We narrowed to 30,195 results for: REP
-
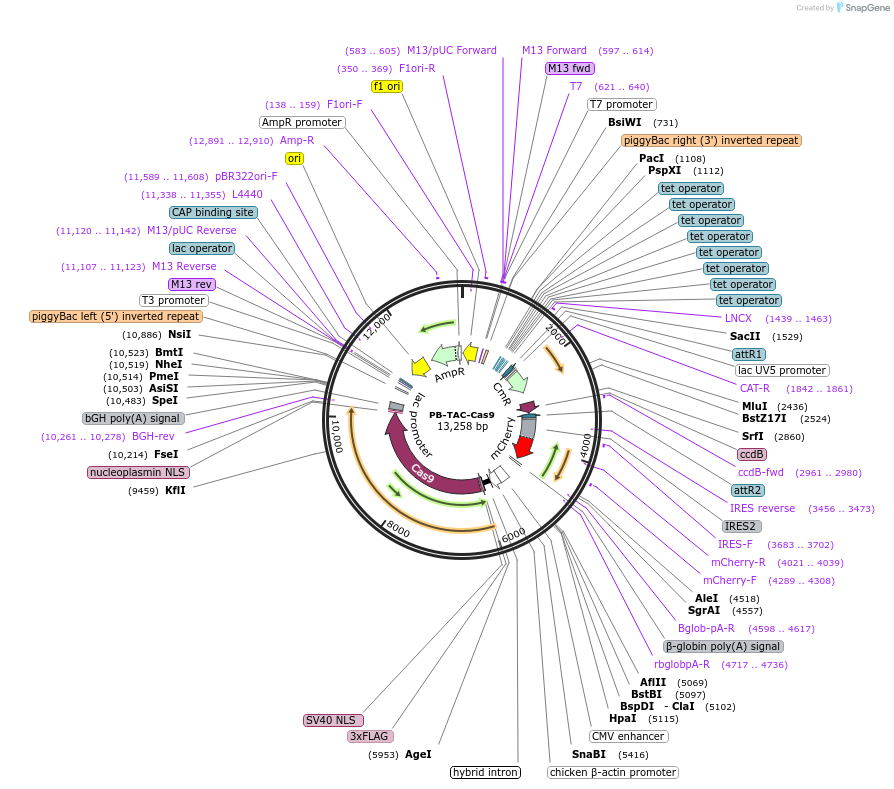
Plasmid#120352PurposepiggyBac transposon Destination vector for dox-inducible expression of Gateway cloned elements (inducible mCherry). SpCas9 is constitutively expressed.DepositorTypeEmpty backboneUseCRISPR; Piggybac transposonExpressionMammalianPromotertet-on and CBhAvailable SinceApril 8, 2021AvailabilityAcademic Institutions and Nonprofits only
-
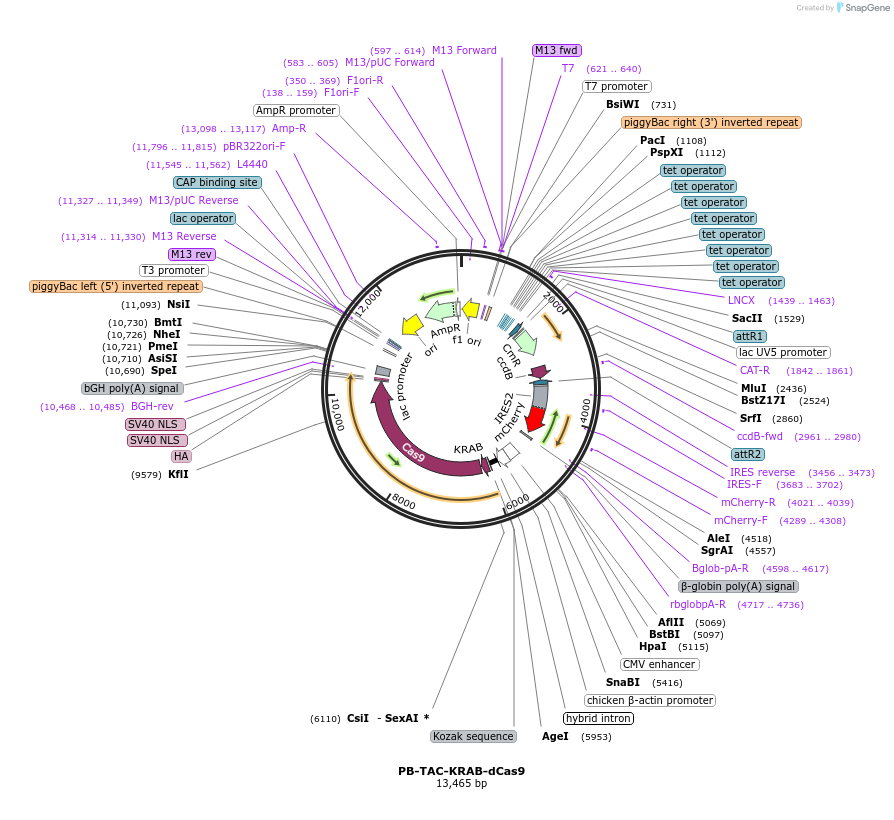
PB-TAC-KRAB-dCas9
Plasmid#120355PurposepiggyBac transposon Destination vector for dox-inducible expression of Gateway cloned elements (inducible mCherry). KRAB-dCas9 is constitutively expressed.DepositorTypeEmpty backboneUseCRISPR; Piggybac transposonExpressionMammalianPromotertet-on and CBhAvailable SinceApril 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
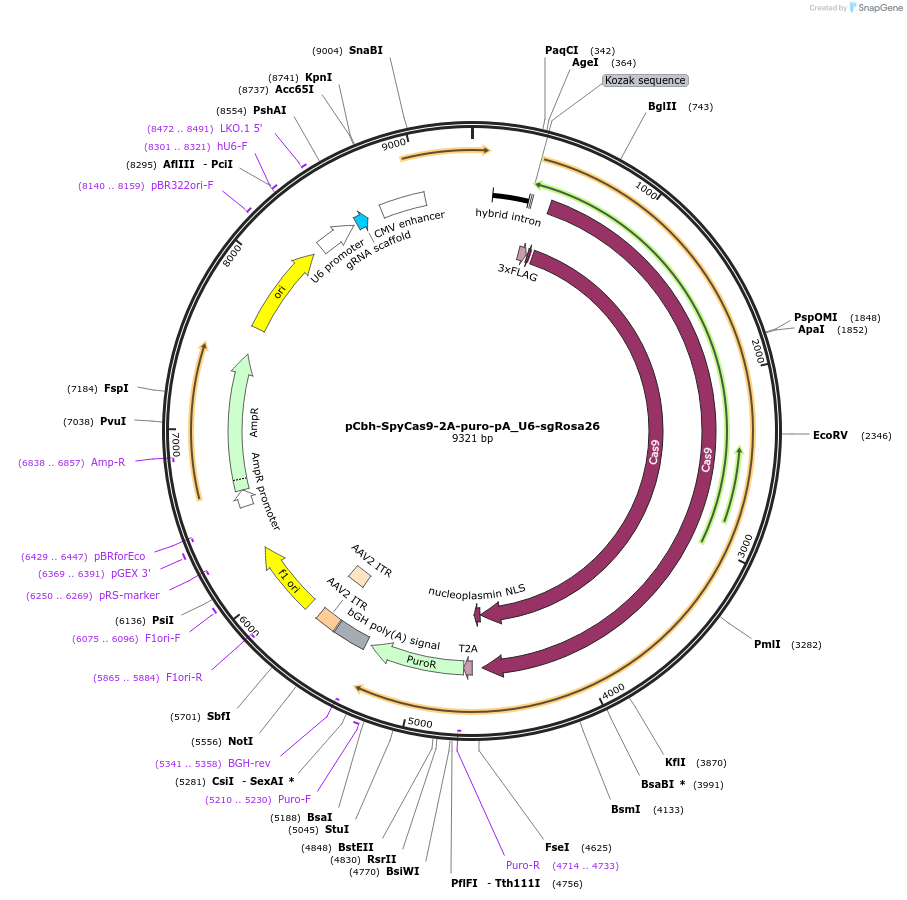
pCbh-SpyCas9-2A-puro-pA_U6-sgRosa26
Plasmid#149346PurposeMammalian expression vector for SpyCas9, puromycin resistance and mouse Rosa26 guide RNADepositorInsertCas9
UseCRISPRTagsFlag tagExpressionMammalianPromoterCbcAvailable SinceJan. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
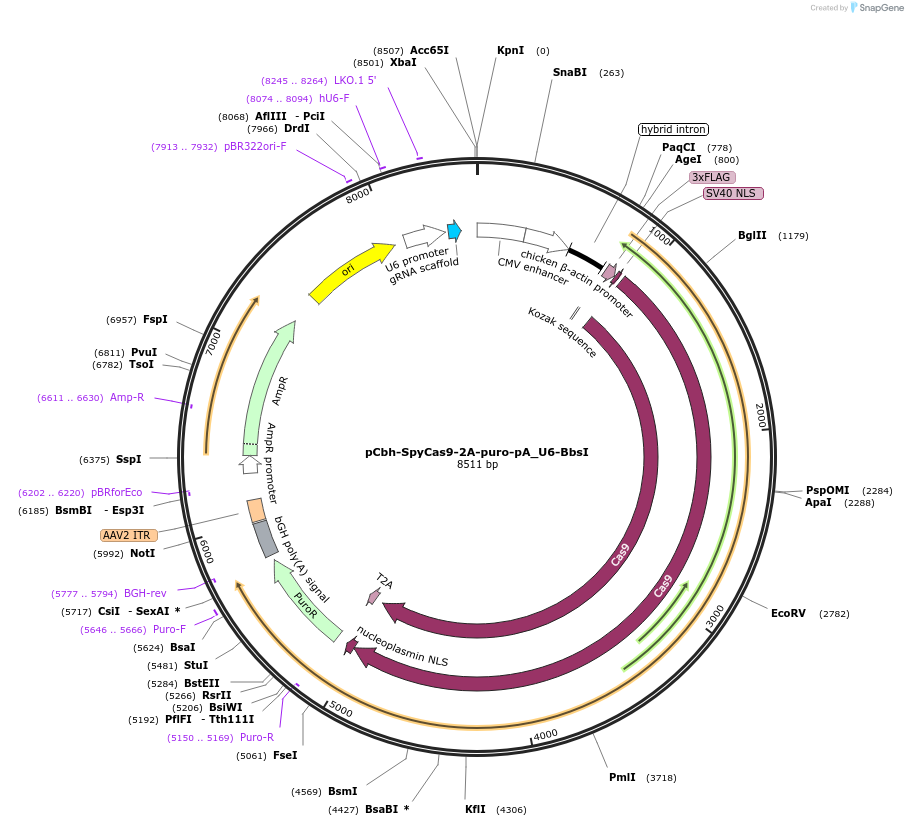
pCbh-SpyCas9-2A-puro-pA_U6-BbsI
Plasmid#149347PurposeMammalian expression vector for SpyCas9 and puromycin resistanceDepositorInsertSpyCas9
UseCRISPRTagsFlag tagExpressionMammalianPromoterCbhAvailable SinceJan. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
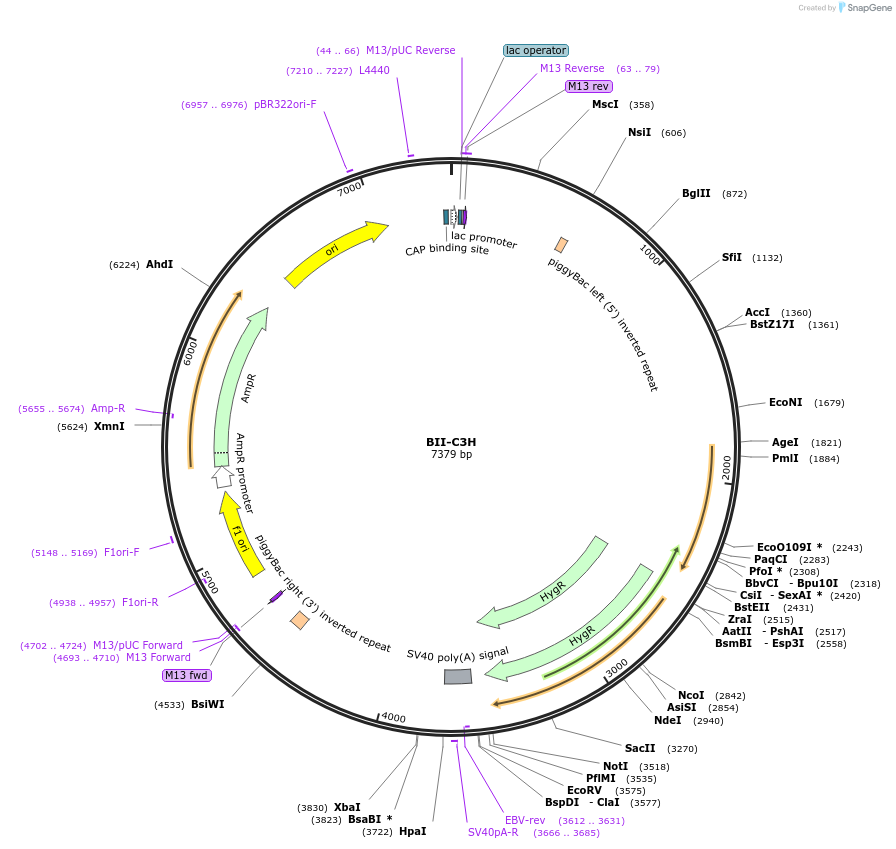
BII-C3H
Plasmid#133386PurposePiggybac vector for SRIRACCHA-mediated mutation enrichment (surrogate target for HDR)DepositorTypeEmpty backboneExpressionMammalianAvailable SinceJan. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
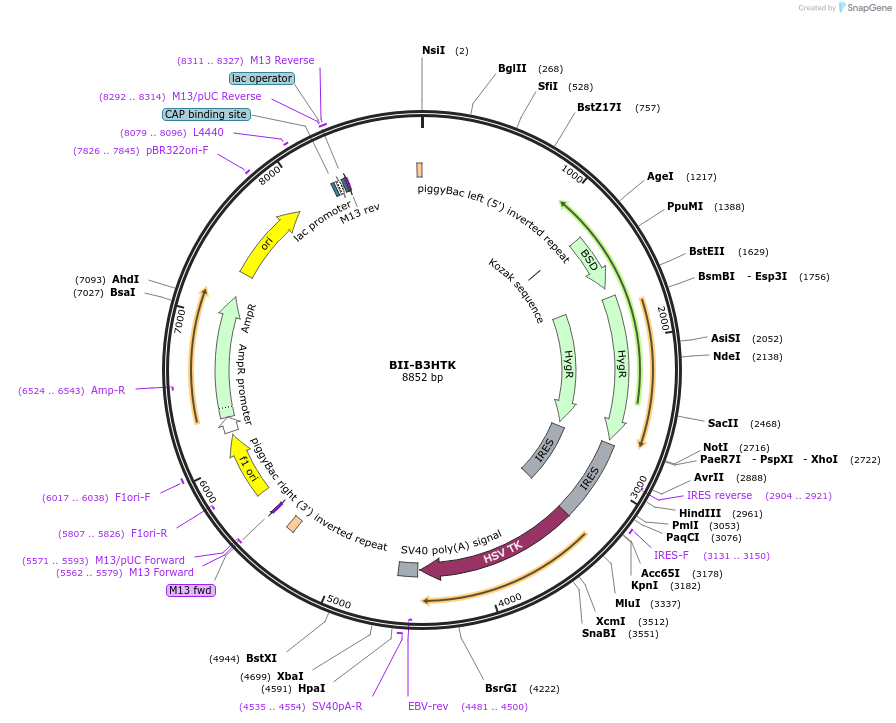
BII-B3HTK
Plasmid#133389PurposePiggybac vector for SRIRACCHA-mediated mutation enrichment (surrogate target for HDR)DepositorTypeEmpty backboneExpressionMammalianAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
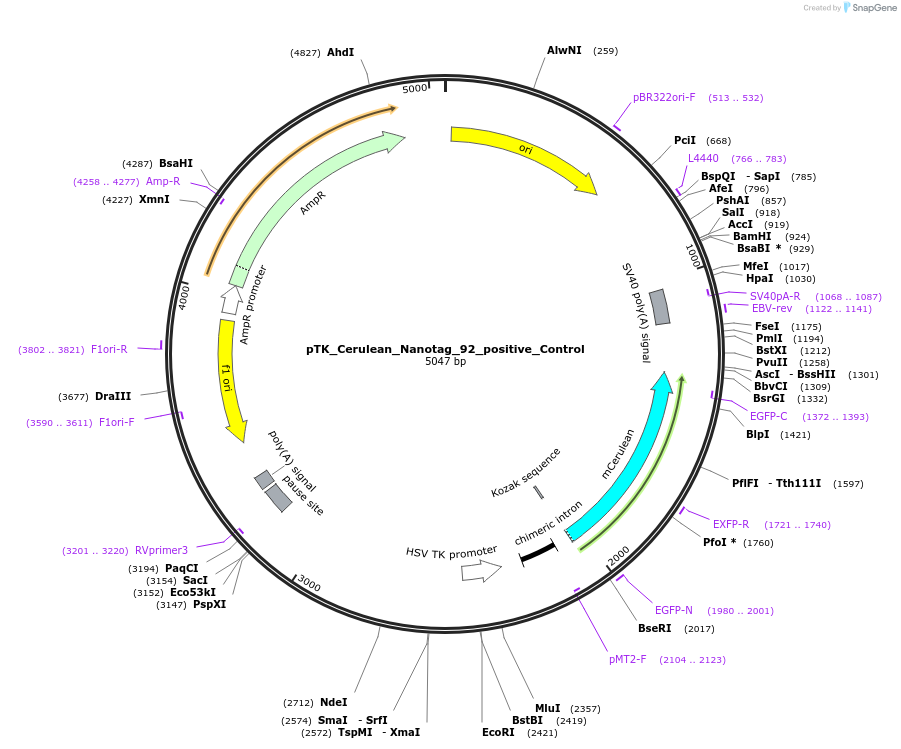
pTK_Cerulean_Nanotag_92_positive_Control
Plasmid#130573PurposeEnhancer cloning nanotag reporter vectorDepositorInsertFoxD3_cranial_neural_crest_enhancer_NC4
UseEnhancer cloning nanotag reporter vectorPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
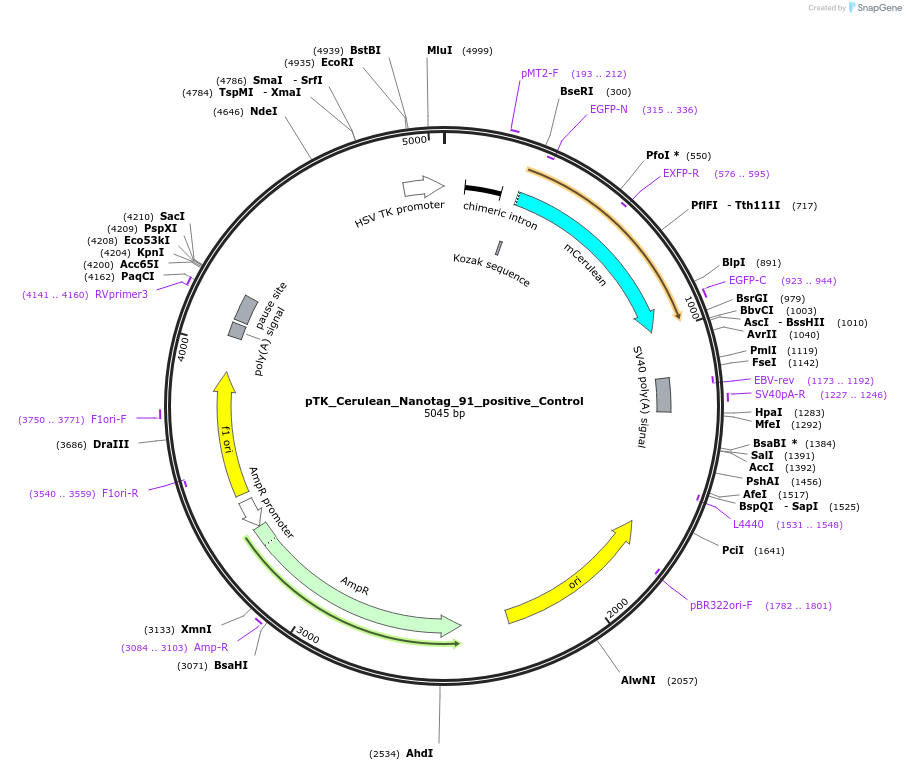
pTK_Cerulean_Nanotag_91_positive_Control
Plasmid#130572PurposeEnhancer cloning nanotag reporter vectorDepositorInsertFoxD3_cranial_neural_crest_enhancer_NC3
UseEnhancer cloning nanotag reporter vectorPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
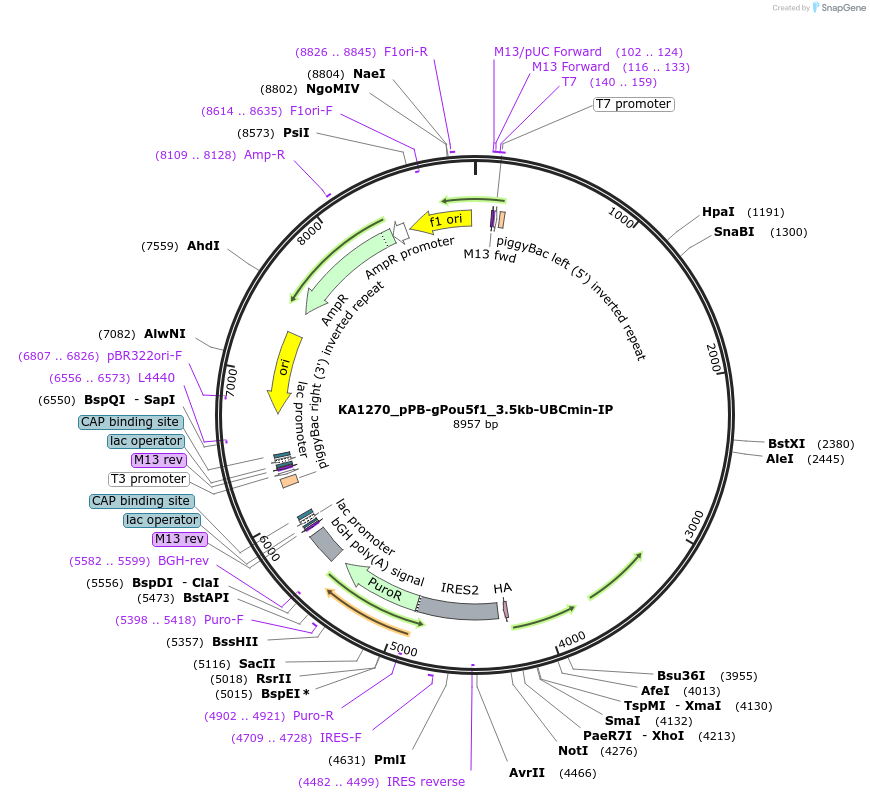
KA1270_pPB-gPou5f1_3.5kb-UBCmin-IP
Plasmid#124181PurposepiggyBac-based reporter (Puro) driven by the Pou5f1 enhancerDepositorInsertIRES-Pac, Pou5f1 enhancer
UseReporterPromoterUBCminAvailable SinceApril 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
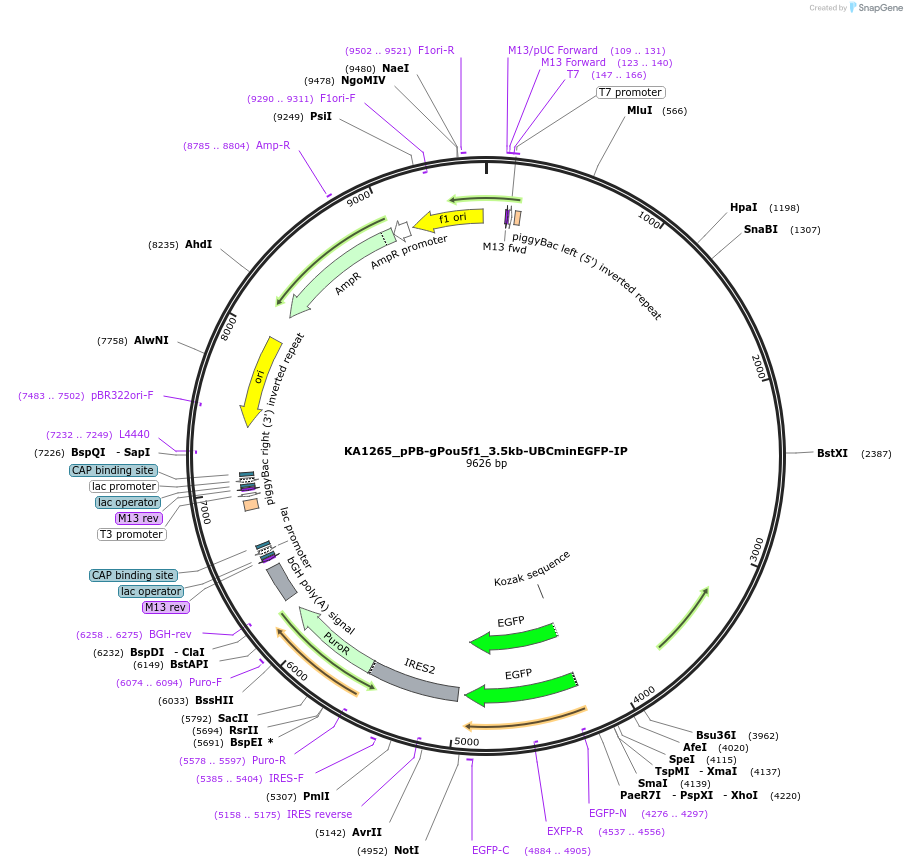
KA1265_pPB-gPou5f1_3.5kb-UBCminEGFP-IP
Plasmid#124180PurposepiggyBac-based reporter (EGFP) driven by the Pou5f1 enhancerDepositorInsertEGFP-IRES-Pac, Pou5f1 enhancer
UseReporterPromoterUBCminAvailable SinceMarch 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
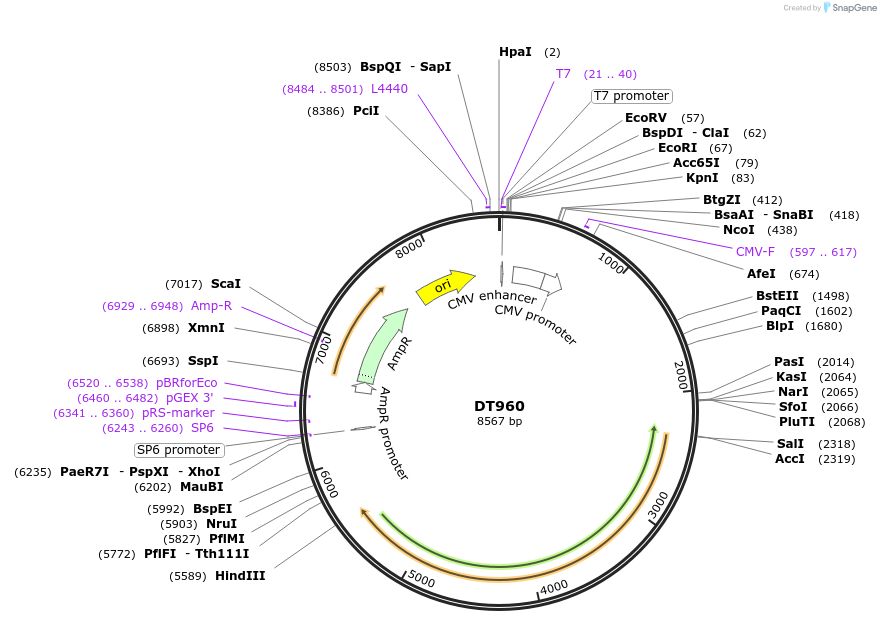
DT960
Plasmid#80412PurposeExpresses 960 interrupted CUG repeats in the context of human DMPK genomic segment containing exons 11-15 with repeats inserted at site in exon 15 that contains the repeatsDepositorInserthuman DMPK exons 11-15 with 960 interrupted CTG repeats (DMPK Human)
ExpressionMammalianAvailable SinceSept. 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
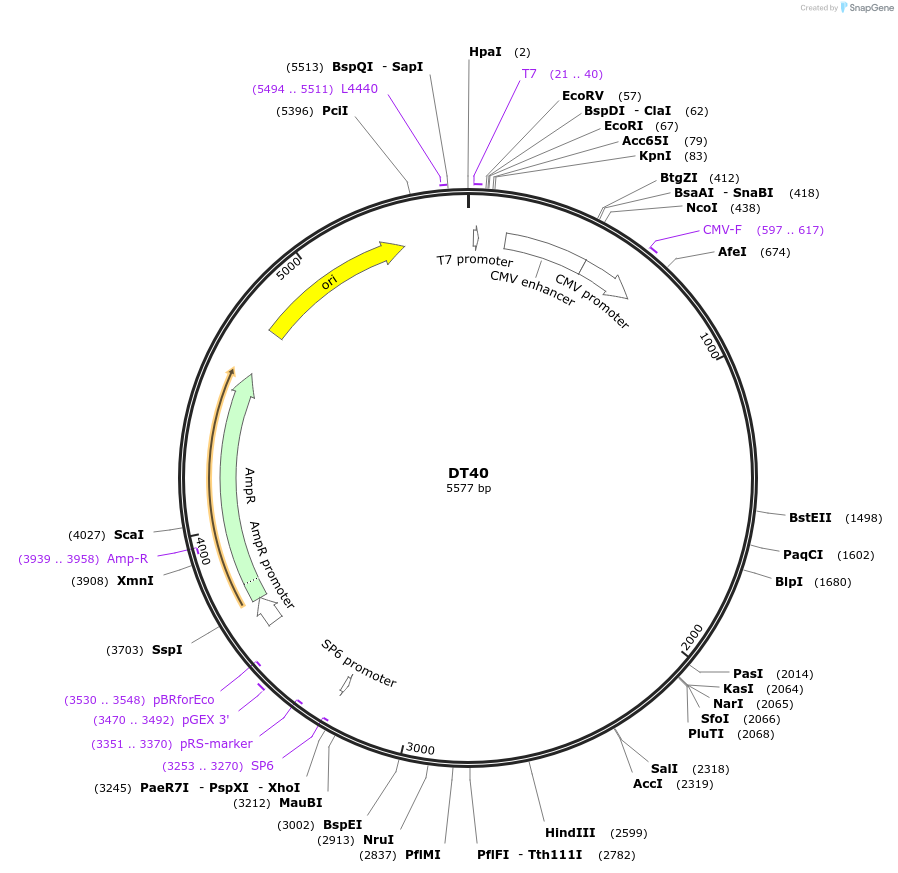
DT40
Plasmid#80415PurposeExpresses 40 interrupted CUG repeats in the context of human DMPK genomic segment containing exons 11-15 with repeats inserted at site in exon 15 that contains the repeatsDepositorInserthuman DMPK exons 11-15 with 40 interrupted CTG repeats (DMPK Human)
ExpressionMammalianAvailable SinceSept. 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
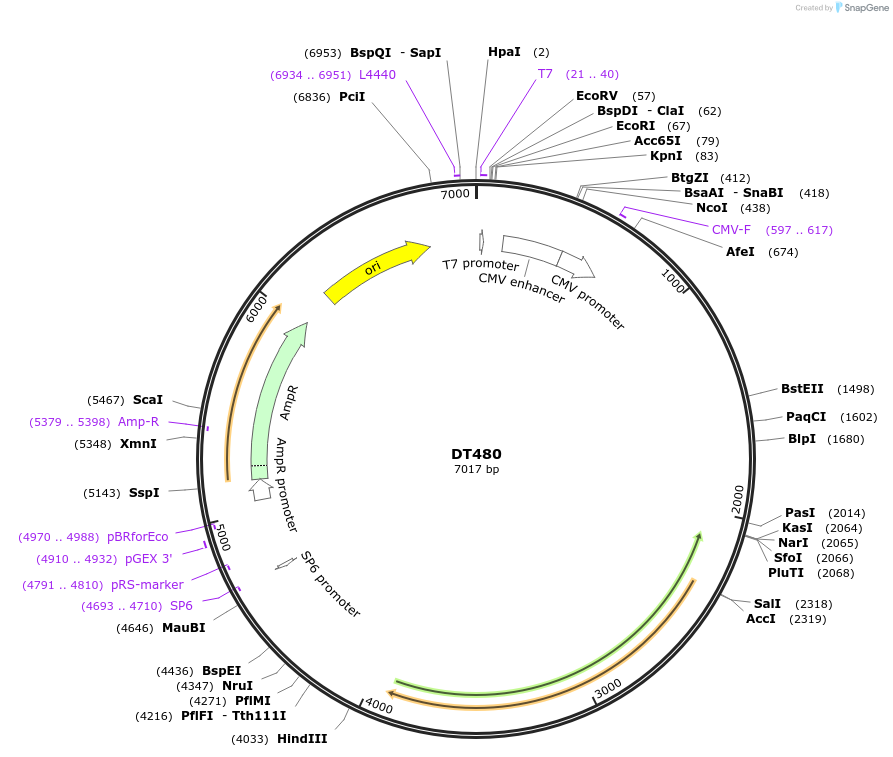
DT480
Plasmid#80413PurposeExpresses 480 interrupted CUG repeats in the context of human DMPK genomic segment containing exons 11-15 with repeats inserted at site in exon 15 that contains the repeatsDepositorInserthuman DMPK exons 11-15 with 480 interrupted CTG repeats (DMPK Human)
ExpressionMammalianAvailable SinceSept. 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
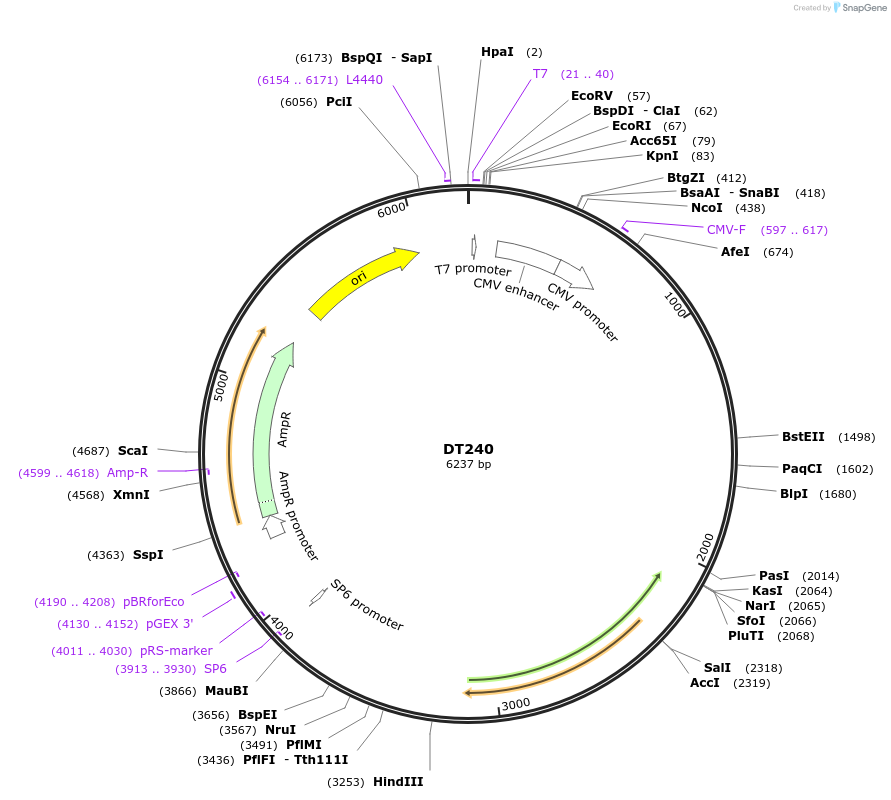
DT240
Plasmid#80414PurposeExpresses 240 interrupted CUG repeats in the context of human DMPK genomic segment containing exons 11-15 with repeats inserted at site in exon 15 that contains the repeatsDepositorInserthuman DMPK exons 11-15 with 240 interrupted CTG repeats (DMPK Human)
ExpressionMammalianAvailable SinceSept. 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
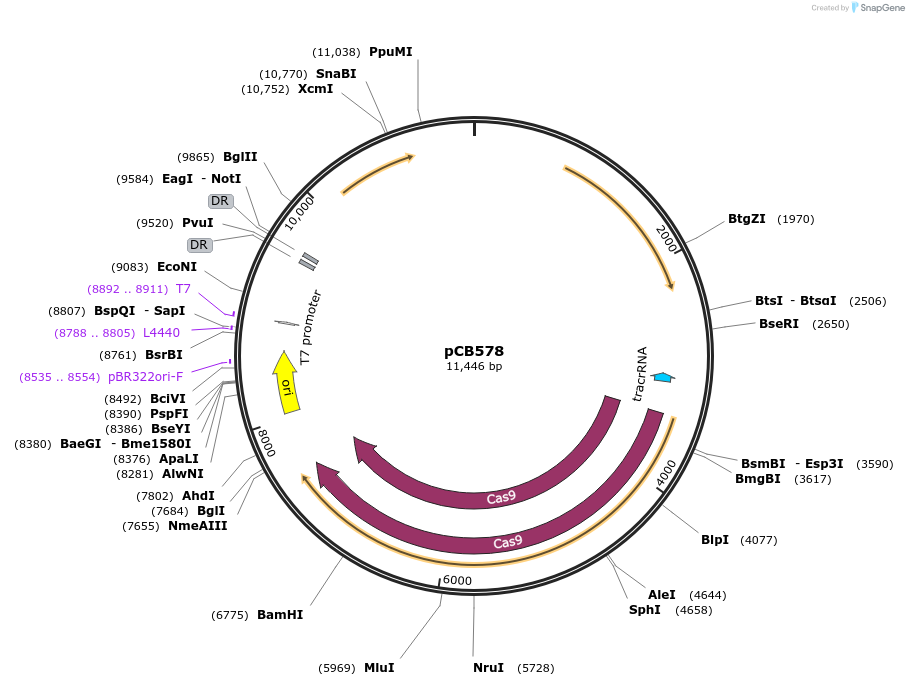
pCB578
Plasmid#141095PurposeE. coli-Lactobacilli shuttle vector containing SpCas9, tracrRNA, and a repeat-spacer-repeat arrayDepositorInsertsCas9 and tracrRNA
repeat-spacer-repeat array
ExpressionBacterialAvailable SinceApril 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
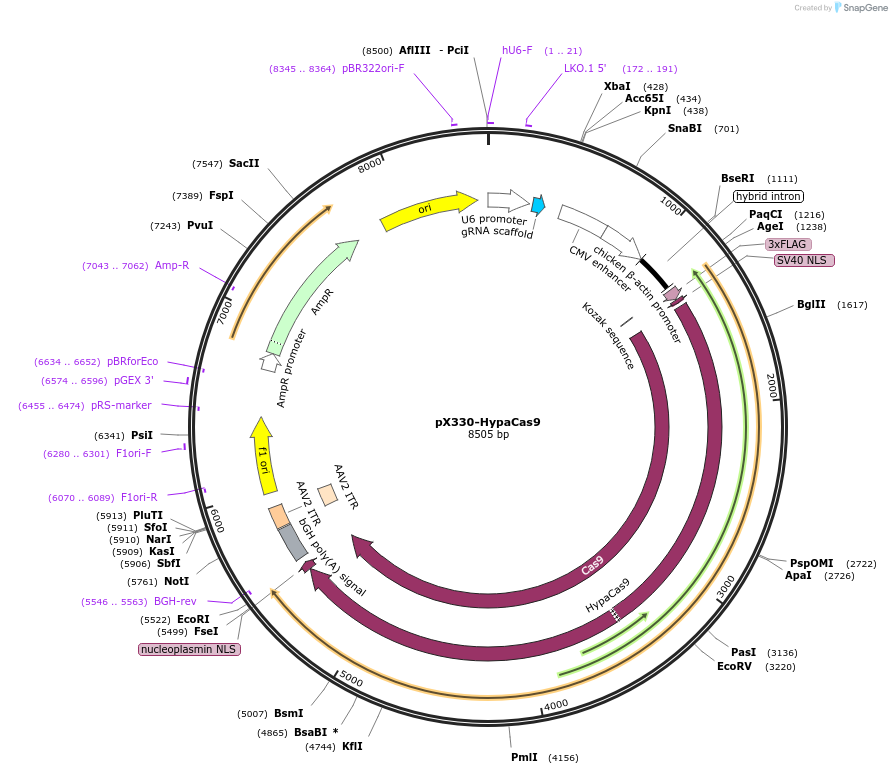
pX330-HypaCas9
Plasmid#108302PurposepX330 (Plasmid #42230) with the N692A, M694A, Q695A, and H698A mutationsDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceMay 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
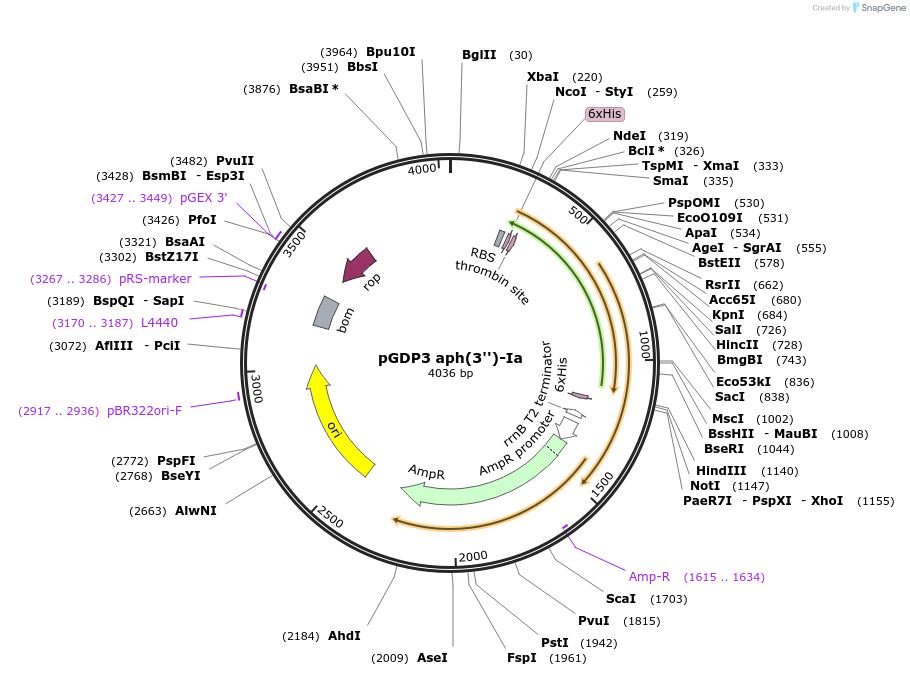
pGDP3 aph(3'')-Ia
Plasmid#112879PurposeThis plasmid is part of the Minimal Antibiotic Resistance Platform (ARP) Kit (Addgene #1000000143). Low copy-number plasmid that constitutively expresses genes using the Pbla promoter.DepositorAvailable SinceDec. 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
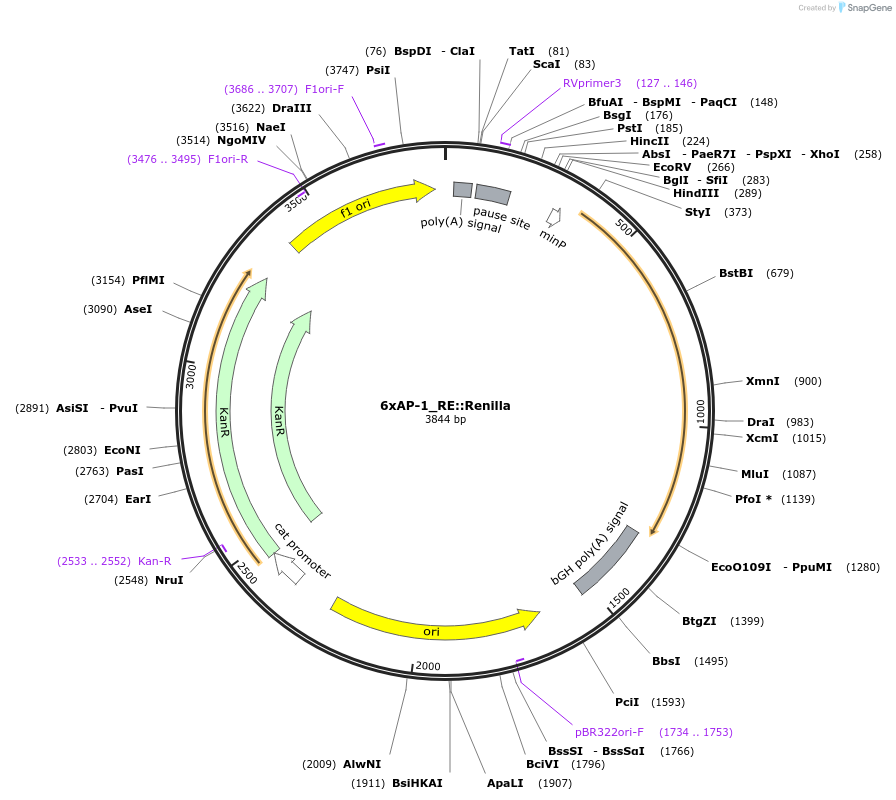
6xAP-1_RE::Renilla
Plasmid#178317PurposeAP-1 Renilla luciferase reporterDepositorInsertAP-1 Renilla Reporter
UseLuciferase and Synthetic BiologyExpressionMammalianAvailable SinceJuly 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
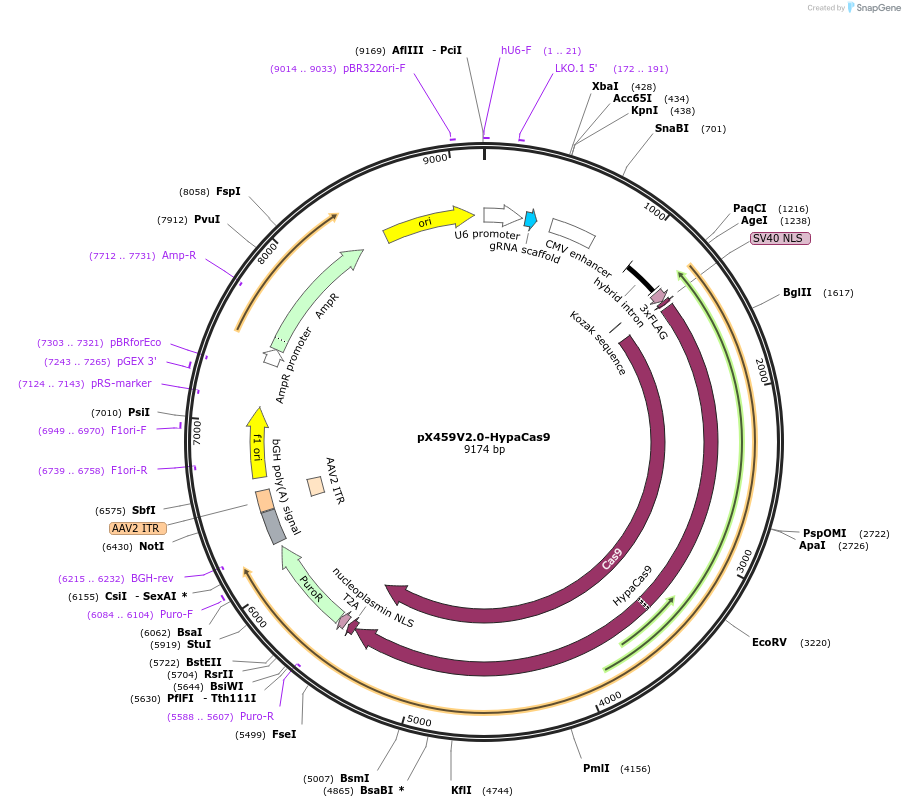
pX459V2.0-HypaCas9
Plasmid#108294PurposepX459 V2.0 (Plasmid #62988) with the N692A, M694A, Q695A, and H698A mutationsDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceJuly 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
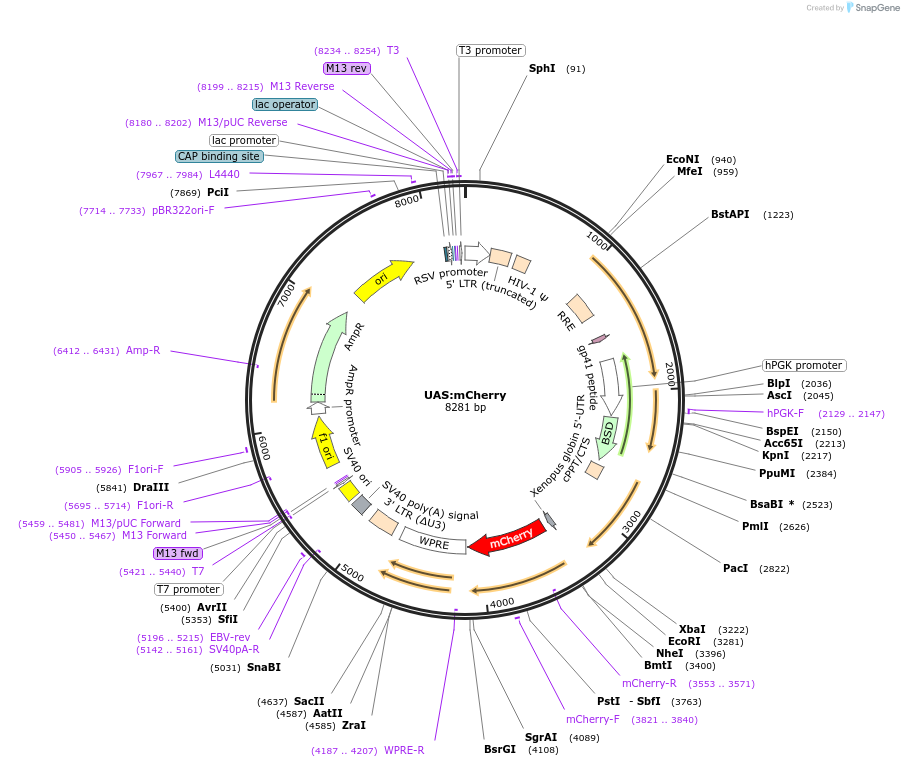
UAS:mCherry
Plasmid#170994PurposeUAS:GAL4 repeat-containing promoter drives simple mCherry reporter gene. Blasticidin selectable.DepositorInsertUAS:mCherry
UseLentiviral and Synthetic BiologyExpressionMammalianPromoterSynthetic 14x UAS:Gal4Available SinceAug. 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
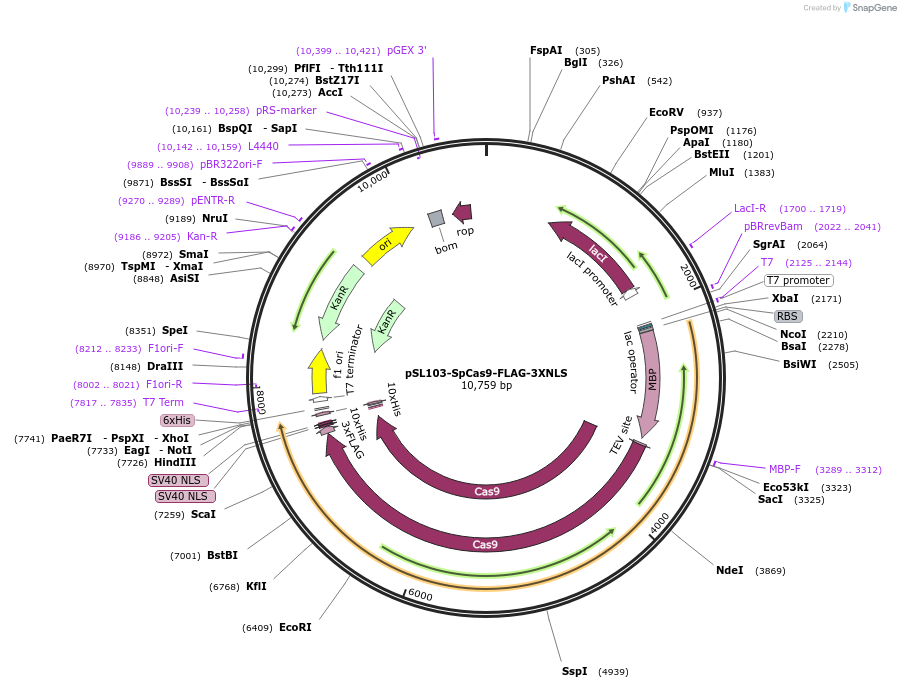
pSL103-SpCas9-FLAG-3XNLS
Plasmid#182032PurposeExpression of recombinant Streptococcus pyogenes Cas9 protein carrying C-terminal FLAG and 3XNLS for genome editing as Cas9 RNPDepositorInsertStreptococcus pyogenes cas9 (cas9 Synthetic)
UseCRISPRTags12X His tag, 3XNLS, FLAG, and MBPExpressionBacterialPromoterT7Available SinceMay 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
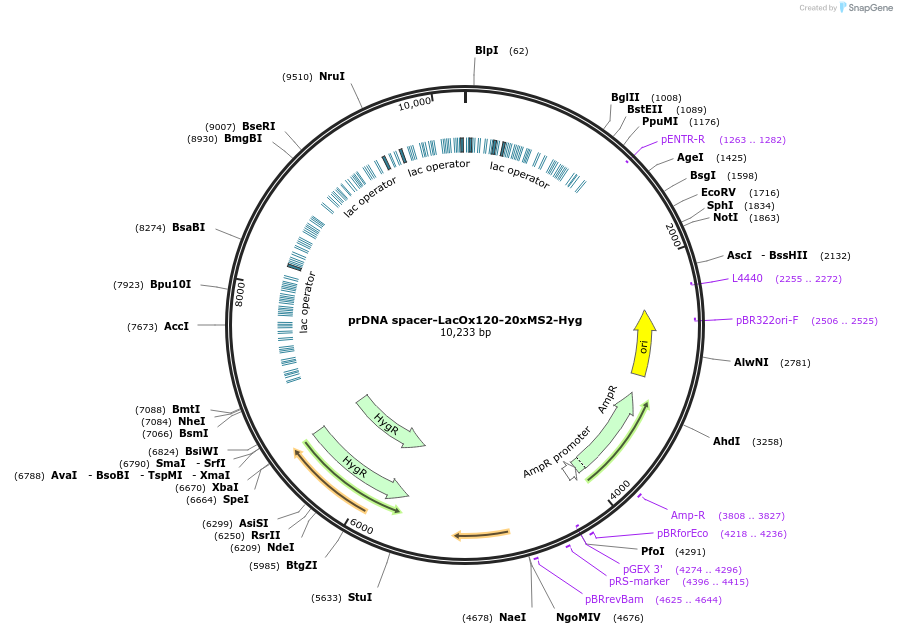
prDNA spacer-LacOx120-20xMS2-Hyg
Plasmid#210220PurposeIntegrates 120xLacO repeats and 20xMS2 repeats with a hygromycin drug selection marker into the rDNA spacer.DepositorInserts120xLacO repeats derived from: PMID: 12864855.
24xMS2 repeats derived from Addgene plasmid # 31865
Available SinceDec. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
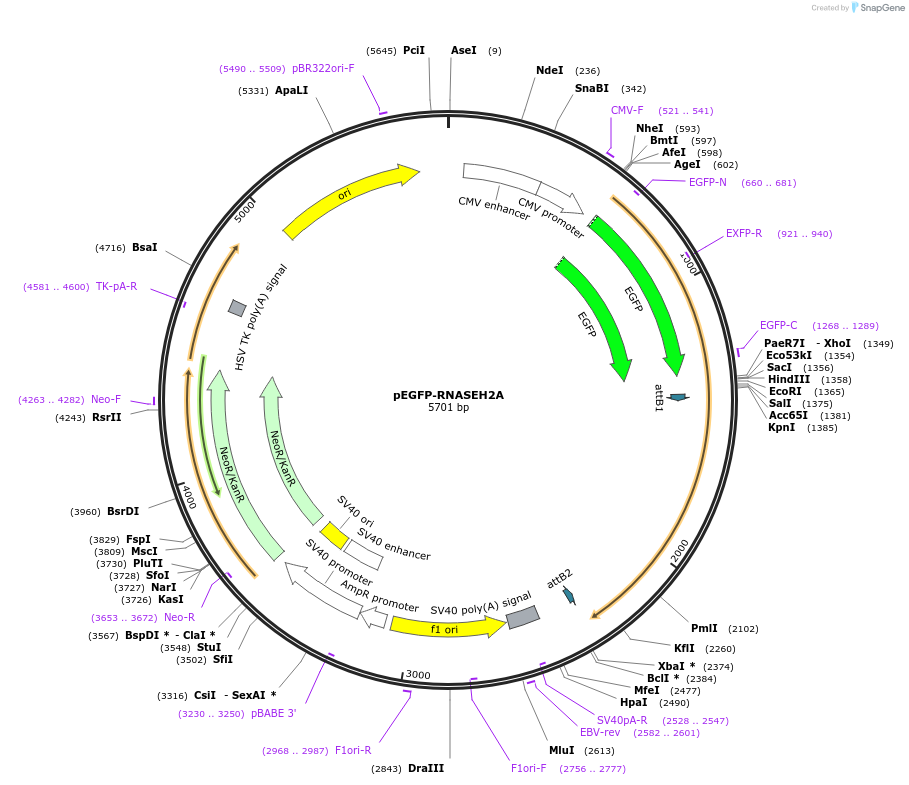
pEGFP‐RNASEH2A
Plasmid#108700PurposeFor expression of N-terminally eGFP-tagged human RNASEH2A in mammalian cellsDepositorAvailable SinceApril 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
pTiGc
Plasmid#78314PurposeMycobacterial dual reporter plasmid for fluorescence dilution. Description: hsp60-gfp, hsp60(ribo)-turboFP635 (inducible TurboFP635 under control of theophylline - inducible riboswitch).DepositorInsertsTurboFP635
GFP
UseSynthetic Biology; Mycobacterial replicating vect…ExpressionBacterialPromoterhsp60 and hsp60(ribo)Available SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
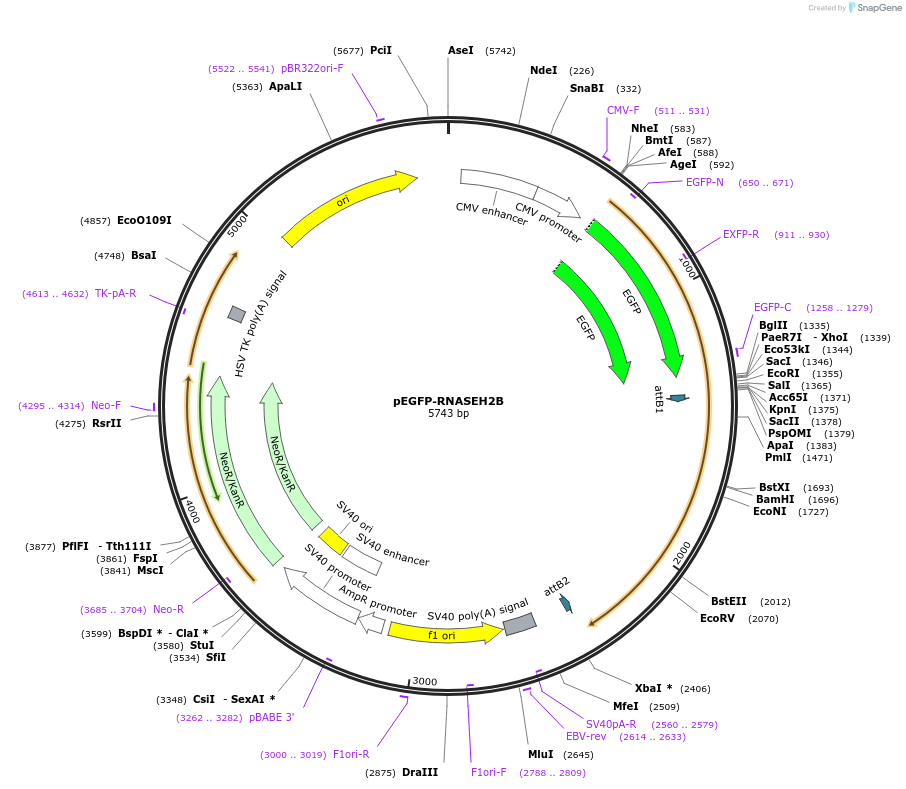
pEGFP‐RNASEH2B
Plasmid#108697PurposeFor expression of N-terminally eGFP-tagged human RNASEH2B in mammalian cellsDepositorAvailable SinceApril 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
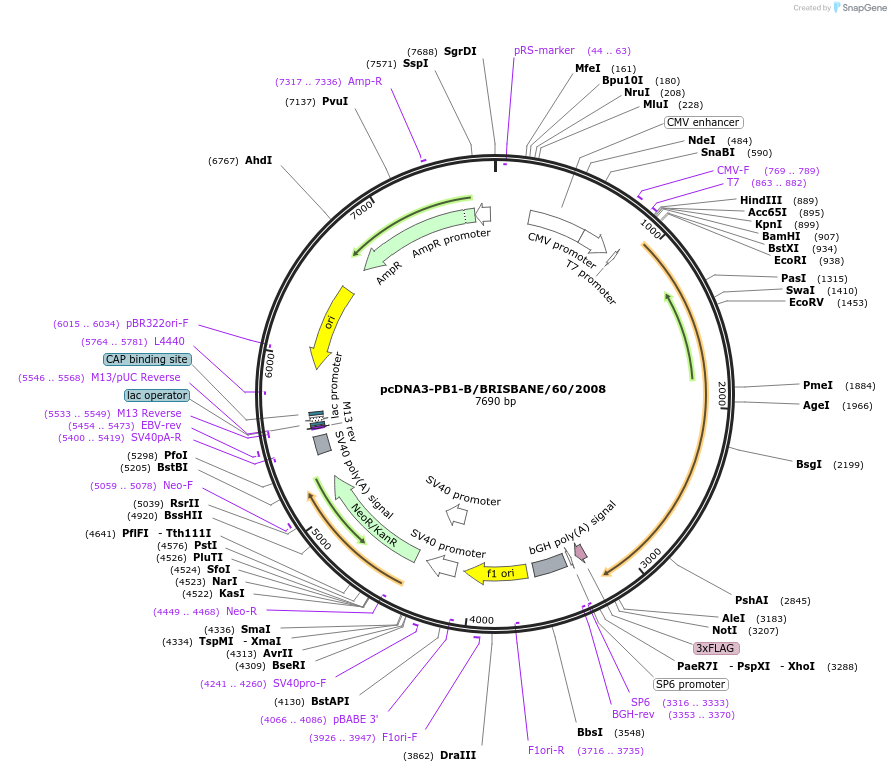
pcDNA3-PB1-B/BRISBANE/60/2008
Plasmid#187919PurposeExpresses PB1 subunit of Influenza B/Brisbane/60/2008 virus polymerase in mammalian cellsDepositorInsertpolymerase basic protein 1
TagsUNTAGGEDPromoterCMV PromoterAvailable SinceOct. 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
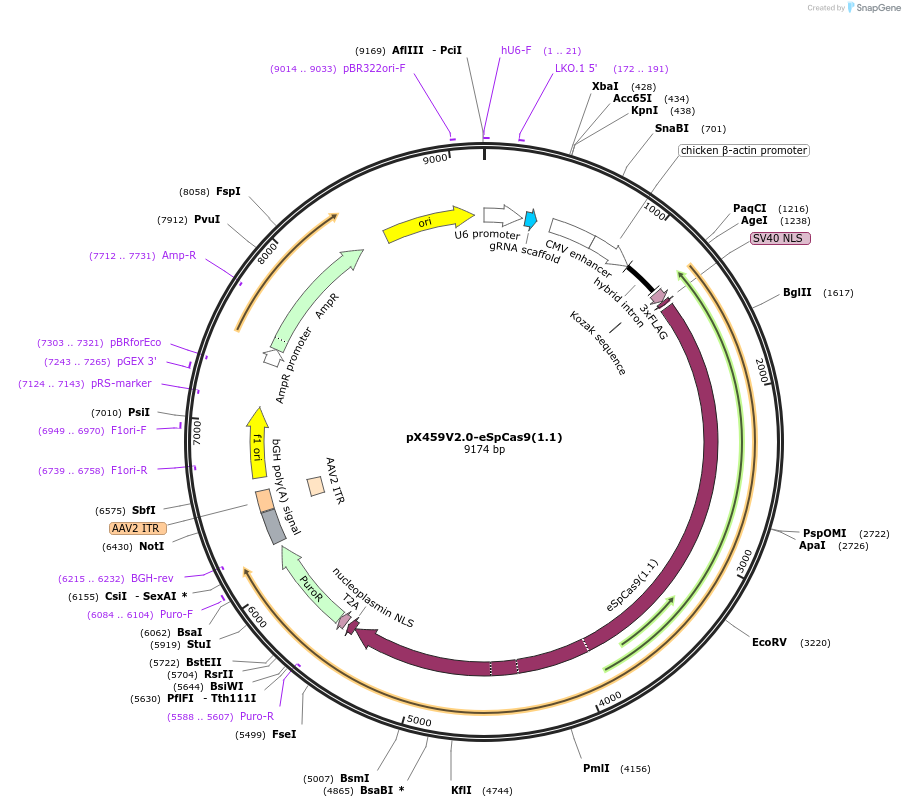
pX459V2.0-eSpCas9(1.1)
Plasmid#108292PurposepX459 V2.0 (Plasmid #62988) with the K848A, K1003A, and R1060A mutationsDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceMay 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
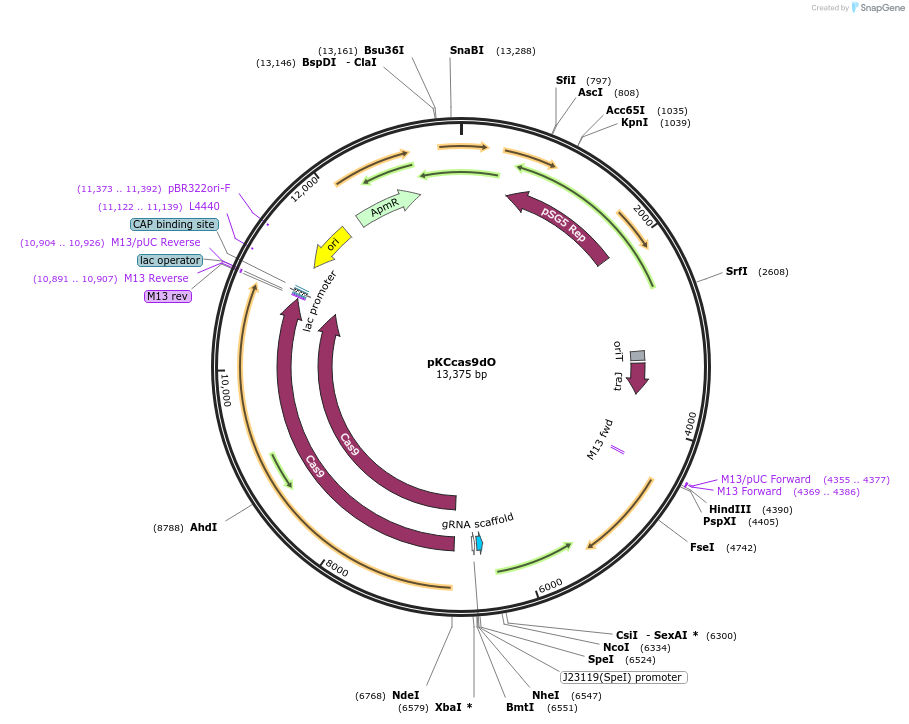
pKCcas9dO
Plasmid#62552PurposeHigh efficiency Streptomyces genome editing by CRISPR/Cas9 systemDepositorInsertsScocas9
sgRNA ()
act-orf4 homology arm
UseCRISPR; Bacterial genome editingExpressionBacterialPromoterj23119 and ptipAAvailable SinceJan. 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
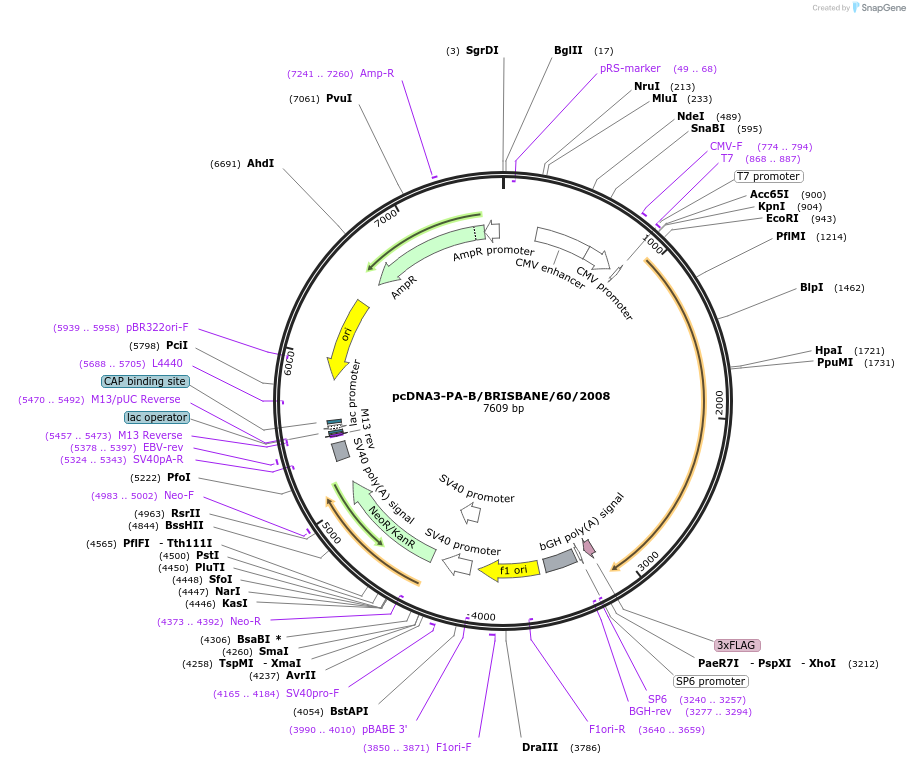
pcDNA3-PA-B/BRISBANE/60/2008
Plasmid#187920PurposeExpresses PA subunit of Influenza B/Brisbane/60/2008 virus polymerase in mammalian cellsDepositorInsertpolymerase acidic protein
TagsUNTAGGEDExpressionMammalianPromoterCMV PromoterAvailable SinceMay 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
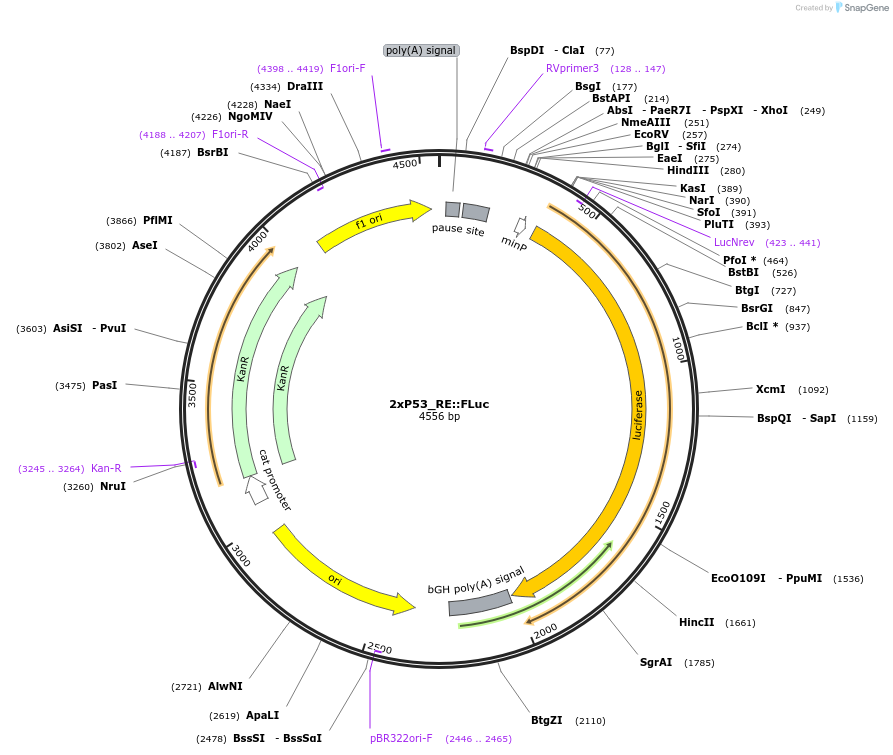
2xP53_RE::FLuc
Plasmid#178316PurposeP53 Firefly luciferase reporterDepositorInsertP53 FLuc Reporter
UseLuciferase and Synthetic BiologyExpressionMammalianAvailable SinceJuly 20, 2022AvailabilityAcademic Institutions and Nonprofits only