8,173 results
-
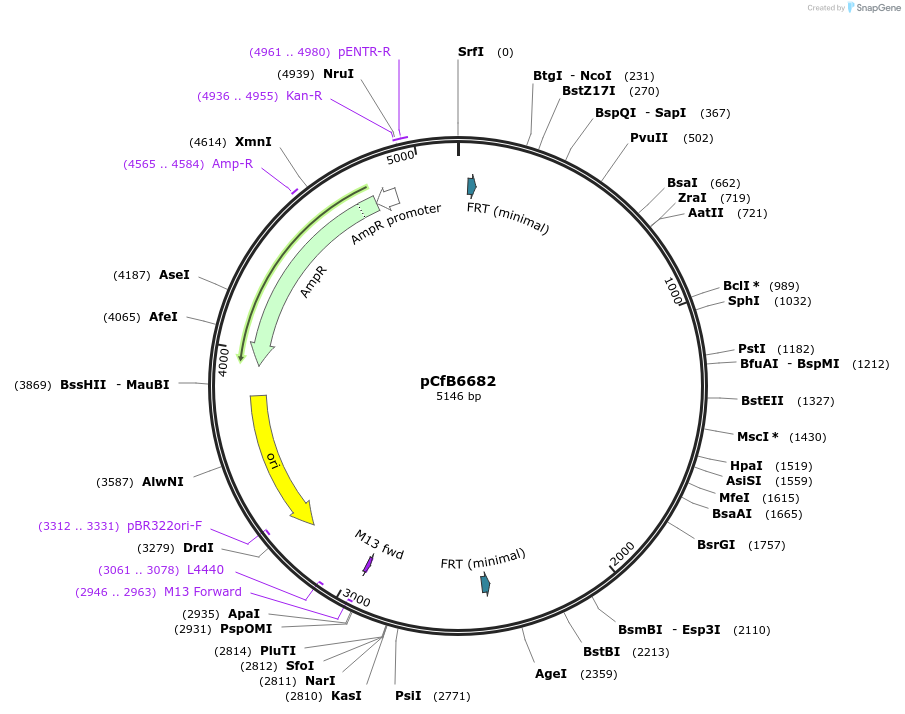
Plasmid#106152PurposeEasyCloneYALI system-based yeast integrative vector for markerfree integration via CRISPR/Cas9 to be used in combination with pCfB6627, integration into Yarrowia lipolytica chromosomal location IntC_2, USER site for cloning, amp resistanceDepositorTypeEmpty backboneExpressionYeastAvailable SinceMay 11, 2018AvailabilityAcademic Institutions and Nonprofits only
-
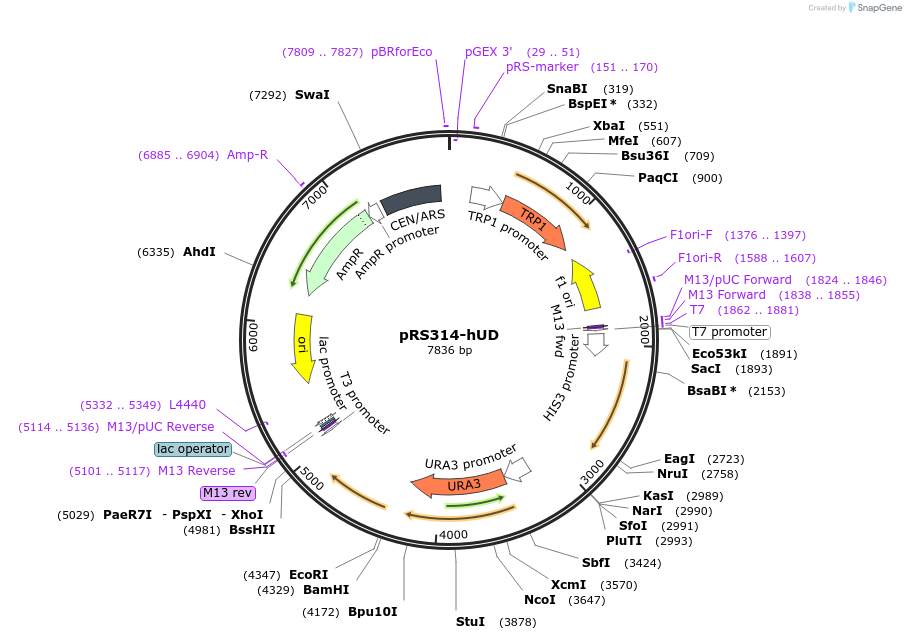
pRS314-hUD
Plasmid#11005DepositorInserthUD recombination system
ExpressionBacterial and YeastMutationDirect repeat recombination system. Recombination…Available SinceApril 28, 2006AvailabilityAcademic Institutions and Nonprofits only -
-
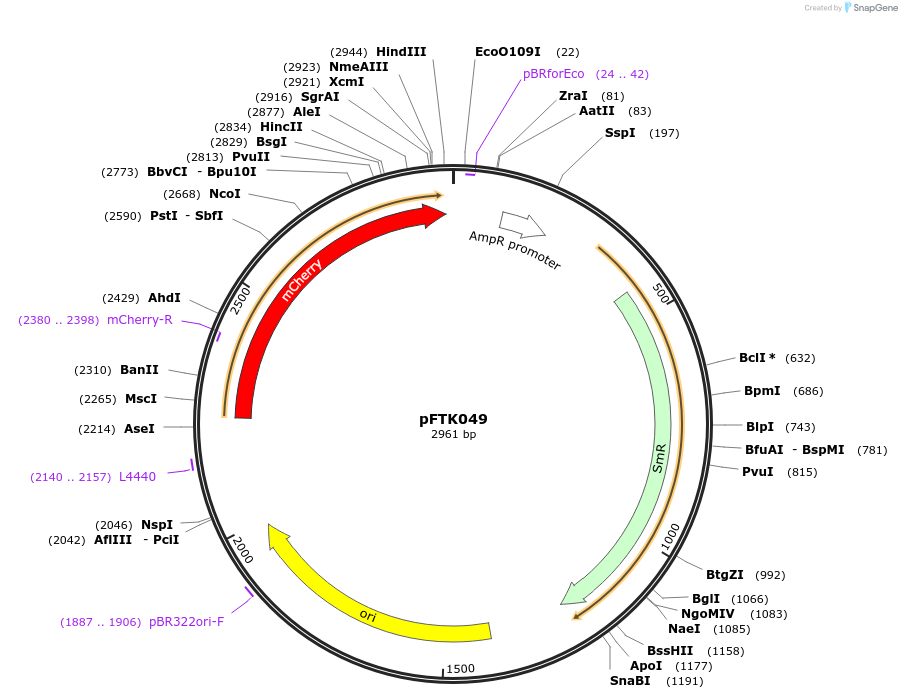
pFTK049
Plasmid#171321PurposePart Plasmid Entry Vector (LVL0) from the Fungal Modular Cloning ToolKit.DepositorInsertmCherry fluorescent reporter
UseSynthetic BiologyExpressionBacterial and YeastMutationn/aAvailable SinceDec. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
LHP336 - HA-Ub-K48R
Plasmid#32164DepositorTagsHAExpressionYeastMutationK48RPromoterCUP1Available SinceSept. 27, 2011AvailabilityAcademic Institutions and Nonprofits only -
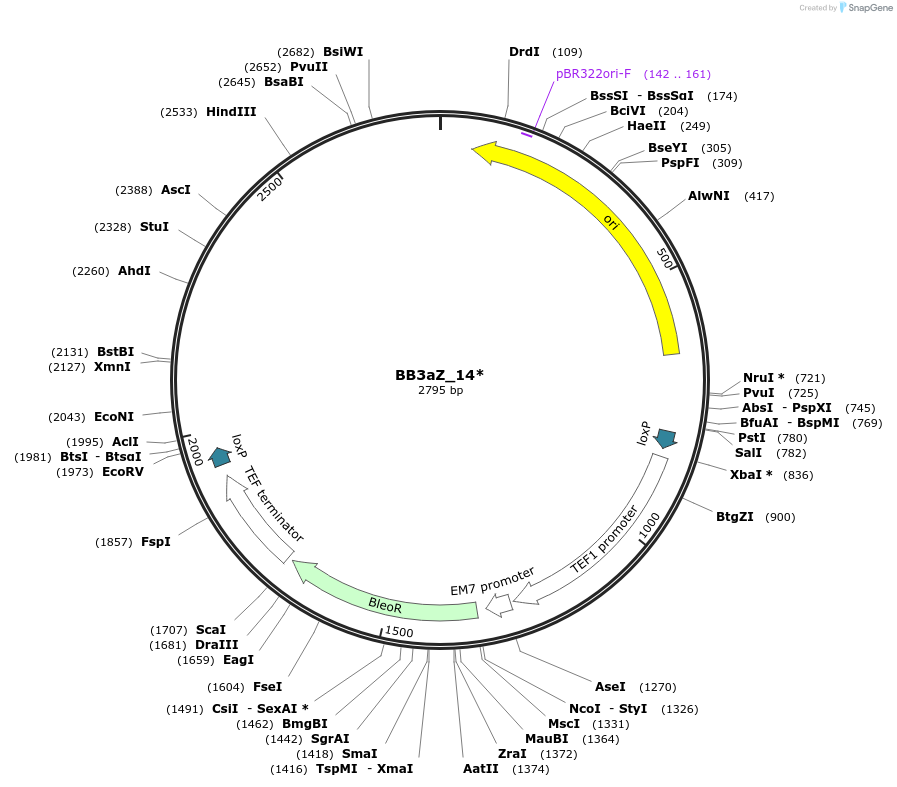
BB3aZ_14*
Plasmid#98542Purposeempty BB3 for insertion and overexpression of 1 gene in P. pastorisDepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
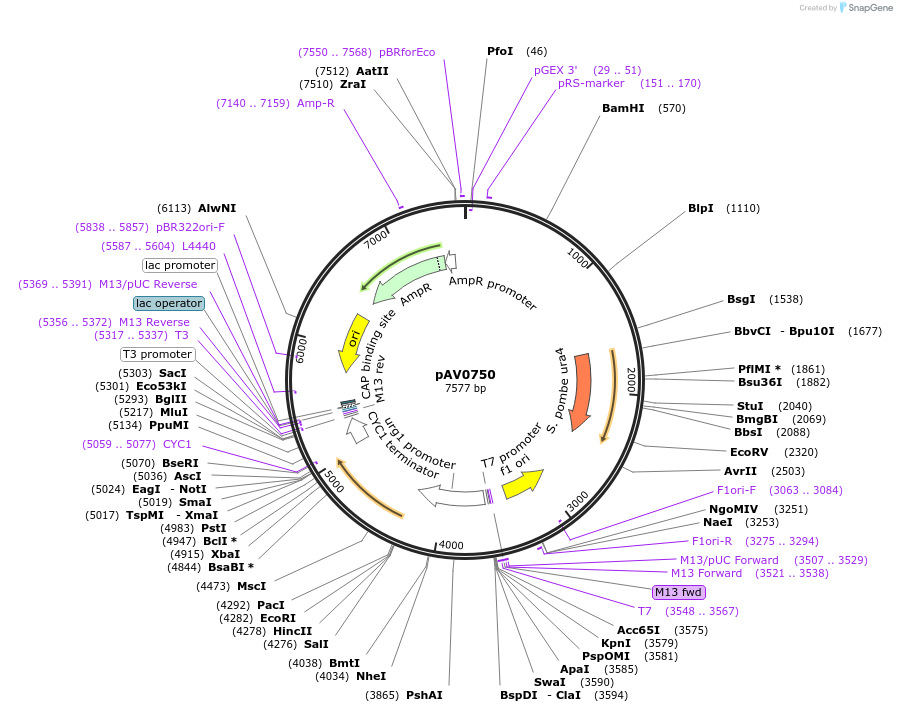
pAV0750
Plasmid#133484PurposeExpression in fission yeast from promoter purg1 , integrated at ura4 locusDepositorInsertpUra4(AfeI)-p(urg1)-sfGFP-terminator(ScCYC1)
ExpressionYeastAvailable SinceNov. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
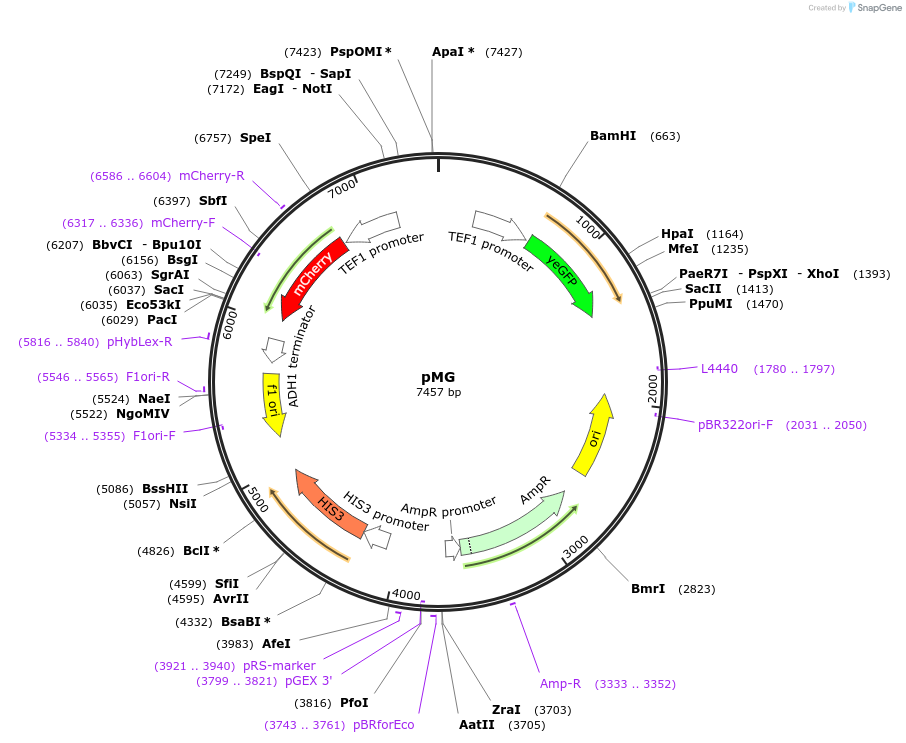
pMG
Plasmid#138264PurposeYeast intergration vector for integration of dual reporter system at the URA3 locus. Dual reporter system contains constitutive mCherry and constitutive eGFP flanked by iCas9 sites.DepositorInsertseGFP flanked by iCas9 sites
mCherry
UseCRISPRExpressionYeastPromoterTef1Available SinceMarch 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
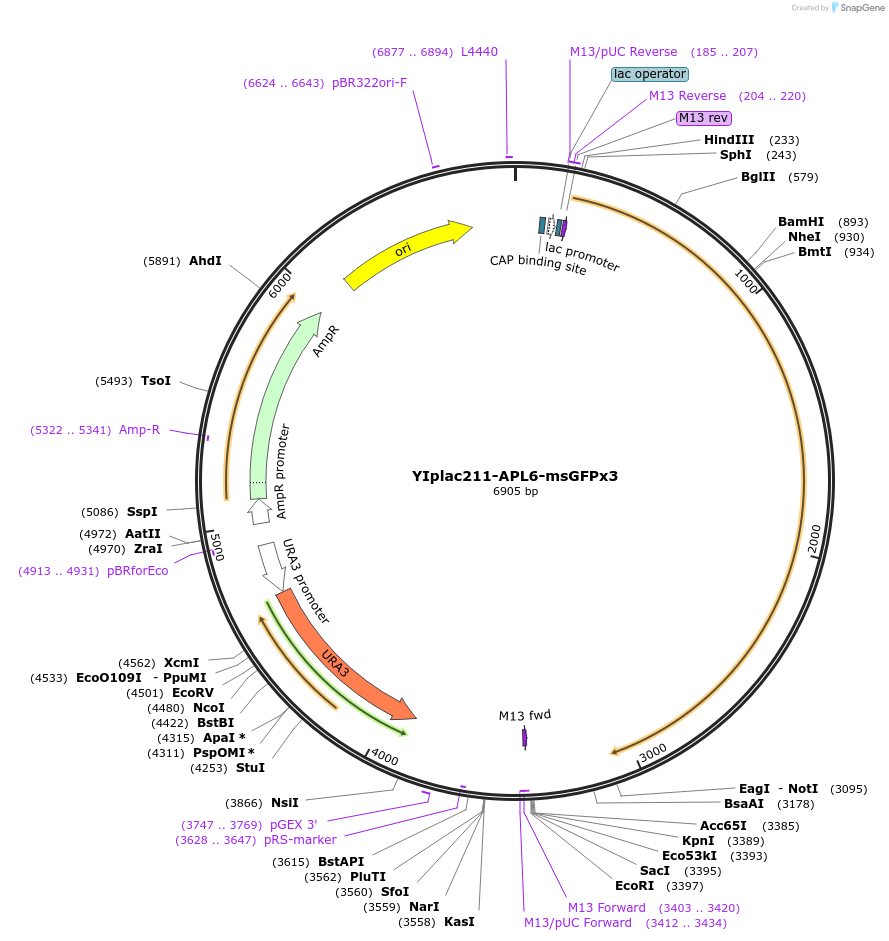
YIplac211-APL6-msGFPx3
Plasmid#105254PurposePlasmid for pop-in/pop-out replacement of Apl6 with Apl6-msGFPx3.DepositorAvailable SinceJan. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
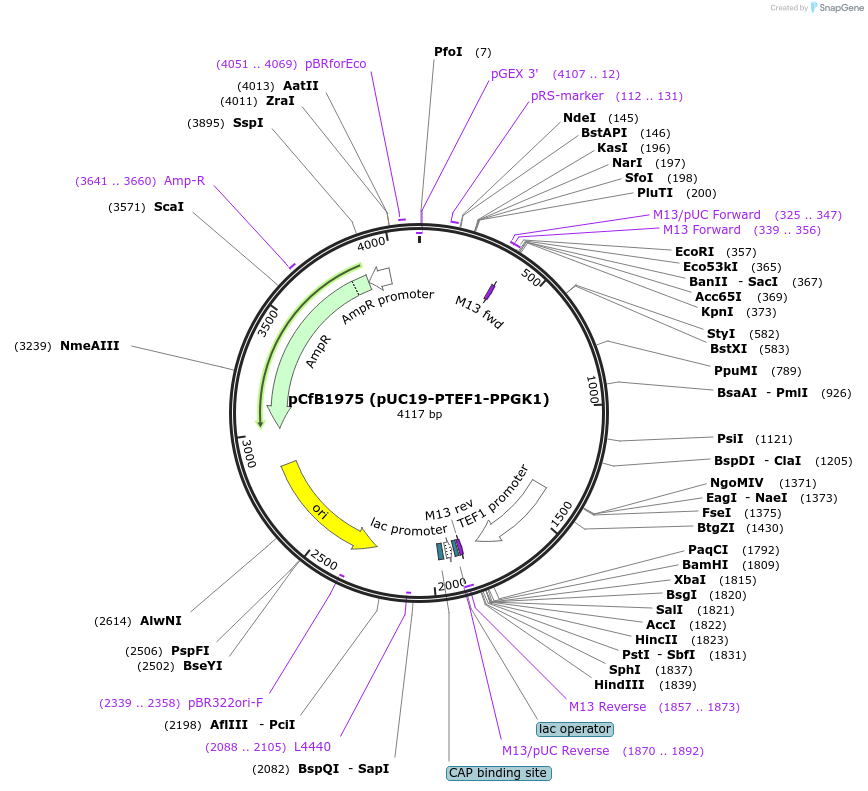
pCfB1975 (pUC19-PTEF1-PPGK1)
Plasmid#73296PurposeExample plasmid containing a bidirectional promoterDepositorInsertpTEF1-pPGK1
UseSynthetic BiologyExpressionBacterial and YeastAvailable SinceNov. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
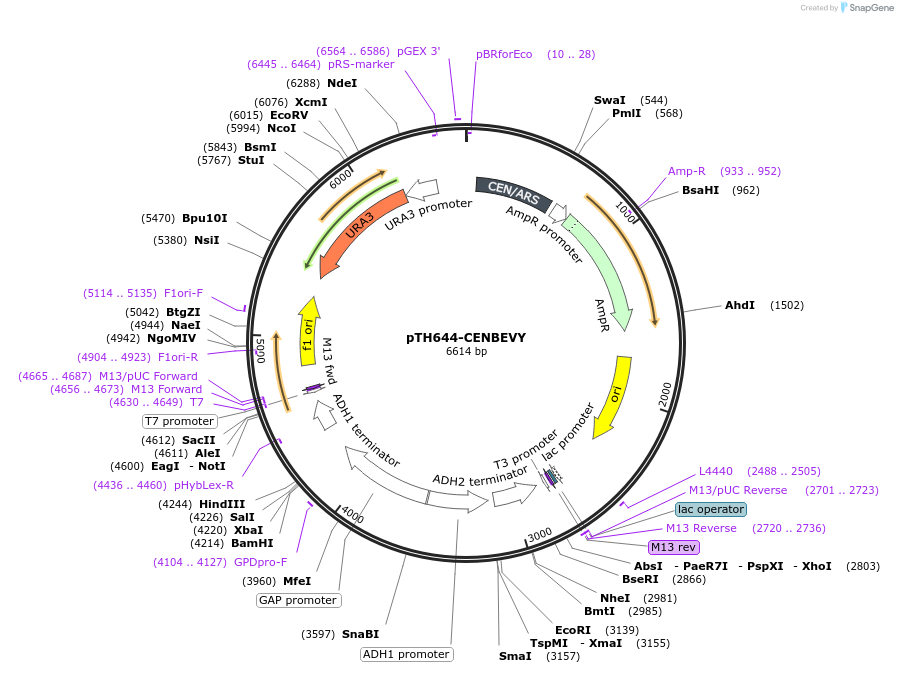
pTH644-CENBEVY
Plasmid#29695DepositorInsertbidirectional promoter cassette from pBEVY-U
ExpressionYeastAvailable SinceJan. 26, 2012AvailabilityAcademic Institutions and Nonprofits only -
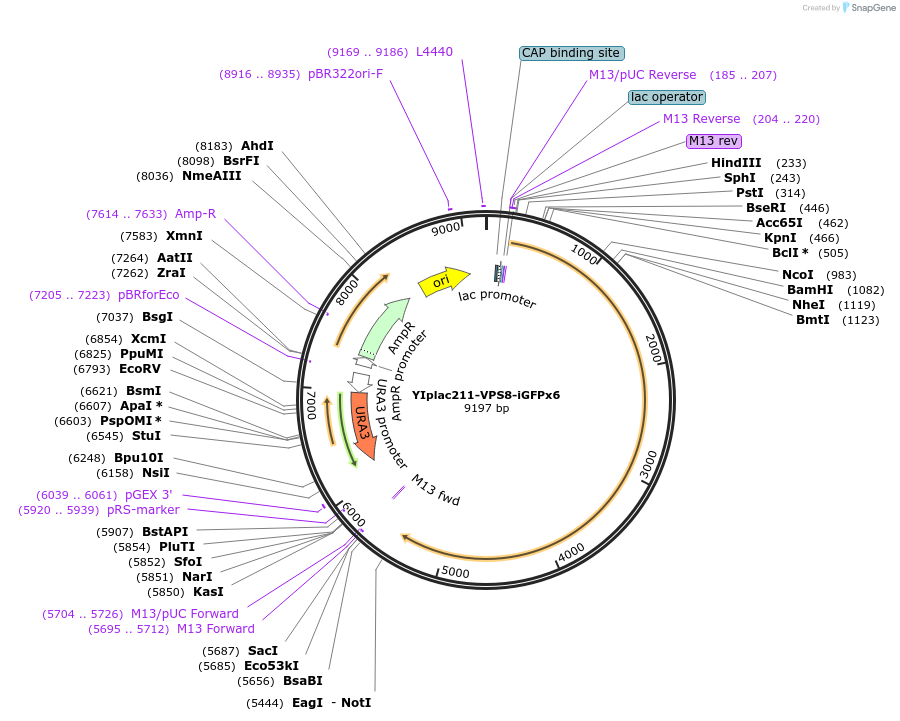
YIplac211-VPS8-iGFPx6
Plasmid#105271PurposePlasmid for pop-in/pop-out replacement of Vps8 with Vps8-iGFPx6.DepositorAvailable SinceJan. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
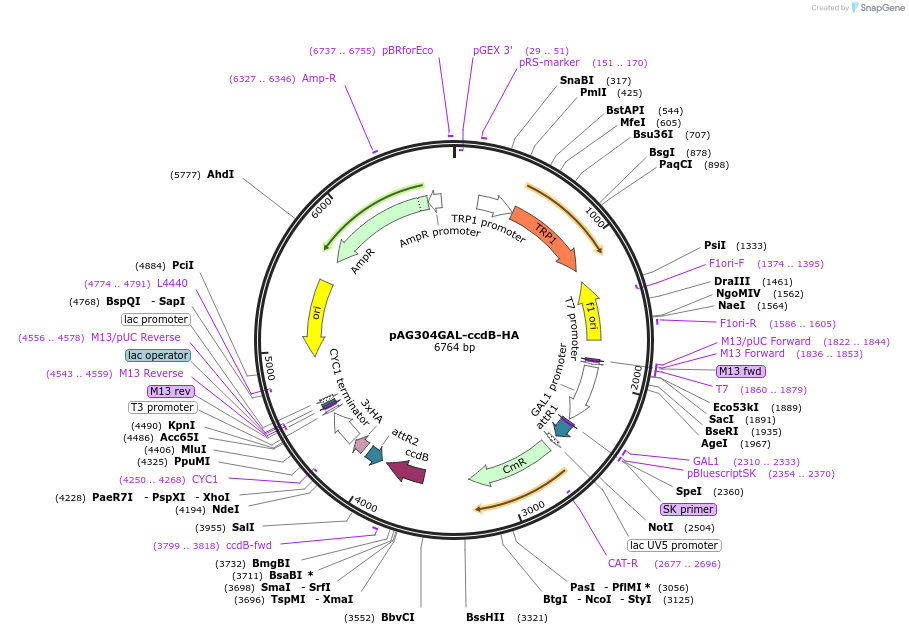
pAG304GAL-ccdB-HA
Plasmid#14231DepositorTypeEmpty backboneUseGateway destinationTags3HAExpressionYeastAvailable SinceMarch 29, 2007AvailabilityAcademic Institutions and Nonprofits only -
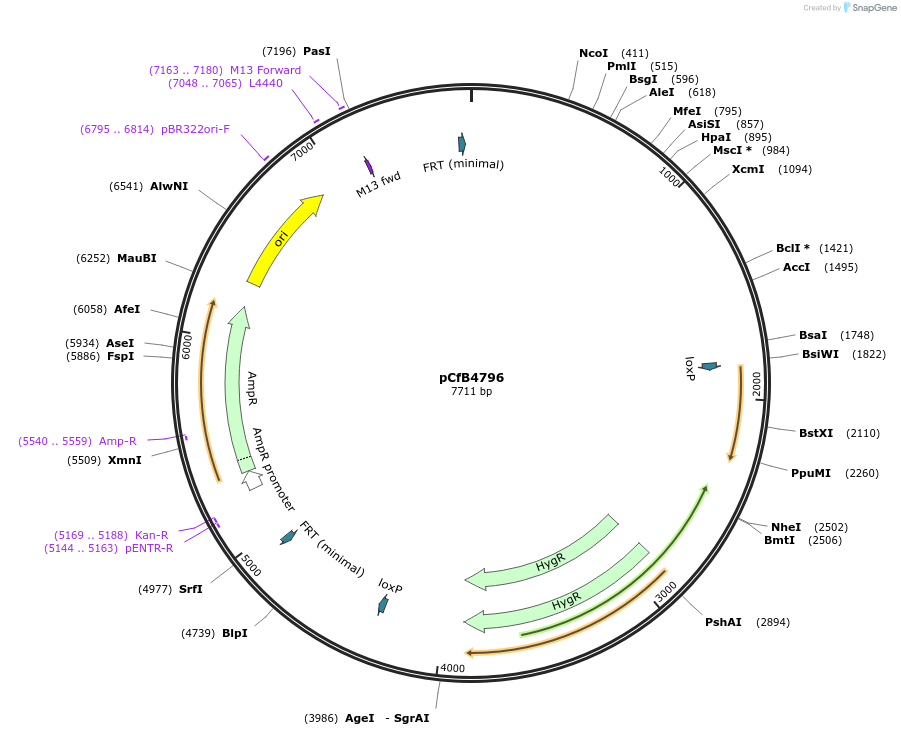
pCfB4796
Plasmid#106131PurposeEasyCloneYALI system-based yeast integrative vector carrying loxP-flanked hygromycin-resistance marker, integration into Yarrowia lipolytica chromosomal location IntD_1, USER site for cloning, amp resistanceDepositorTypeEmpty backboneExpressionYeastAvailable SinceApril 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
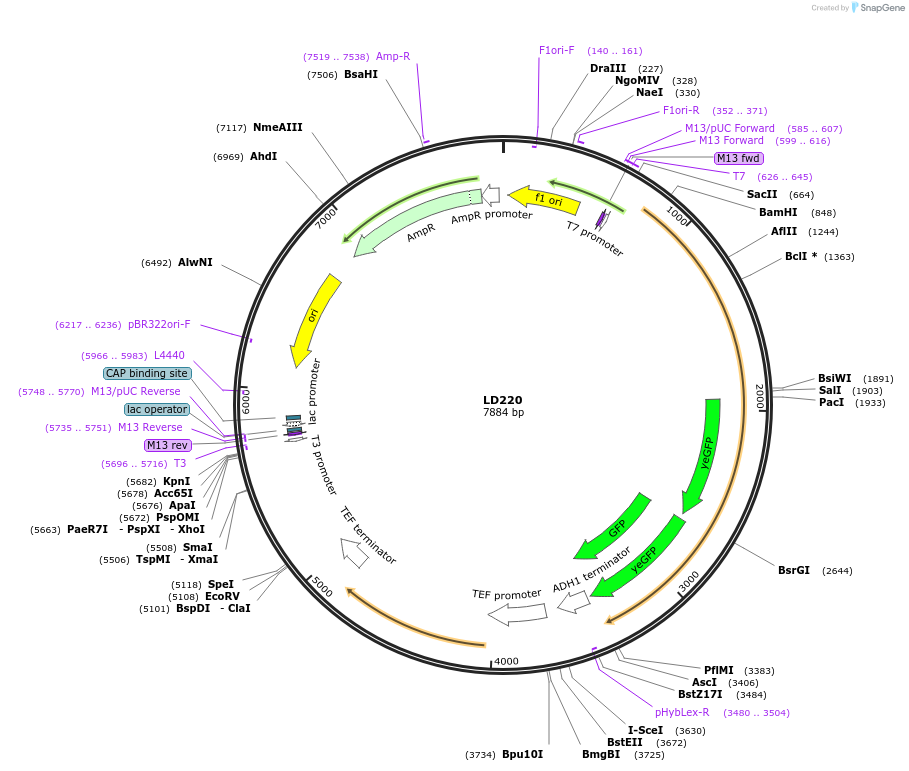
LD220
Plasmid#69188PurposeIntegration of 2GFP tag at Atg20 C terminus, use auxotrophic marker URA3(Candida albicans)DepositorInsertAtg20-2EGFP
Tags2xEGFPExpressionYeastAvailable SinceOct. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
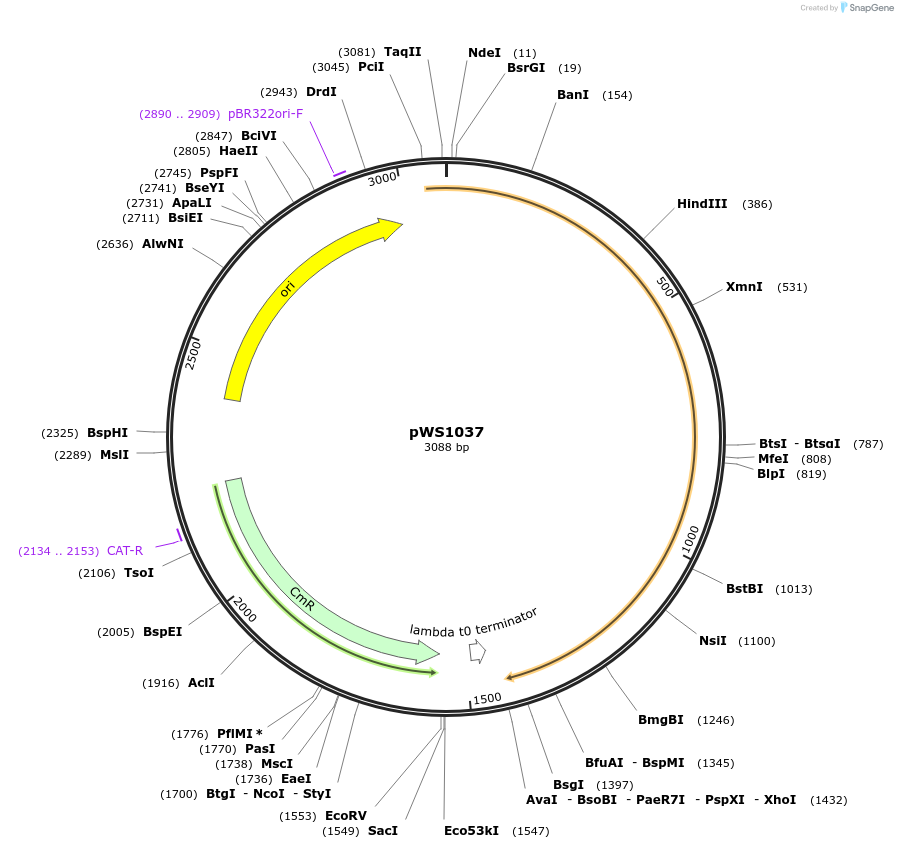
pWS1037
Plasmid#123049PurposeG_ subunitDepositorInsertGpa1 [3-4a]
ExpressionYeastAvailable SinceApril 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
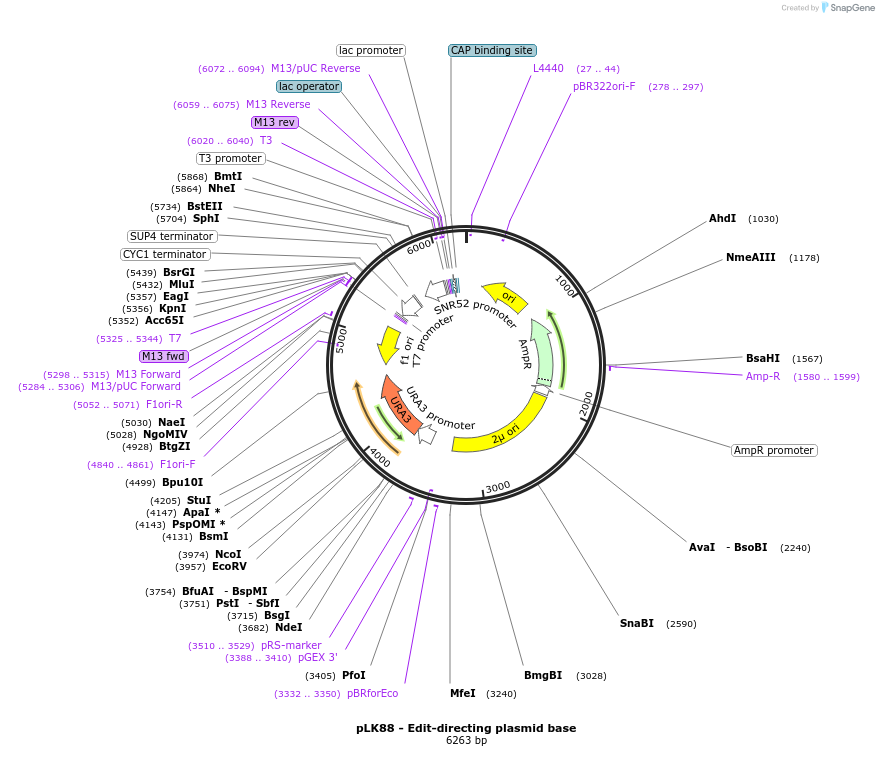
pLK88 - Edit-directing plasmid base
Plasmid#98814PurposeEdit-directing plasmid baseDepositorInsertCAN1.y gRNA
UseCRISPRExpressionYeastAvailable SinceJuly 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
-
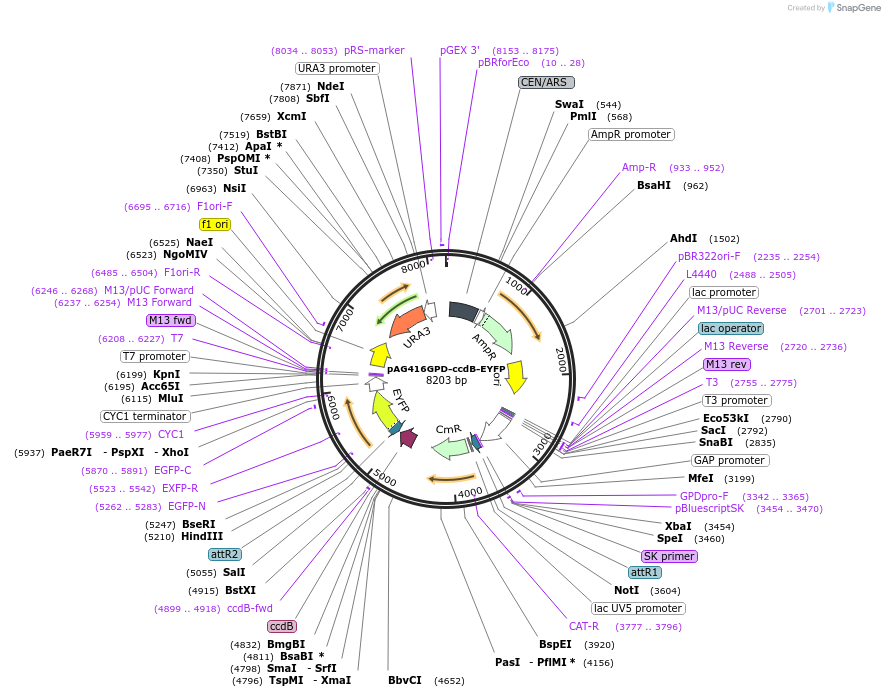
pAG416GPD-ccdB-EYFP
Plasmid#14220DepositorTypeEmpty backboneUseGateway destinationTagsEYFPExpressionYeastAvailable SinceMarch 16, 2007AvailabilityAcademic Institutions and Nonprofits only -
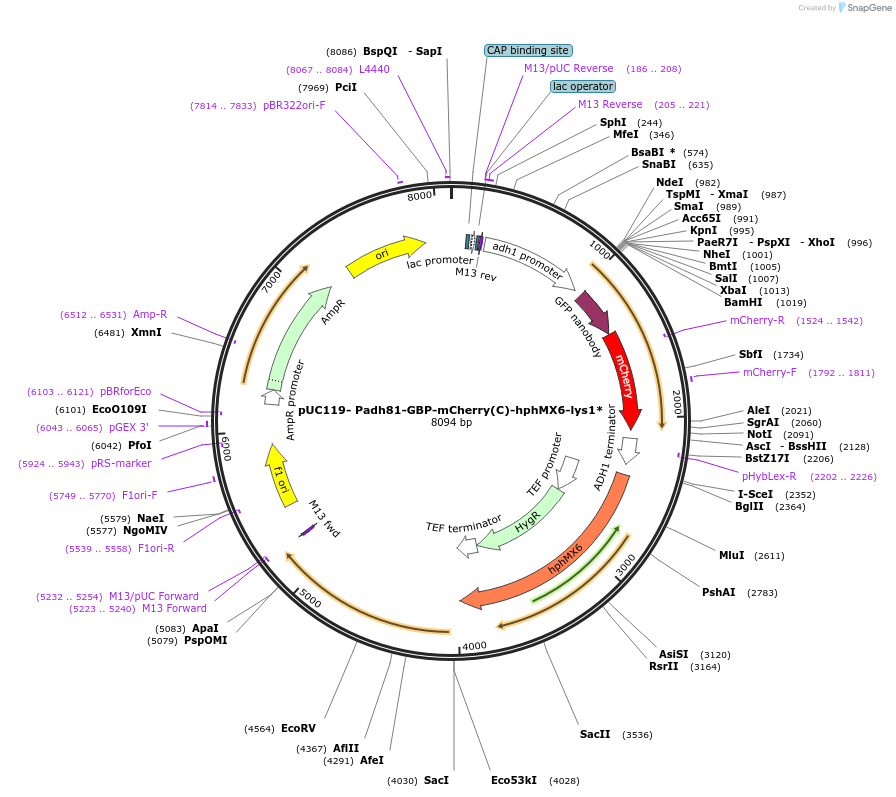
pUC119- Padh81-GBP-mCherry(C)-hphMX6-lys1*
Plasmid#89071Purposegenomic GBP-mCherry epitope C-terminal tagging at lys1+ locus in S. pombeDepositorTypeEmpty backboneUseGenomic integration in s. pombeTagsGBP-mCherryExpressionYeastAvailable SinceJune 14, 2017AvailabilityAcademic Institutions and Nonprofits only -
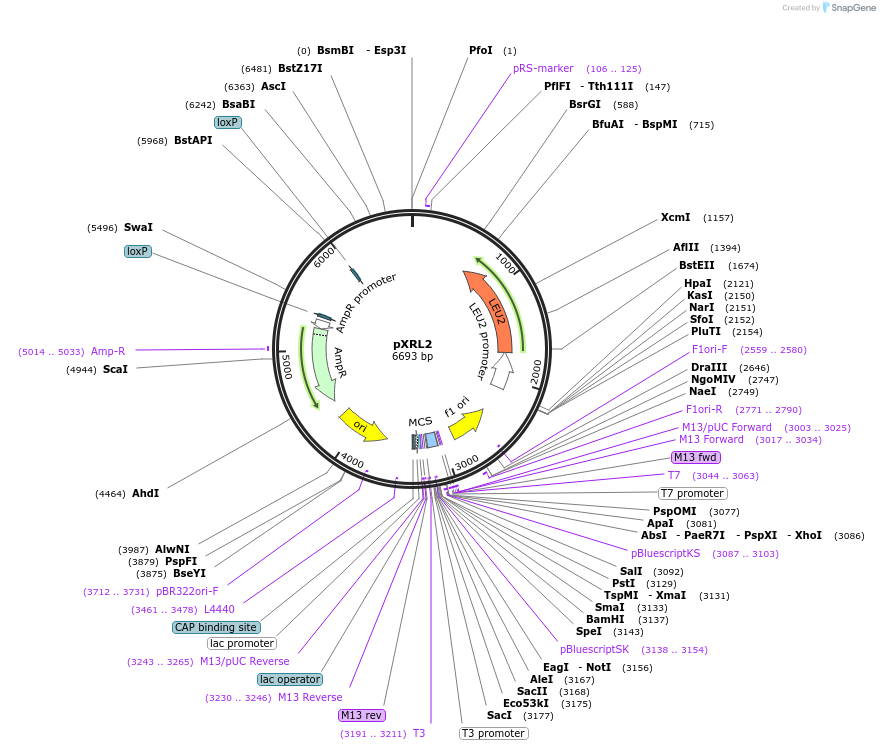
pXRL2
Plasmid#63148PurposeLeu2 marker pXR vector uses for both plasmid assembly in vivo, as well as targeted genomic integration.DepositorTypeEmpty backboneUseCre/LoxExpressionBacterial and YeastAvailable SinceApril 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
pPS808
Plasmid#8856DepositorTypeEmpty backboneTagsGFPExpressionYeastAvailable SinceAug. 3, 2005AvailabilityAcademic Institutions and Nonprofits only -
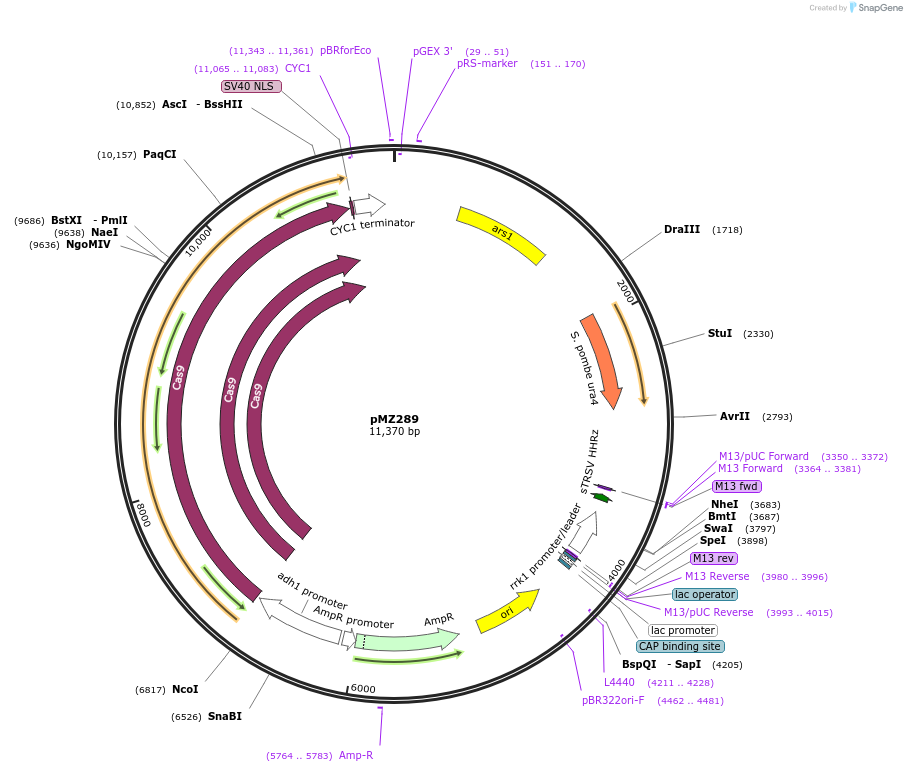
pMZ289
Plasmid#59898PurposeCombination adh1:cas9/rrk1:sgRNA for CRISPR genome editing in fission yeast: sgRNA targeted against ade6-M210DepositorInsertsCas9
rrk1:sgRNA
UseCRISPRExpressionYeastMutationSilent muation of the CspCI siteAvailable SinceOct. 29, 2014AvailabilityAcademic Institutions and Nonprofits only -
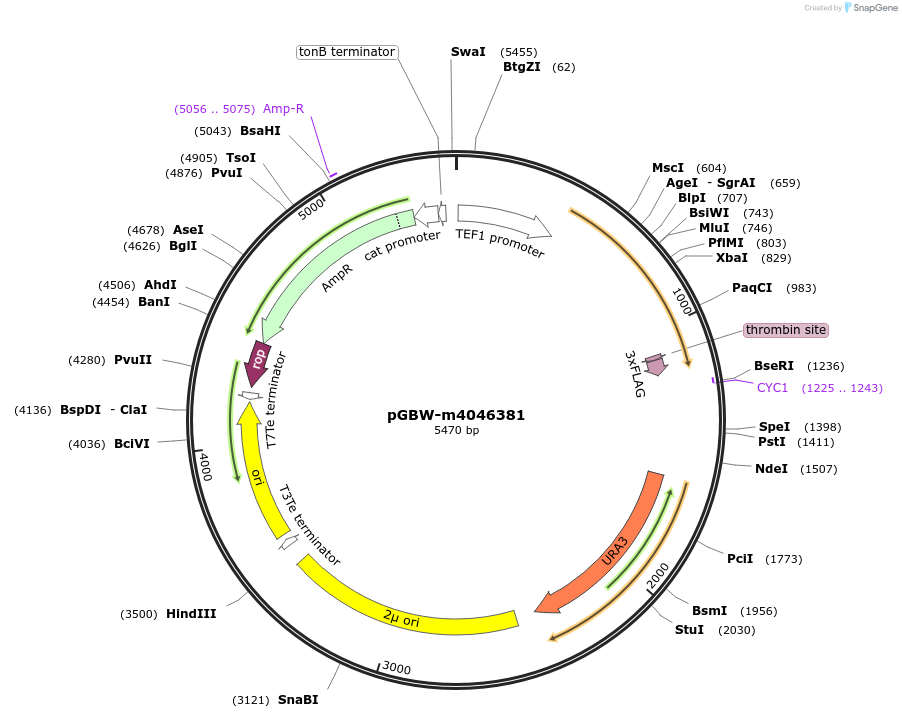
pGBW-m4046381
Plasmid#149010PurposeYeast Expression plasmid for SARS-CoV-2 M (membrane)DepositorInsertSARS-CoV-2 M (membrane) (M Severe acute respiratory syndrome coronavirus 2)
TagsCleavable thrombin;3xFLAGExpressionYeastMutationwtPromoterpTEF1Available SinceMay 14, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
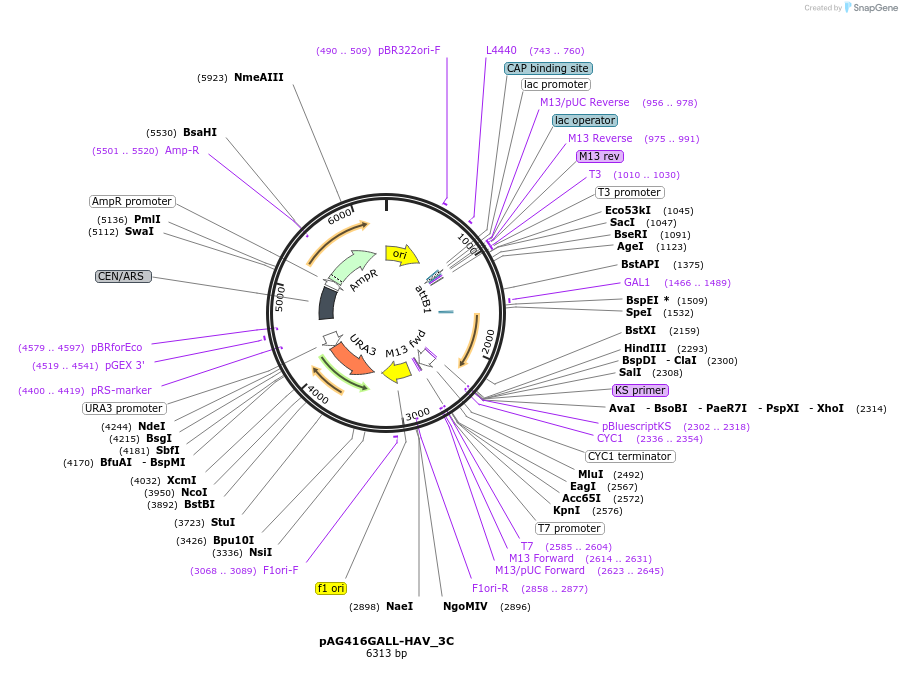
pAG416GALL-HAV_3C
Plasmid#203481PurposeExpresses HAV 3C protease from a GALL promoter with a URA3 markerDepositorInsertHAV 3C protease
ExpressionYeastPromoterGALLAvailable SinceJuly 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
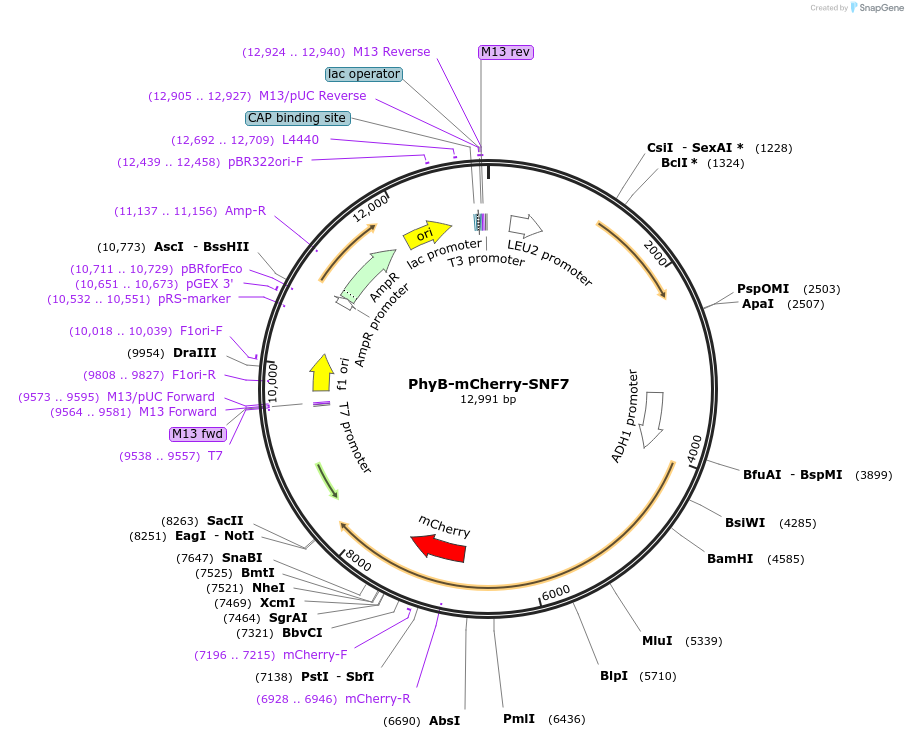
PhyB-mCherry-SNF7
Plasmid#51574PurposeConstitutively express endosome localized PhyB in Saccharomyces cerevisiaeDepositorInsertPhyB (PHYB Mustard Weed)
TagsSNF7 and mCherryExpressionYeastMutation1-908aa onlyPromoterADH1prAvailable SinceAug. 25, 2014AvailabilityAcademic Institutions and Nonprofits only -
KAP95-FLU (113)
Plasmid#24045DepositorAvailable SinceMarch 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
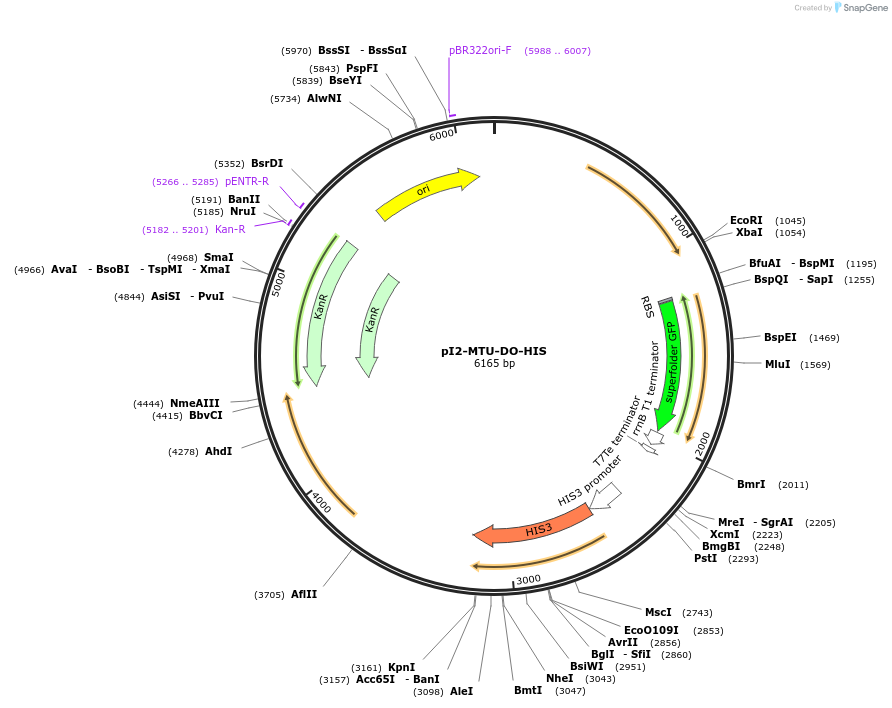
pI2-MTU-DO-HIS
Plasmid#160016PurposeIntegrative vector for K. marxianus, targeting integration site I2 described in (Rajkumar et al.,2019). Histidine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
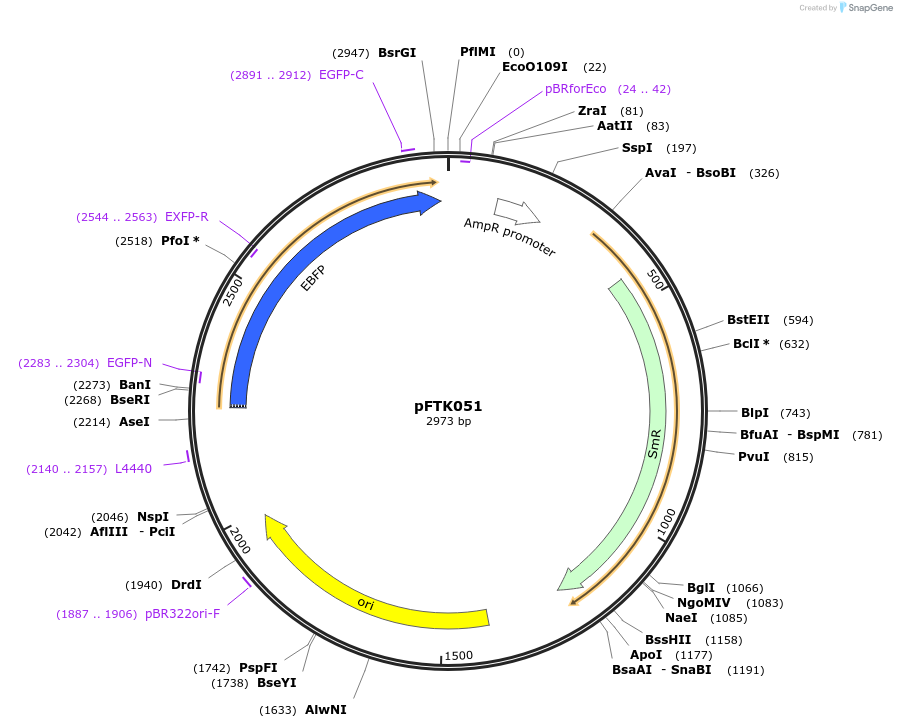
pFTK051
Plasmid#171323PurposePart Plasmid Entry Vector (LVL0) from the Fungal Modular Cloning ToolKit.DepositorInserteBFP fluorescent reporter
UseSynthetic BiologyExpressionBacterial and YeastMutationn/aAvailable SinceJan. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
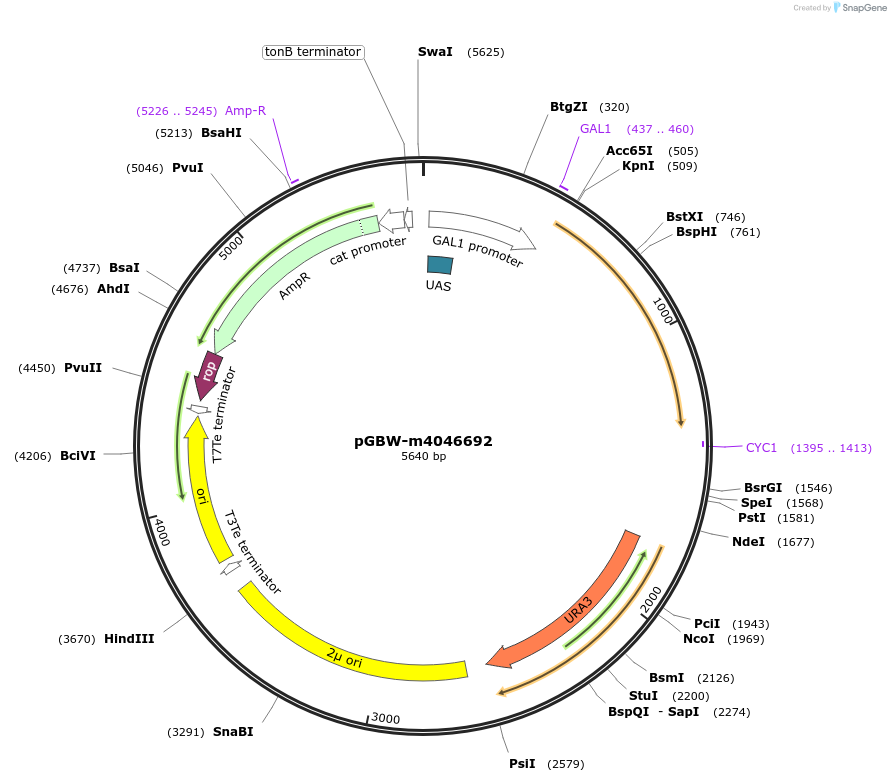
pGBW-m4046692
Plasmid#149195PurposeYeast Expression plasmid for SARS-CoV-2 nsp6DepositorInsertSARS-CoV-2 nsp6 (ORF1ab Severe acute respiratory syndrome coronavirus 2, Budding Yeast)
ExpressionYeastMutationSaccharomyces cerevisiae recode 1PromoterpGAL1Available SinceMay 14, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
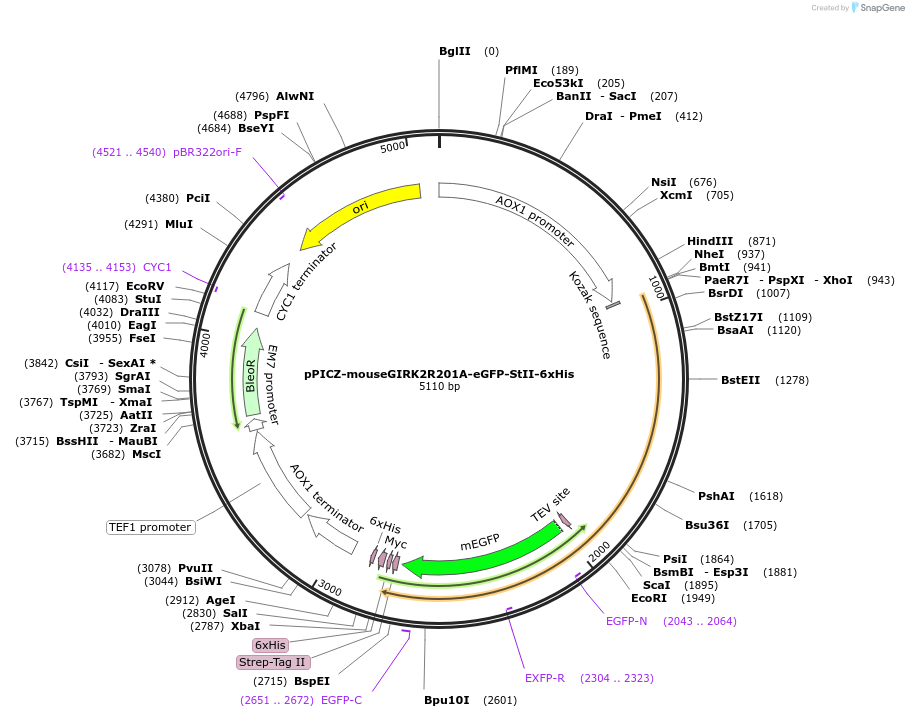
pPICZ-mouseGIRK2R201A-eGFP-StII-6xHis
Plasmid#124278PurposeExpresses mouse GIRK2 R201A as a TEV protease cleavable C-terminal fusion to eGFP, strep tag II, and 6x His tagDepositorAvailable SinceApril 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
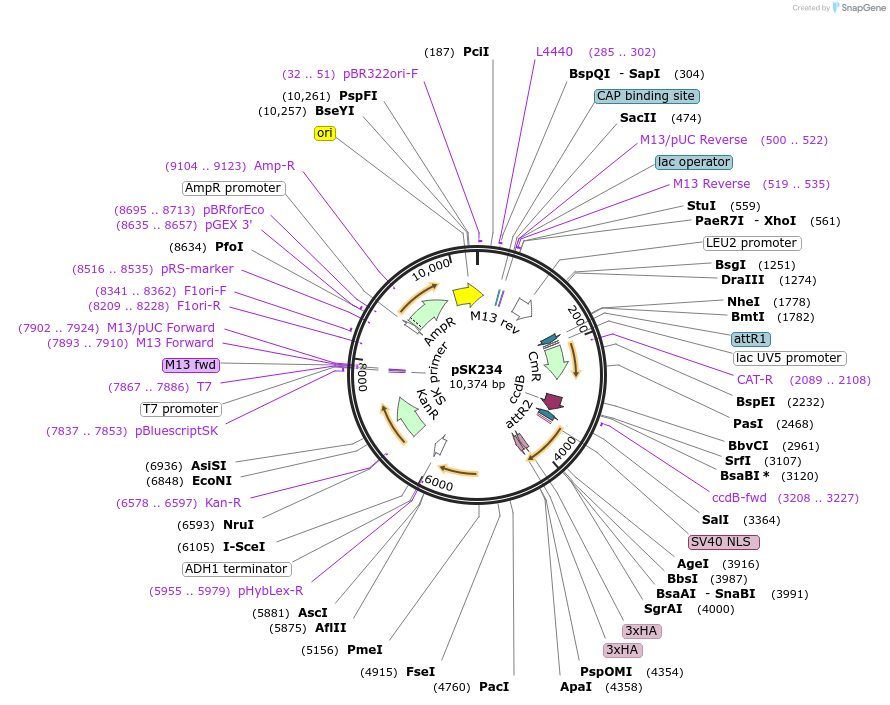
pSK234
Plasmid#131149Purposedual yTRAP, Gateway destination vector of aTF2 fusion reporting on mKate2DepositorTypeEmpty backboneUseGateway destination vectorExpressionYeastAvailable SinceOct. 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
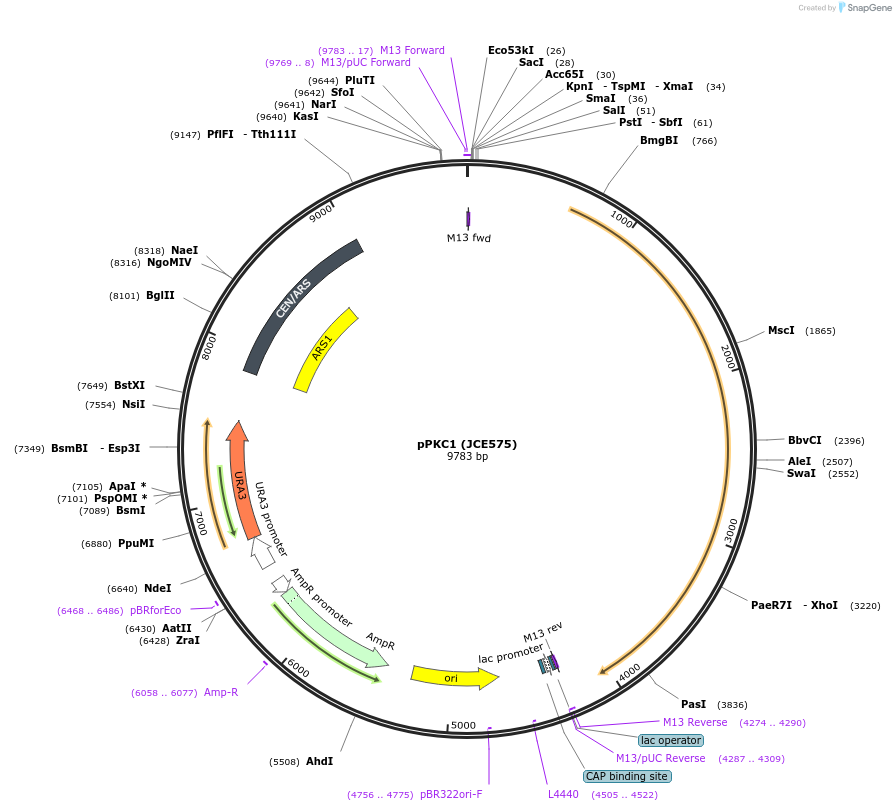
pPKC1 (JCE575)
Plasmid#89773Purposeyeast expression of PKC1DepositorAvailable SinceApril 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
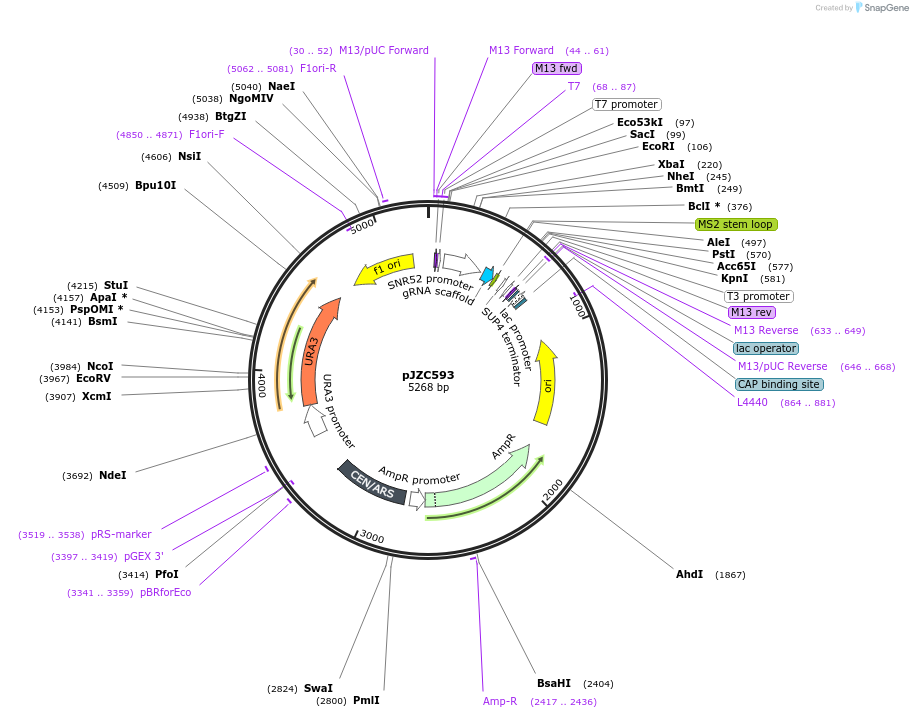
pJZC593
Plasmid#62319PurposesgRNA with MS2-PP7 for yeast cellsDepositorInsertsgRNA + MS2-PP7 RNA binding module
ExpressionYeastPromoterSNR52Available SinceMarch 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
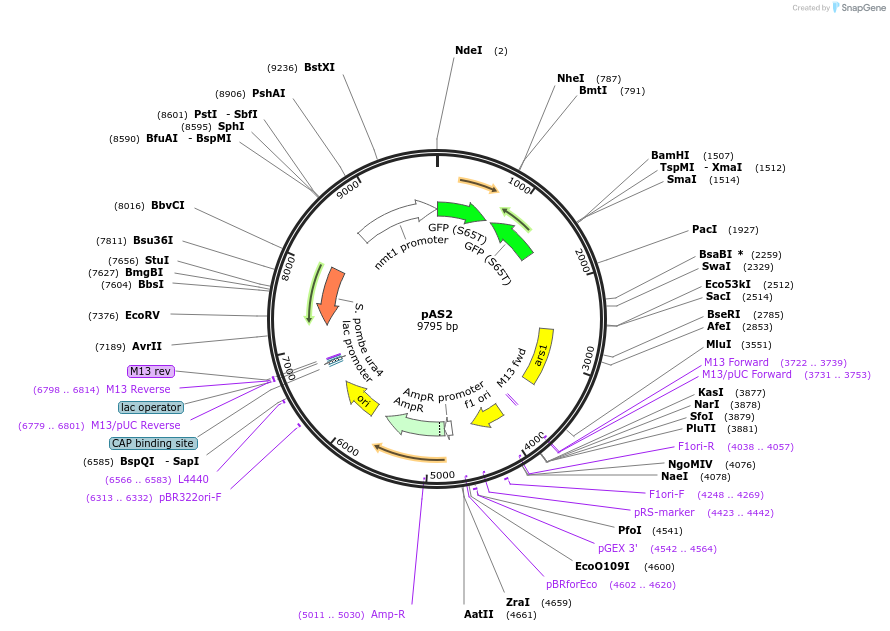
pAS2
Plasmid#12389DepositorInsertgreen fluorescent protein
ExpressionYeastAvailable SinceAug. 24, 2006AvailabilityAcademic Institutions and Nonprofits only -
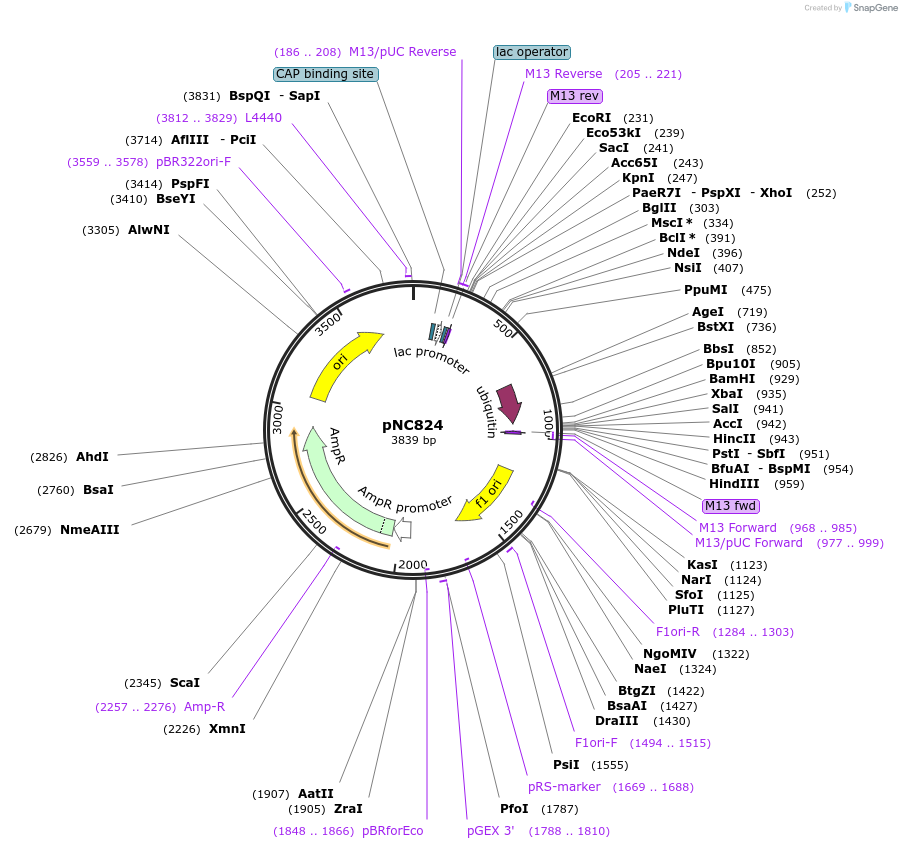
pNC824
Plasmid#20384DepositorInsertUAS cassette (FUS1)
ExpressionYeastMutationX=UbiY (see schematic)Available SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
GAL 103Q+ProCFPp303
Plasmid#15575DepositorAvailable SinceAug. 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
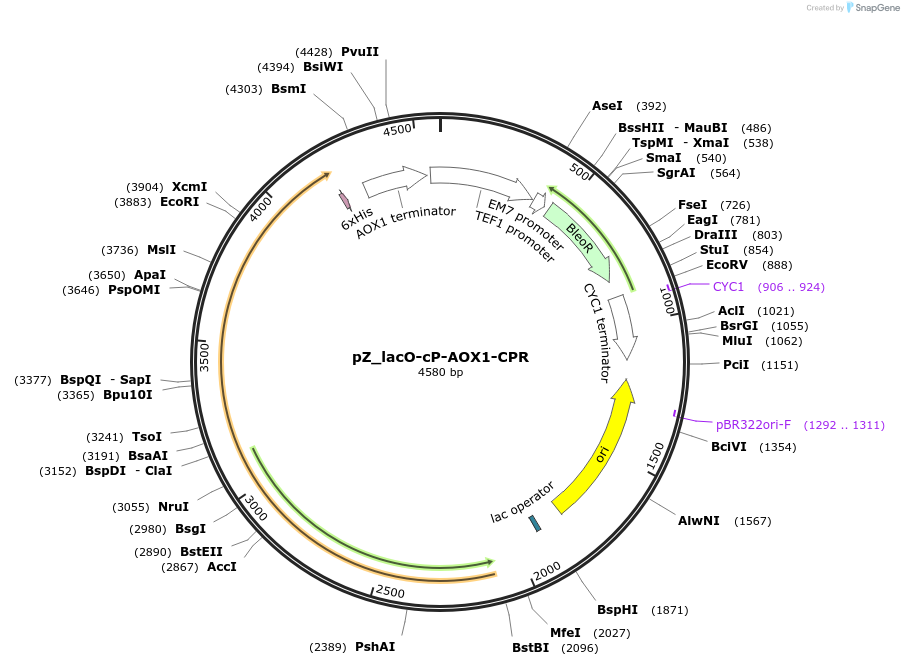
pZ_lacO-cP-AOX1-CPR
Plasmid#126742Purposeyeast plasmid overexpressing CPRDepositorInsertcpr
Tags6xHisExpressionYeastAvailable SinceJuly 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
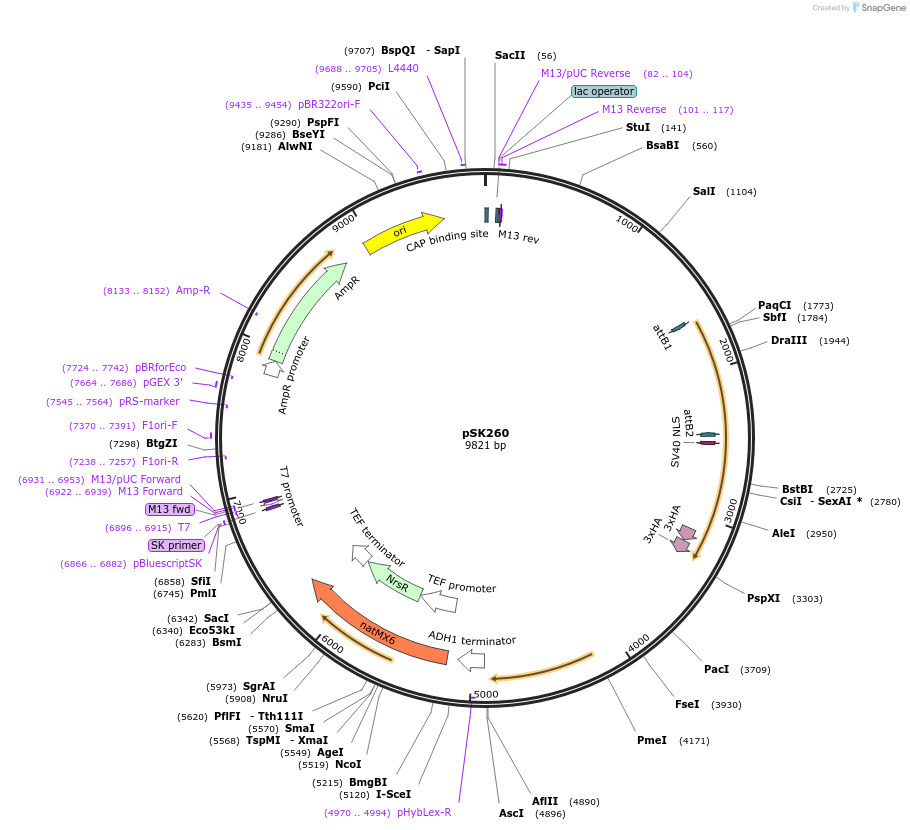
pSK260
Plasmid#129252Purposedual yTRAP, NM fusion to aTF1 reporting on [PSI+] state with mKate2 reporterDepositorInsertSup35 NM yTRAP sensor reporting on mKate2
ExpressionYeastAvailable SinceOct. 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
pUB23-L-beta-gal
Plasmid#8820DepositorInsertbeta-gal
TagsUbiquitinExpressionYeastMutationL (leucine) at amino terminus of beta-galAvailable SinceDec. 28, 2005AvailabilityAcademic Institutions and Nonprofits only