We narrowed to 22,610 results for: ESC
-
Plasmid#186520PurposeVisualization of the ER in cells using hyperfolder YFP (compatible with OSER assay)DepositorInsertHyperfolder YFP
TagsCytochrome p450 first 29 animo acids (Oryctolagus…ExpressionMammalianMutationmGreenLantern-F46L/C48S/V68L/A69M/C70V/K101E/T105…PromoterCMVAvailable SinceNov. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
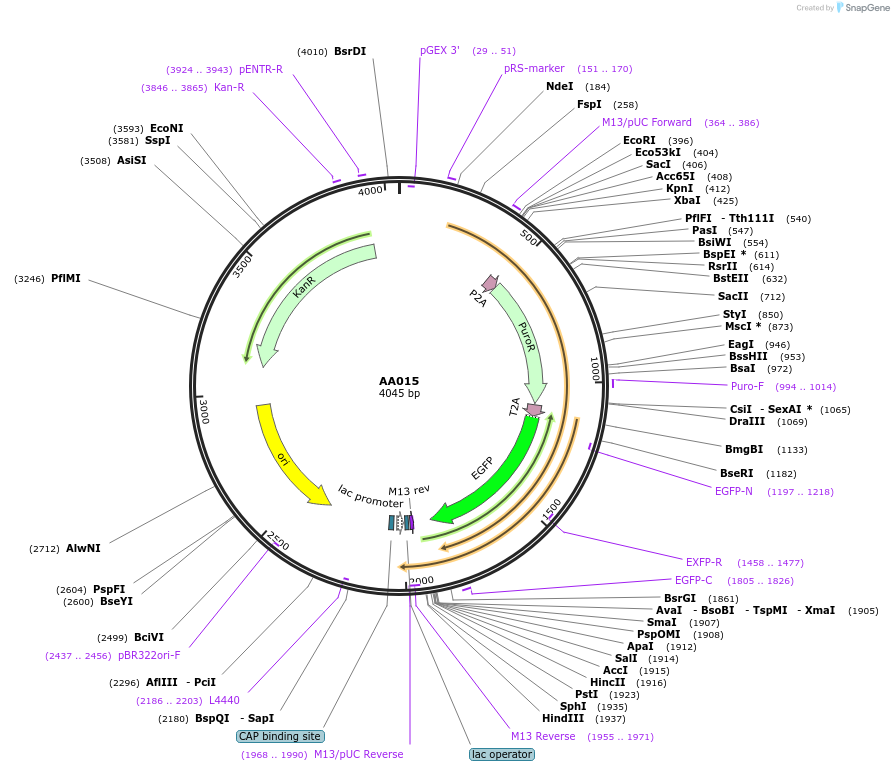
AA015
Plasmid#216058PurposeFragmid fragment: (2A Selection) Puromycin resistance, T2A, EGFPDepositorHas ServiceCloning Grade DNAInsertP2A_v1.1; PuroR_v1.1; T2A_v1.2; EGFP_v1.1; Stop
UseFragmentAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
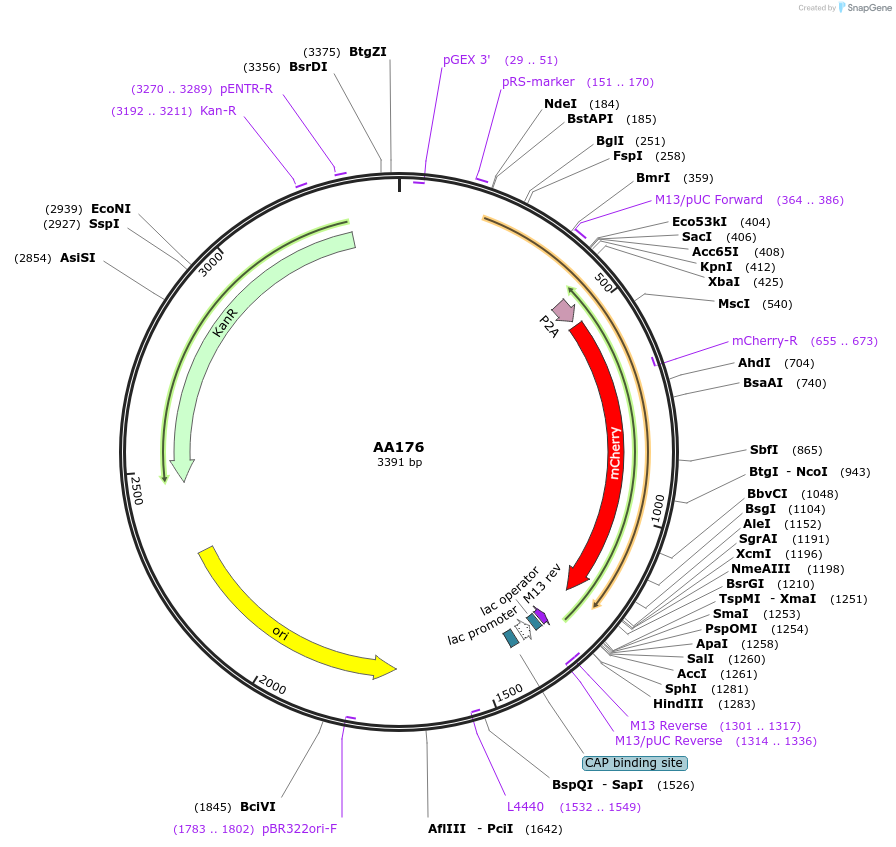
AA176
Plasmid#216063PurposeFragmid fragment: (2A Selection) mCherryDepositorHas ServiceCloning Grade DNAInsertP2A_v1.1; mCherry_v1.1; Stop
UseFragmentAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
pYNL-N1_TUBB5
Plasmid#65678PurposeExpresses beta-tubulin-YNL in mammalian cellsDepositorInsertbeta-tubulin class V
UseLuciferaseTagsYNLExpressionMammalianPromoterCMVAvailable SinceJune 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
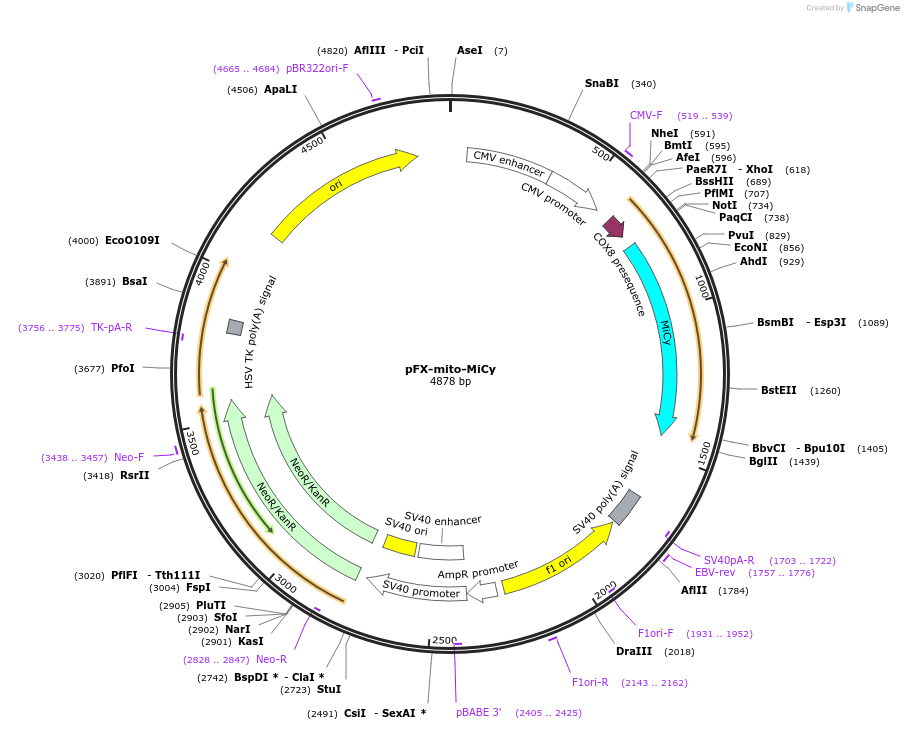
pFX-mito-MiCy
Plasmid#174177PurposeExpress MiCy with mitochondria-targeting sequence in mammalian cellsDepositorInsertMiCy with mitochondria-targeting sequence
Tagsthe mitochondria-targeting sequence of the cytoch…ExpressionMammalianPromoterCMVAvailable SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
mOrange2-Tubulin-6
Plasmid#57974PurposeLocalization: Microtubules, Excitation: 549, Emission: 565DepositorAvailable SinceOct. 24, 2014AvailabilityAcademic Institutions and Nonprofits only -
mIFP12-sEspin-C-18
Plasmid#56262PurposeLocalization: Actin Bundling, Excitation: 683, Emission: 704DepositorInsertsEspin
TagsmIFP12ExpressionMammalianPromoterCMVAvailable SinceApril 28, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
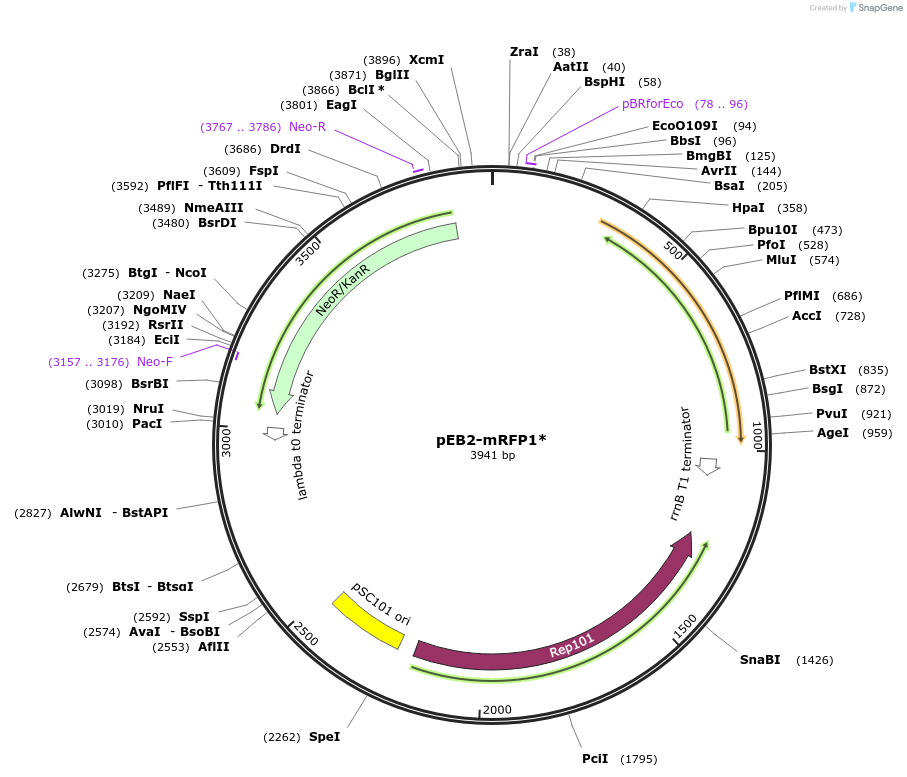
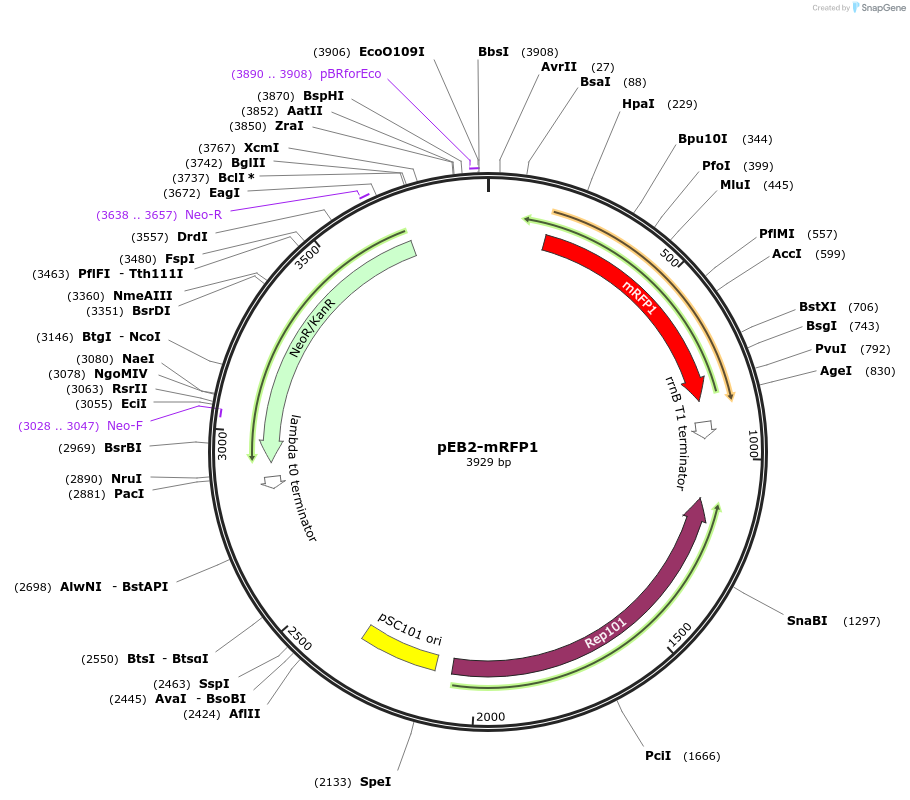
pEB2-mRFP1*
Plasmid#104000PurposePlasmid encoding mRFP1*DepositorInsertmRFP1*
UseLow copyExpressionBacterialMutationMutations relative to DsRed (MSKGEE NNLAVIKEF...T…PromoterproCAvailable SinceDec. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
pETM41-EcLonA
Plasmid#38330DepositorInsertLon protease fragment (lon Escherichia coli)
TagsHis, MBP, and TEVExpressionBacterialMutationPolyphosphate binding fragment of Lon protease. …PromoterT7Available SinceAug. 10, 2012AvailabilityAcademic Institutions and Nonprofits only -
mEos2-Alpha-5-Integrin-12
Plasmid#57344PurposeLocalization: Focal Adhesions, Excitation: 505 / 569, Emission: 516 / 581DepositorAvailable SinceFeb. 5, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
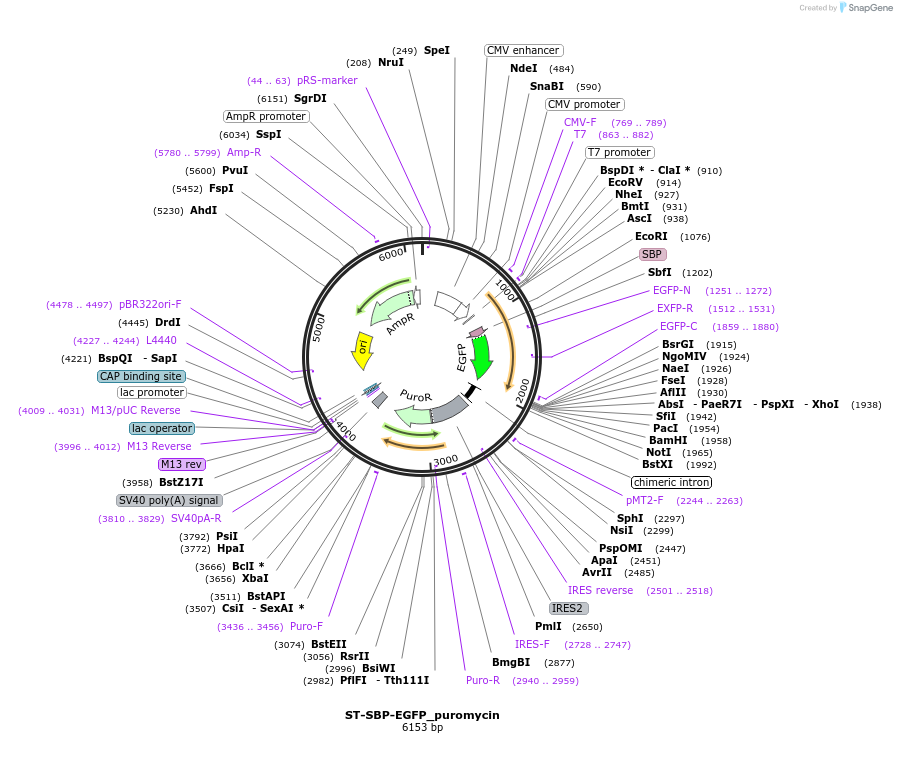
ST-SBP-EGFP_puromycin
Plasmid#65270PurposeST reporter only ( no hook) (RUSH system)DepositorInserttruncated sialyltransferase (ST6GAL1 Human)
TagsEGFP and Streptavidin Binding Protein (SBP)ExpressionMammalianMutationcontains only amino acids 1 to 43 of STPromoterCMVAvailable SinceAug. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
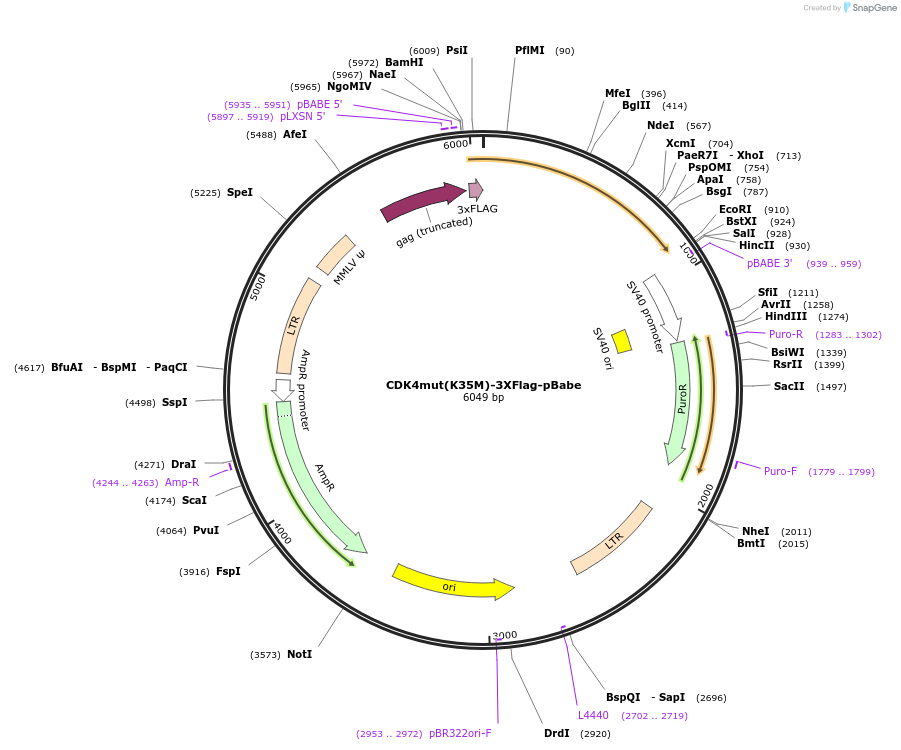
CDK4mut(K35M)-3XFlag-pBabe
Plasmid#73559Purposevector encoding a fusion protein between a CDK4 mutant (K35M) and 3 Flag tagsDepositorInsertCDK4mut(K35M)
UseRetroviralTags3X-FLAGExpressionMammalianPromoterSV40Available SinceMarch 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
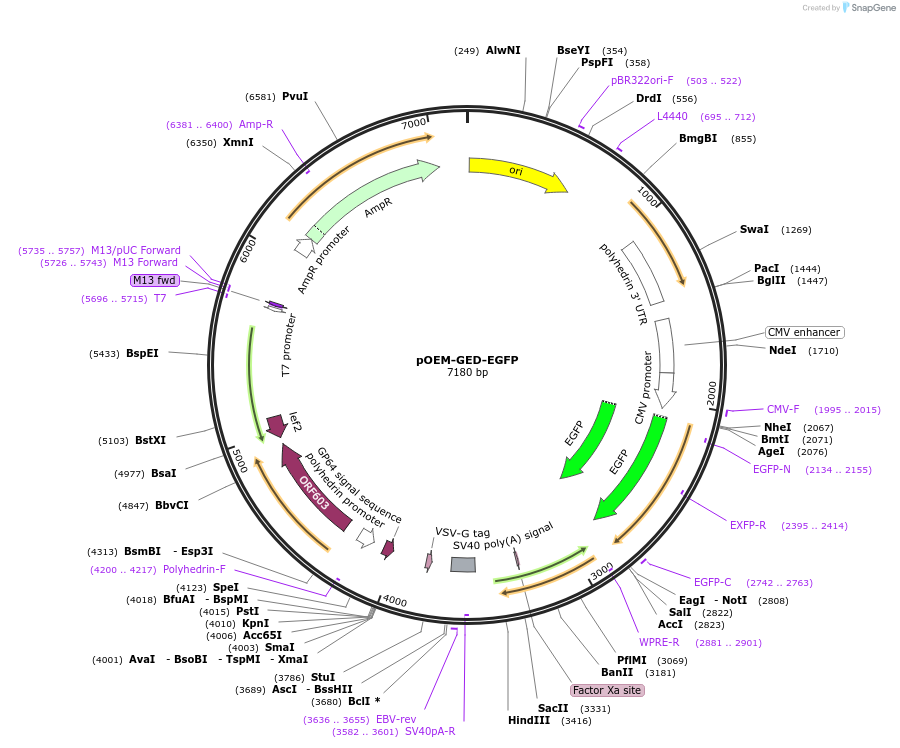
pOEM-GED-EGFP
Plasmid#102782PurposeBaculovirus rescue vector, VSVGED pseudotype, eGFPDepositorInsertEGFP
UseBaculoviral rescue shuttle vectorExpressionInsect and MammalianPromoterCMVAvailable SinceDec. 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
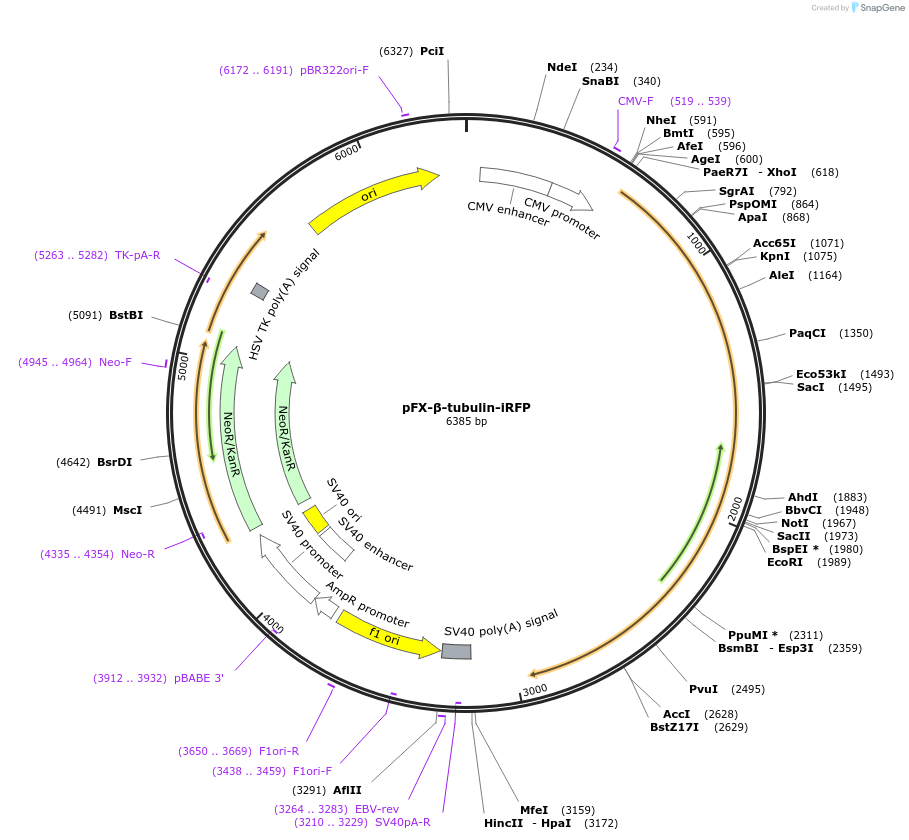
pFX-β-tubulin-iRFP
Plasmid#175830PurposeExpress iRFP with microtubule-targeting sequence in mammalian cellsDepositorInsertiRFP with microtuble-targeting sequence
Tagshuman tubulin beta 4B class IVb;TUBB4BExpressionMammalianPromoterCMVAvailable SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
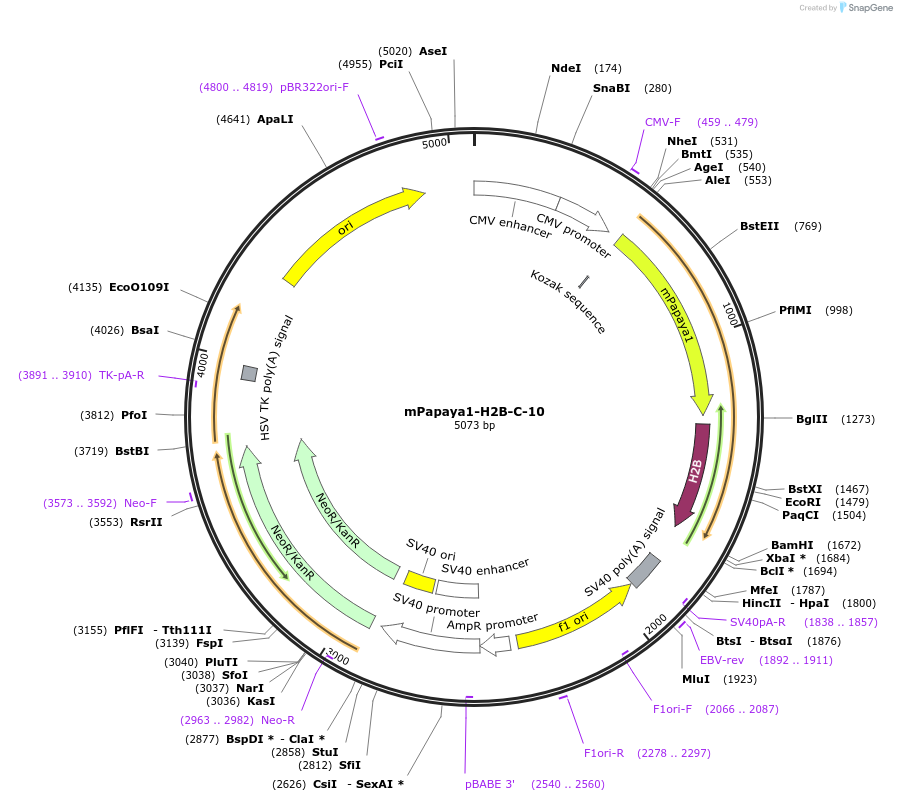
mPapaya1-H2B-C-10
Plasmid#56401PurposeLocalization: Nucleus/Histones, Excitation: 530, Emission: 541DepositorInsertH2B (H2BC11 Human)
TagsmPapaya1ExpressionMammalianMutationD26G and V119I in H2BPromoterCMVAvailable SinceDec. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
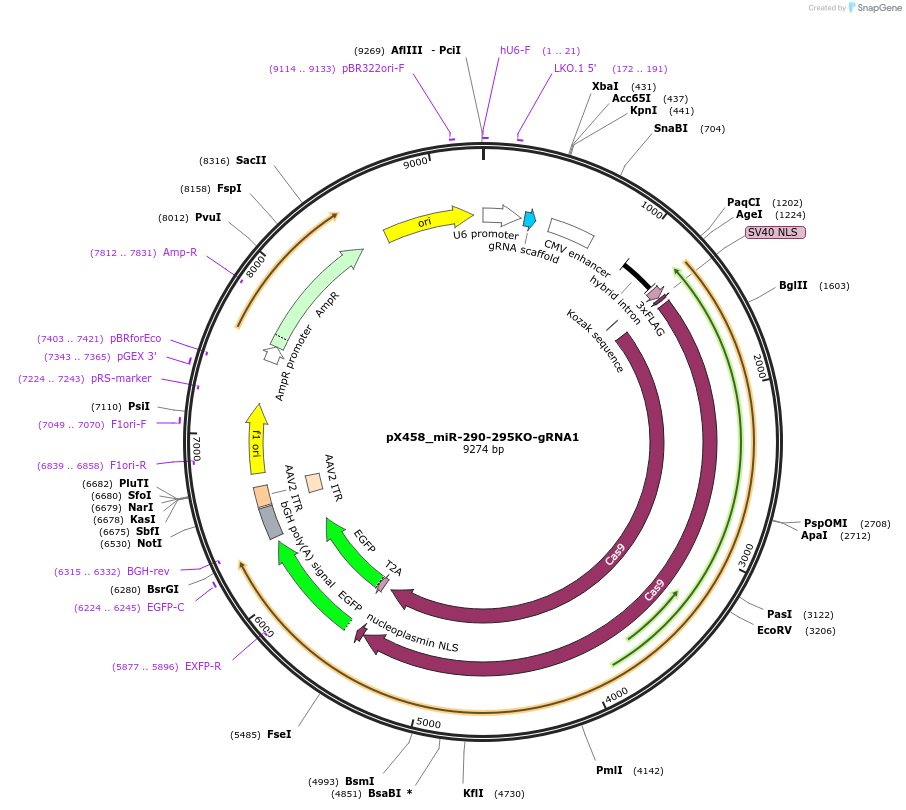
pX458_miR-290-295KO-gRNA1
Plasmid#172709PurposeCas9 with 5' gRNA for paired CRISPR KO of miR-290-295 clusterDepositorInsertmiR-290-295
UseCRISPRAvailable SinceAug. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
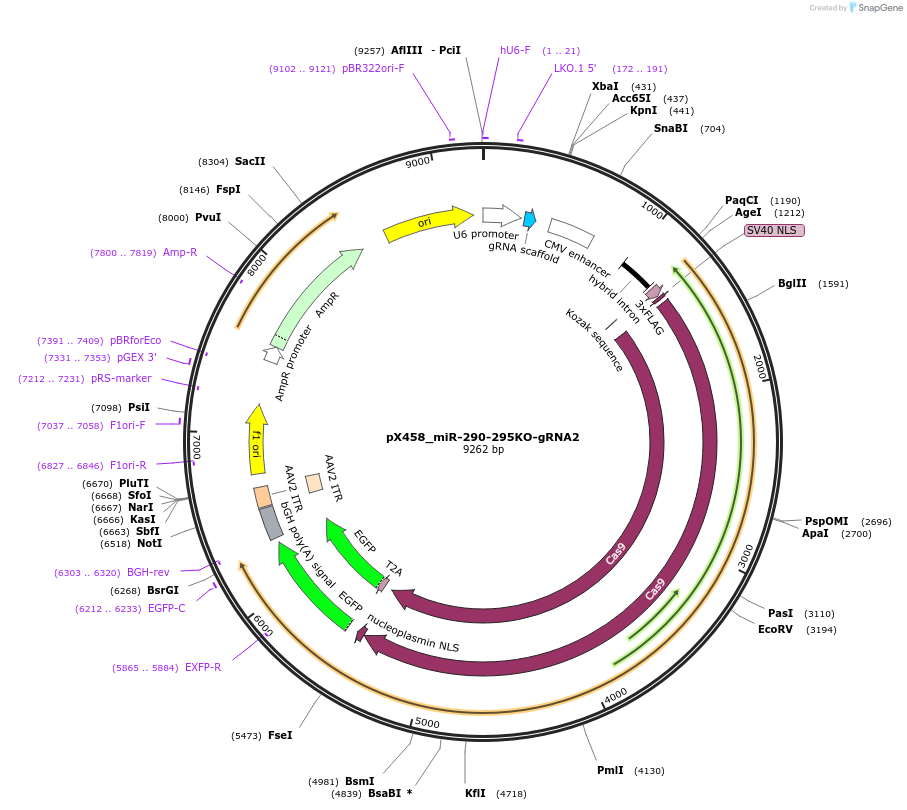
pX458_miR-290-295KO-gRNA2
Plasmid#172710PurposeCas9 with 3' gRNA for paired CRISPR KO of miR-290-295 clusterDepositorInsertmiR-290-295
UseCRISPRAvailable SinceAug. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
BAS292 C terminal SF-GFP kanR
Plasmid#74087PurposeS.pombe recoded super-folder GFP in universal C-terminal tagging plasmid, G418 resistanceDepositorInsertSF-GFP recoded for S.pombe
Available SinceAug. 2, 2016AvailabilityAcademic Institutions and Nonprofits only -
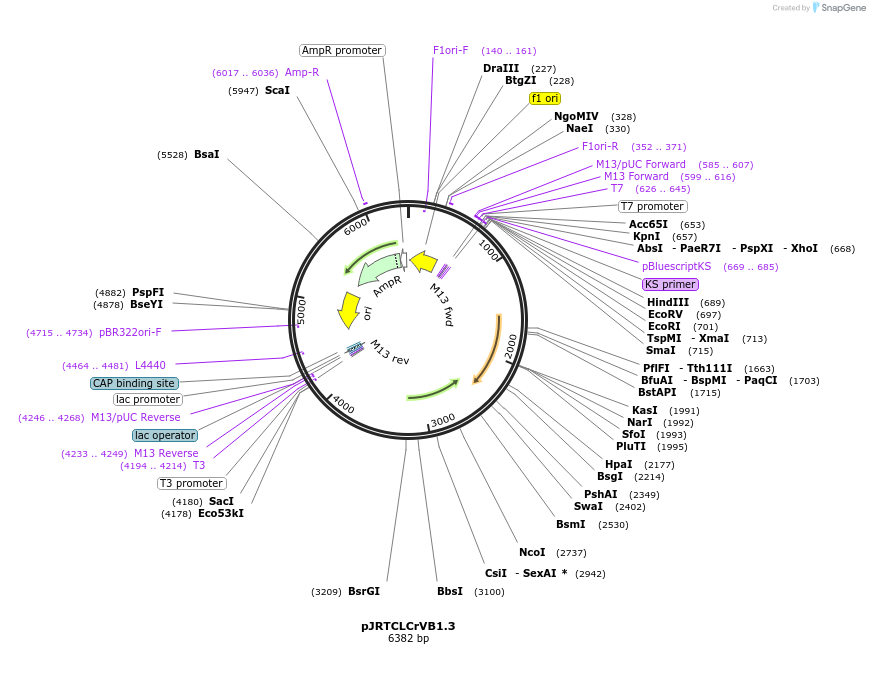
pJRTCLCrVB1.3
Plasmid#22844DepositorInsertB DNA
UseCotton vigs vectorExpressionBacterialMutation1.3 unit B componet length inserted into pBlueScr…Available SinceMarch 16, 2010AvailabilityAcademic Institutions and Nonprofits only -
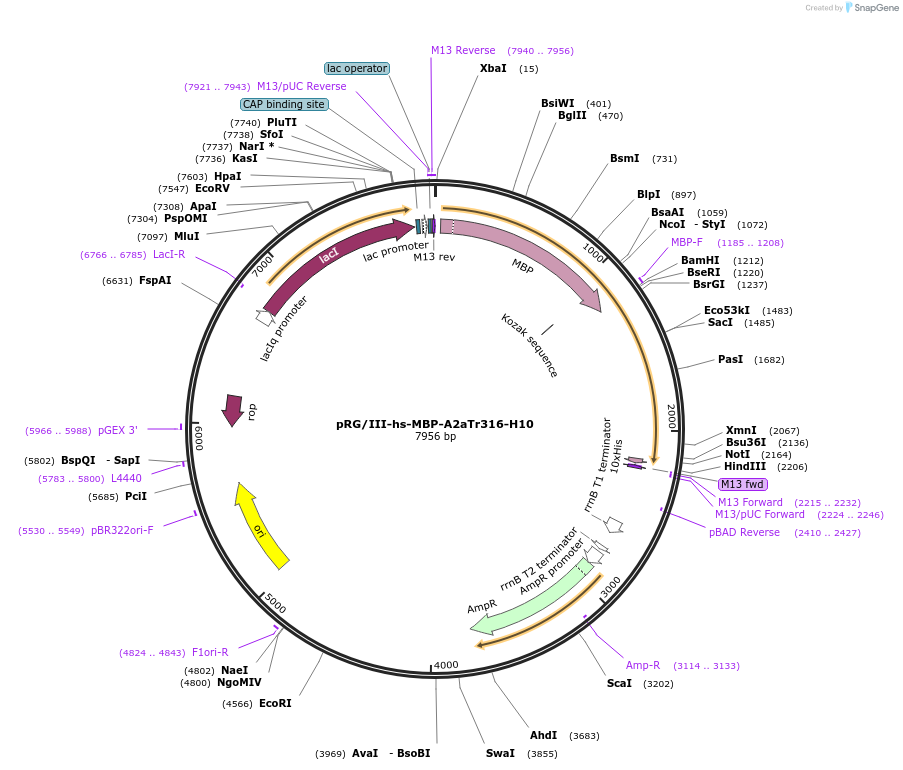
pRG/III-hs-MBP-A2aTr316-H10
Plasmid#121683PurposeBacterial expression of adenosine A2a receptor fusion proteinDepositorAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
mTagBFP2-FYN-N-10
Plasmid#55297PurposeLocalization: Membrane Associated Kinase, Excitation: 399, Emission: 456DepositorAvailable SinceOct. 10, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
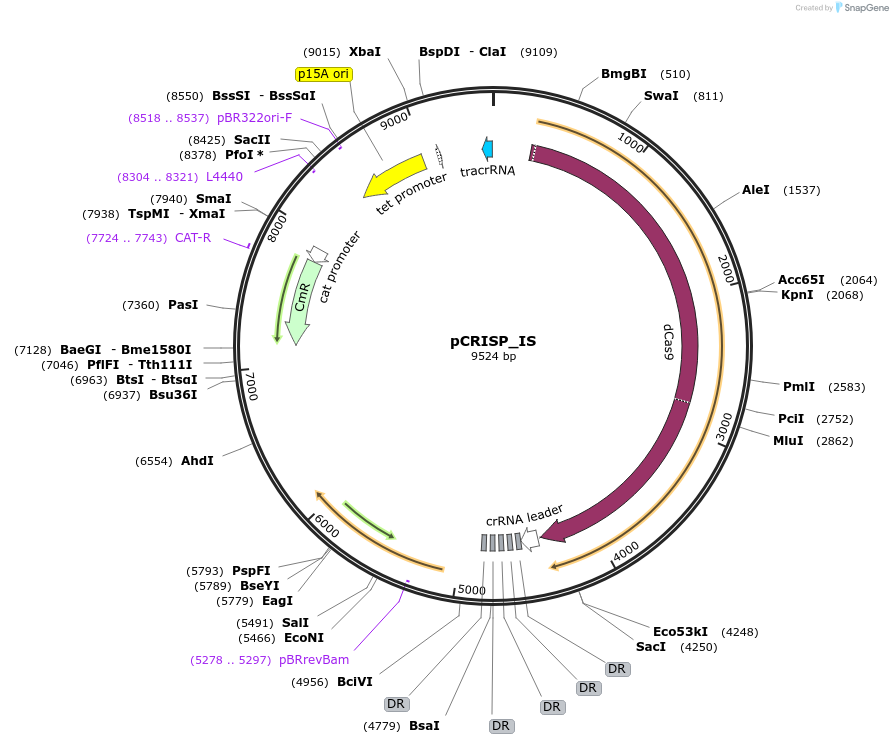
pCRISP_IS
Plasmid#120425PurposeThis plasmid encodes the complete CRISPR/dCas9 machinery for repressing transposition of the following bacterial Insertion Sequences: IS1, IS3 and IS5.DepositorInsertCRISPR spacers targeting IS1, IS5, IS3
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterconstitutiveAvailable SinceJan. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
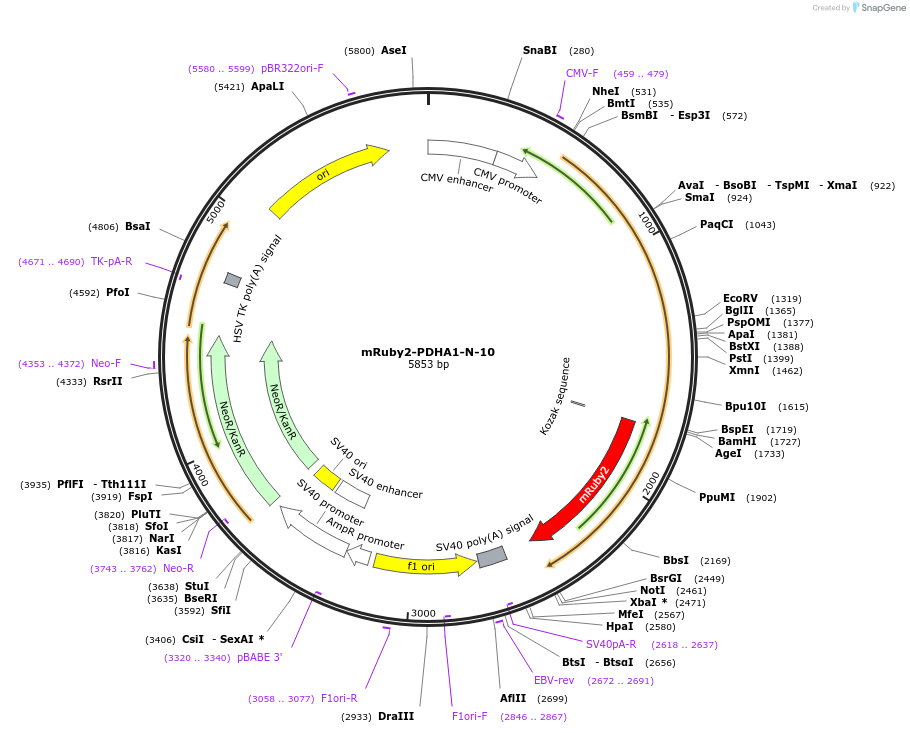
mRuby2-PDHA1-N-10
Plasmid#55909PurposeLocalization: Mitochondria, Excitation: 559, Emission: 600DepositorAvailable SinceMarch 12, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
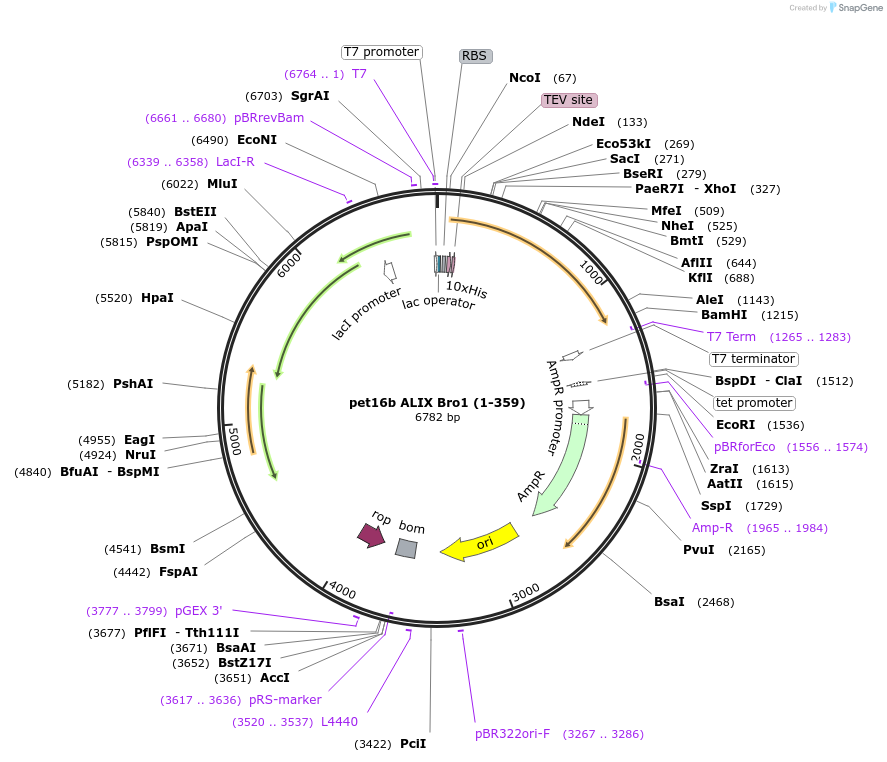
pet16b ALIX Bro1 (1-359)
Plasmid#80641PurposeResidues 1-359 of ALIX; Bacterial Expression Vector; N-term His-tag; Sundquist Lab Internal ID: WISP11-428DepositorInsertALIX Bro1, residues 1-359 (PDCD6IP Human)
TagsHis TagExpressionBacterialMutationResidues 1-359PromoterT7Available SinceSept. 30, 2016AvailabilityAcademic Institutions and Nonprofits only -
pEB2-mRFP1
Plasmid#104001PurposePlasmid encoding mRFP1DepositorInsertmRFP1
UseLow copyExpressionBacterialMutationMutations relative to DsRed (R2A, K5E, N6D, T21S,…PromoterproCAvailable SinceDec. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
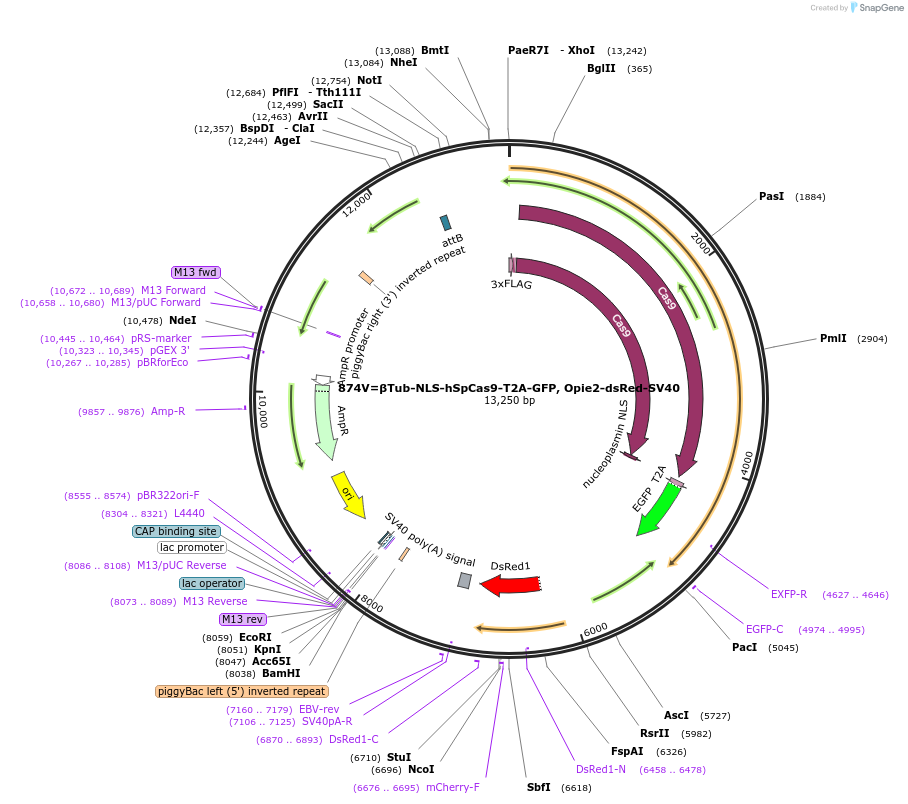
874V=βTub-NLS-hSpCas9-T2A-GFP, Opie2-dsRed-SV40
Plasmid#159672PurposePlasmid supports Cas9 expression only in males, Opie2-dsRed tagged, and can be integrated with pBac or phC31.DepositorInsertβTub-NLS-hSpCas9-T2A-GFP
TagsT2A-GFPExpressionInsectAvailable SinceMarch 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
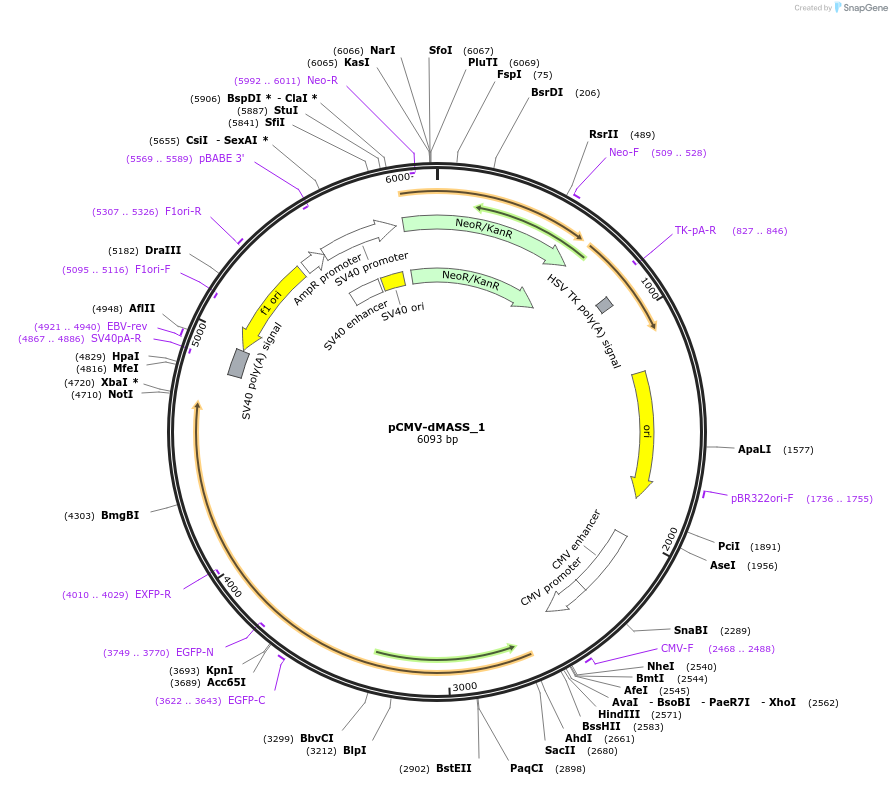
pCMV-dMASS_1
Plasmid#209764PurposeExpression of dMASS_1 in mammalian cells under CMV promoterDepositorInsertdMASS_1
ExpressionMammalianAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
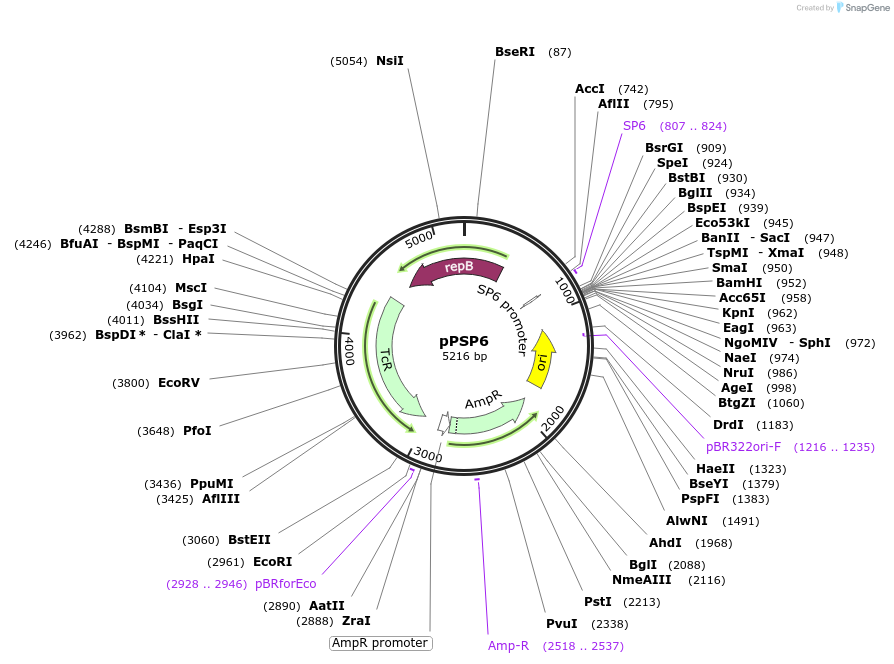
pPSP6
Plasmid#48150PurposeShuttle vector E. coli/B.meg., encoding a promoter for RNA polymerase of the phage SP6DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceOct. 11, 2013AvailabilityAcademic Institutions and Nonprofits only -
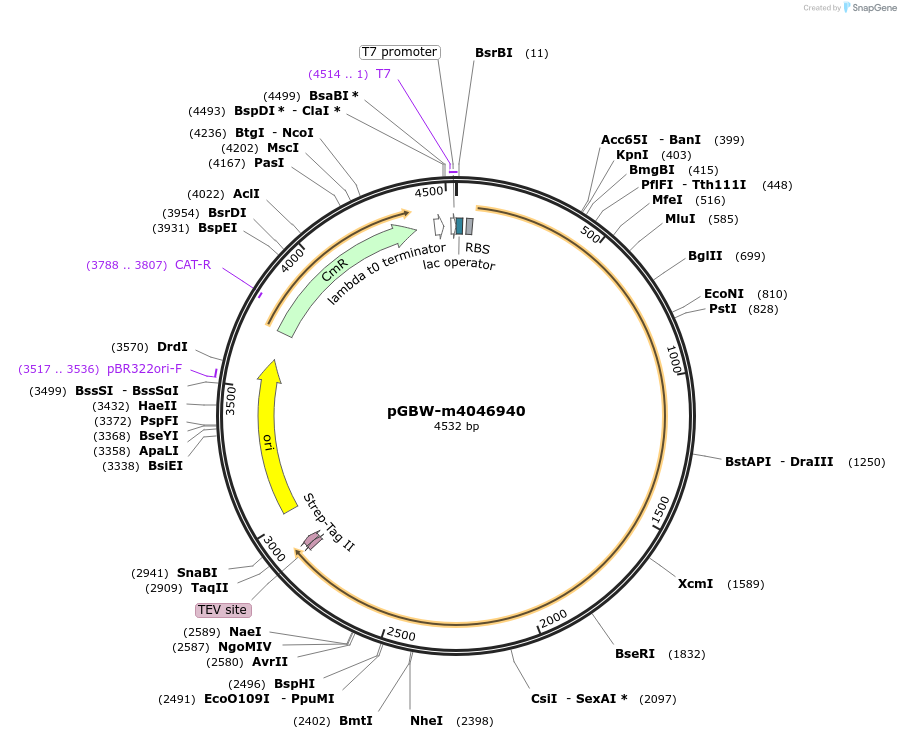
pGBW-m4046940
Plasmid#149261PurposeBacterial expression plasmid for SARS-CoV-2 RNA-dependent RNA polymeraseDepositorInsertSARS-CoV-2 RNA-dependent RNA polymerase (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceMay 12, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
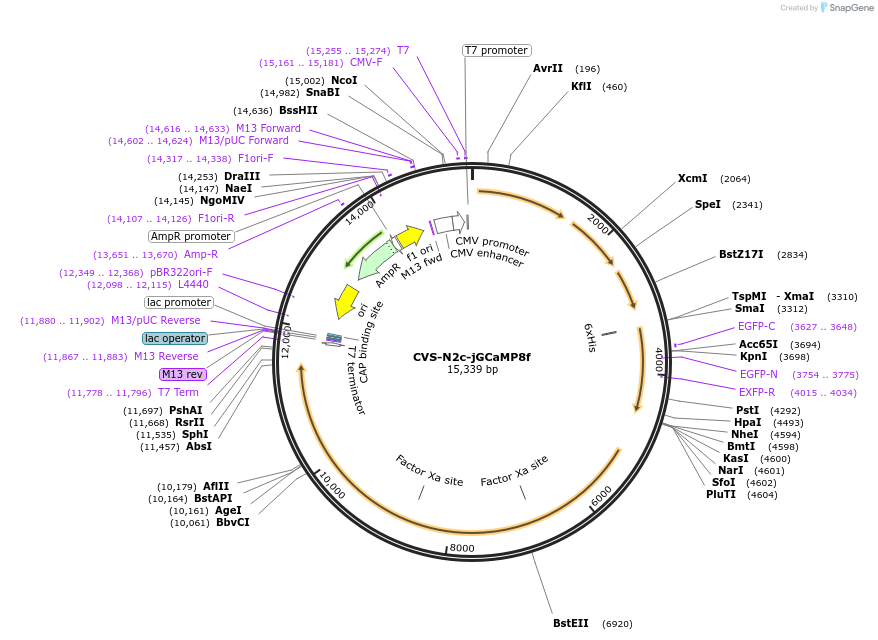
CVS-N2c-jGCaMP8f
Plasmid#172387PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertGCaMP8f
UseNeurotropic virusAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
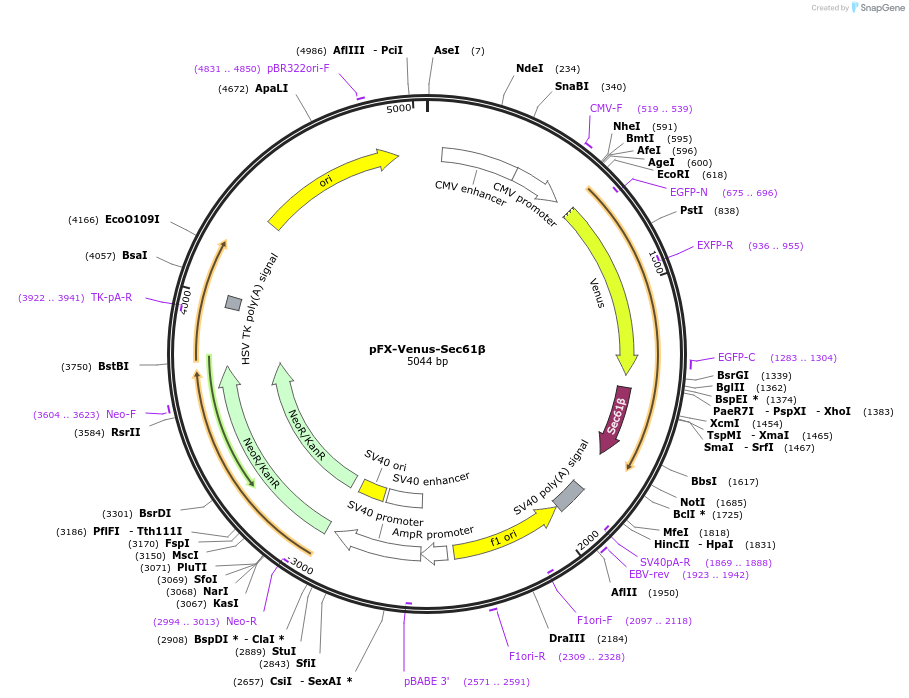
pFX-Venus-Sec61β
Plasmid#174467PurposeExpress Venus with endoplasmic reticulum-targeting sequence in mammalian cellsDepositorInsertVenus with endoplasmic reticulum-targeting sequence
TagsSec61 subunit betaExpressionMammalianPromoterCMVAvailable SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
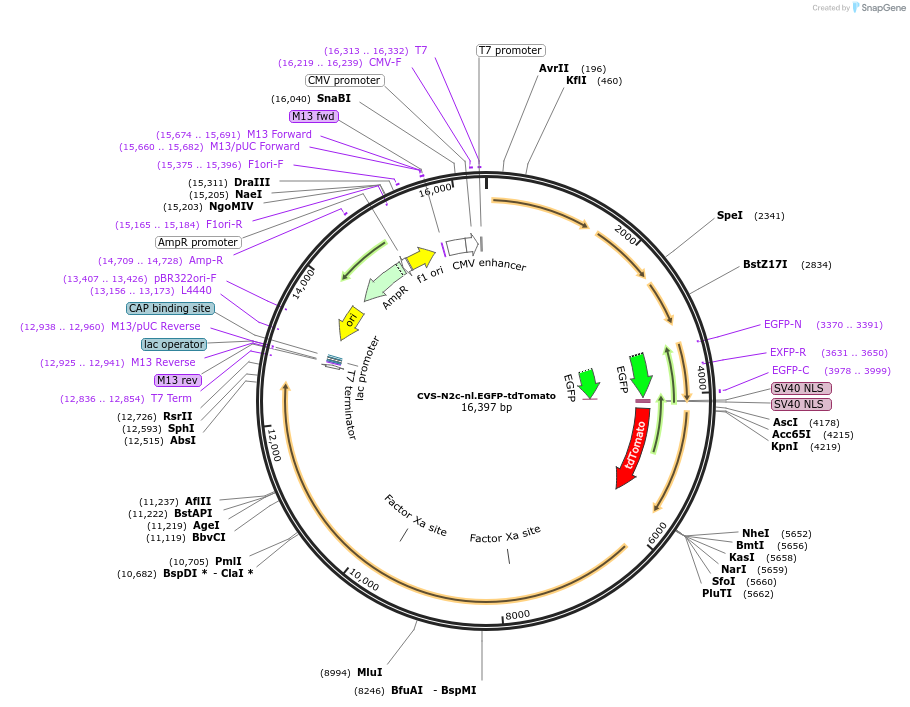
CVS-N2c-nl.EGFP-tdTomato
Plasmid#172382PurposeProduction of RVdG-CVS-N2c viral vectors for cell-type specific trans-synaptic retrograde labeling.DepositorInsertnuclear-localized EGFP + tdTomato
UseNeurotropic virusAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
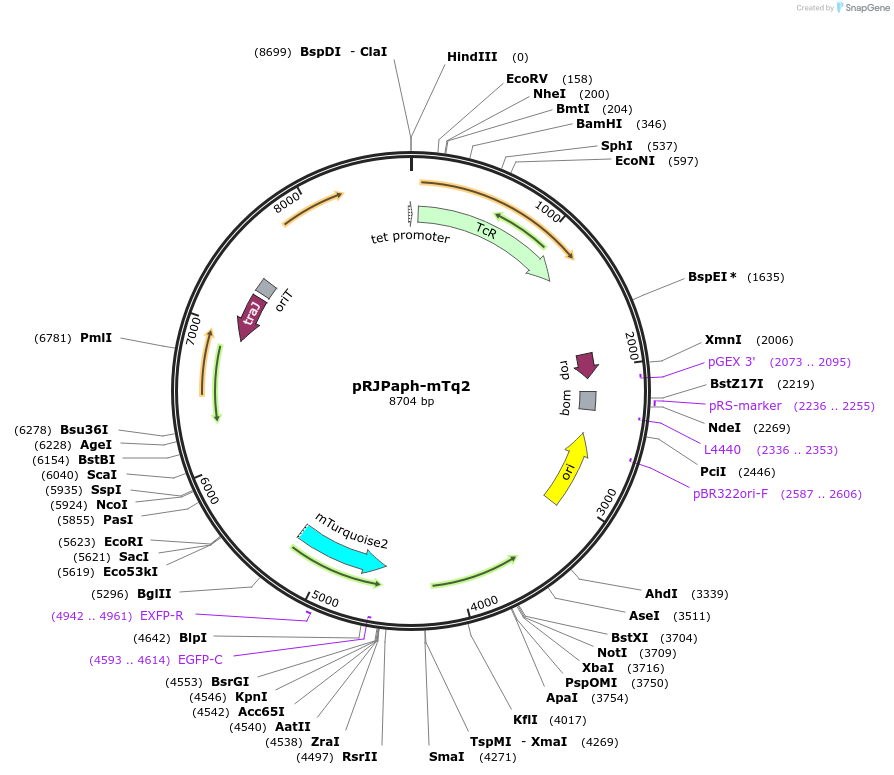
pRJPaph-mTq2
Plasmid#67031PurposePlasmid for Paph-mTurqoise2 integration downstream of B. diazoefficiens (B. japonicum) scoIDepositorInsertsB. diazoefficiens DNA from scoI downstream region (comprising blr1132')
mTurqoise2
UseConjugative bacterial vectorAvailable SinceOct. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
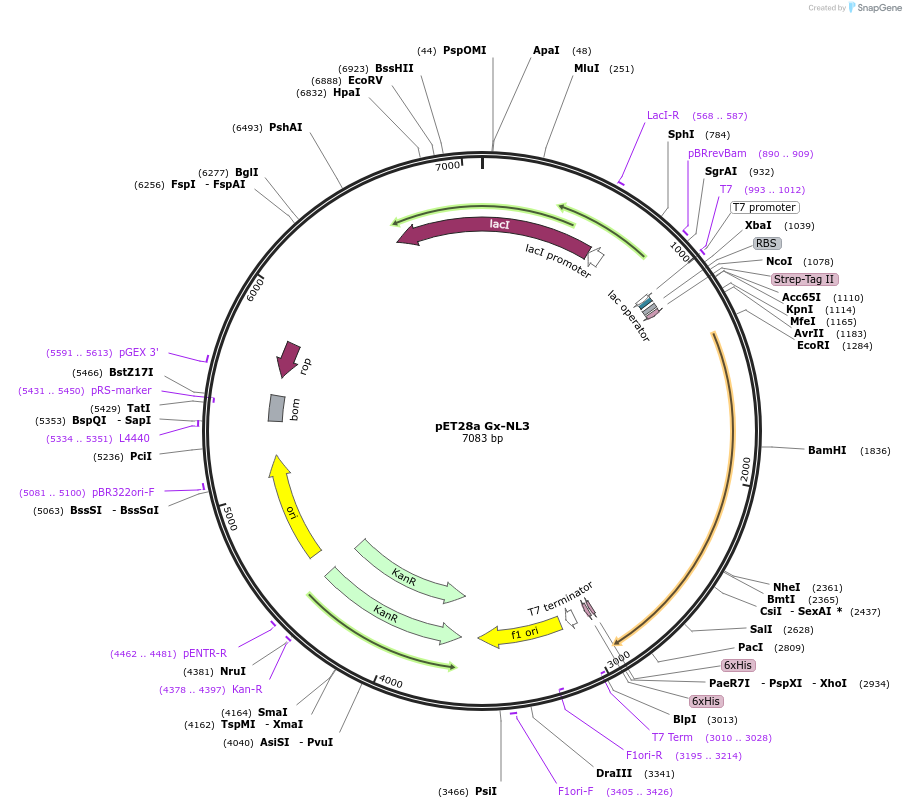
pET28a Gx-NL3
Plasmid#169108PurposeExpresses protein G fused to 3 copies of NanoLuc. The protein G domain contains an amber stop-codon at position 24 for incorporation of the unnatural amino acid para-benzoylphenylalanine.DepositorInsertprotein G - NanoLuc - NanoLuc - NanoLuc
Tagshis-tag and strep-tagExpressionBacterialMutationAla 24 stopPromoterT7Available SinceJune 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
mCerulean3-EB1-C-18
Plasmid#55415PurposeLocalization: MT End Binding Protein, Excitation: 433, Emission: 475DepositorAvailable SinceOct. 10, 2014AvailabilityAcademic Institutions and Nonprofits only -
BAS287 C terminal E2C hygBR
Plasmid#74082PurposeS.pombe recoded E2Crimson in universal C-terminal tagging plasmid, hygB resistance.DepositorInsertE2Crimson
Available SinceAug. 2, 2016AvailabilityAcademic Institutions and Nonprofits only -
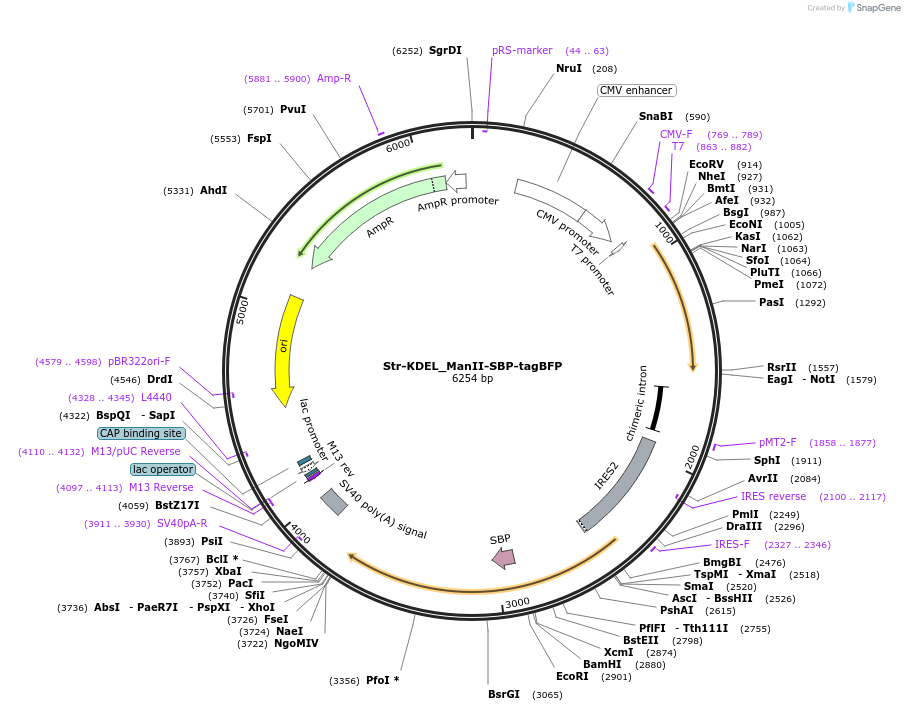
Str-KDEL_ManII-SBP-tagBFP
Plasmid#65254Purposesynchronize trafficking of ManII from the ER (RUSH system)DepositorInsertStreptavidin-KDEL and truncated Mannosidase II
TagsStreptavidin Binding Protein (SBP) and tagBFPExpressionMammalianMutationcontains only amino acids 1 to 116 of ManIIPromoterCMVAvailable SinceJune 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
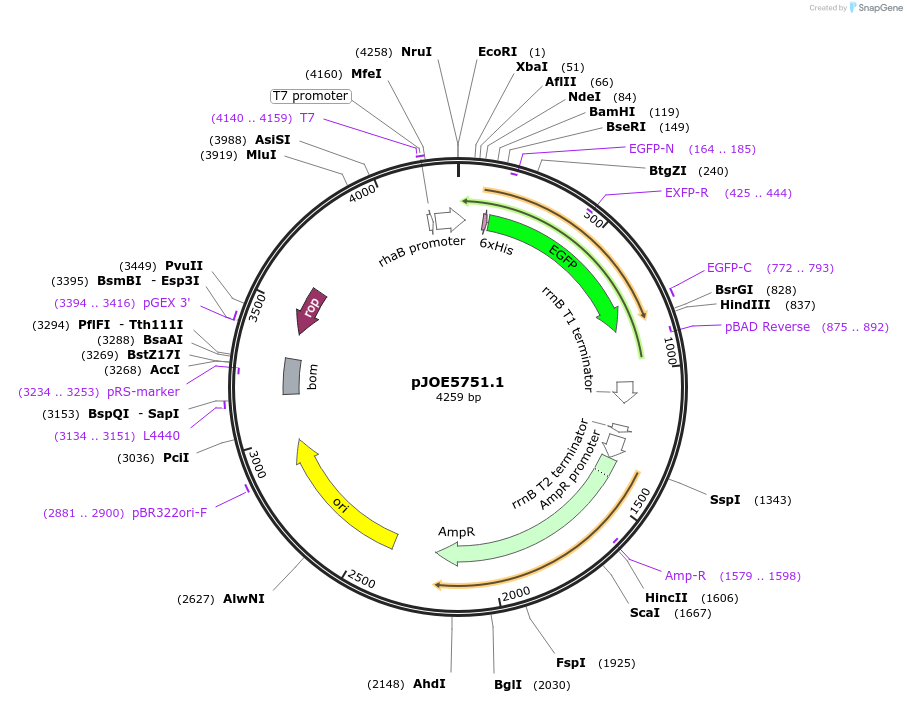
pJOE5751.1
Plasmid#135083PurposepBR322-based L-rhamnose-inducible vector.DepositorInsertP-rhaP-BAD-His tag-eGFP
ExpressionBacterialAvailable SinceFeb. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
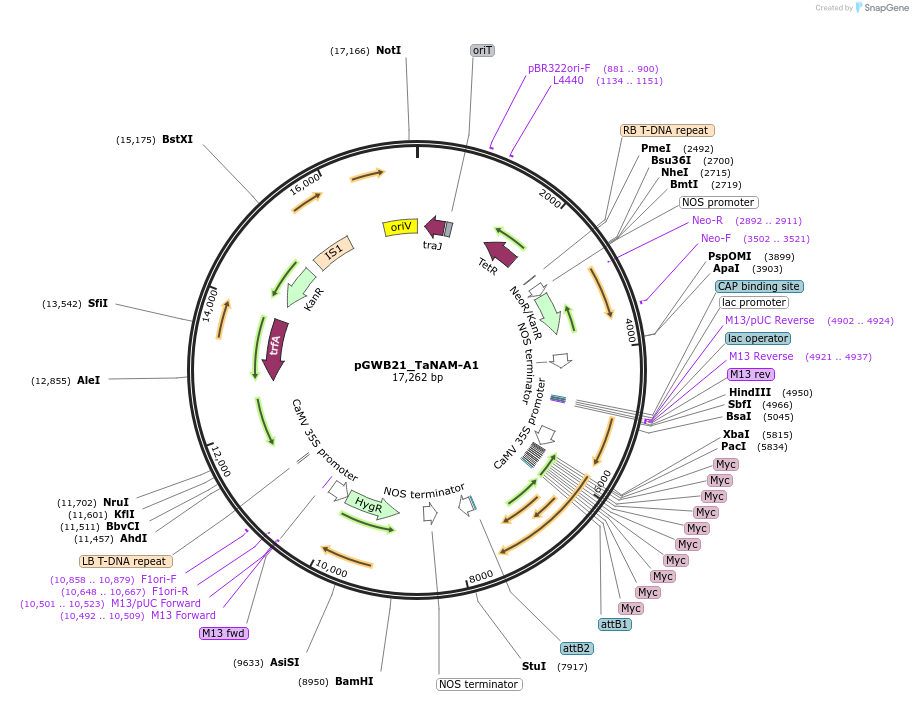
pGWB21_TaNAM-A1
Plasmid#124572PurposeExpresses full-length TaNAM-A1 with an N-terminal 10x Myc tag under the 35s promoter.DepositorInsertTaNAM-A1
Tags10xMycExpressionPlantPromoter35sAvailable SinceAug. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
mTagBFP2-Dectin1A-N-10
Plasmid#55291PurposeLocalization: Membrane, Excitation: 399, Emission: 456DepositorAvailable SinceOct. 7, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
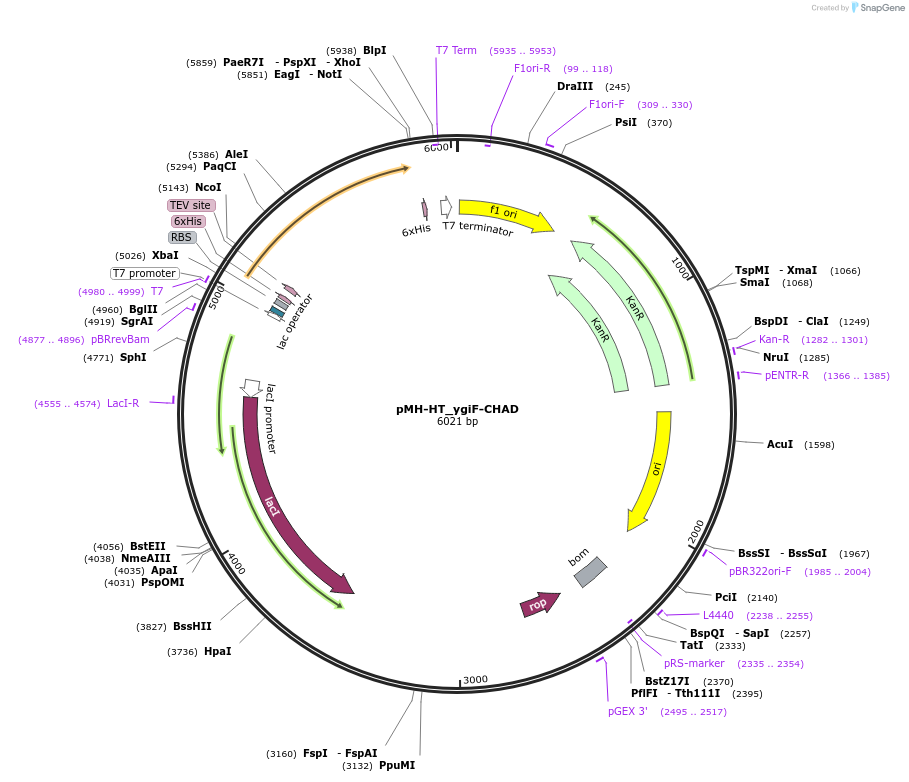
pMH-HT_ygiF-CHAD
Plasmid#127793PurposeExpresses the CHAD domain of ygiF in E. coli cellsDepositorInsertygiF-CHAD (ygiF Escherichia coli)
Tags6xHis tag + TEV cleavage siteExpressionBacterialMutationN-terminal TTM domain of ygiF (residues 201-433)Available SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
pENTR4_FLAG/HA_FUS_His6
Plasmid#26365DepositorAvailable SinceOct. 18, 2010AvailabilityAcademic Institutions and Nonprofits only -
Clover-Nup50-N-10
Plasmid#56303PurposeLocalization: Nuclear Pore, Excitation: 505, Emission: 515DepositorAvailable SinceDec. 18, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
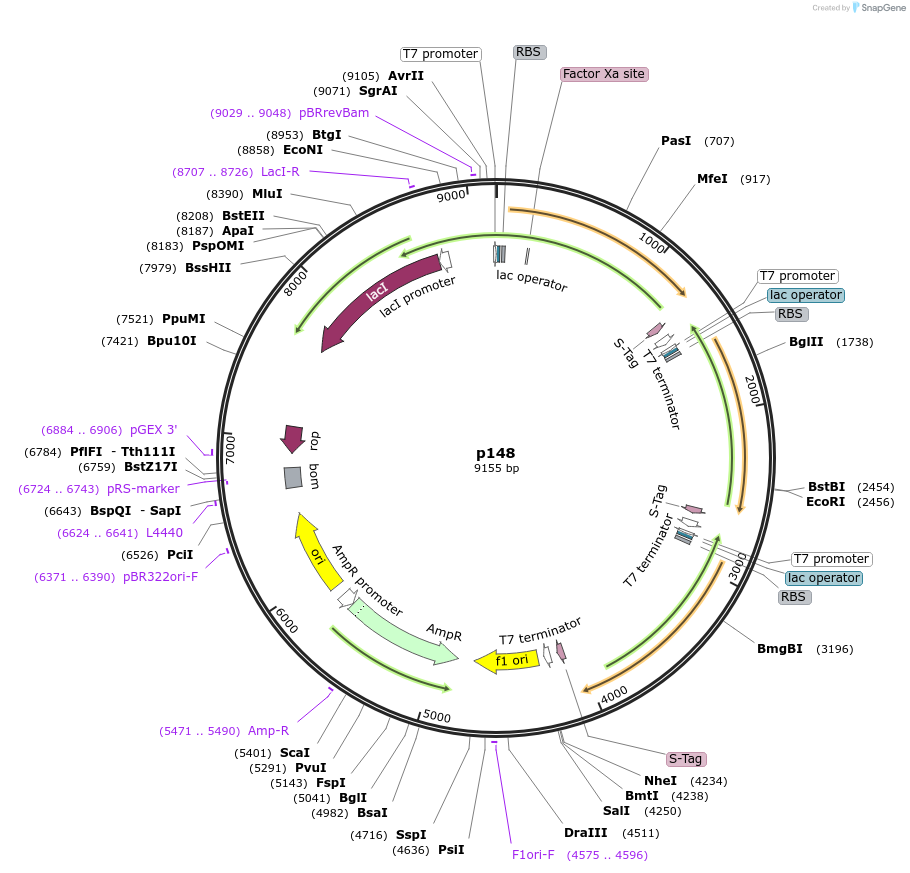
p148
Plasmid#62616PurposeePathBrick pETM6 vector encoding CsF3H, AaDFR, and DuLARDepositorInsertsflavaone 3B-hydroxylase
dihydroflavonol 4-reductase
leucoanthocyanidin reductase
UseSynthetic BiologyTagsGBD, PDZ, and SH3ExpressionBacterialPromoterT7Available SinceJuly 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
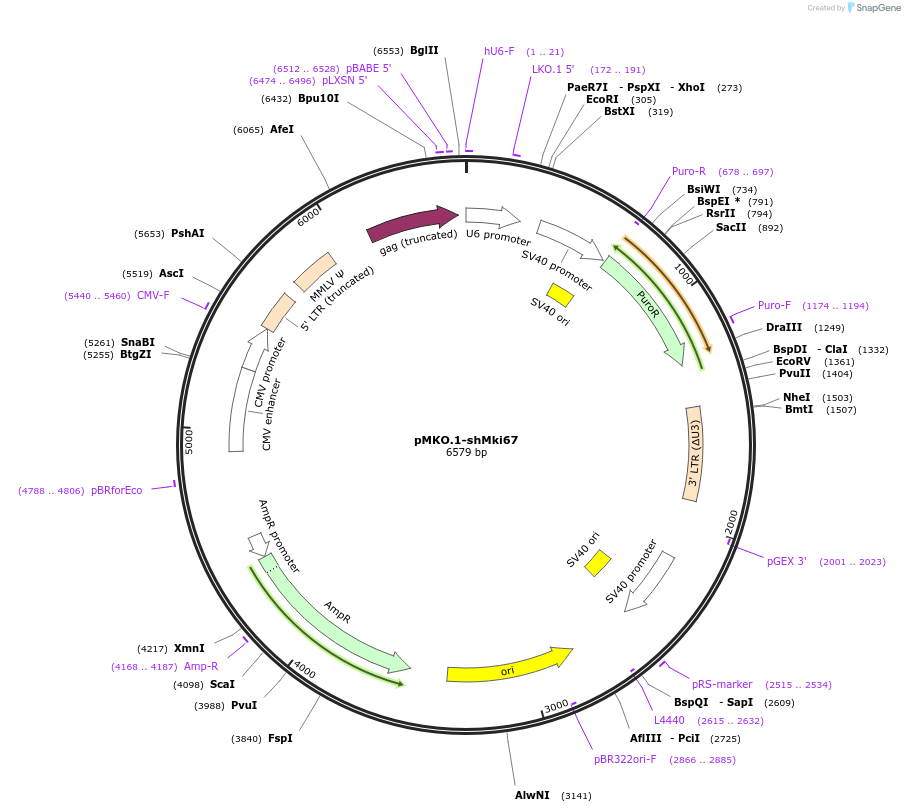
pMKO.1-shMki67
Plasmid#160958PurposeMki67 shRNA in pMKO.1 retroviral vectorDepositorAvailable SinceDec. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
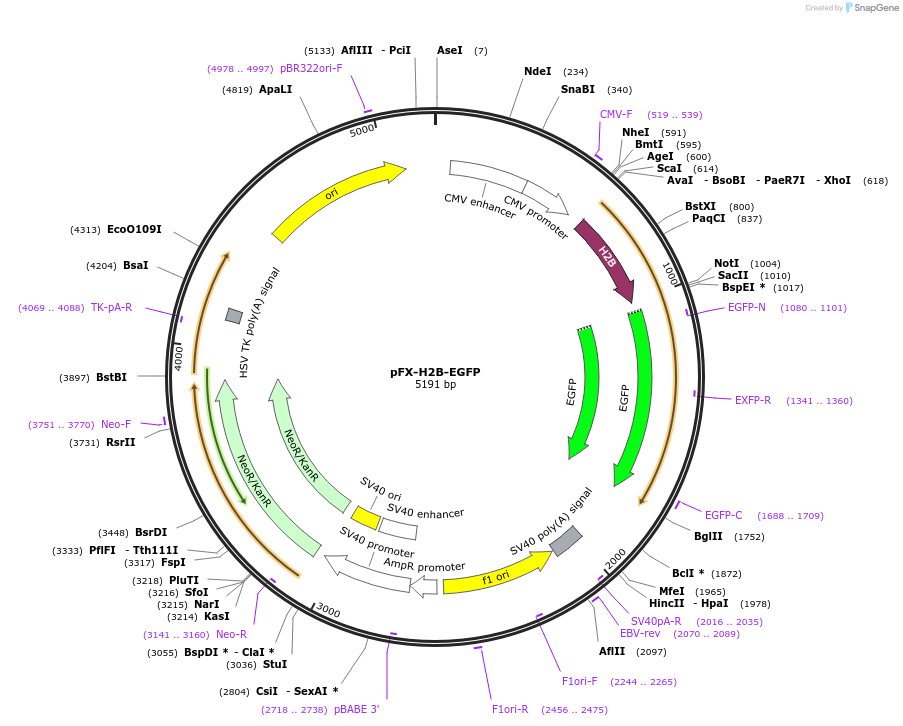
pFX-H2B-EGFP
Plasmid#175839PurposeExpress EGFP with nucleus-targeting sequence in mammalian cellsDepositorInsertEGFP with nucleus-targeting sequence
TagsH2B (human histone 2B)ExpressionMammalianPromoterCMVAvailable SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
E0030 (YFP)_CD
Plasmid#66031PurposeMoClo Basic Part: CDS - Fluorescent protein. Yellow. [C:E0030:D]DepositorInsertFluorescent reporter - YFP
UseSynthetic BiologyAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
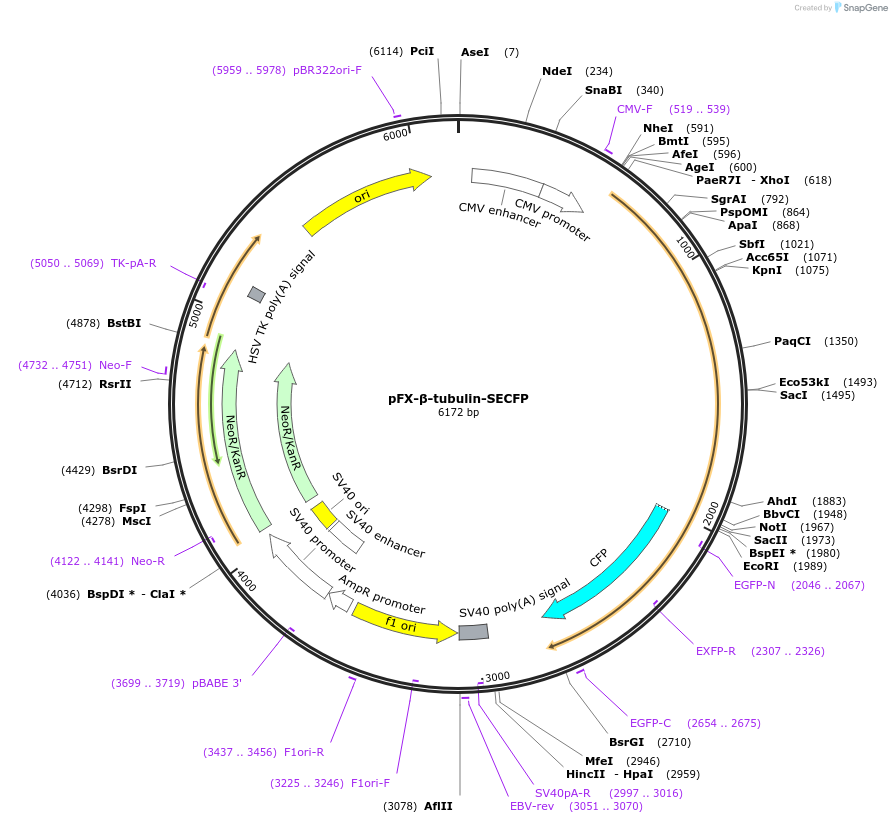
pFX-β-tubulin-SECFP
Plasmid#175826PurposeExpress SECFP with microtubule-targeting sequence in mammalian cellsDepositorInsertSECFP with microtuble-targeting sequence
Tagshuman tubulin beta 4B class IVb;TUBB4BExpressionMammalianPromoterCMVAvailable SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
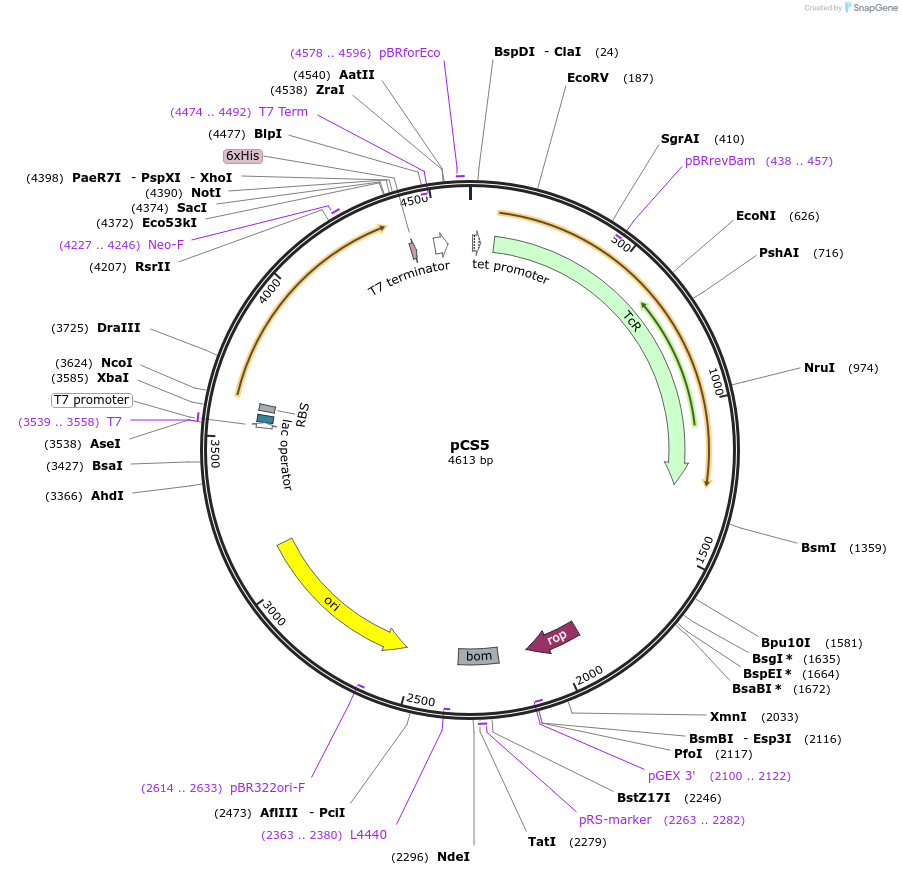
pCS5
Plasmid#55750PurposeProduces one half (beta) of aminoglycoside phosphotransferase (3′)-IIa when in the presence of T7 RNA Polymerase.DepositorInsertKanR
ExpressionBacterialMutationResidues 59-264 of KanRPromoterT7-lacAvailable SinceJuly 15, 2014AvailabilityAcademic Institutions and Nonprofits only -
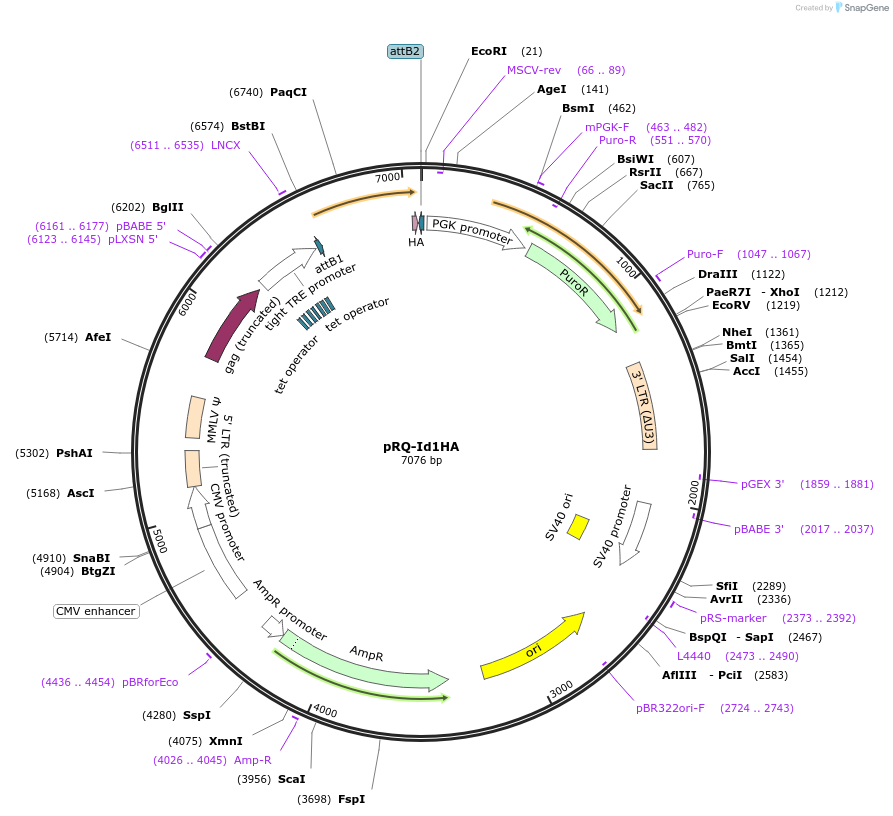
pRQ-Id1HA
Plasmid#18112DepositorAvailable SinceJan. 29, 2009AvailabilityAcademic Institutions and Nonprofits only