We narrowed to 2,834 results for: Star;
-
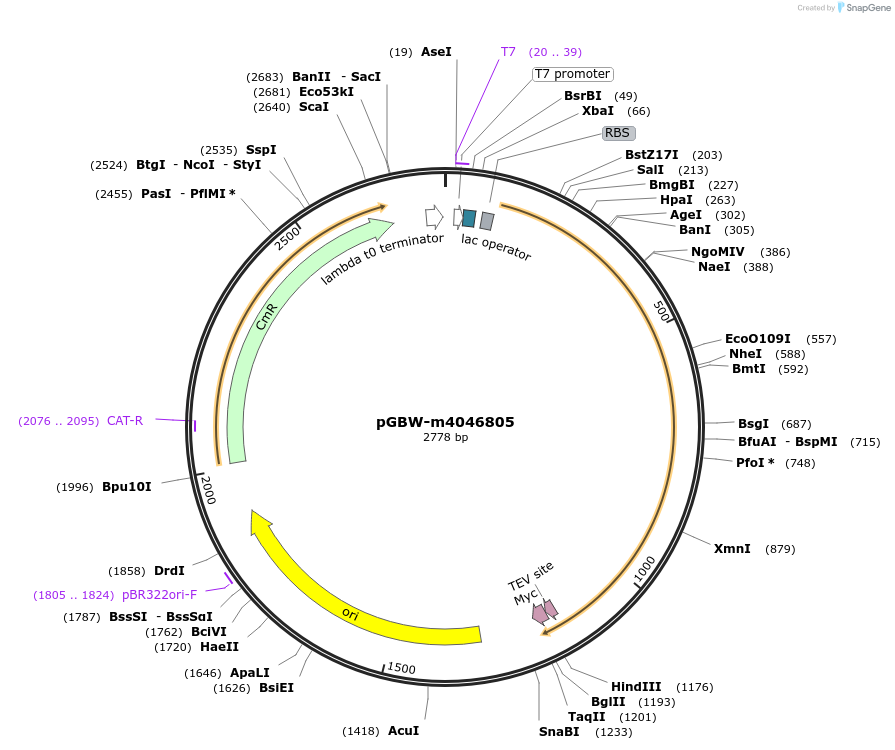
Plasmid#145589PurposeBacterial Expression plasmid for SARS-CoV-2 endoRNAseDepositorInsertSARS-CoV-2 endoRNAse (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;MycExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGBW-m4046935
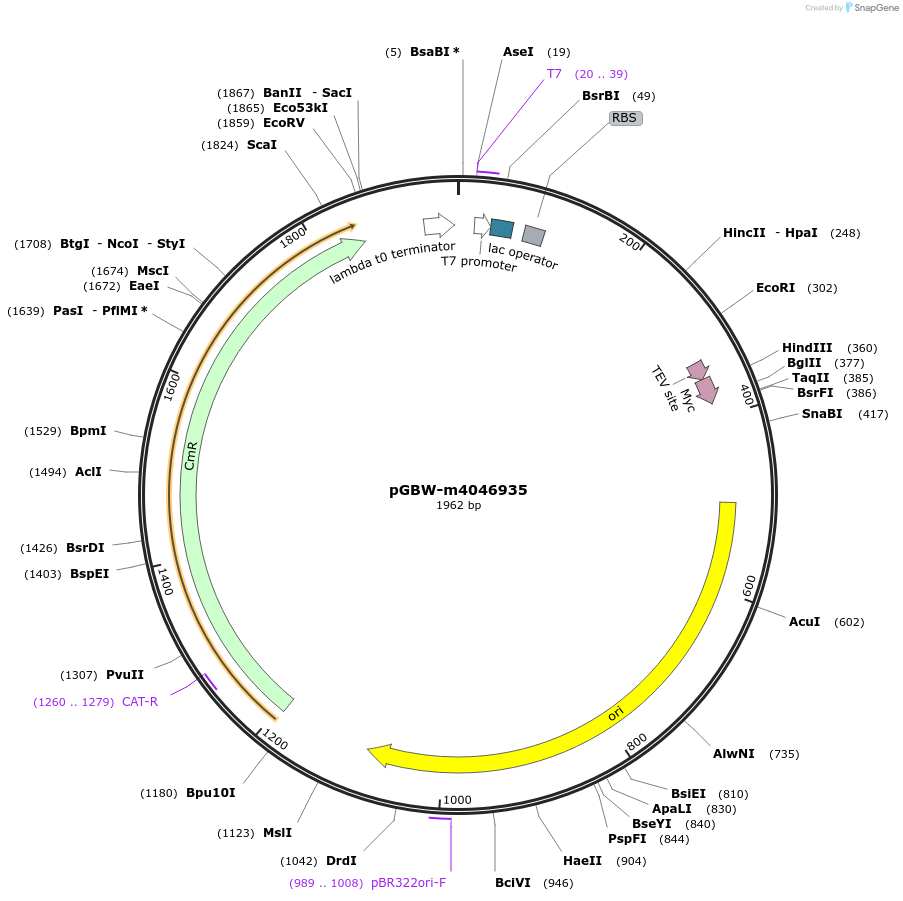
Plasmid#145580PurposeBacterial Expression plasmid for SARS-CoV-2 E (envelope)DepositorInsertSARS-CoV-2 E (envelope) (E Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;MycExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGBW-m4046725
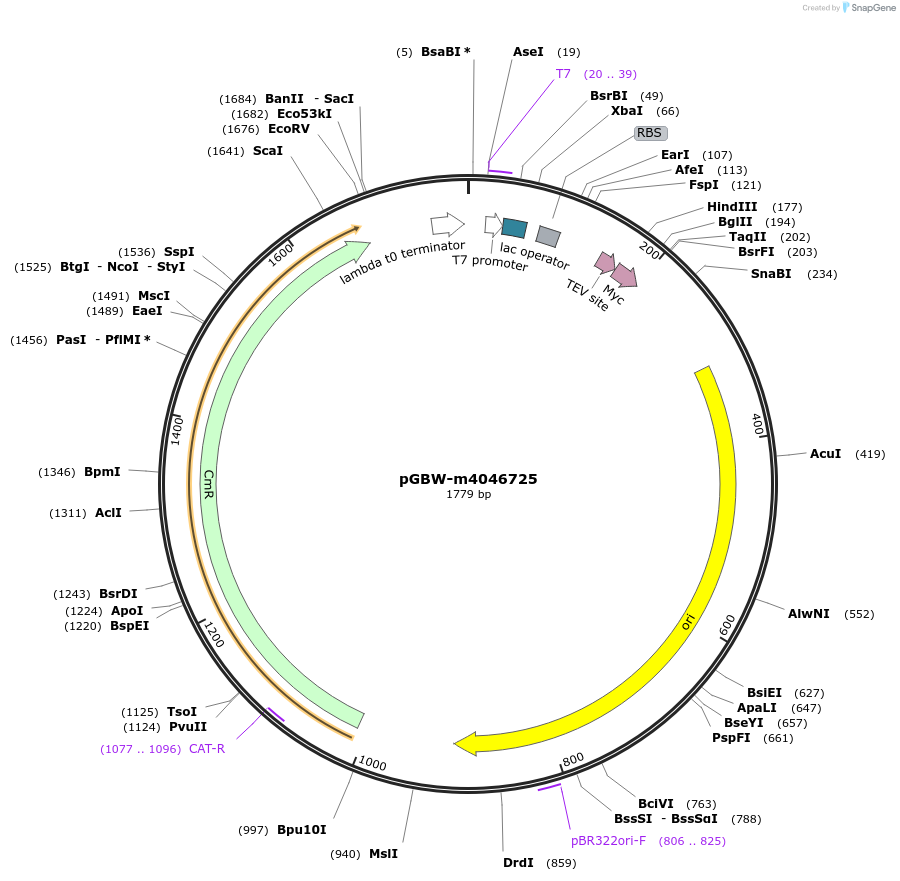
Plasmid#145581PurposeBacterial Expression plasmid for SARS-CoV-2 nsp11DepositorInsertSARS-CoV-2 nsp11 (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;MycExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGBW-m4046761
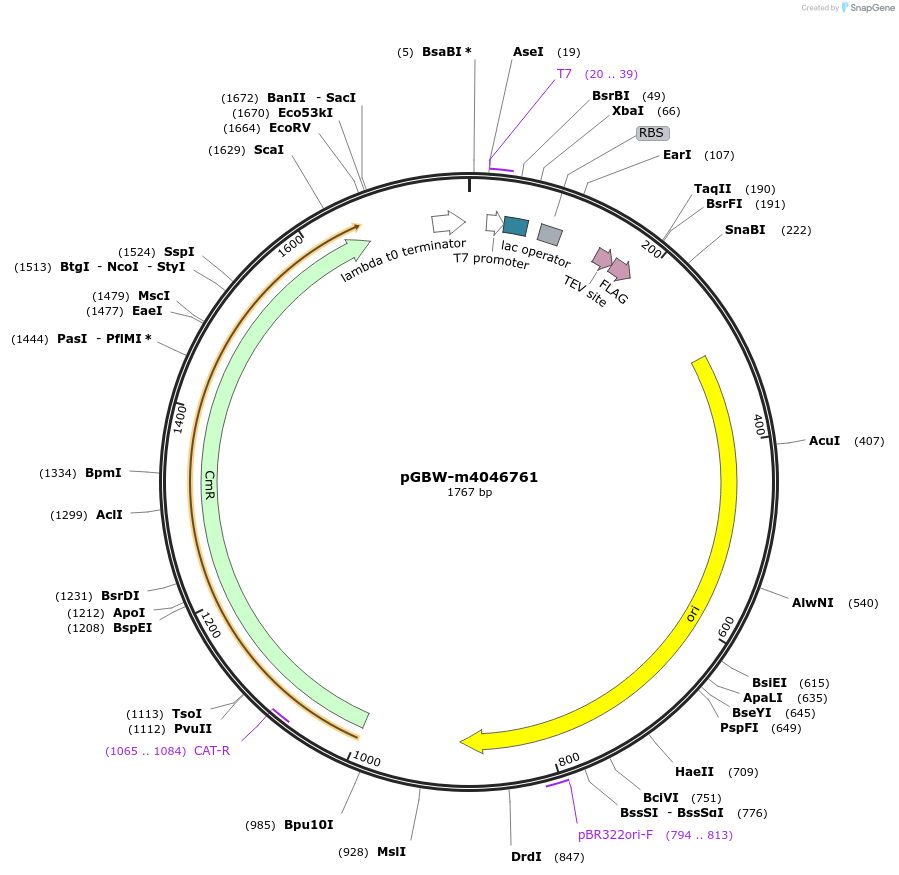
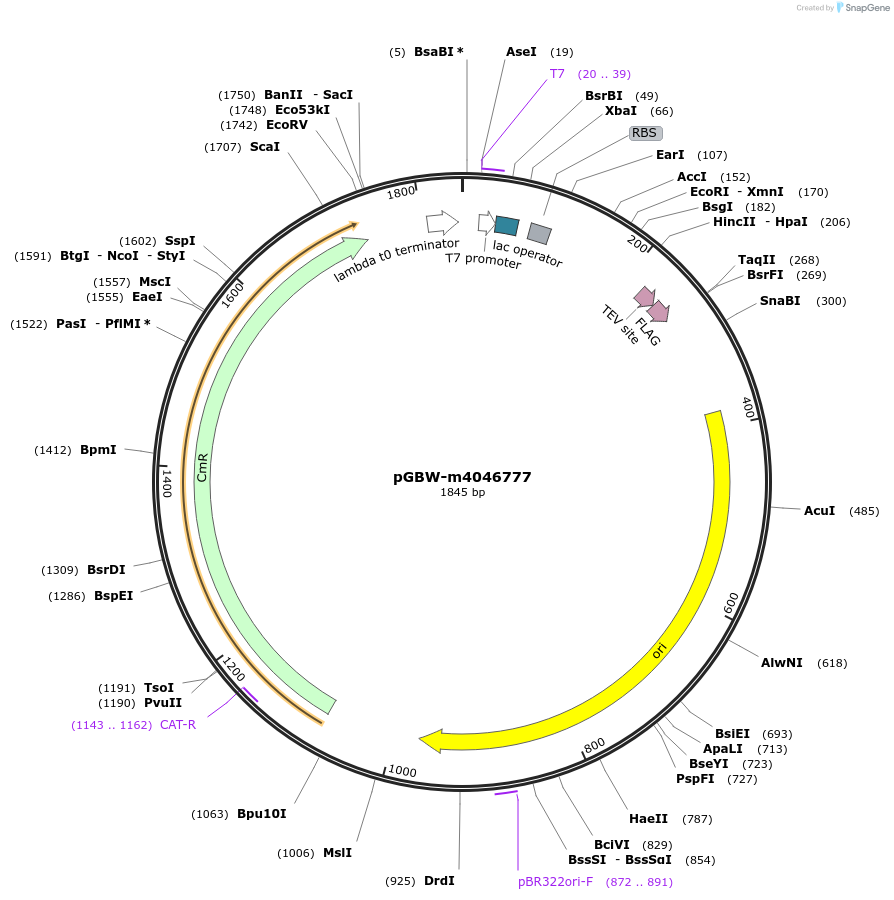
Plasmid#145582PurposeBacterial Expression plasmid for SARS-CoV-2 nsp11DepositorInsertSARS-CoV-2 nsp11 (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;FLAG_2ExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGBW-m4046777
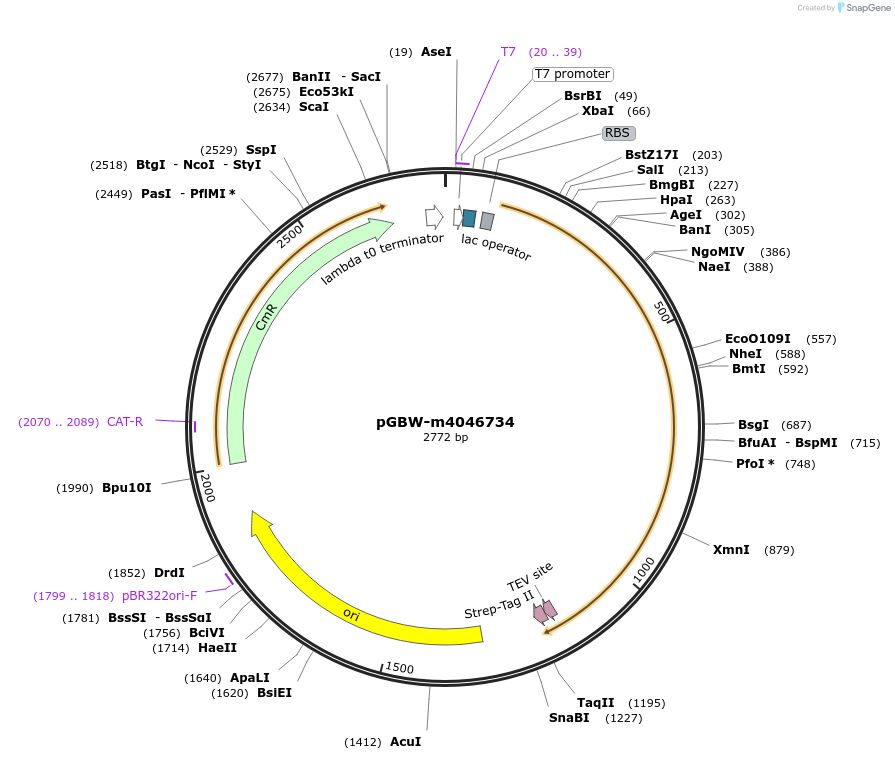
Plasmid#145583PurposeBacterial Expression plasmid for SARS-CoV-2 ORF10 proteinDepositorInsertSARS-CoV-2 ORF10 protein (ORF10 Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;FLAG_2ExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGBW-m4046734
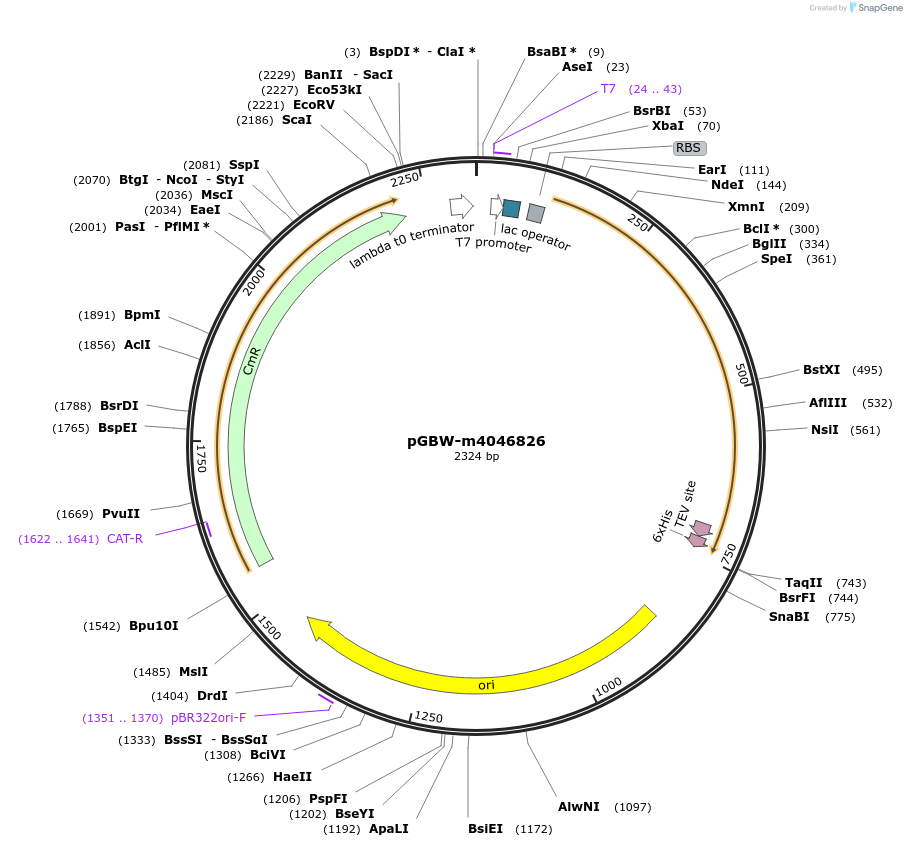
Plasmid#145576PurposeBacterial Expression plasmid for SARS-CoV-2 endoRNAseDepositorInsertSARS-CoV-2 endoRNAse (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGBW-m4046826
Plasmid#145687PurposeBacterial Expression plasmid for SARS-CoV-2 nsp8DepositorInsertSARS-CoV-2 nsp8 (ORF1ab Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;6xHISExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 27, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
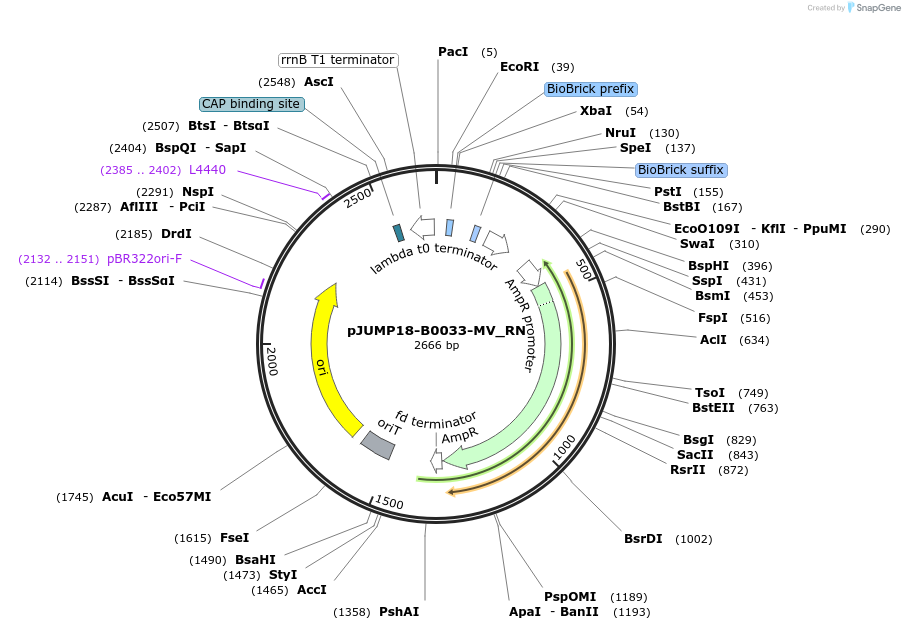
pJUMP18-B0033-MV_RN
Plasmid#127016PurposeBasic Part RN- RBS+N-terminus; B0034m Ribosome Binding Site + Met-Val N-terminus start.DepositorInsertPart B0033m-MV_RN
UseSynthetic BiologyExpressionBacterialMutationNoneAvailable SinceJuly 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
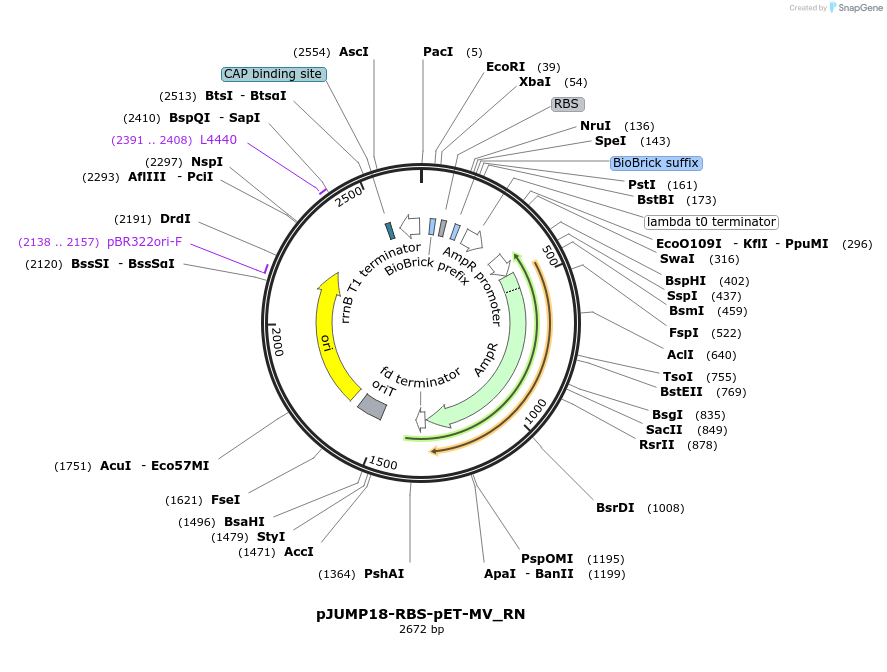
pJUMP18-RBS-pET-MV_RN
Plasmid#127014PurposeBasic Part RN- RBS+N-terminus; B0032m Ribosome Binding Site + Met-Val N-terminus start.DepositorInsertPart RBS-pET-MV_RN
UseSynthetic BiologyExpressionBacterialMutationNoneAvailable SinceJuly 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
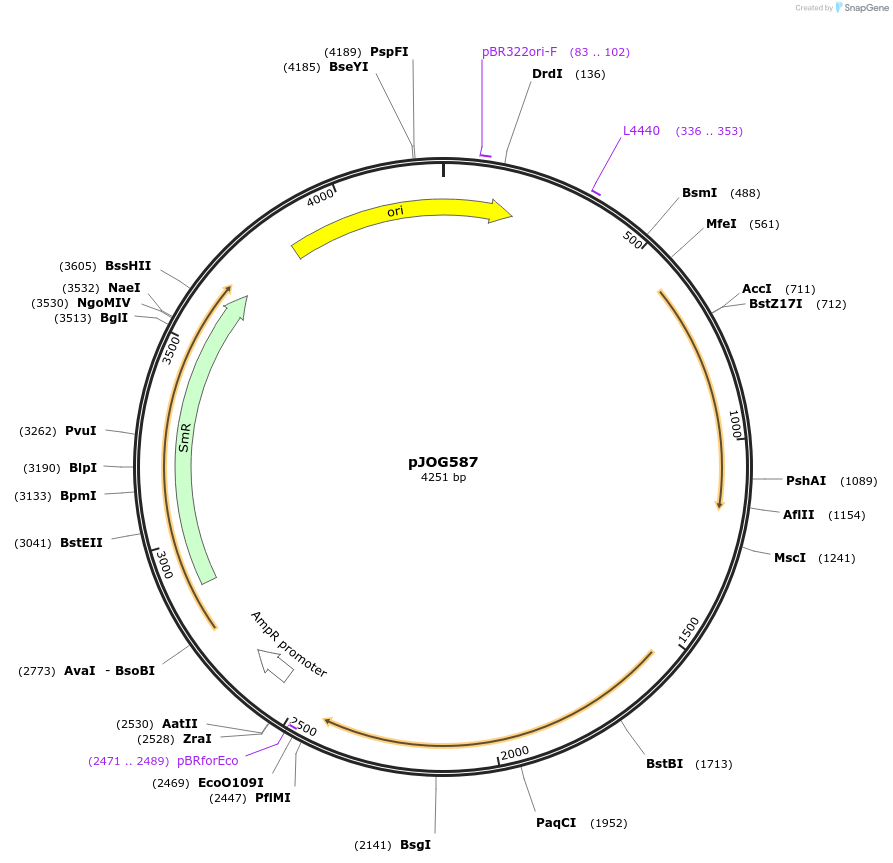
pJOG587
Plasmid#105431PurposeSynthetic biologyDepositorInsertbeta-glucuronidase (2 introns; no ATG-Start codon)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
pHL 802
Plasmid#52991PurposepLtetO-1:OxyS-mCherry, pLlacO-1:ompC::gfpDepositorInsertsOxyS sRNA
ompC
UseSynthetic BiologyTagsGFP and RBS-mCherryExpressionBacterialMutationcontains −81 to +36 bp relative to start codonPromoterpLlacO-1 and pLtetO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL 1148
Plasmid#53006PurposepCon:OxyS, pLlacO-1:sodB::gfpDepositorInsertsOxyS sRNA
sodB
UseSynthetic BiologyTagsGFPExpressionBacterialMutationcontains −56 to +141 bp relative to start codonPromoterpCon and pLlacO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL 1070
Plasmid#53002PurposepCon:DsrA, pLlacO-1:sodB::gfpDepositorInsertsDsrA sRNA
sodB
UseSynthetic BiologyTagsGFPExpressionBacterialMutationcontains −56 to +141 bp relative to start codonPromoterpCon and pLlacO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL 1057
Plasmid#53001PurposepCon:RhyB, pLlacO-1:rpoS::gfpDepositorInsertsRhyB sRNA
rpoS
UseSynthetic BiologyTagsGFPExpressionBacterialMutationcontainsn −150 to +30 bp relative to start codonPromoterpCon and pLlacO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pMSCV-HA-shift-IRES-GFP(KaganE21)
Plasmid#52004PurposeExpresses the human MAVS CDS containing an N-terminal HA tag and a frameshift mutation inserting TA at bp number 254 of the MAVS CDS . And a GFP markerDepositorInsertMAVS-Shift
UseRetroviralTagsHAExpressionMammalianMutationA 2 nucleotide insertion was introduced between t…Available SinceApril 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
pGL4
Plasmid#48744PurposeExpression of luciferase driven by truncated mPer2 promoter region (bases -1128 to +1536 with respect to transcription start site at +1)DepositorInserttruncated mPer2 promoter/enhancer region (-1128 to +1536)
UseLuciferaseMutationtruncated promoter region (see pGL6)Available SinceJan. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
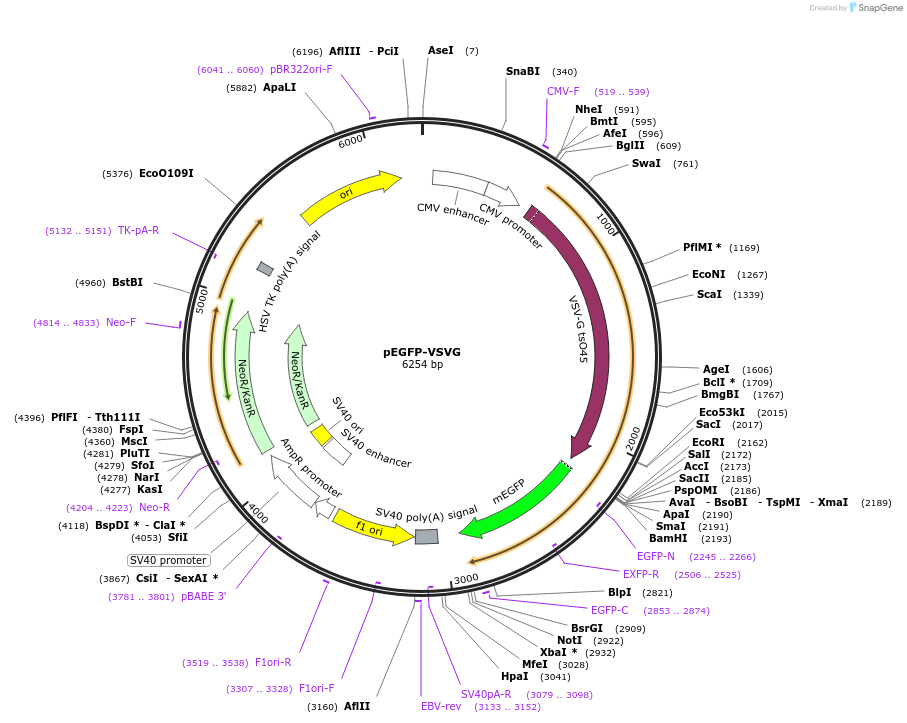
pEGFP-VSVG
Plasmid#11912DepositorInsertVSVG
ExpressionMammalianMutationF64L, S65T, A206K relative the wild type GFP sequ…Available SinceSept. 13, 2006AvailabilityAcademic Institutions and Nonprofits only -
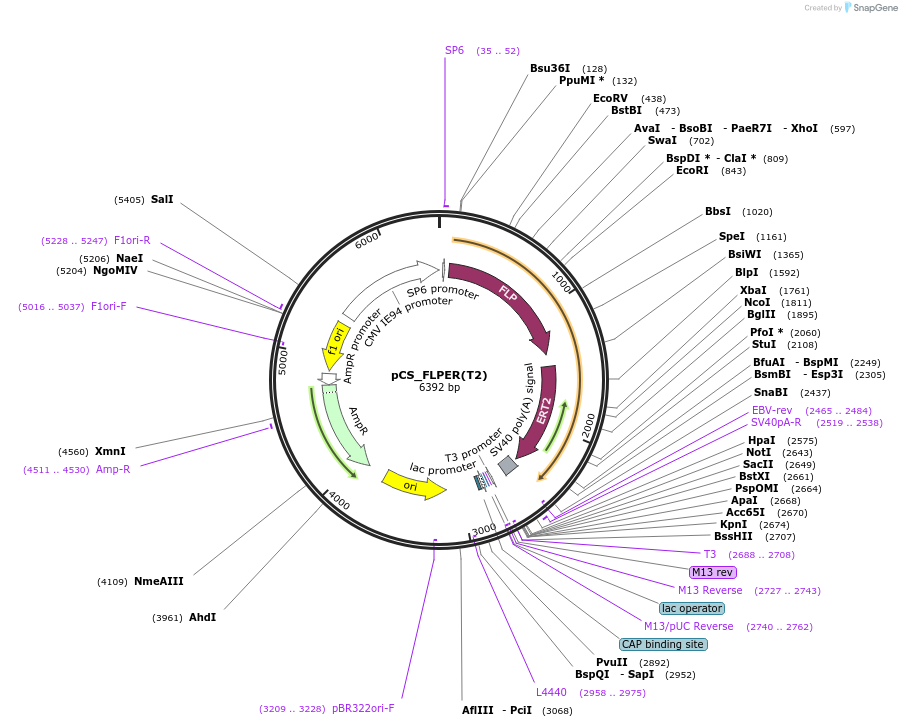
pCS_FLPER(T2)
Plasmid#31303DepositorInsertFLP-ER ligand binding domain (ESR1 Budding Yeast, Human)
ExpressionMammalianMutationfull length FLP linked to ligand binding domain o…Available SinceOct. 17, 2011AvailabilityAcademic Institutions and Nonprofits only -
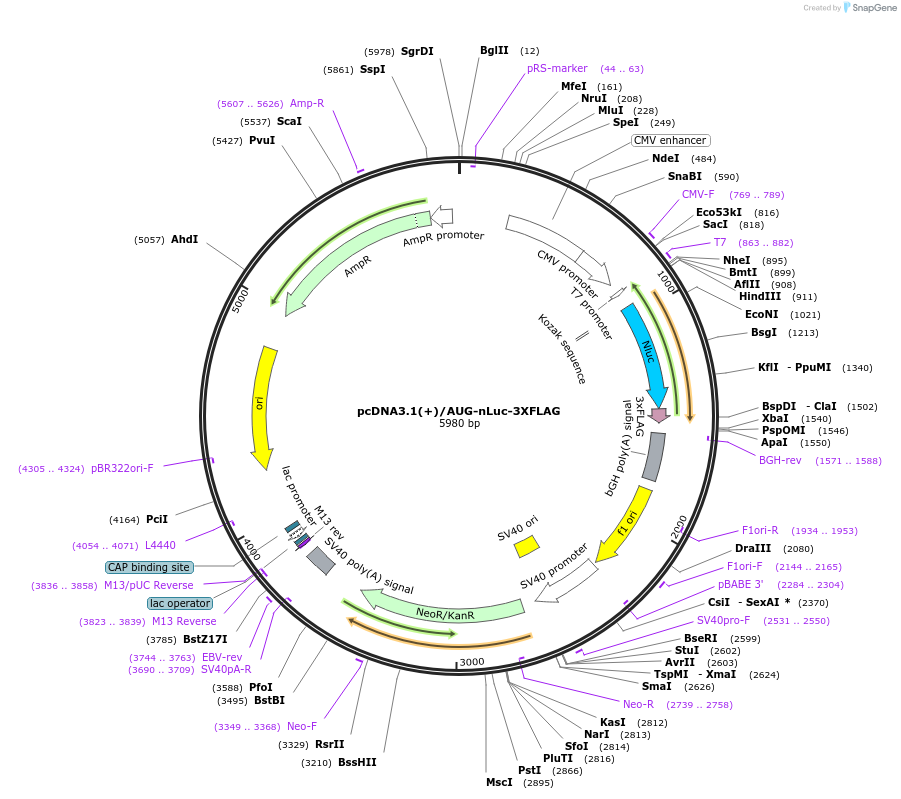
pcDNA3.1(+)/AUG-nLuc-3XFLAG
Plasmid#127299PurposeExpress 3xFLAG tagged nanoLuciferase (nLuc) from AUG start codonDepositorInsertnanoLuciferase
Tags3xFLAGExpressionMammalianPromoterCMVAvailable SinceJune 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
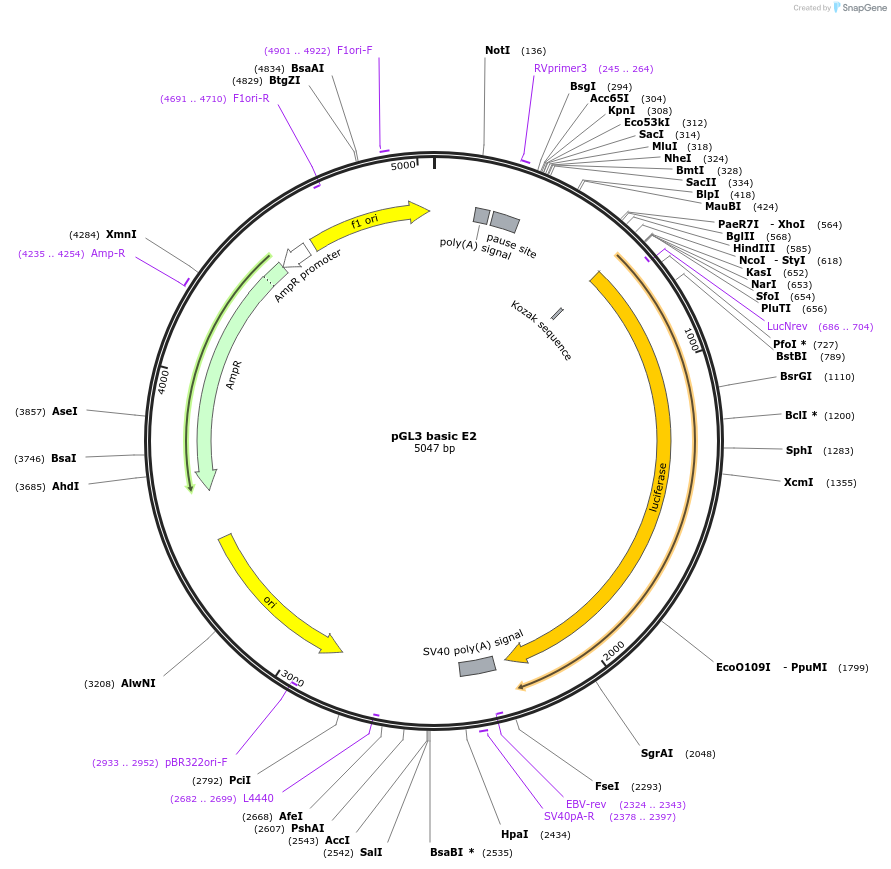
pGL3 basic E2
Plasmid#48747PurposeExpression of luciferase driven by mPer2 promoter fragment containing E-box2 (bases -112 to +98 with respect to transcription start site at +1)DepositorInsertmPer2 promoter/enhancer region containing E-box2 (-112 to +98)
UseLuciferaseAvailable SinceDec. 5, 2013AvailabilityAcademic Institutions and Nonprofits only -
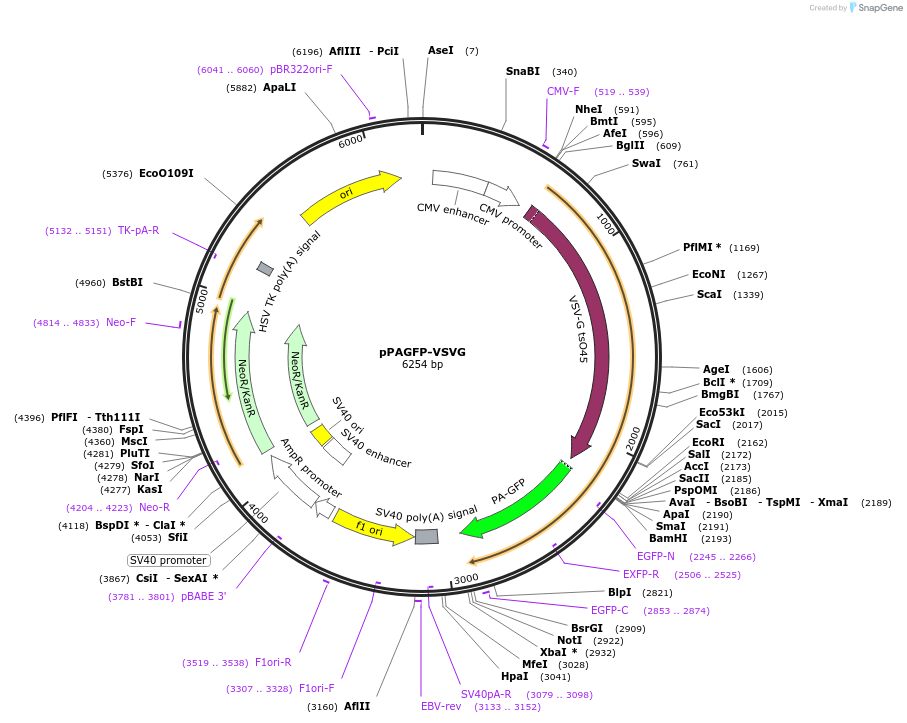
pPAGFP-VSVG
Plasmid#11915DepositorInsertVSVG
ExpressionMammalianMutation(L64F/T65S/V163A/T203H). The PAGFP also contains …Available SinceJuly 3, 2008AvailabilityAcademic Institutions and Nonprofits only -
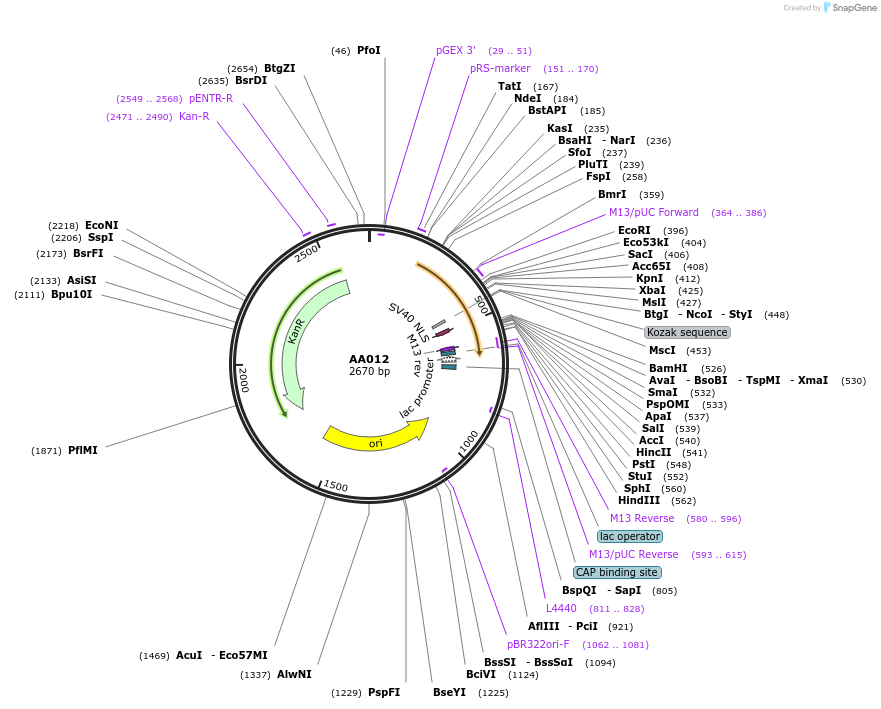
AA012
Plasmid#215974PurposeFragmid fragment: (N' terminus) Bipartite Nuclear Localization SignalDepositorHas ServiceCloning Grade DNAInsertStart; BPNLS_v1.1
UseFragmentAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
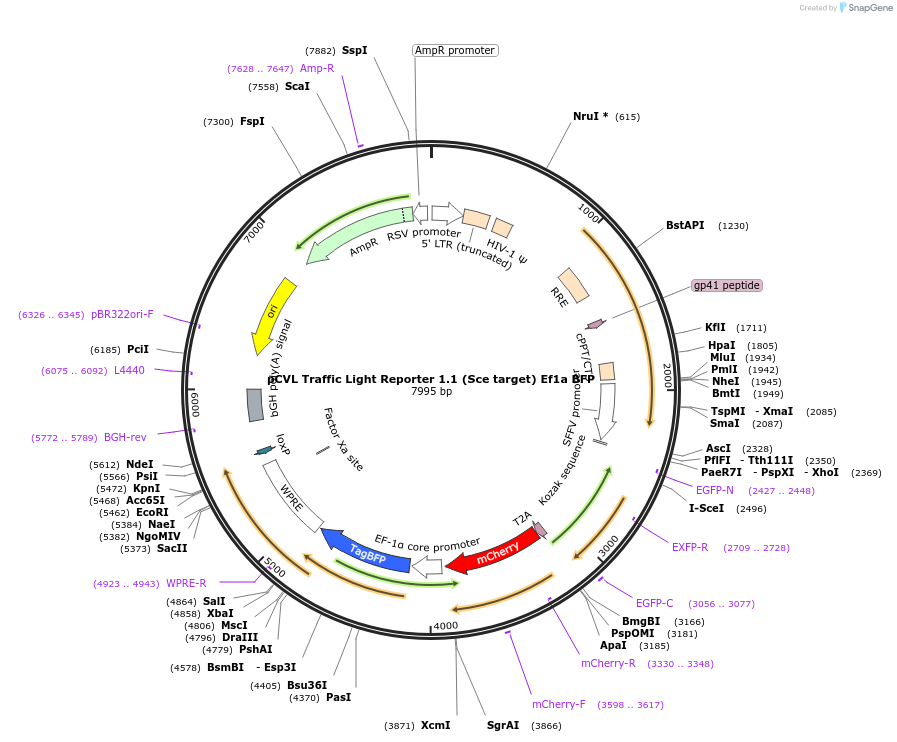
pCVL Traffic Light Reporter 1.1 (Sce target) Ef1a BFP
Plasmid#31481DepositorInsertTraffic Light Reporter 1.1 (Sce target) EF BFP
UseLentiviralMutationmCherry contains M9S and M16L to reduce backgroun…Available SinceAug. 22, 2011AvailabilityAcademic Institutions and Nonprofits only -
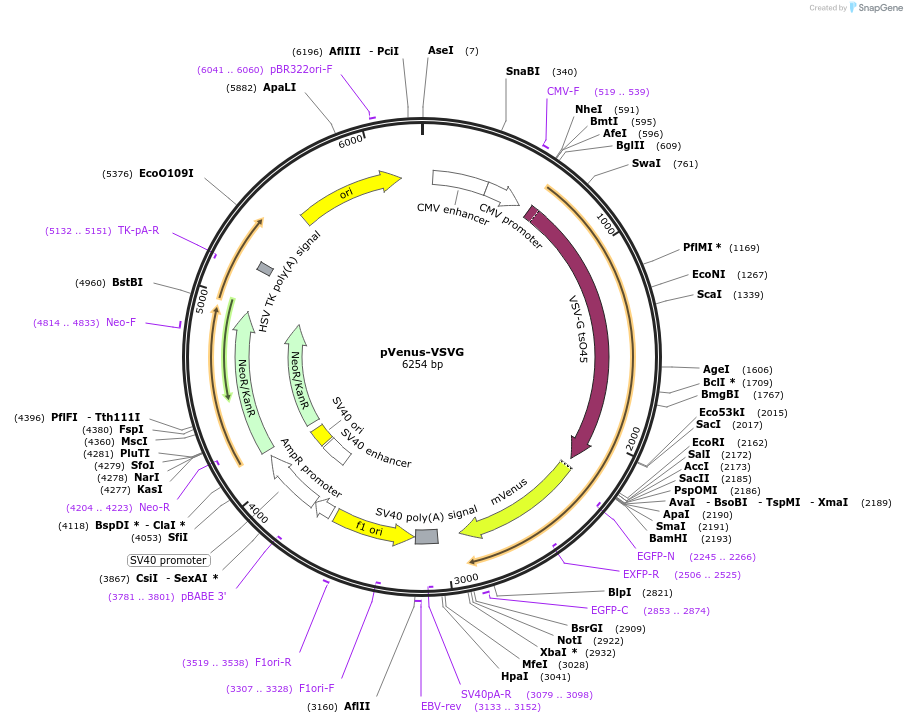
pVenus-VSVG
Plasmid#11914DepositorInsertVSVG
ExpressionMammalianMutationF46L, F64L, M153T, V163A, S175G, T203Y, A206K rel…Available SinceSept. 13, 2006AvailabilityAcademic Institutions and Nonprofits only -
pCMV4-NIK-HA
Plasmid#27554DepositorInsertNIK (MAP3K14 Human)
TagsHAExpressionMammalianMutationNIK starts at amino acid 3; C52R and R139H mutati…PromoterCMVAvailable SinceAug. 17, 2011AvailabilityAcademic Institutions and Nonprofits only -
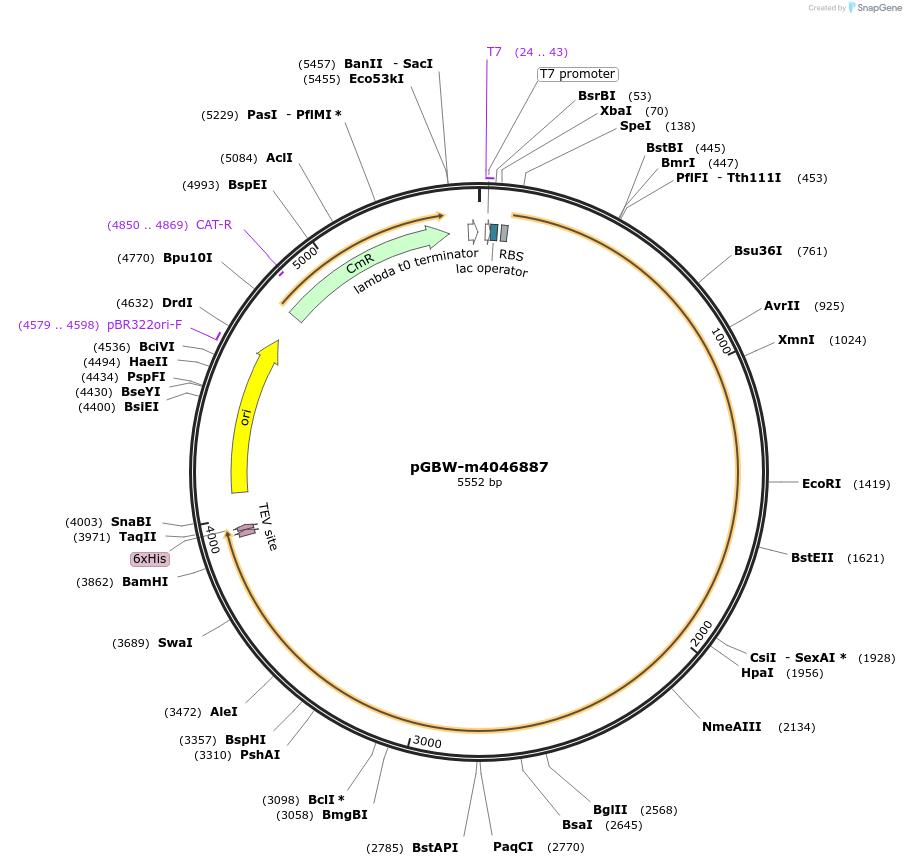
pGBW-m4046887
Plasmid#145730PurposeBacterial Expression plasmid for SARS-CoV-2 surface glycoprotein (Spike protein)DepositorInsertSARS-CoV-2 S (surface glycoprotein) (S Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;6xHISExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 28, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pHL 1405
Plasmid#53010PurposepLtetO-1:RhyB, pCon:DsrA, pLtetO-1:hfq, pLlacO-1:sodB::gfpDepositorInsertsRhyB sRNA
DsrA sRNA
hfq
sodB
UseSynthetic BiologyTagsgfpExpressionBacterialMutationcontains -56 to +141 relative to start codonPromoterpCon, pLlacO-1, and pLtetO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
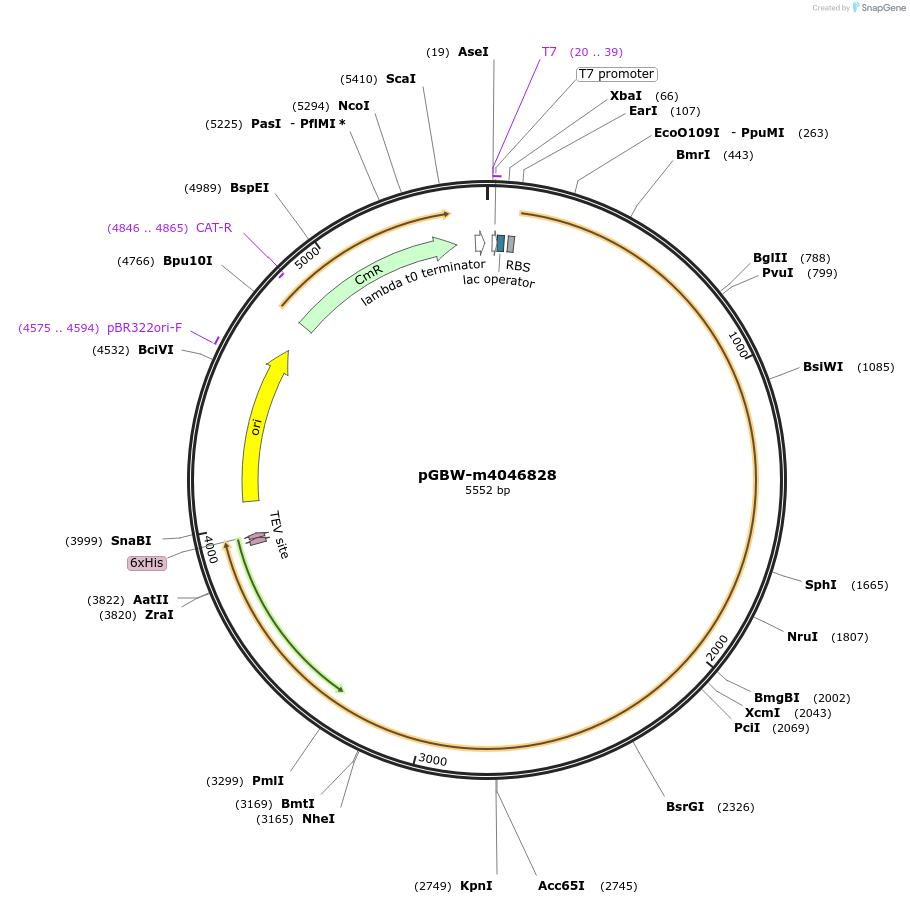
pGBW-m4046828
Plasmid#145742PurposeBacterial Expression plasmid for SARS-CoV-2 surface glycoprotein (Spike protein)DepositorInsertSARS-CoV-2 S (surface glycoprotein) (S Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;6xHISExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pAAV-nef-CIAO2-TVA-mCherry
Plasmid#149297PurposeCre-dependent expression of TVA:mCherry fusion protein for rabies tracing.DepositorInsertTVA:mCherry
UseAAVMutationStart codon removed.Available SinceFeb. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL2-Skp2 promoter-luciferase
Plasmid#81119PurposeReporter activity of SKP2 gene promoterDepositorInsertSKP2 (Skp2 Mouse)
UseLuciferaseTagsluciferaseMutationcontains the region from 1_kb upstream of transcr…PromoterSKP2Available SinceSept. 27, 2016AvailabilityAcademic Institutions and Nonprofits only -
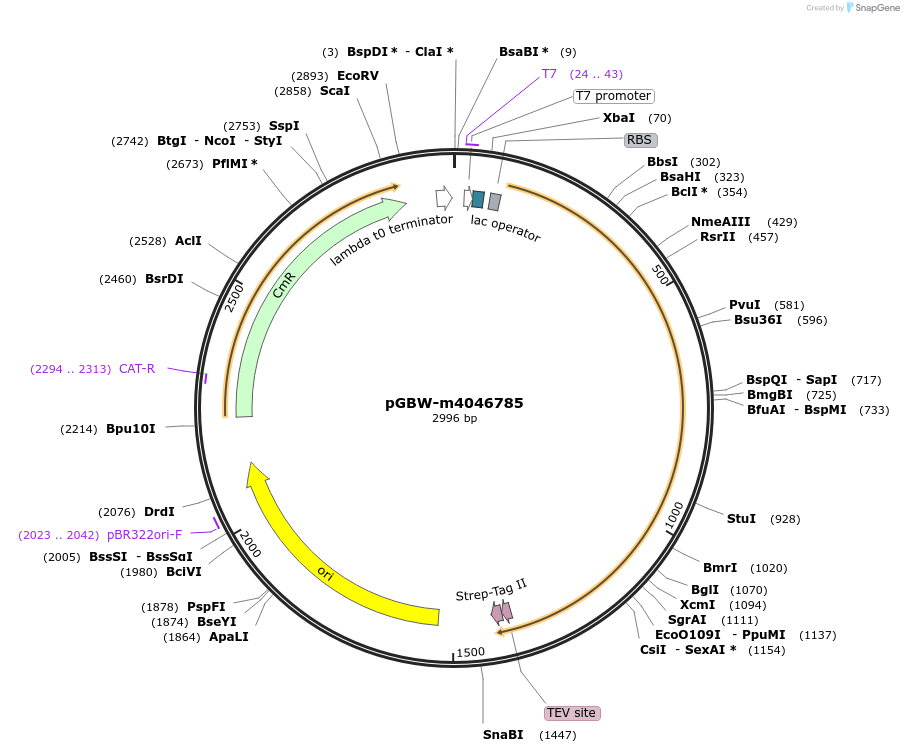
pGBW-m4046785
Plasmid#145684PurposeBacterial Expression plasmid for SARS-CoV-2 N (nucleocapsid)DepositorInsertSARS-CoV-2 N (nucleocapsid) (N Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGL3
Plasmid#48743PurposeExpression of luciferase driven by truncated mPer2 promoter region (bases -1128 to +1060 with respect to transcription start site at +1)DepositorInserttruncated mPer2 promoter/enhancer region (-1128 to +1060)
UseLuciferaseMutationtruncated promoter region (see pGL6)Available SinceDec. 19, 2013AvailabilityAcademic Institutions and Nonprofits only -
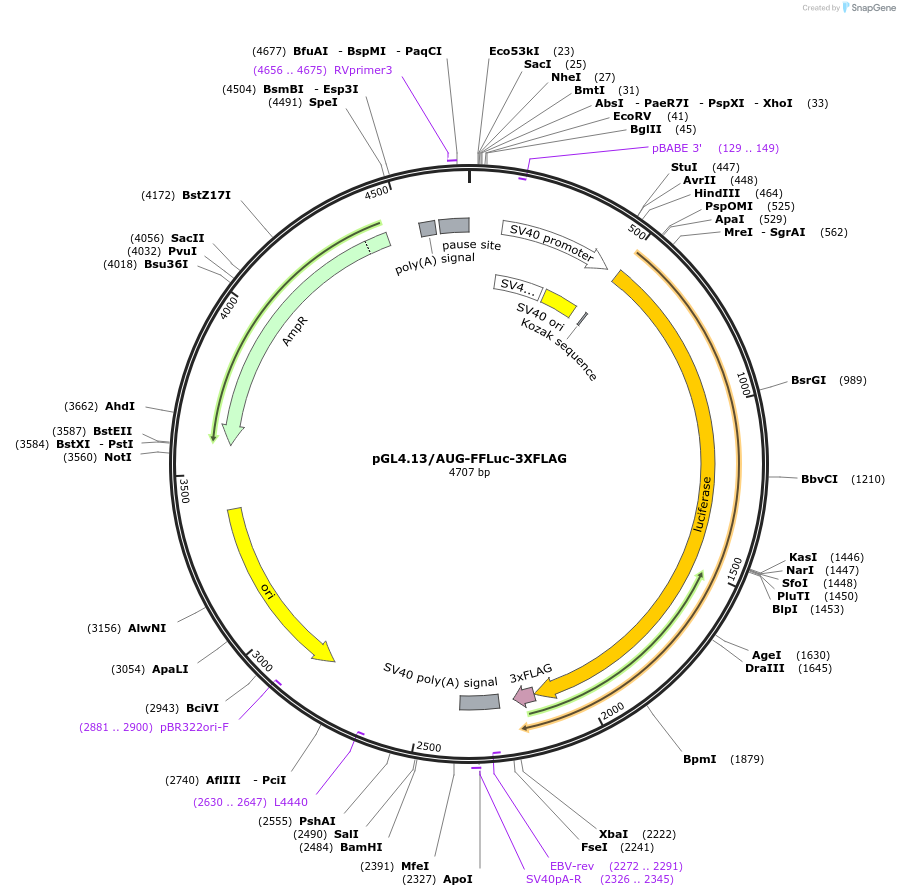
pGL4.13/AUG-FFLuc-3XFLAG
Plasmid#127333PurposeExpress 3xFLAG tagged firefly luciferase (FFLuc) from AUG start codonDepositorInsertFirefly luciferase
Tags3xFLAGExpressionMammalianPromoterCV40Available SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
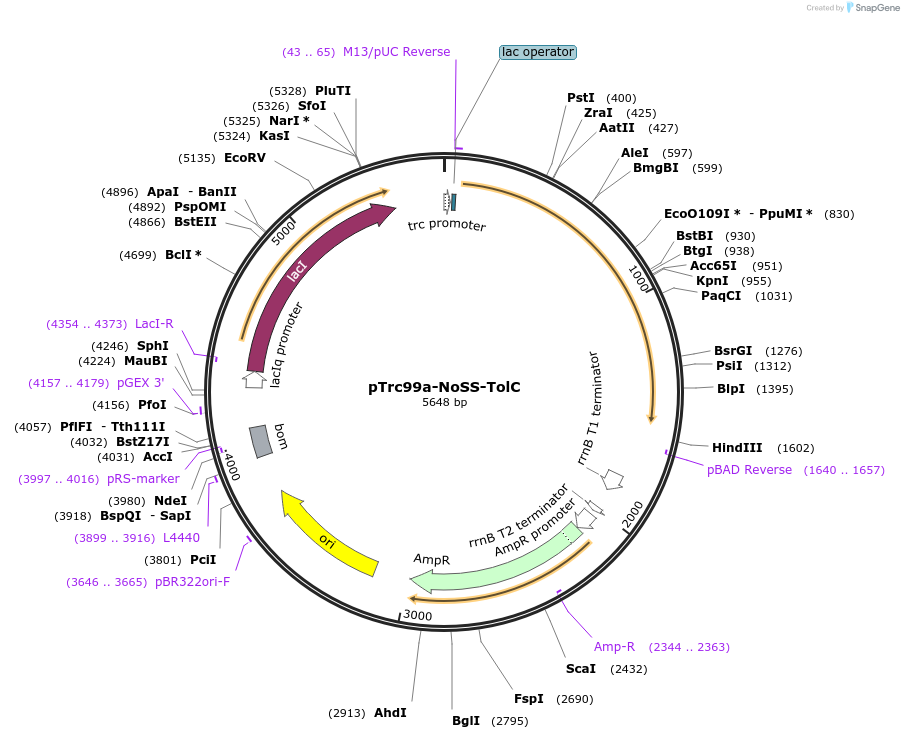
pTrc99a-NoSS-TolC
Plasmid#155179PurposeExpresses signal sequence-less TolC as inclusion bodiesDepositorInsertTolC (tolC )
ExpressionBacterialMutationdeleted residues 1-20, residue 21 mutated to Met …PromotertrcAvailable SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
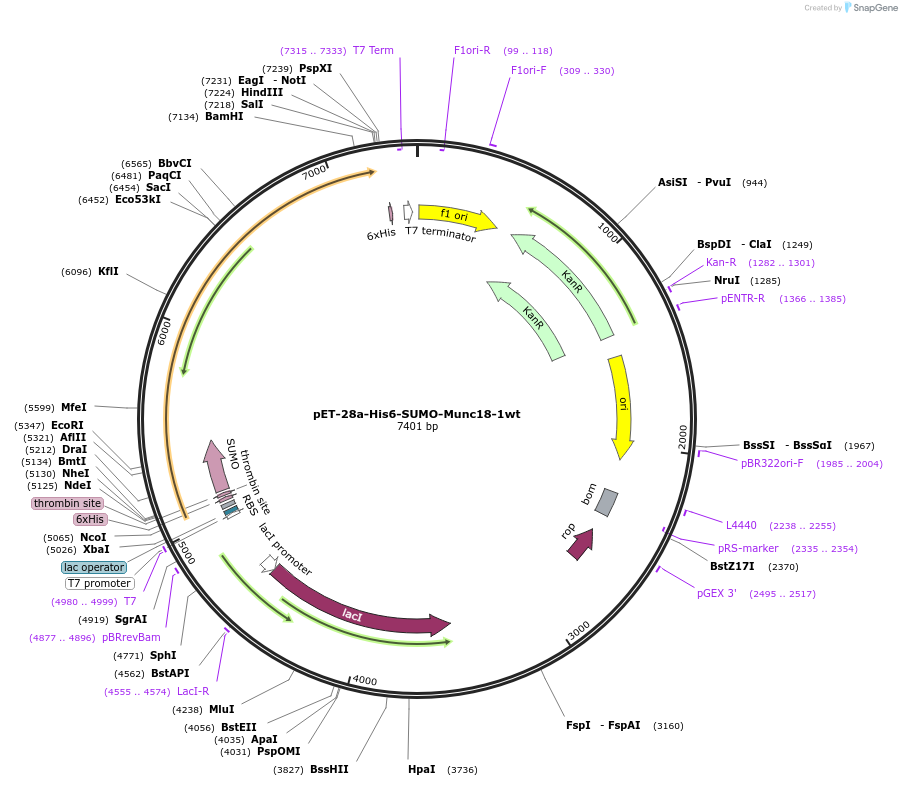
pET-28a-His6-SUMO-Munc18-1wt
Plasmid#135550PurposeExpress rat Munc18-1 in E. coli BL21, purified via HisPur Ni-NTA ResinDepositorInsertMunc18-1wt
Tags6xHis tag, thrombin recognition and cleavage site…ExpressionBacterialMutationNo ATG start codonPromoterT7 promoterAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
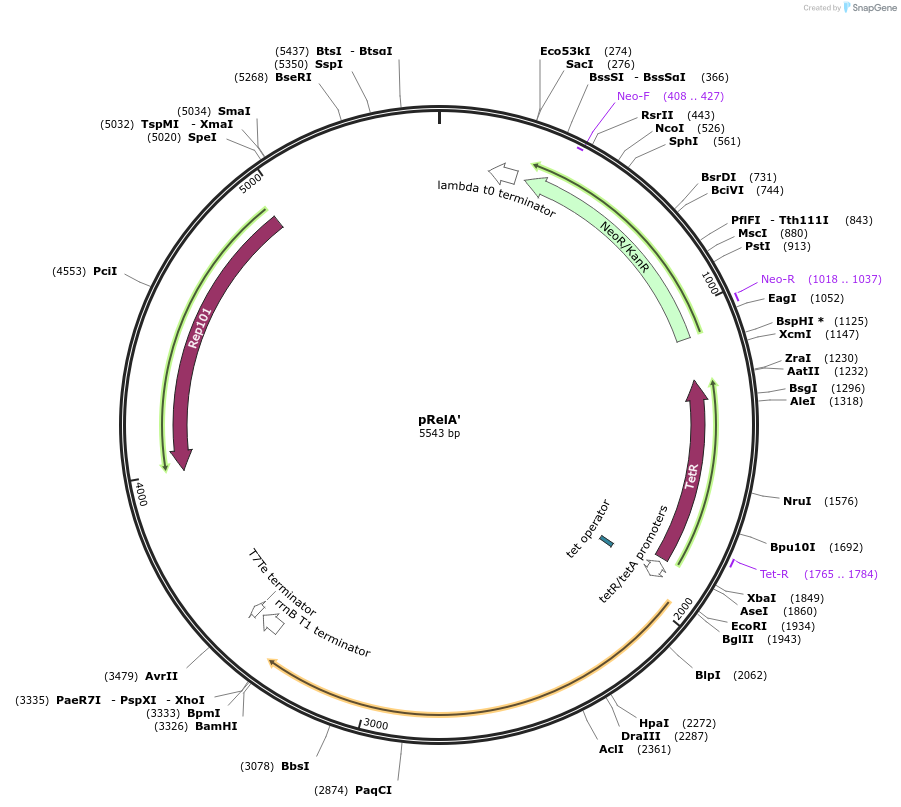
pRelA'
Plasmid#175595PurposeTet inducible, KAN Resistant, non labelled synthesis domain of the E. coli relA gene on a low copy backbone. Used to induce controllable synthesis of ppGpp in E. coli.DepositorInsertCatalytic domain of E.coli relA gene.
ExpressionBacterialMutationOnly the catalytic domain of the enzyme is used (…Available SinceNov. 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
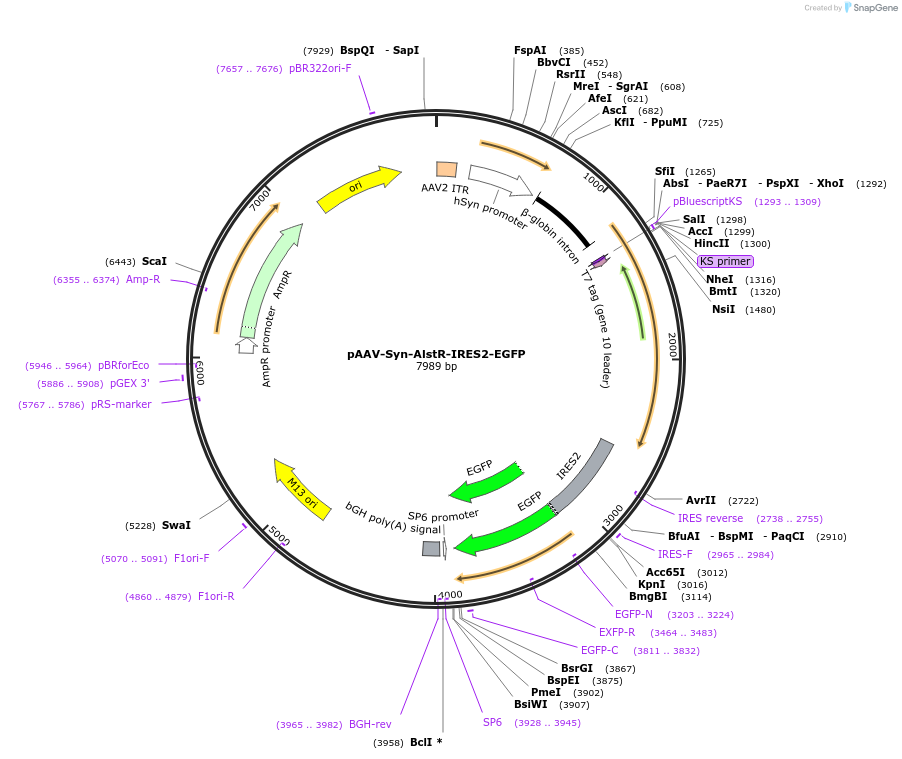
pAAV-Syn-AlstR-IRES2-EGFP
Plasmid#14895DepositorAvailable SinceMay 11, 2007AvailabilityAcademic Institutions and Nonprofits only -
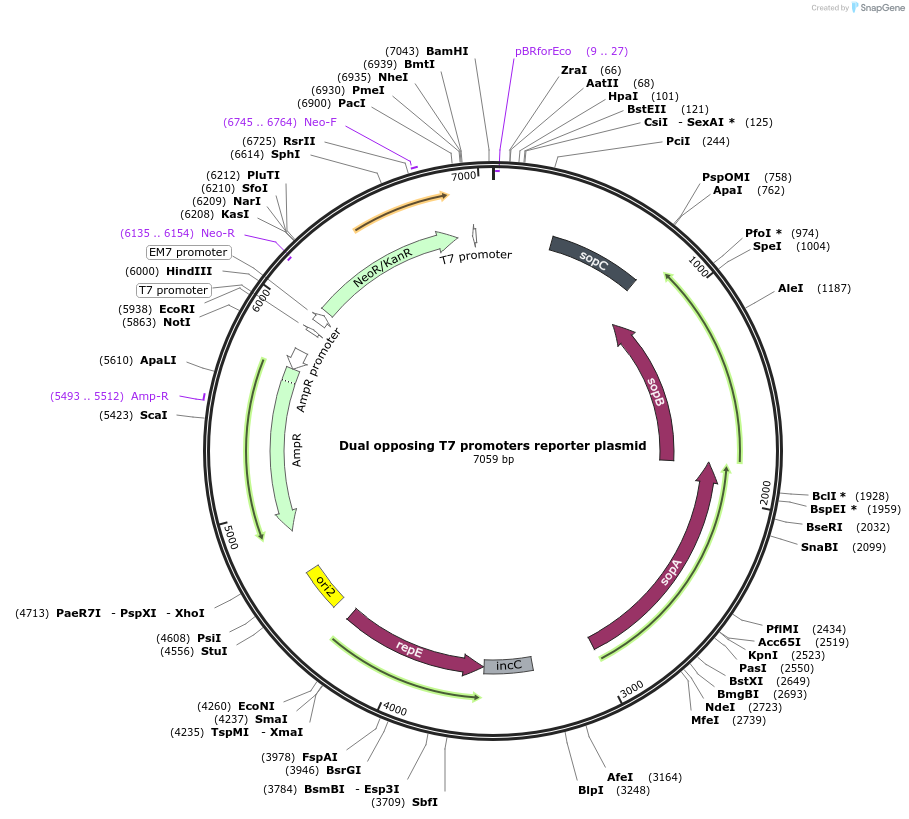
Dual opposing T7 promoters reporter plasmid
Plasmid#156457PurposeA mutational reporter plasmid that can gain Kanamycin resistance upon mutation. Contains two T7 promoters flanking the kanamycin resistance gene.DepositorInsertNeoR/KanR
ExpressionBacterialMutationATG start codon mutated to ACGPromoterEm7Available SinceOct. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
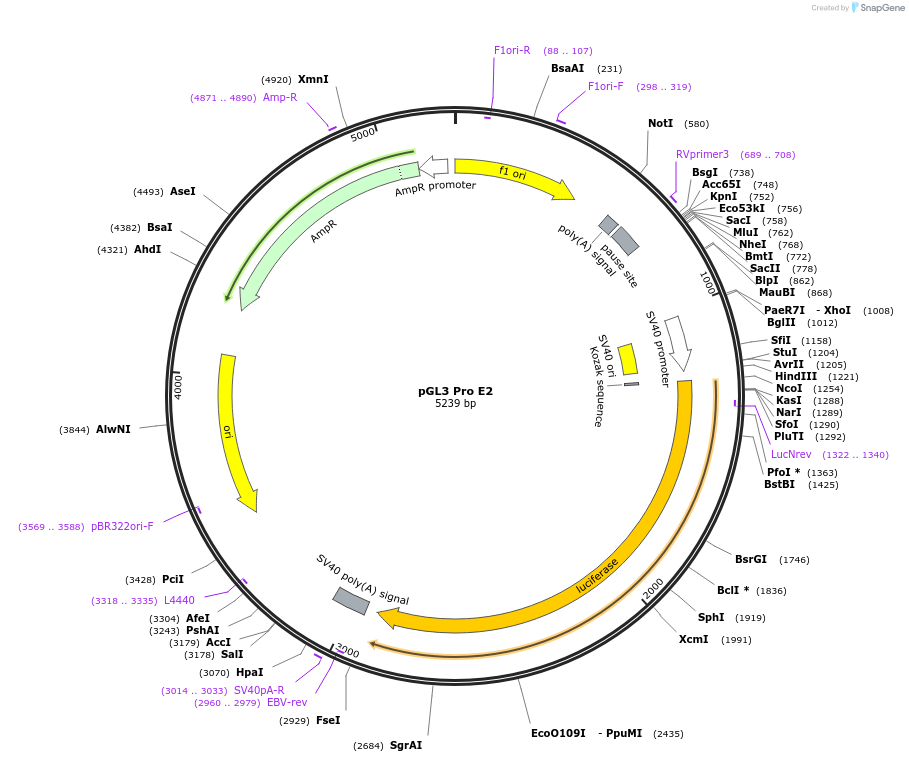
pGL3 Pro E2
Plasmid#48749PurposeExpression of luciferase driven by mPer2 promoter fragment containing E-box2 (bases -112 to +98 with respect to transcription start site at +1) and SV40 promoter (backbone)DepositorInsertmPer2 promoter/enhancer region containing E-box2 (-112 to +98)
UseLuciferaseAvailable SinceDec. 5, 2013AvailabilityAcademic Institutions and Nonprofits only -
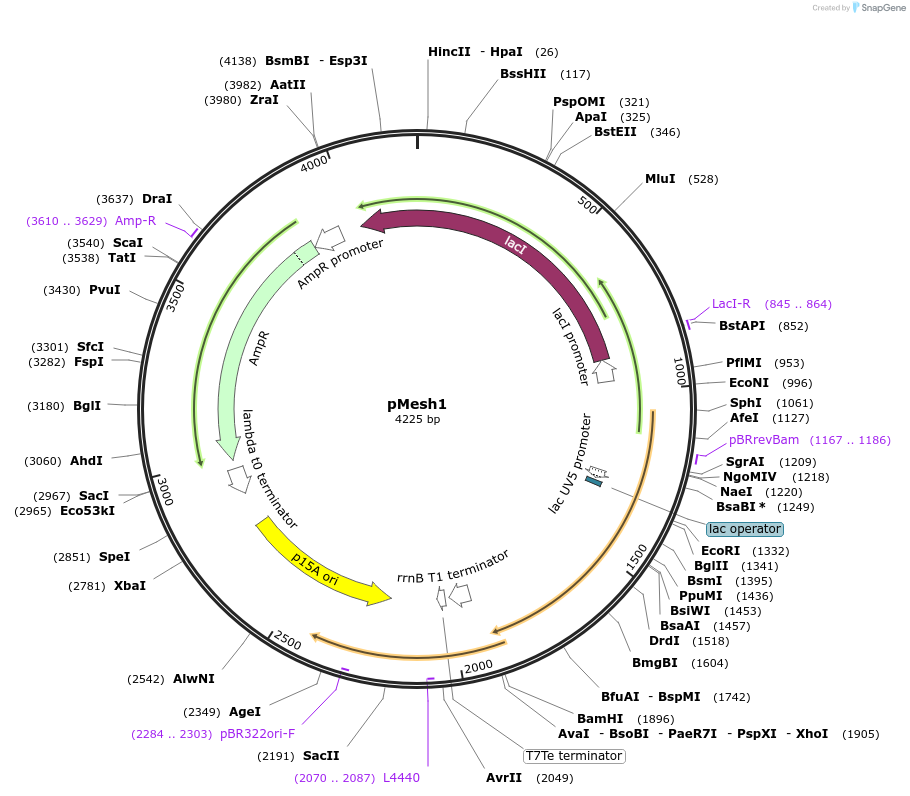
pMesh1
Plasmid#175594PurposeLac inducible, AMP Resistant, non labelled gene mesh1 from D. melanogaster on a low copy backbone. Used to induce controllable hydrolysis of ppGpp in E. coli.DepositorInsertMesh1 (Mesh1 Fly)
ExpressionBacterialMutationCodon optimized for translation in E.coli. AGGAGG…Available SinceNov. 19, 2021AvailabilityAcademic Institutions and Nonprofits only