We narrowed to 18,624 results for: RAN-1
-
Plasmid#159449PurposeInsert: Pfus1tet-citrine-tCyc1. The fluorescent protein Citrine is inducible by alpha-factor and repressible by TetR. Backbone: pRG207 (addGene). Marker:Lys2.DepositorInsertPfus1tet-citrine-tCyc1
UseSynthetic BiologyTagsExpressionYeastMutationPromoterPfus1tetAvailable SinceDec. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
pGEX-delN-EcGlpG
Plasmid#158537PurposeBacterial expression of rhomboid protease EcGlpG without N terminusDepositorInsertdelta N-terminus EcGlpG
UseTagsGST tagExpressionBacterialMutationN-terminal deletion (1-86)PromoterAvailable SinceSept. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
pFH39
Plasmid#126013PurposeLevel0 Rice OsU3 Pol III promoter with transcription Start-ADepositorInsertLevel0 Rice OsU3 Pol III promoter with transcription Start-A
UseCRISPR and Synthetic BiologyTagsExpressionPlantMutationPromoterAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
YIplac211-HSE1-iGFPx6
Plasmid#105259PurposePlasmid for pop-in/pop-out replacement of Hse1 with Hse1-iGFPx6.DepositorAvailable SinceJan. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
YIplac211-MUP1-iYFP
Plasmid#105266PurposePlasmid for pop-in/pop-out replacement of Mup1 with Mup1-iYFP.DepositorAvailable SinceJan. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
CLYBL-(Ef1a-SBP-LNGFR-T2A-mApple)-(CAG-rtTA)-(TRE-hNIL))
Plasmid#105842PurposeDonor construct for introduction of hNIL factors and SBP-LNGFR for streptavidin bead enrichmentDepositorInsertsUseCRISPR and TALENTagsExpressionMutationPromoterCAG, EF-1alpha, and TRE3GAvailable SinceApril 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
pSOUP
Plasmid#165419PurposeThe pSOUP helper plasmid needed for replication of all pGreen (Hellens et al., 2000) based vectors in AgrobacteriumInsertRep A
UseHelper plasmid required for replication for all p…TagsExpressionMutationPromoterAvailable SinceApril 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
-
pDV002
Plasmid#191823PurposePlasmid for the transformation of the cyanobacterium Arthrospira platensis at the NS1 locus. No promoter. No terminator. Golden Gate compatible. Selection: in A. platensis: aadA; E. coli: AmpRDepositorTypeEmpty backboneUseUnspecifiedTagsExpressionMutationPromoterAvailable SinceJan. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
pDV044
Plasmid#191824PurposePlasmid for the transformation of the cyanobacterium Arthrospira platensis at the KmR locus. pCPC600 promoter. With terminator. Golden Gate compatible. Selection: in A. platensis: KmR; E. coli: AmpRDepositorTypeEmpty backboneUseUnspecifiedTagsExpressionMutationPromoterAvailable SinceJan. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
pDV052
Plasmid#191825PurposePlasmid for the transformation of the cyanobacterium Arthrospira platensis at the KmR locus. No promoter. No terminator. Golden Gate compatible. Selection: in A. platensis: KmR; E. coli: AmpRDepositorTypeEmpty backboneUseUnspecifiedTagsExpressionMutationPromoterAvailable SinceJan. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3-AKAR4-Kras
Plasmid#61621PurposeNon-raft/plasma membrane-targeted FRET biosensor for monitoring PKA activityDepositorInsertAKAR4-Kras
UseTagsC-terminal targeting sequence from Kras, Cerulean…ExpressionMammalianMutationPromoterCMVAvailable SinceMarch 13, 2015AvailabilityAcademic Institutions and Nonprofits only -
pXD68Kan2-AphA
Plasmid#191248PurposeRSF1010-based broad host-range plasmid engineered from pAM5409, kanamycin resistanceDepositorTypeEmpty backboneUseTagsExpressionBacterialMutationPromoterAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
pSC101-GFPmut2-mScarlet-I TAA
Plasmid#208187Purposenonsense mistranslation, TAA stopDepositorInsertsGFPmut2
mScarlet-I
UseReporterTagsExpressionBacterialMutationInsertion 6TAA*(stop) 7AGT(Ser)PromoterPtet+dnaK P1Available SinceMarch 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pBTSG
Plasmid#159420PurposeTransient expression of proteins with a C-terminal thermostable GFP (TGP) tag in mammalian cellsDepositorInsertthermostable GFP (TGP)
UseTagspolyhistidine (poly-His)ExpressionMammalianMutationPromoterAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLux76-LasI
Plasmid#73444PurposeHSL relay device: produces 3O-C12-HSL in response to 3O-C6-HSLDepositorInsertLasI
UseSynthetic BiologyTagsExpressionMutationPromoterpLux76Available SinceApril 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
pLas81-LuxI
Plasmid#73445PurposeHSL relay device: produces 3O-C6-HSL in response to 3O-C12-HSLDepositorInsertLuxI
UseSynthetic BiologyTagsExpressionMutationPromoterpLas81Available SinceApril 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
pSC101-GFPmut2-mScarlet-I TAG
Plasmid#208188Purposenonsense mistranslation, TAG stopDepositorInsertsGFPmut2
mScarlet-I
UseReporterTagsExpressionBacterialMutationInsertion 6TAG*(stop) 7AGT(Ser)PromoterPtet+dnaK P1Available SinceMarch 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pSC101-GFPmut2-mScarlet-I TGA
Plasmid#208189Purposenonsense mistranslation, TGA stopDepositorInsertsGFPmut2
mScarlet-I
UseReporterTagsExpressionBacterialMutationInsertion 6TGA*(stop) 7AGT(Ser)PromoterPtet+dnaK P1Available SinceMarch 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pSC101-GFPmut2-mScarlet-I TGATC
Plasmid#208190Purposenonsense mistranslation, TGA stop variant 2DepositorInsertsGFPmut2
mScarlet-I
UseReporterTagsExpressionBacterialMutationInsertion 6TGA*(stop) 7TCT(Ser)PromoterPtet+dnaK P1Available SinceMarch 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pSC101-GFPmut2-mScarlet-I 6Trp7Ser
Plasmid#208186Purposemistranslation GFP and mScarlet-I positive control with Trp insertionDepositorInsertsGFPmut2
mScarlet-I
UseReporterTagsExpressionBacterialMutationInsertion 6Trp7SerPromoterPtet+dnaK P1Available SinceMarch 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
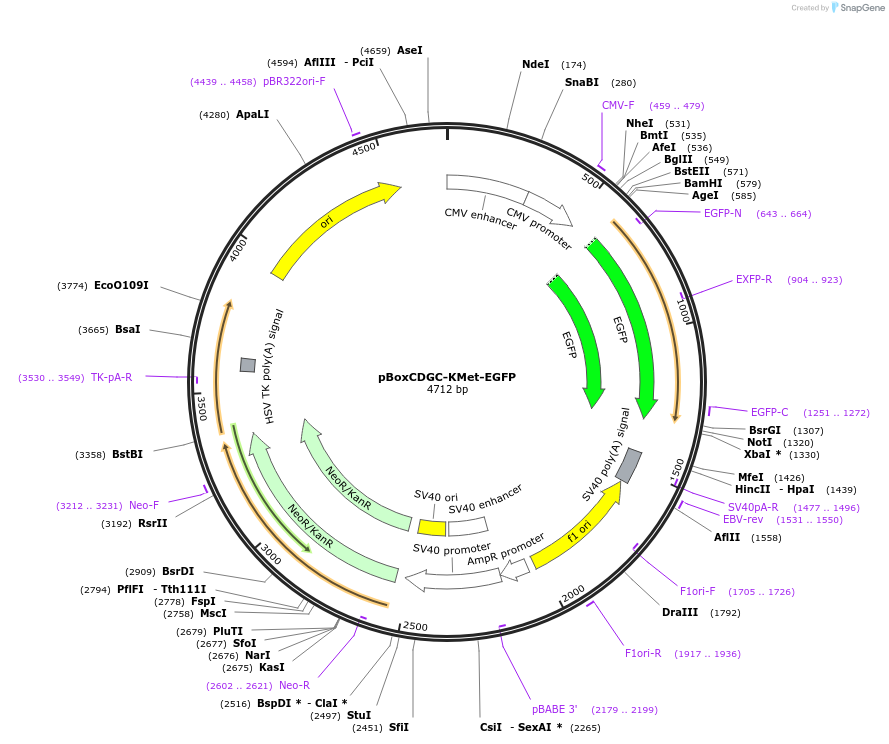
pBoxCDGC-KMet-EGFP
Plasmid#140272PurposeL7Ae-dependent translational repressionDepositorInsertBoxCD
UseSynthetic BiologyTagsExpressionMammalianMutationPromoterCMVAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
pF(UG) CamKII sf iGluSnFR
Plasmid#158769PurposeiGluSnFR glutamate sensor containing a Golgi export sequence and and an ER exit motif for improved trafficking to the plasma membrane.DepositorInsertiGluSnFR
UseLentiviralTagsExpressionMutationAdditional ER and Golgi export sequencesPromoterAvailable SinceFeb. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
TrmD_HaeIn/pET28b(+) N,C-tag
Plasmid#12665DepositorInserttRNA (guanine-N(1)-)-methyltransferase (trmD Haemophilus influenzae)
UseTagsHisExpressionBacterialMutationPromoterAvailable SinceJan. 5, 2007AvailabilityAcademic Institutions and Nonprofits only -
pED7x26
Plasmid#134813PurposeStringent phage propagation reporter containing pIII-16-29-34-TAGADepositorInsertpIII 16-TAGA 29-TAGA 34-TAGA
UseSynthetic BiologyTagsExpressionBacterialMutation16-TAGA 29-TAGA 34-TAGAPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pMIR-R/Tac1/SG
Plasmid#16296DepositorAvailable SinceMarch 12, 2008AvailabilityAcademic Institutions and Nonprofits only -
pZA68
Plasmid#174781PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-L17E/D481V-eGFP, Kmr plant selection marker
UseTagseGFPExpressionPlantMutationL17E/D481VPromoterAvailable SinceOct. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
pZA64
Plasmid#174777PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-L17E/D481V-6xHA, Kmr plant selection marker
UseTags6xHAExpressionPlantMutationL17E/D481VPromoterAvailable SinceOct. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
pZA60
Plasmid#174773PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-L17E/D481V-4xMyc, Kmr plant selection marker
UseTags4xMycExpressionPlantMutationL17E/D481VPromoterAvailable SinceOct. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
pZA22
Plasmid#158500PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V. Km plant selection marker.DepositorInsertZAR1
UseTagsExpressionPlantMutationD481VPromoterAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
pZA45
Plasmid#158523PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
UseTagseGFPExpressionPlantMutationPromoterAvailable SinceSept. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
pZA37
Plasmid#158515PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
UseTags6xHAExpressionPlantMutationPromoterAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
pZA21
Plasmid#158499PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1. Km plant selection marker.DepositorInsertZAR1
UseTagsExpressionPlantMutationPromoterAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
pZA29
Plasmid#158507PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1-4xMyc. Km plant selection marker.DepositorInsertZAR1-4xMyc
UseTags4xMycExpressionPlantMutationPromoterAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
pZA23
Plasmid#158501PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 L17E. Km plant selection marker.DepositorInsertZAR1
UseTagsExpressionPlantMutationL17EPromoterAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
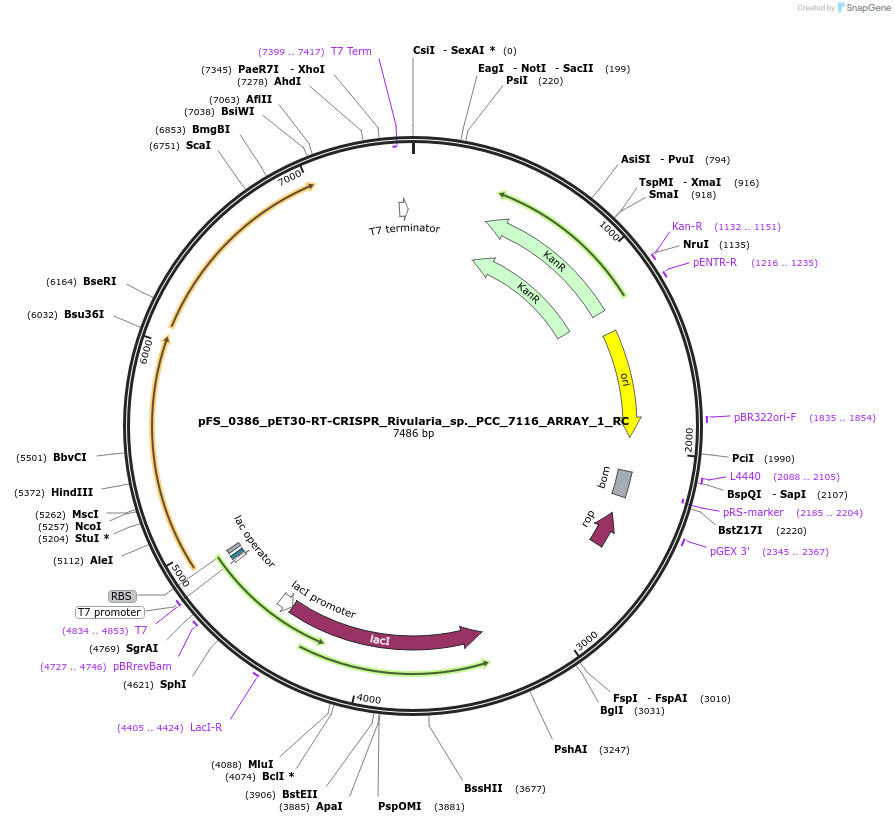
pFS_0386_pET30-RT-CRISPR_Rivularia_sp._PCC_7116_ARRAY_1_RC
Plasmid#116998PurposeExpresses RPCC7116RT-Cas1-Cas2 under pT7lac promoter, encodes RPCC7116CRISPR array 1 RC, compatible with SENECA acquisition readoutDepositorInsertRivularia sp. PCC 7116 RT-Cas1-Cas2
UseTagsExpressionBacterialMutationPromoterT7Available SinceNov. 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
ninaE-stops m3-myc
Plasmid#26855DepositorInsertSlow Termination of Phototransduction (stops Fly)
UseTagsmycExpressionInsectMutationAmino acids 296-299 (SLRH in wild-type) are mutat…PromoterAvailable SinceApril 5, 2011AvailabilityAcademic Institutions and Nonprofits only -
pmTurquoise2-Golgi
Plasmid#36205PurposeIn vivo visualization of golgi apparatus (can be used for colocalization studies)DepositorInsertGolgi targeting sequence (B4GALT1 Human)
UseTagsmTurquoise2ExpressionMammalianMutationPromoterCMVAvailable SinceAug. 15, 2012AvailabilityAcademic Institutions and Nonprofits only -
pCAGGS-hPhyBY276H-mCherry-HRasCT
Plasmid#100540PurposeExpresses Phytochrome B-Y276H (1-908 aa) fused with mCherry and Hras C-terminus in mammalian cellsDepositorInsertPhytochrome B (1-908 aa, Y276H mutation) (PHYB Mustard Weed)
UseTagsmCherry fused with HRas C-terminus for plasma mem…ExpressionMammalianMutationHuman codon optimized PhyB (1-908 aa), Y276H muta…PromoterCAG promoterAvailable SinceOct. 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
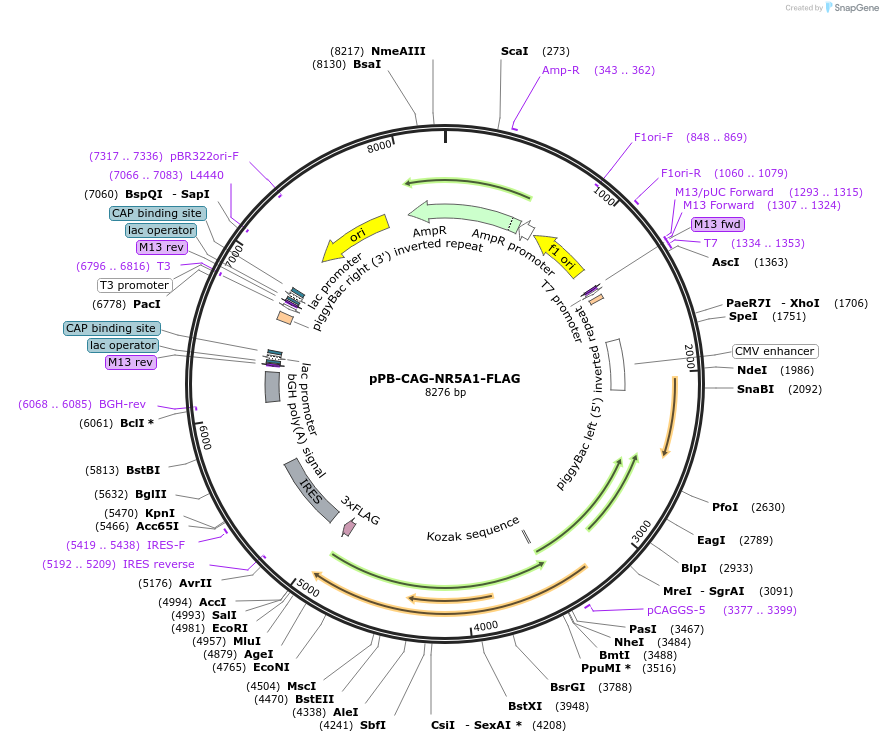
pPB-CAG-NR5A1-FLAG
Plasmid#242227PurposeExpression of full-length human SF-1 using piggyBac transpositionDepositorAvailable SinceSept. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
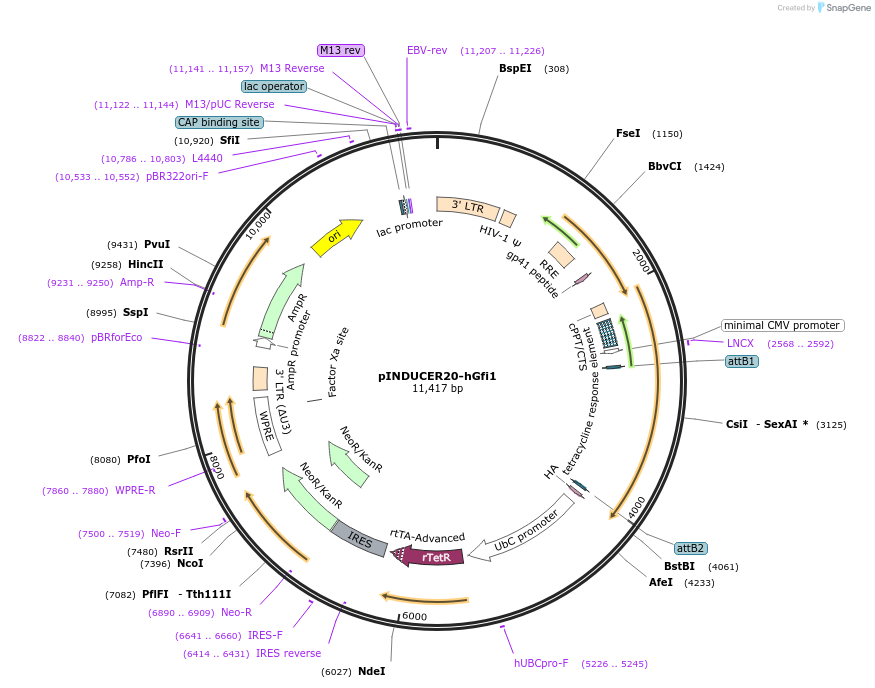
pINDUCER20-hGfi1
Plasmid#121200Purposeinducible expression of hGfi1 in colorectal cancer cellsDepositorInsertGrowth factor independent 1 transcriptional (GFI1 Human)
UseLentiviralTagsExpressionMammalianMutationP2LPromoterAvailable SinceJuly 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
pMSCV-FKBP-GMIP(trunc C1)
Plasmid#221279PurposeExpression of GMIP C-terminal truncation 1 (1-64) in mammalian cells by retroviral transductionDepositorInsertGMIP C-terminal truncation 1 (1-64) (Gmip Mouse)
UseRetroviralTags3xFLAG-4xFKBPExpressionMutationPromoterAvailable SinceJuly 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
pVW296
Plasmid#139496PurposeExpression vector for the PP7-mCherry protein in S. cerevisiaeDepositorInsertpADH1 PP7∆FG-mCherry tCYC1
UseTagsExpressionYeastMutation∆FGPromoterpADH1Available SinceJune 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
pKL06
Plasmid#104430Purposesuper Ecliptic pHluorin with C terminal mRuby2 fusion tag in yeast expression vectorDepositorInsertSuper Ecliptic pHluorin-mRuby2
UseTagsExpressionYeastMutationPromoterTef1Available SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSEVA451-Cpf1
Plasmid#192362PurposeCRISPR-based vector for genome editingDepositorInsertCas12a, gRNA scaffold
UseCRISPRTagsExpressionMutationPromoterAvailable SinceSept. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
pSEVA351-Cpf1
Plasmid#192361PurposeCRISPR-based vector for genome editingDepositorInsertCas12a, gRNA scaffold
UseCRISPRTagsExpressionMutationPromoterAvailable SinceSept. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
pRS42H_PTEF1_dCas9_TCYC1
Plasmid#177155PurposeExpresses dCas9 in yeast cellsDepositorInsertNuclease-null Cas9
UseTagsSV40 NLSExpressionYeastMutationPromoterTEF1 promoterAvailable SinceFeb. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
pCS2+ YFP-Smo
Plasmid#41086DepositorInsertsmoothened homolog (Smo Mouse)
UseTagsYFPExpressionMammalianMutationYFP inserted after Smo signal sequencePromoterAvailable SinceNov. 16, 2012AvailabilityAcademic Institutions and Nonprofits only -
pCL66-decoder
Plasmid#159453PurposeInsert: Pfus1mut-tetR-nls-malE-tCyc1. Same insert as pCL33-decoder in a multiple integration shuttle vector. Backbone: pRG235 (addgene). Marker: Leu2.DepositorInsertPfus1mut-tetR-nls-malE-tCyc1
UseSynthetic BiologyTagsExpressionYeastMutationPromoterPfus1mutAvailable SinceDec. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPB-R1R2-EM7neoPheS
Plasmid#112923PurposepiggyBac Transposon vector with a Gateway destination cassette (neoPheS selection)DepositorInsertneoPheS
UsePiggybac transposon gateway destination vectorTagsExpressionMammalianMutationPromoterAvailable SinceSept. 4, 2019AvailabilityAcademic Institutions and Nonprofits only