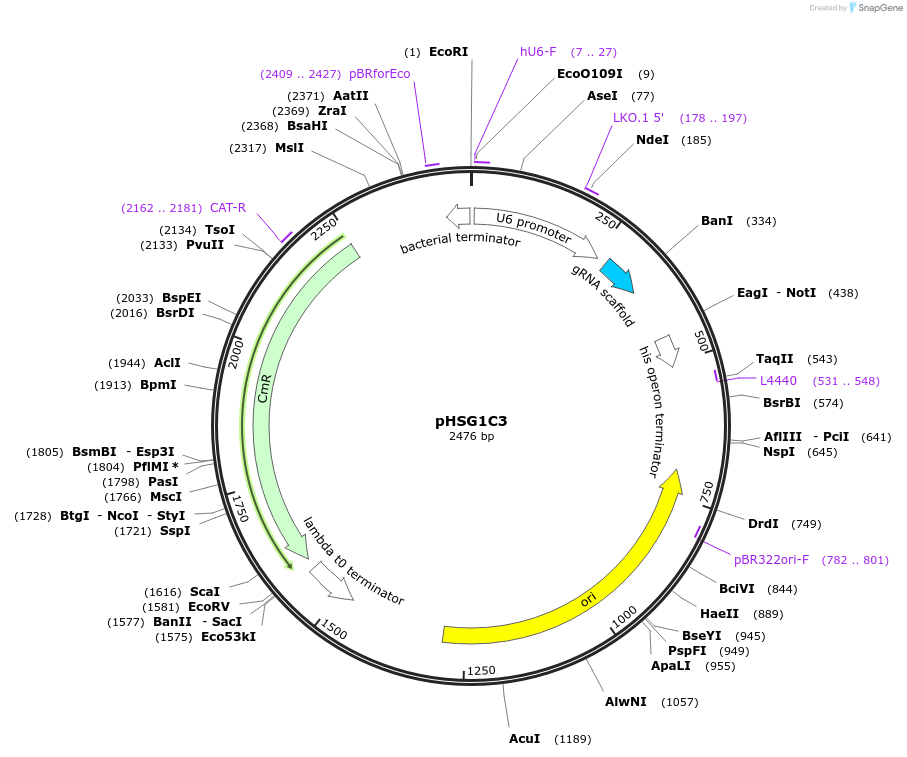
pHSG1C3
Plasmid#164423Purposesingle guide RNA (sgRNA) and Prime editing guide RNA (pegRNA) cloning and mammalian cell expression. BbsI cloning for sgRNAs and BbsI/PstI for pegRNAs.DepositorInsertsgRNA
UseCRISPR and Synthetic BiologyExpressionMammalianPromoterU6Available SinceAug. 4, 2021AvailabilityAcademic Institutions and Nonprofits only -
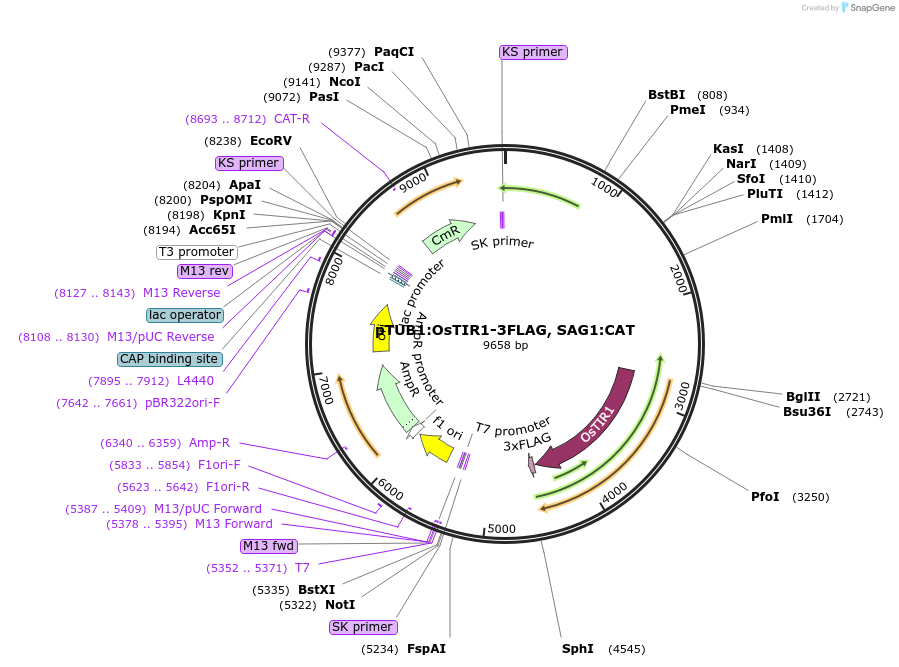
pTUB1:OsTIR1-3FLAG, SAG1:CAT
Plasmid#87258PurposeTIR1-3FLAG fusion driven by a TUB1 promoter with a CAT drug selectable marker. The TIR1 CDS from Oryza sativa was codon optimized for T. gondii expression.DepositorInsertsTIR1
CAT
UseToxoplasma expressionTags3FLAGMutationCodon optimized for Toxoplasma gondii expressionPromoterTgSAG1 361 bp and TgTUB1 2717 bpAvailable SinceMay 4, 2017AvailabilityAcademic Institutions and Nonprofits only -
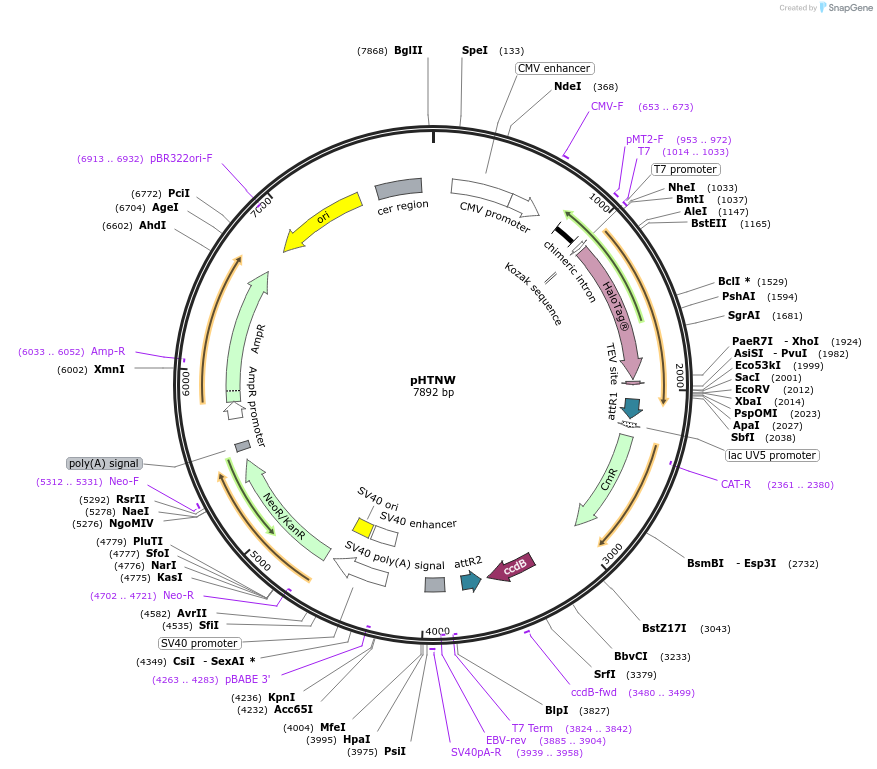
pHTNW
Plasmid#136403PurposeExpression of N-terminal HaloTag-fused proteins in mammalian cells. Genes can be cloned using Gateway cloning system. Cloned vectors can be used for NanoBRET assay.DepositorTypeEmpty backboneTagsHaloTagExpressionMammalianPromoterCMVAvailable SinceFeb. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
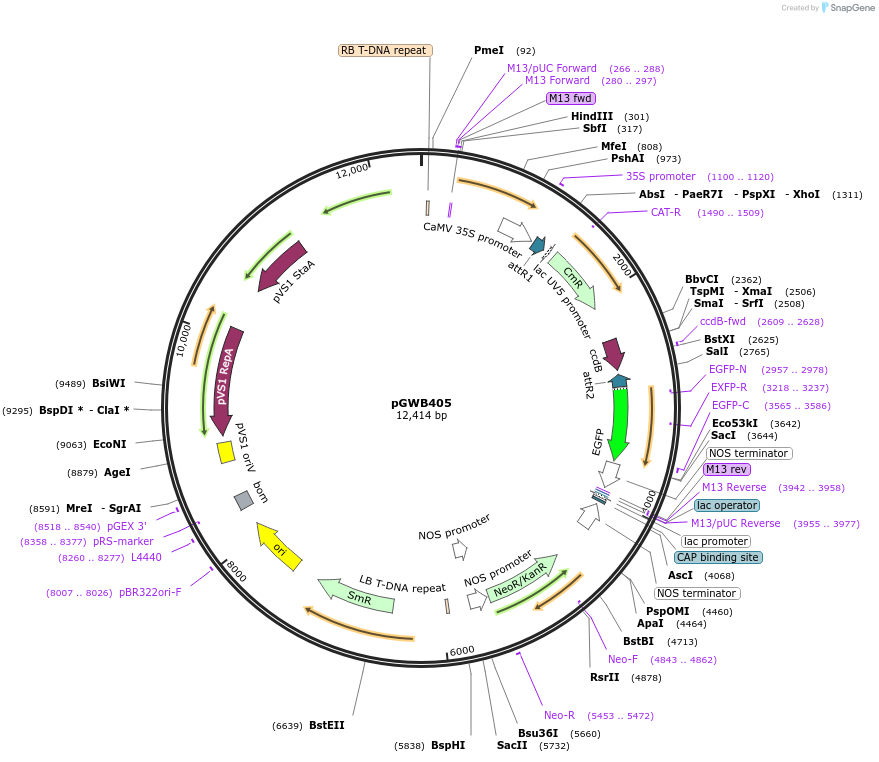
pGWB405
Plasmid#74799PurposeGateway cloning compatible binary vector for C-terminal fusion with sGFP (CaMV35S promoter).DepositorTypeEmpty backboneExpressionPlantPromoterCaMV35SAvailable SinceJuly 18, 2016AvailabilityAcademic Institutions and Nonprofits only -
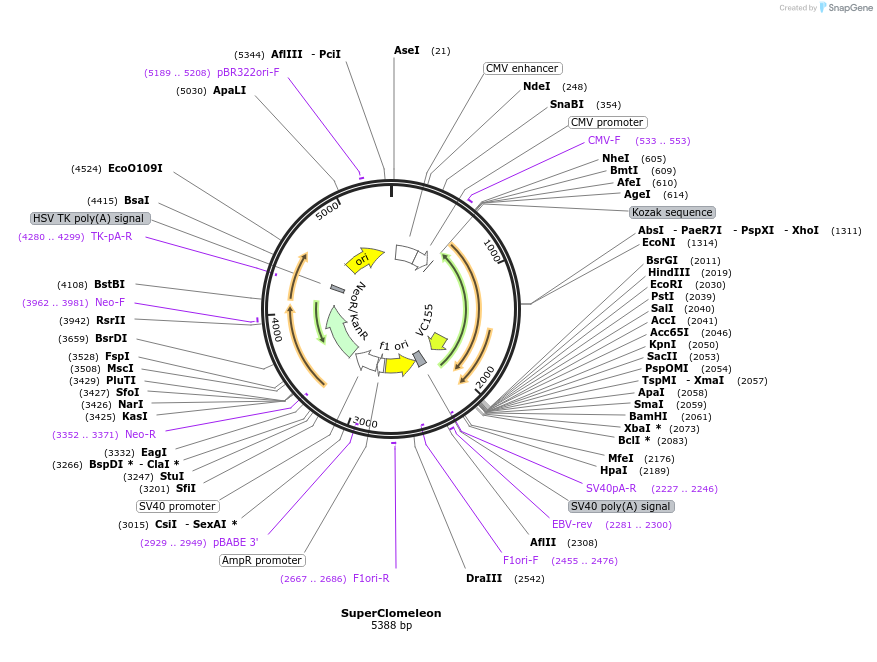
SuperClomeleon
Plasmid#203724PurposeAn improved engineered fluorescent ratiometric optogenetic sensor to detect inhibitory synaptic activity via changes in intracellular chlorideDepositorInsertCerulean linked topaz fluorescent protein
ExpressionBacterial and MammalianMutationCerulean, modified Topaz Fluorescent Protein with…Available SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
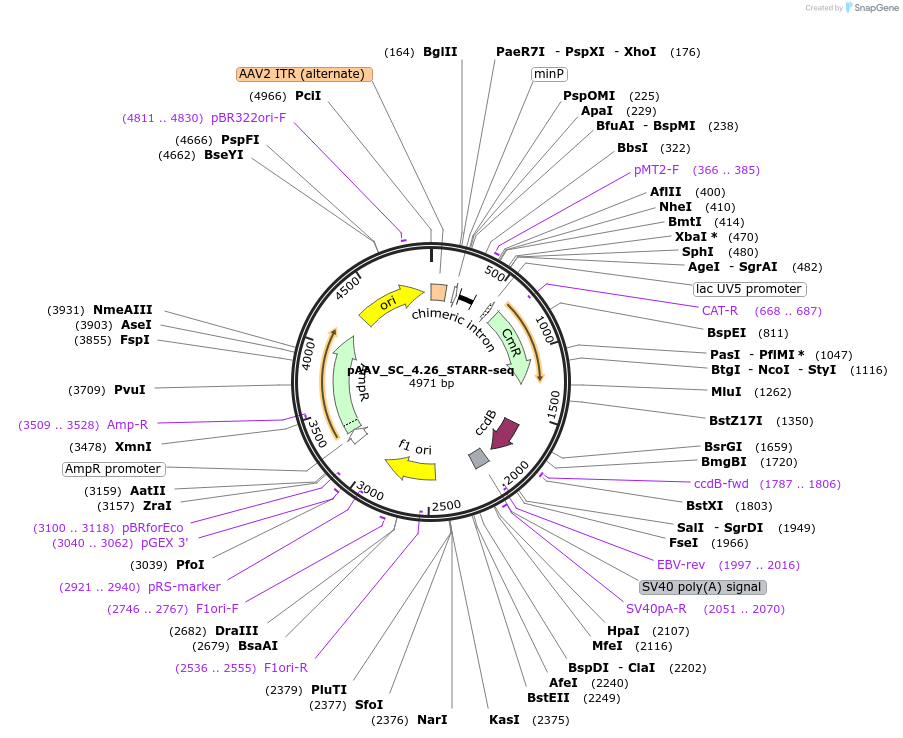
pAAV_SC_4.26_STARR-seq
Plasmid#220220PurposeAAV-STARR-seq screening vector with 4.26 promoteDepositorTypeEmpty backboneUseAAVExpressionMammalianPromoter4.26Available SinceAug. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
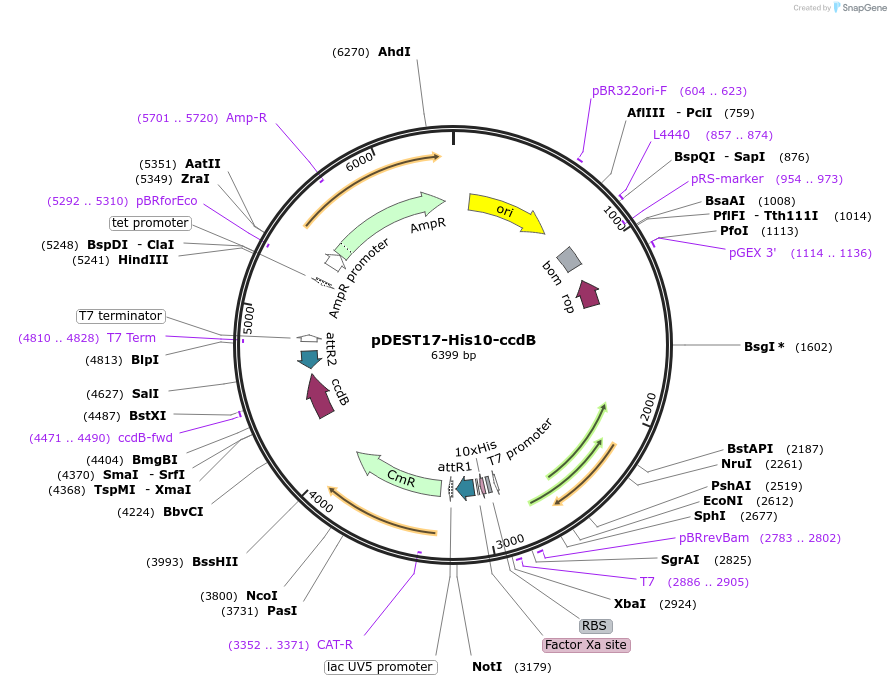
pDEST17-His10-ccdB
Plasmid#166145PurposeGateway destination vector for generation of N-terminal His10-tagged proteins in E. coli.DepositorTypeEmpty backboneTagsHis10ExpressionBacterialAvailable SinceMay 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
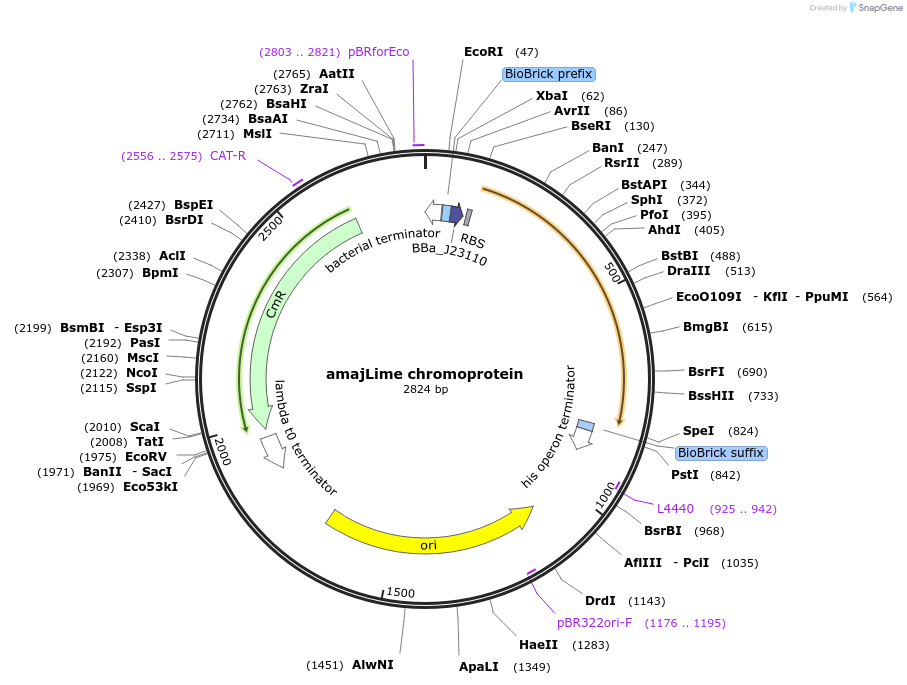
amajLime chromoprotein
Plasmid#117843PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amajLime chromoprotein in E. coliDepositorInsertpromoter, RBS, amajLime
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
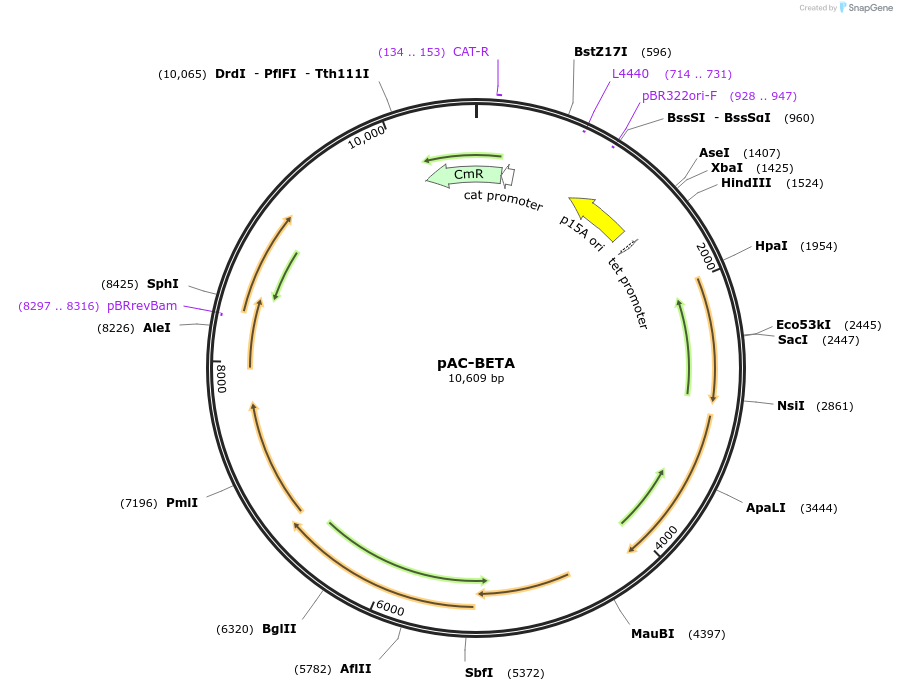
pAC-BETA
Plasmid#53272PurposeContains crtE, crtB, crtI, and crtY carotenoid pathway genes of Erwinia herbicola (Pantoea agglomerans) Eho10 and thereby produces beta-carotene in Escherichia coliDepositorInsertcrtE, crtY, crtI, crtB
UseLow copy number bacterial cloning vectorPromoterendogenous promotersAvailable SinceJan. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
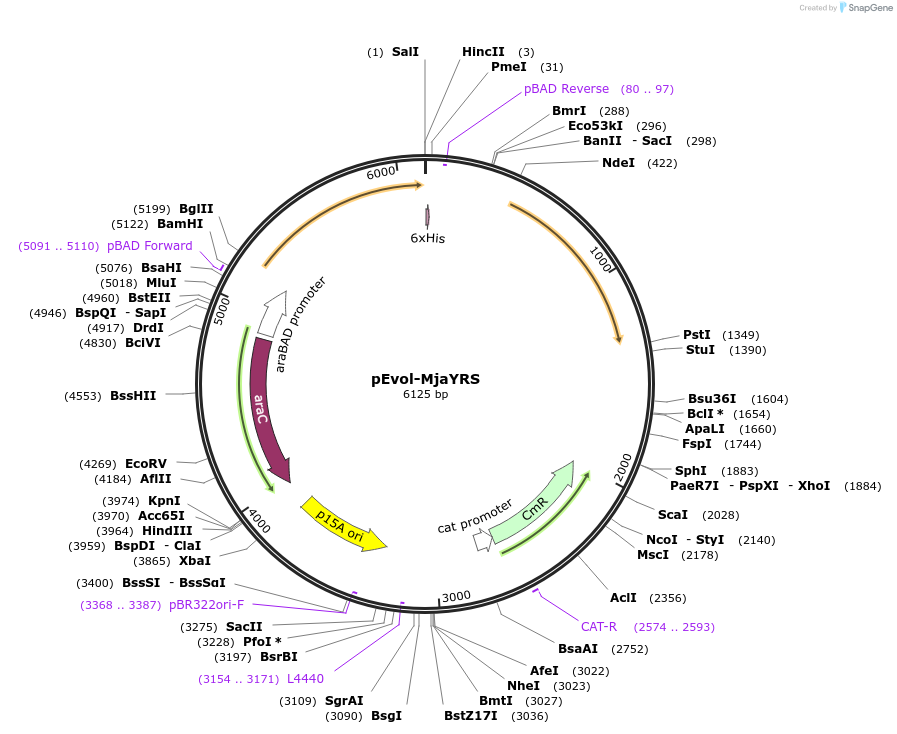
pEvol-MjaYRS
Plasmid#153557PurposeAmber suppression for incorporating 3-aminotyrosine to proteins in E. coliDepositorInsertsMJaYRS (first copy)
MJaYRS (second copy)
amber suppression tRNA under proK promoter
ExpressionBacterialPromoterglnS and pBADAvailable SinceJuly 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
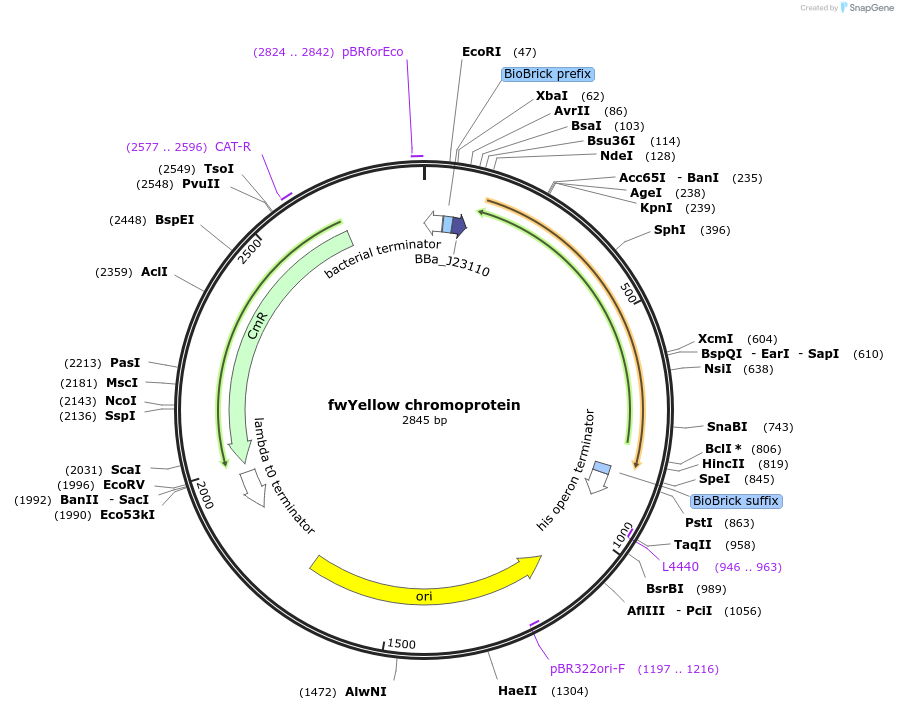
fwYellow chromoprotein
Plasmid#117841PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses fwYellow chromoprotein in E. coliDepositorInsertpromoter, RBS, fwYellow
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
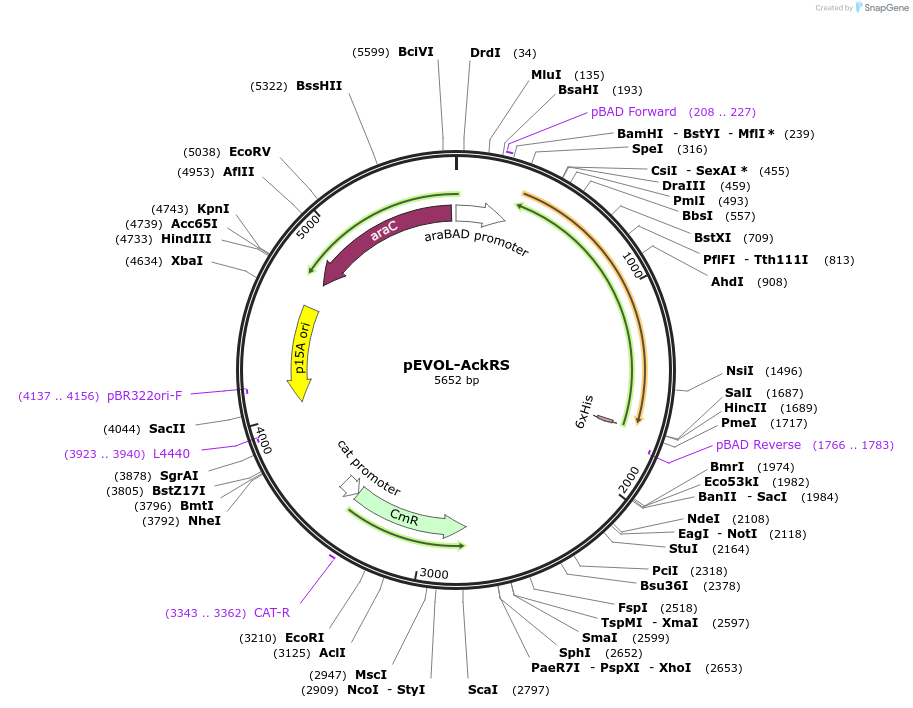
pEVOL-AckRS
Plasmid#137976Purposesequence optimized N‐acetyl lysyl‐tRNA synthetase with cognate tRNA for genetic code expansionDepositorInsertAckRS and pylTcua
ExpressionBacterialPromoteraraBADAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
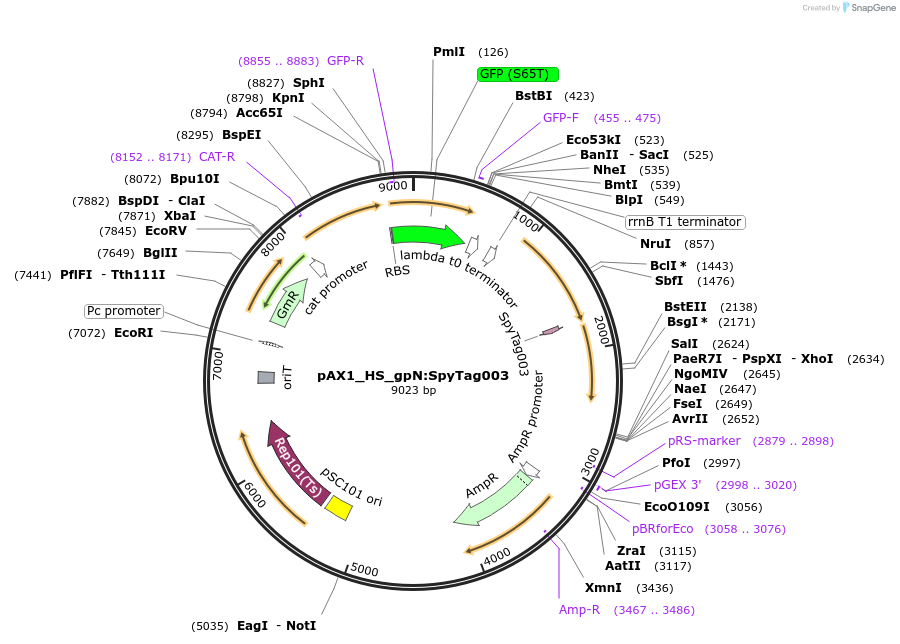
pAX1_HS_gpN:SpyTag003
Plasmid#225194PurposeUsed for adding C-terminal SpyTag003 to the E. coli HS P2 phage major capsid proteinDepositorInsertgpN homology arms and SpyTag003
ExpressionBacterialAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only