CRISPR Plasmids: RNA Targeting
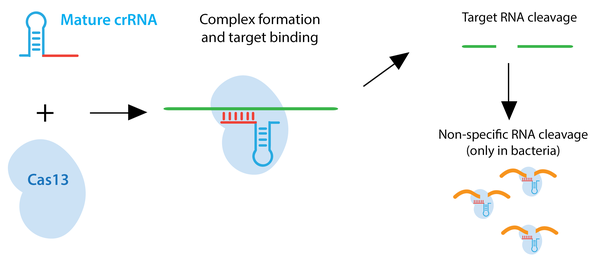
Type VI CRISPR systems, including the enzymes Cas13a/C2c2 and Cas13b, target RNA rather than DNA. In bacteria, once they have recognized and cleaved the target RNA sequence, they adopt an enzymatically active state and can bind and cleave additional RNAs regardless of homology to the crRNA. This activity provides a stark contrast to Cas9 and Cpf1, which require that each DNA target have high sequence identity to the spacer sequence and contain a PAM sequence just downstream of the sequence to be cleaved. This non-specific cleavage is thought to activate programmed cell death or dormancy for phage-infected bacterial cells so as to limit the spread of infection throughout the entire population.
Type VI enzymes that function in mammalian cells can be used to attentuate RNA levels. In mammalian systems, Cas13a does not exhibit the collateral RNA degradation seen in bacteria.
Want more information on the wide variety of Cas enzymes? CasPEDIA is an encyclopedia of CRISPR systems with wiki entries describing enzyme activity, experimental considerations, and more, created and maintained by the Doudna Lab.
Browse, sort, or search the tables below for CRISPR RNA targeting plasmids.Plasmids are available for expression in mammalian systems, bacteria, and plants.
Mammalian
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Bacterial
| ID | Plasmid | Gene/Insert | PI | Publication |
|---|
Plant
| ID | Plasmid | Gene/Insert | PI | Publication |
|---|

Do you have suggestions for other plasmids that should be added to this list?
Fill out our Suggest a Plasmid form or e-mail [email protected] to help us improve this resource!
