We narrowed to 6,964 results for: GFP expression plasmids
-
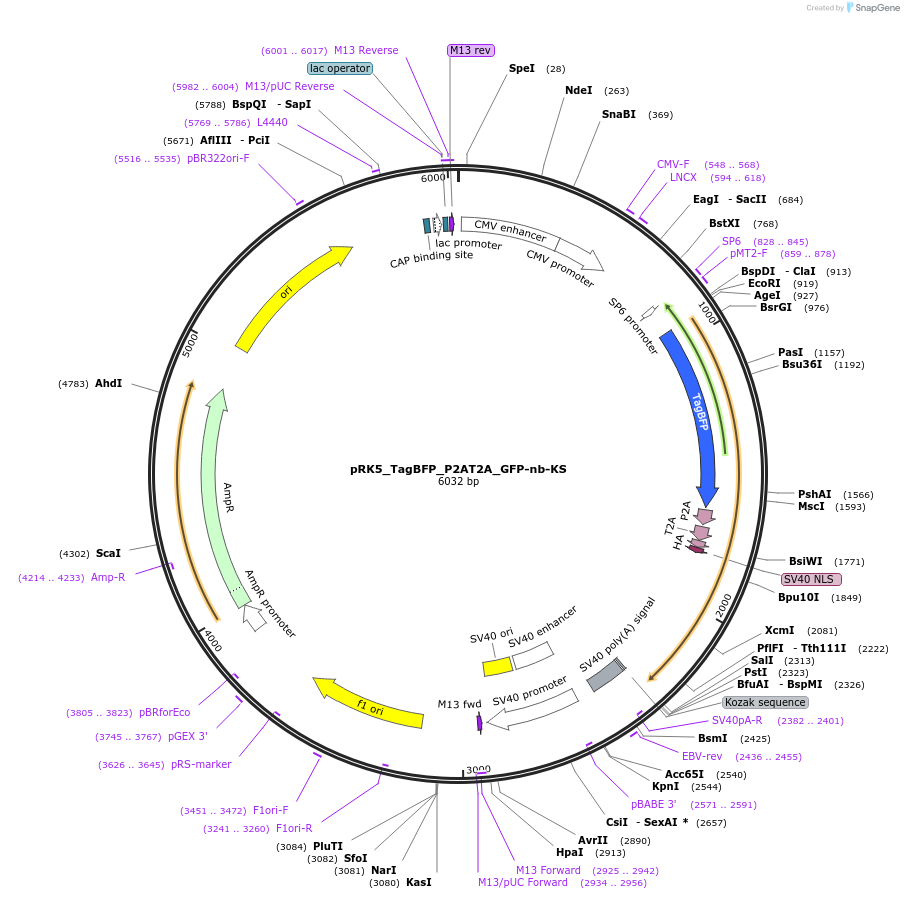
Plasmid#238274PurposeFor overexpression of TagBFP_P2AT2A_GFP-nb-KSDepositorInsertTagBFP_P2AT2A_GFP-nb-KS
ExpressionMammalianAvailable SinceJune 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
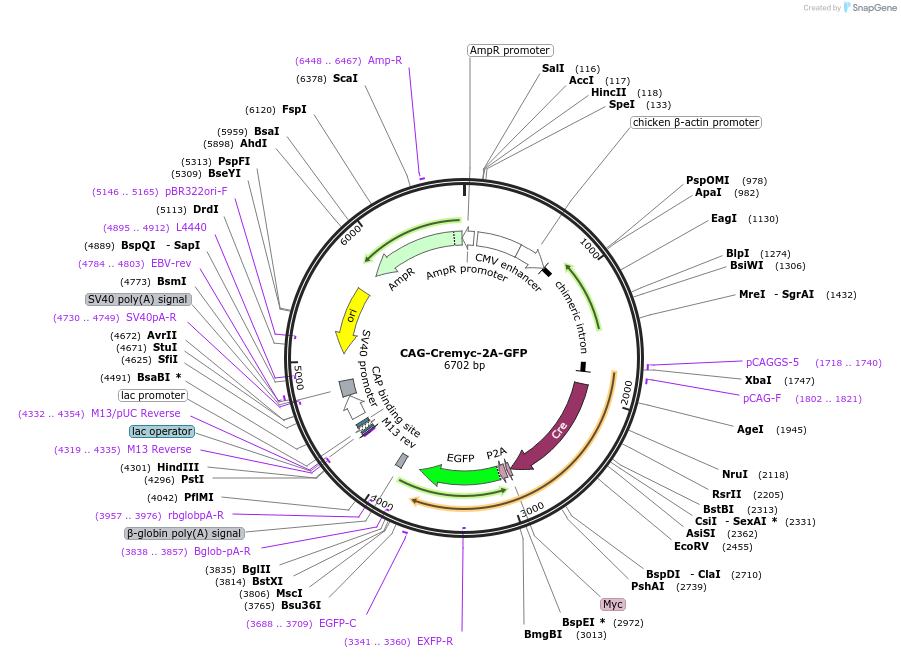
CAG-Cremyc-2A-GFP
Plasmid#116879PurposeExpress Cre enzyme in mammalian cells with floxed genes to achieve somatic knockout. GFP is simultaneously expressed for convenient tracking the transfected cells.DepositorInsertCremyc-2A-GFP
ExpressionMammalianAvailable SinceMay 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
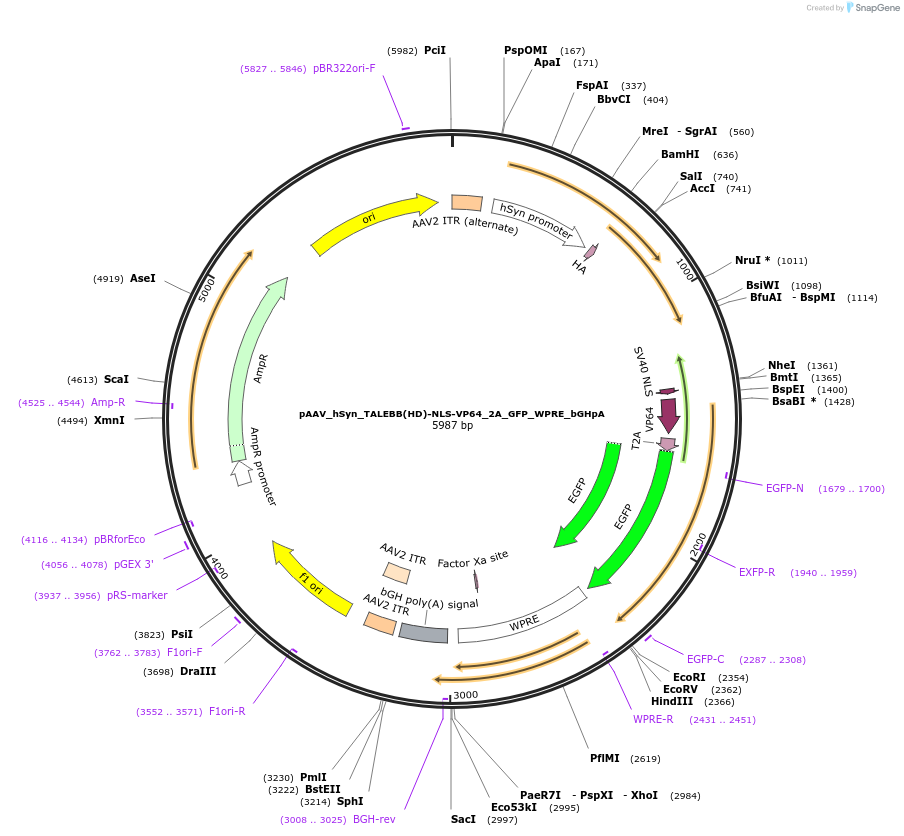
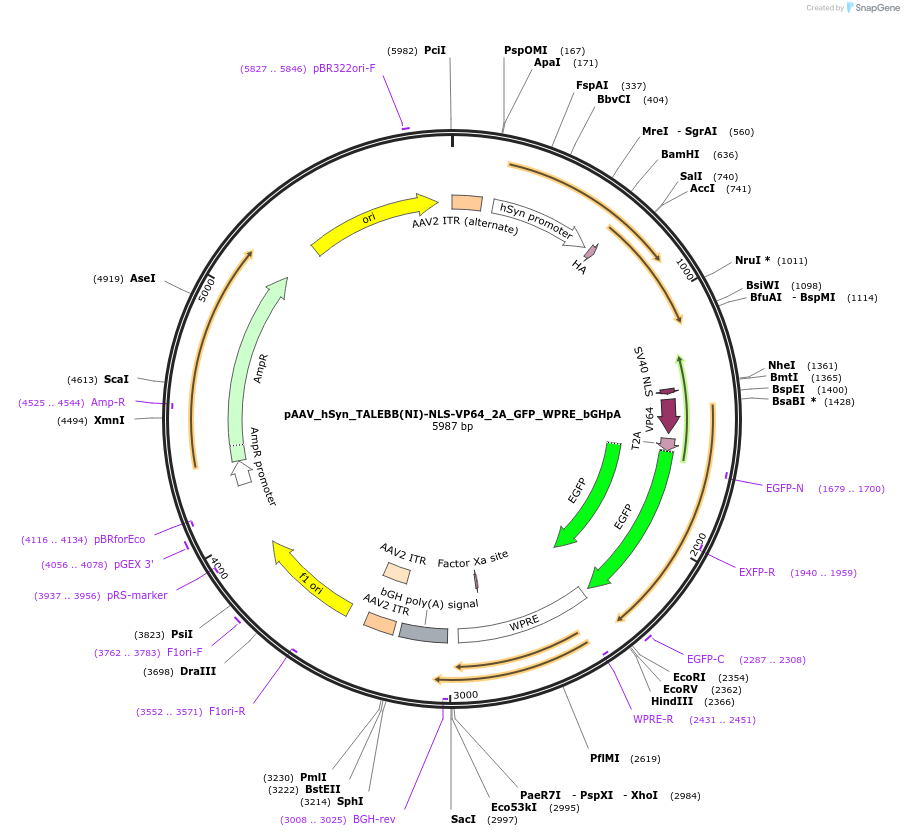
pAAV_hSyn_TALEBB(HD)-NLS-VP64_2A_GFP_WPRE_bGHpA
Plasmid#47446PurposeAAV TALE synthesis backbone with HD half repeat; fused to nuclear localization sequence linker and VP64 transcriptional activator effector domain. Synapsin promoter for neuronal expression.DepositorInsertTALE BB(N136_0.5HD_C63)-VP64
UseAAV; TaleTags2A_GFPExpressionMammalianPromoterhSyn human synapsin (mammalian neuronal expressio…Available SinceOct. 15, 2013AvailabilityAcademic Institutions and Nonprofits only -
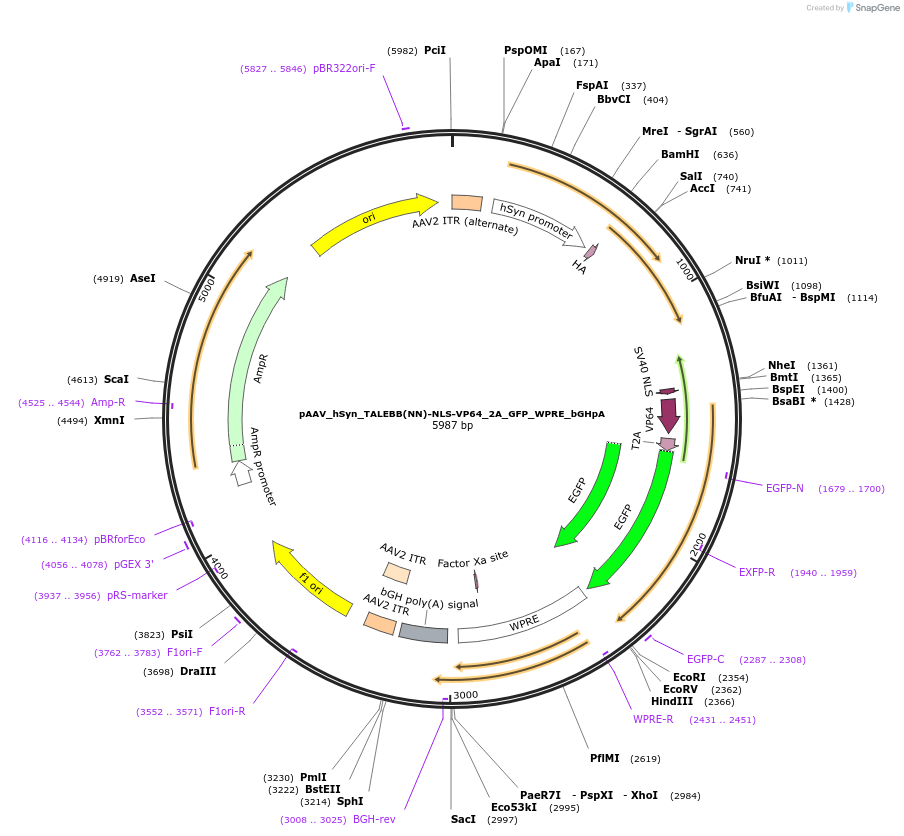
pAAV_hSyn_TALEBB(NN)-NLS-VP64_2A_GFP_WPRE_bGHpA
Plasmid#47445PurposeAAV TALE synthesis backbone with NN half repeat; fused to nuclear localization sequence linker and VP64 transcriptional activator effector domain. Synapsin promoter for neuronal expression.DepositorInsertTALE BB(N136_0.5NN_C63)-VP64
UseAAV; TaleTags2A_GFPExpressionMammalianPromoterhSyn human synapsin (mammalian neuronal expressio…Available SinceOct. 22, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAAV_hSyn_TALEBB(NI)-NLS-VP64_2A_GFP_WPRE_bGHpA
Plasmid#47448PurposeAAV TALE synthesis backbone with NI half repeat; fused to nuclear localization sequence linker and VP64 transcriptional activator effector domain. Synapsin promoter for neuronal expression.DepositorInsertTALE BB(N136_0.5NI_C63)-VP64
UseAAV; TaleTags2A_GFPExpressionMammalianPromoterhSyn human synapsin (mammalian neuronal expressio…Available SinceOct. 15, 2013AvailabilityAcademic Institutions and Nonprofits only -
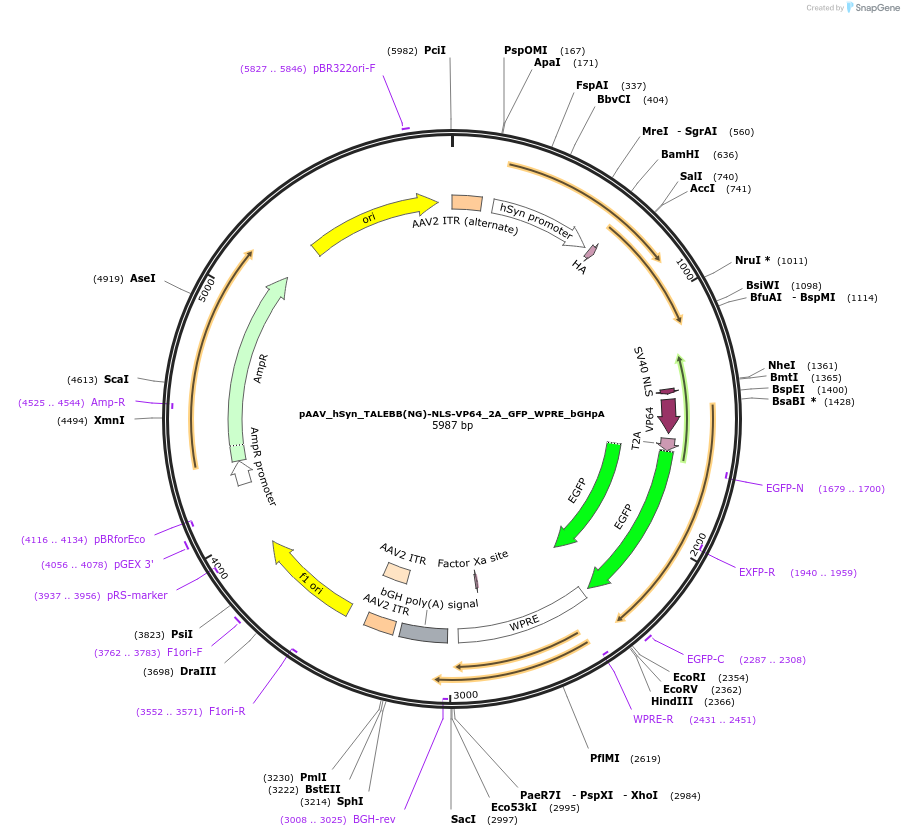
pAAV_hSyn_TALEBB(NG)-NLS-VP64_2A_GFP_WPRE_bGHpA
Plasmid#47447PurposeAAV TALE synthesis backbone with NG half repeat; fused to nuclear localization sequence linker and VP64 transcriptional activator effector domain. Synapsin promoter for neuronal expression.DepositorInsertTALE BB(N136_0.5NG_C63)-VP64
UseAAV; TaleTags2A_GFPExpressionMammalianPromoterhSyn human synapsin (mammalian neuronal expressio…Available SinceOct. 22, 2013AvailabilityAcademic Institutions and Nonprofits only -
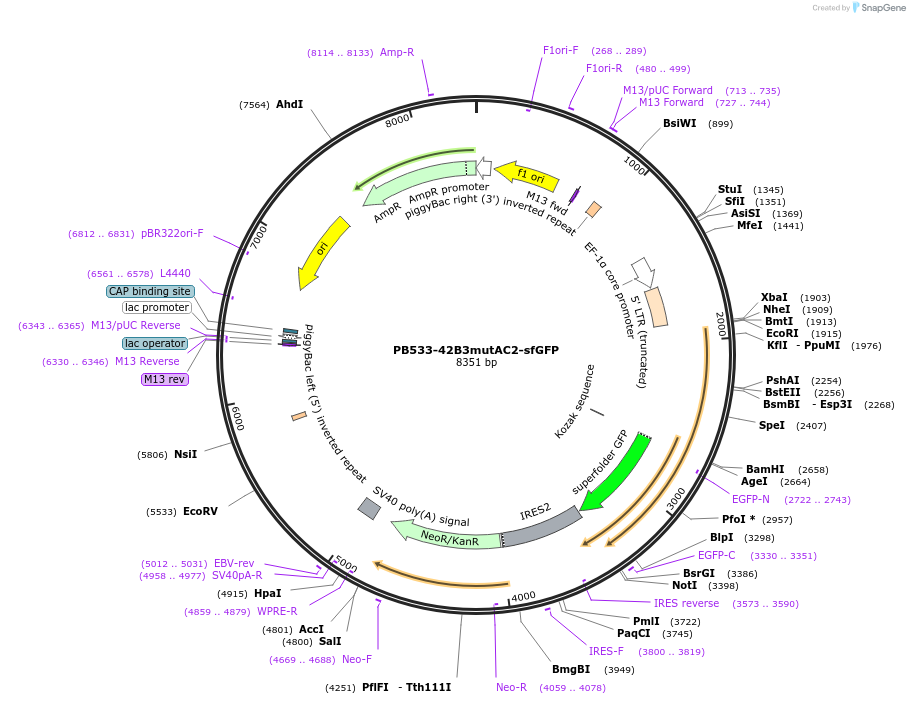
PB533-42B3mutAC2-sfGFP
Plasmid#186777PurposeRNAP2 Ser2ph-mintbody [42B3(R78K/A80T/M95V)-scFv] expression vector for mammalian cellsDepositorInsert42B3mutAC2-scFv
TagssfGFPExpressionMammalianAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
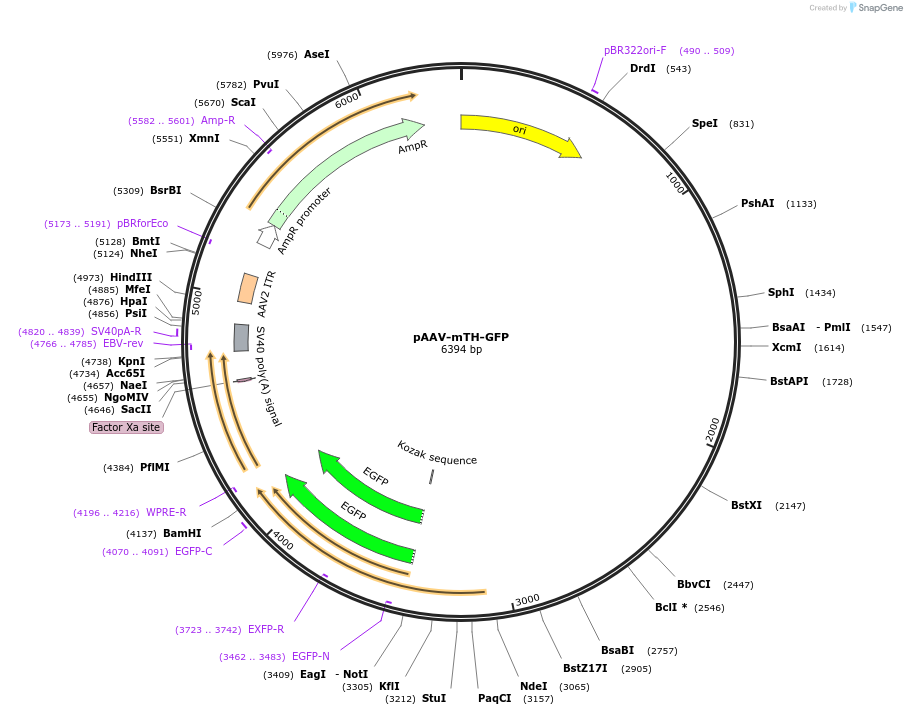
pAAV-mTH-GFP
Plasmid#99128PurposeAn AAV genome that expresses GFP from the mouse TH promoterDepositorInsertGFP
UseAAVExpressionMammalianPromotermTHAvailable SinceAug. 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
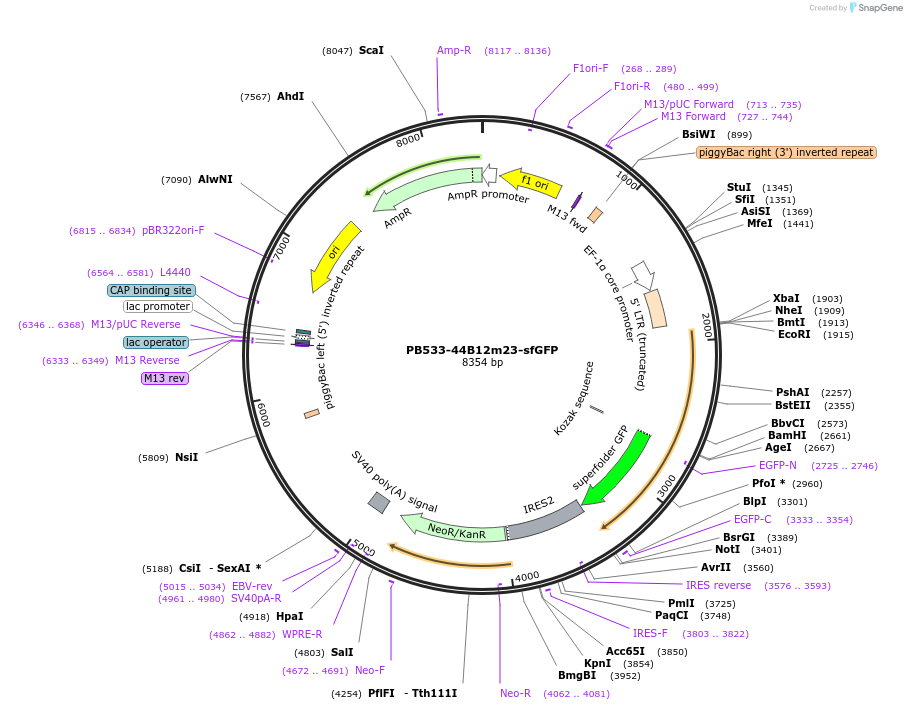
PB533-44B12m23-sfGFP
Plasmid#186778PurposeRNAP2 Ser5ph-mintbody expression vector for mammalian cellsDepositorInsert44B12m23-scFv
TagssfGFPExpressionMammalianAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
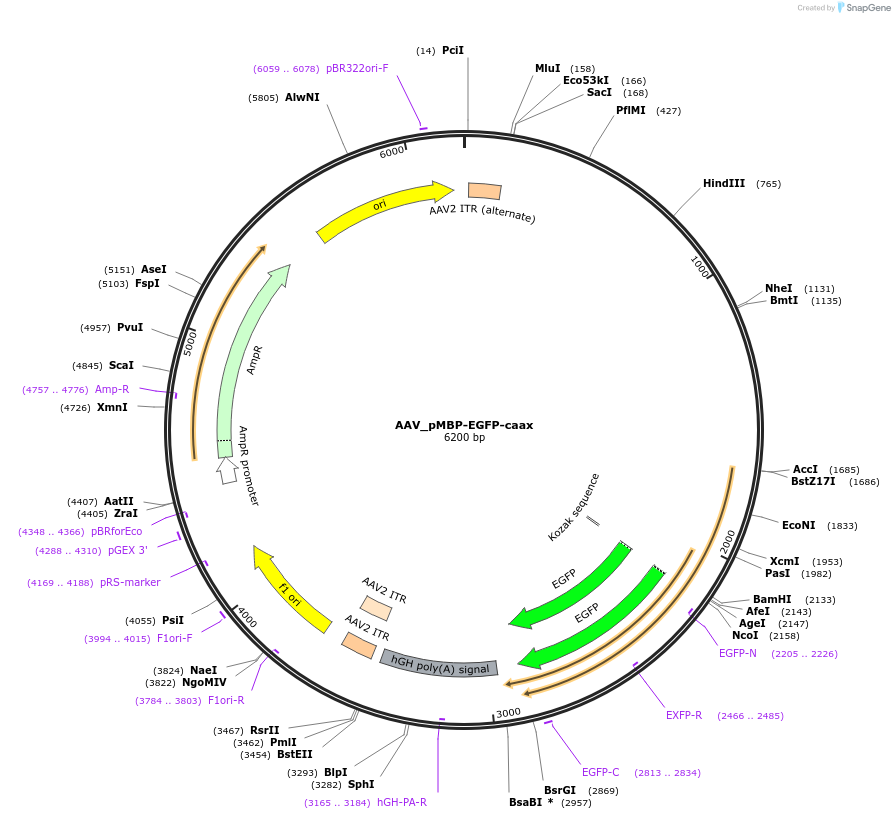
AAV_pMBP-EGFP-caax
Plasmid#190155PurposeExpresses membrane-targeted EGFP in oligodendrocytes; AAV vectorDepositorInsertEGFP-caax
UseAAVExpressionMammalianPromoterMBPAvailable SinceDec. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
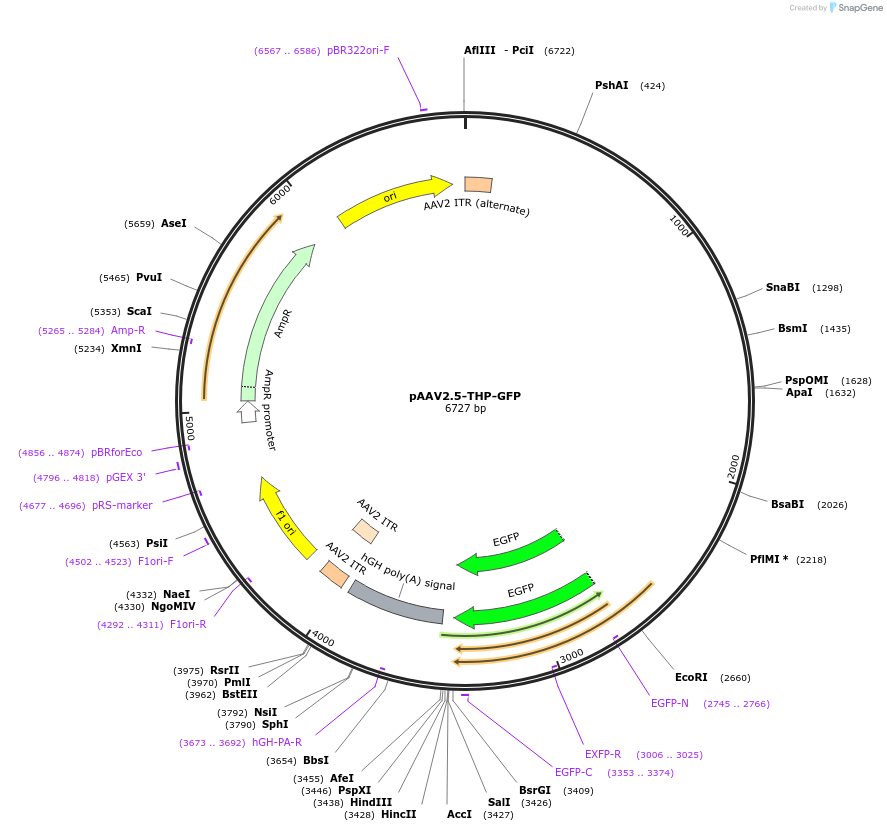
pAAV2.5-THP-GFP
Plasmid#80336PurposeFluorescent Reporter for Dopaminergic Neuron DifferentiationDepositorAvailable SinceOct. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
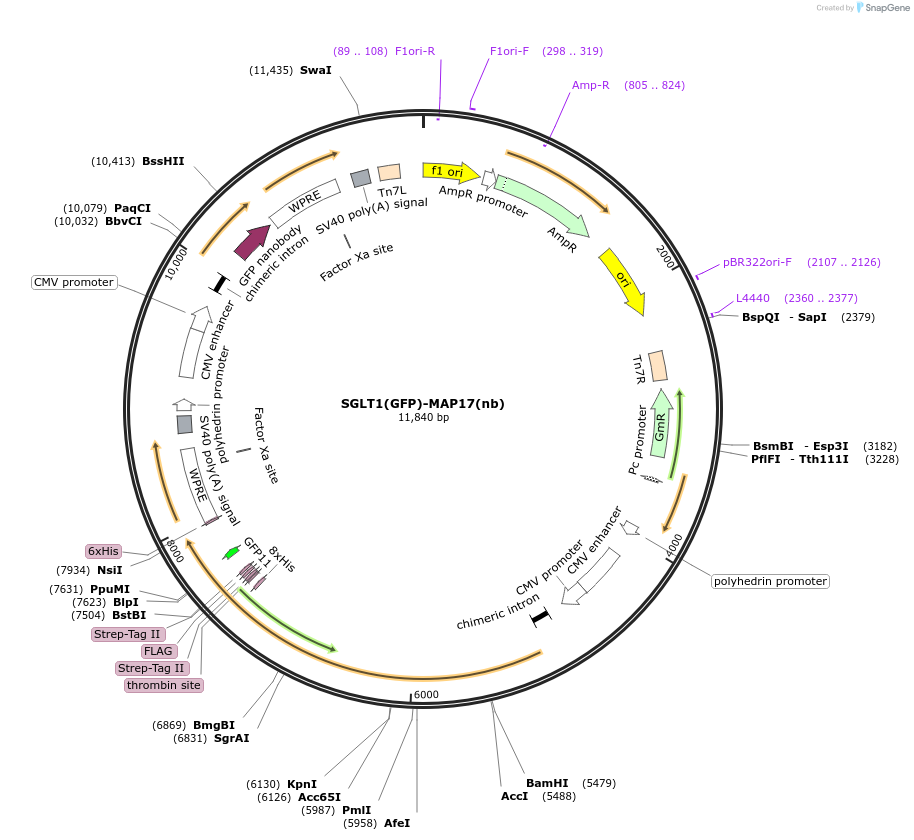
SGLT1(GFP)-MAP17(nb)
Plasmid#216210Purposefor expression of human GFP-tagged SGLT1-MAP17 nanobody complex in BacMam systemDepositorTagsGFPExpressionMammalianAvailable SinceMay 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
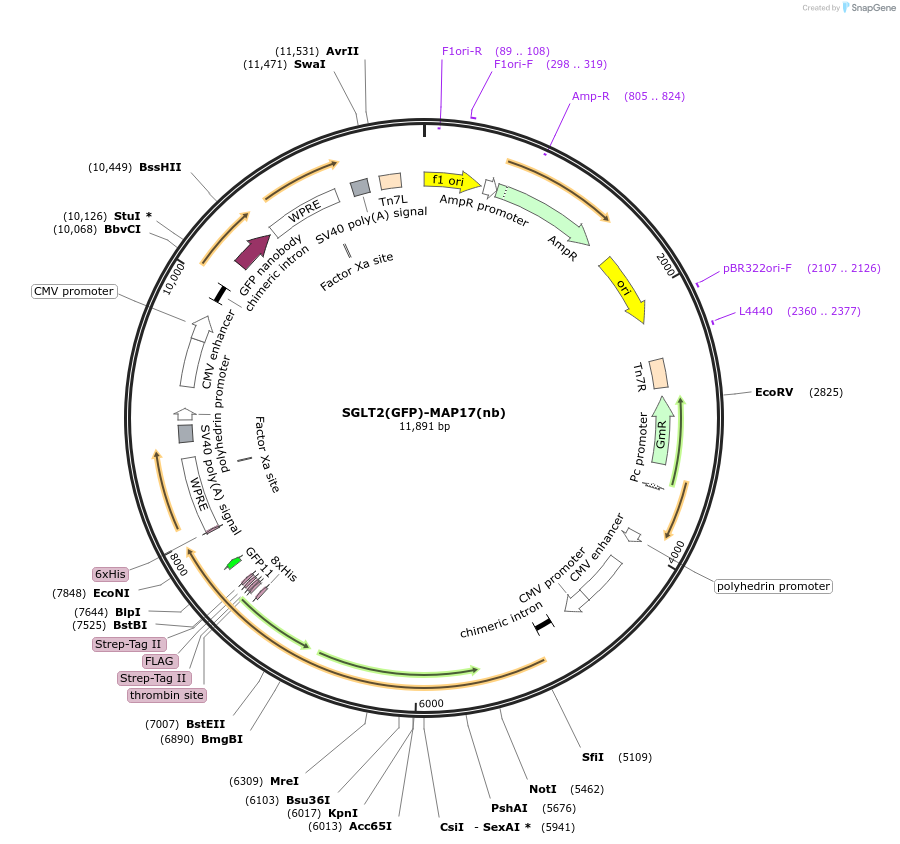
SGLT2(GFP)-MAP17(nb)
Plasmid#216211Purposefor expression of human GFP-tagged SGLT2-MAP17 nanobody complex in BacMam systemDepositorTagsGFPExpressionMammalianAvailable SinceMay 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
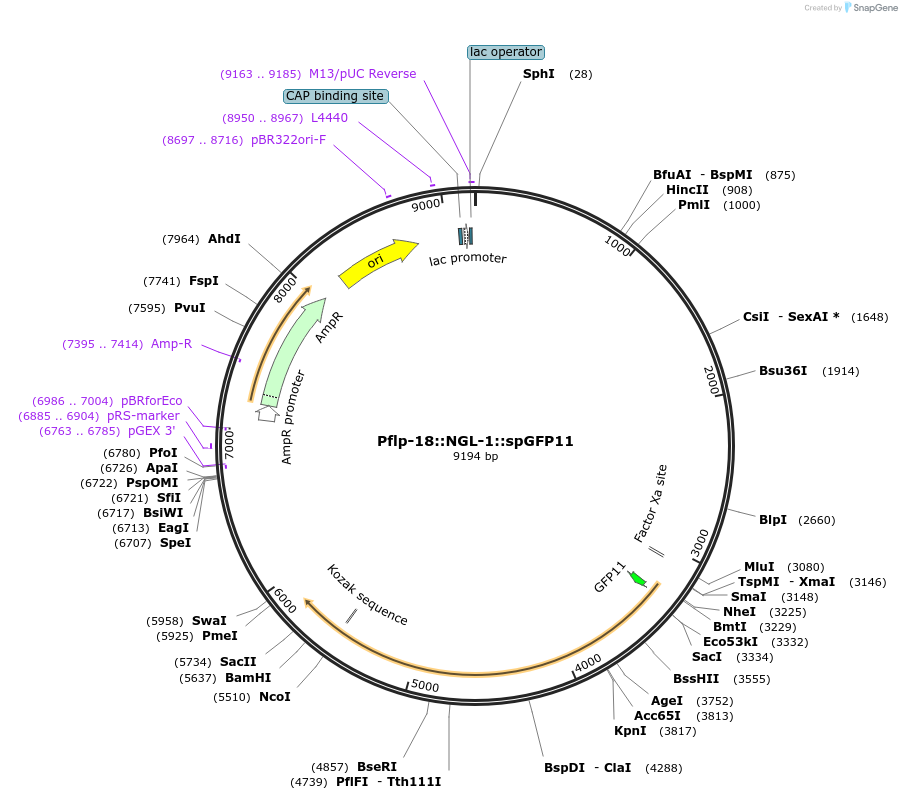
Pflp-18::NGL-1::spGFP11
Plasmid#65828PurposeFor trans-synaptic labelingDepositorAvailable SinceNov. 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
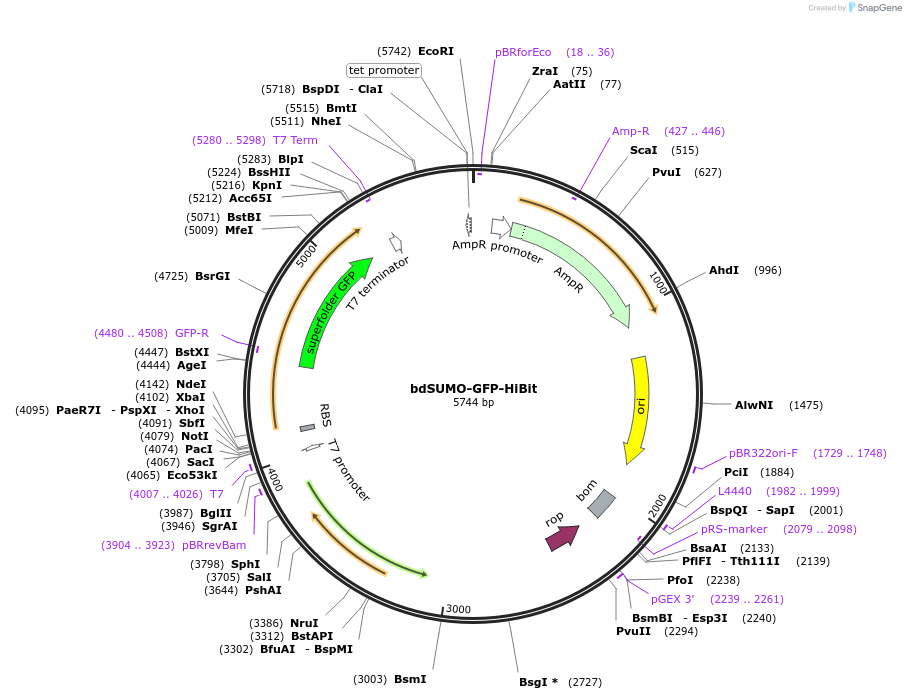
bdSUMO-GFP-HiBit
Plasmid#170989PurposeFor bacterial expression of bdSUMO tagged muGFP with NanoLuc complementation peptide variant #86DepositorInsert14x-His-bdSUMO-muGFP-HiBiT
UseLuciferaseExpressionBacterialPromoterT7Available SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
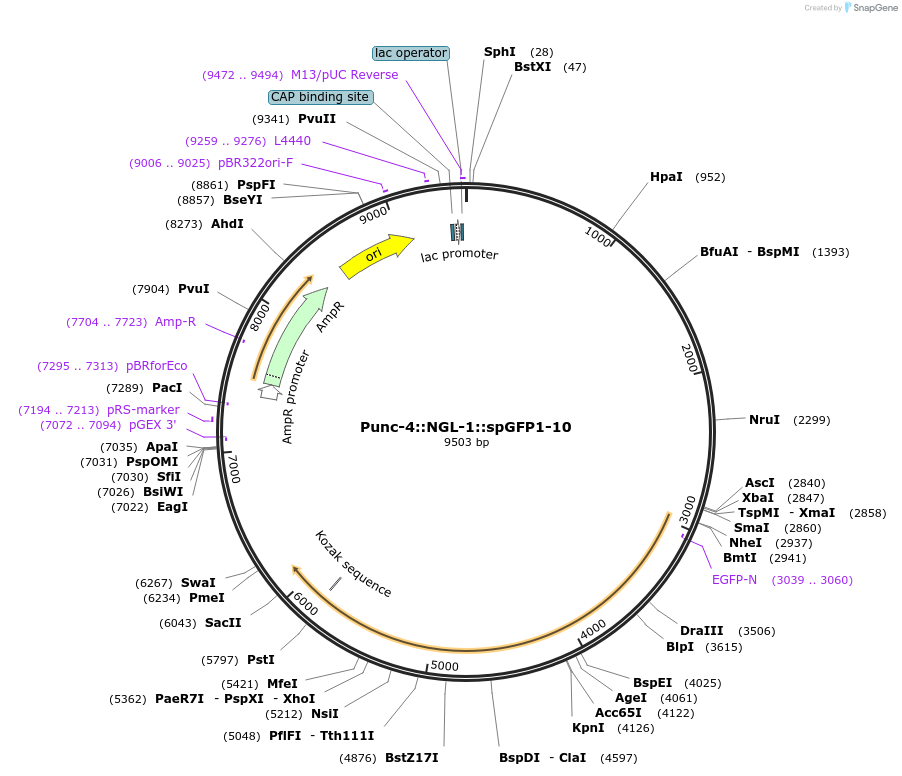
Punc-4::NGL-1::spGFP1-10
Plasmid#65827PurposeFor trans-synaptic labelingDepositorAvailable SinceSept. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
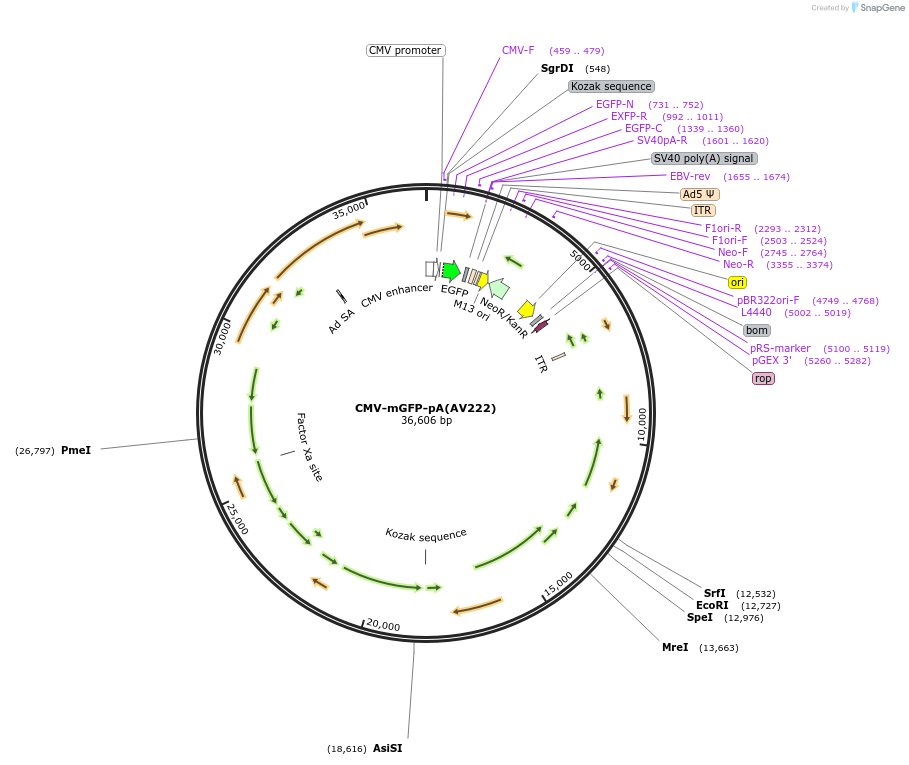
CMV-mGFP-pA(AV222)
Plasmid#117858PurposeThis plasmid expresses membrane anchored GFP to facility axon morphology study.DepositorInsertmGFP
UseAdenoviralExpressionMammalianPromoterCMVAvailable SinceMay 30, 2019AvailabilityAcademic Institutions and Nonprofits only -
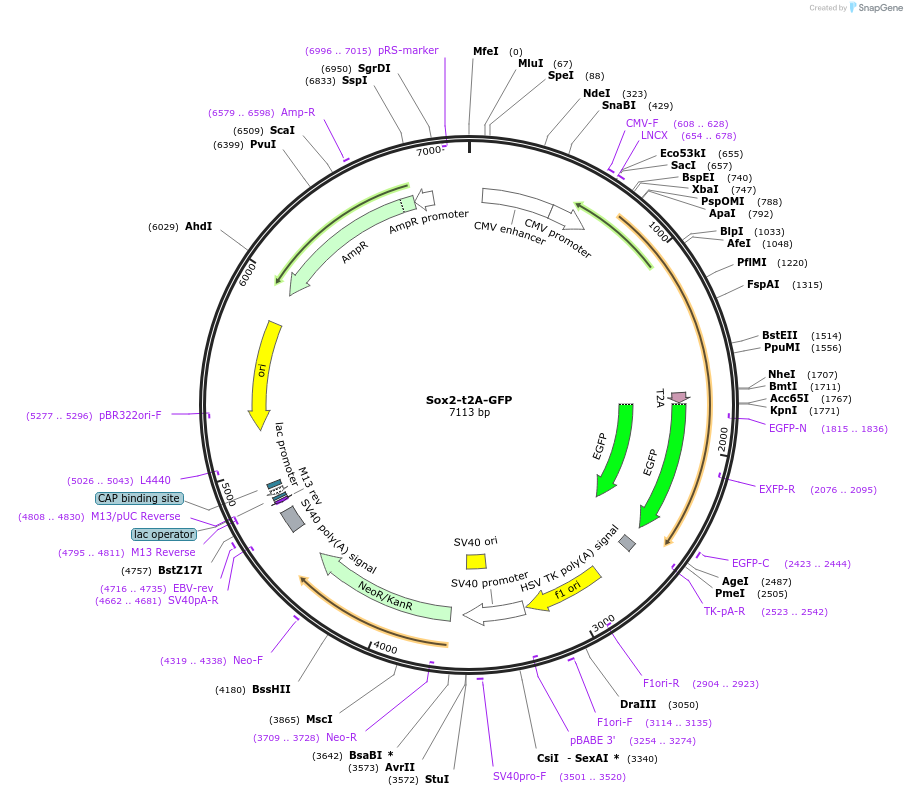
Sox2-t2A-GFP
Plasmid#127537PurposeExpresses human SOX2 and eGFP via t2A linker.DepositorAvailable SinceJuly 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
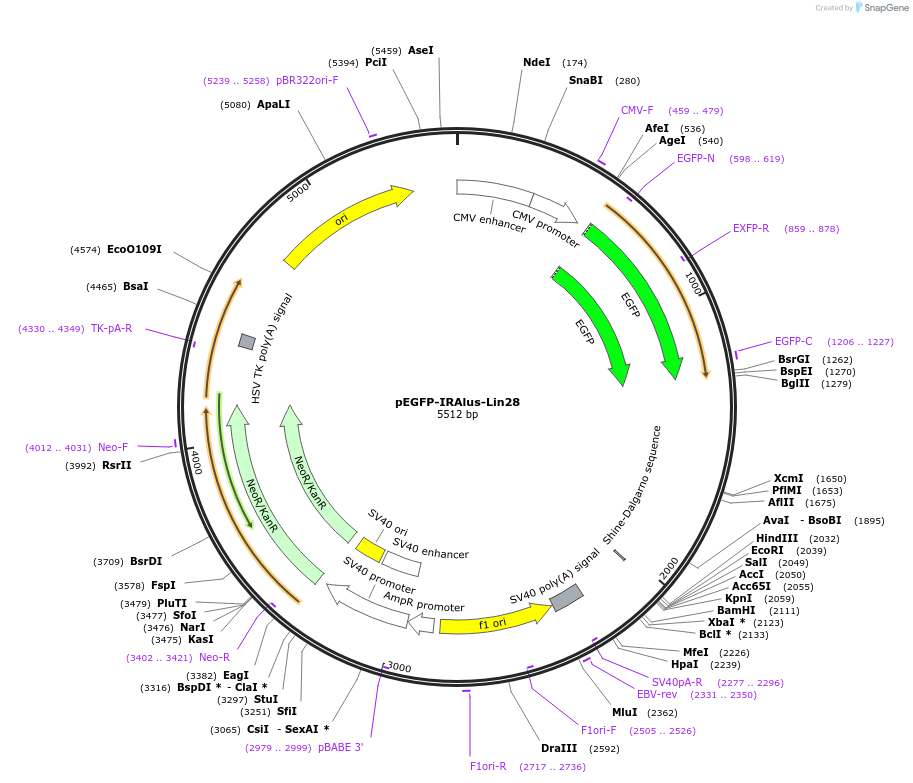
pEGFP-IRAlus-Lin28
Plasmid#92347PurposePlasmid for expression of GFP gene with a Lin28 gene derived IR Alu element containing 3'UTR.DepositorInsertLin28 gene derived IR Alu element containing 3'UTR (LIN28A Human)
ExpressionMammalianPromoterCMVAvailable SinceJune 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
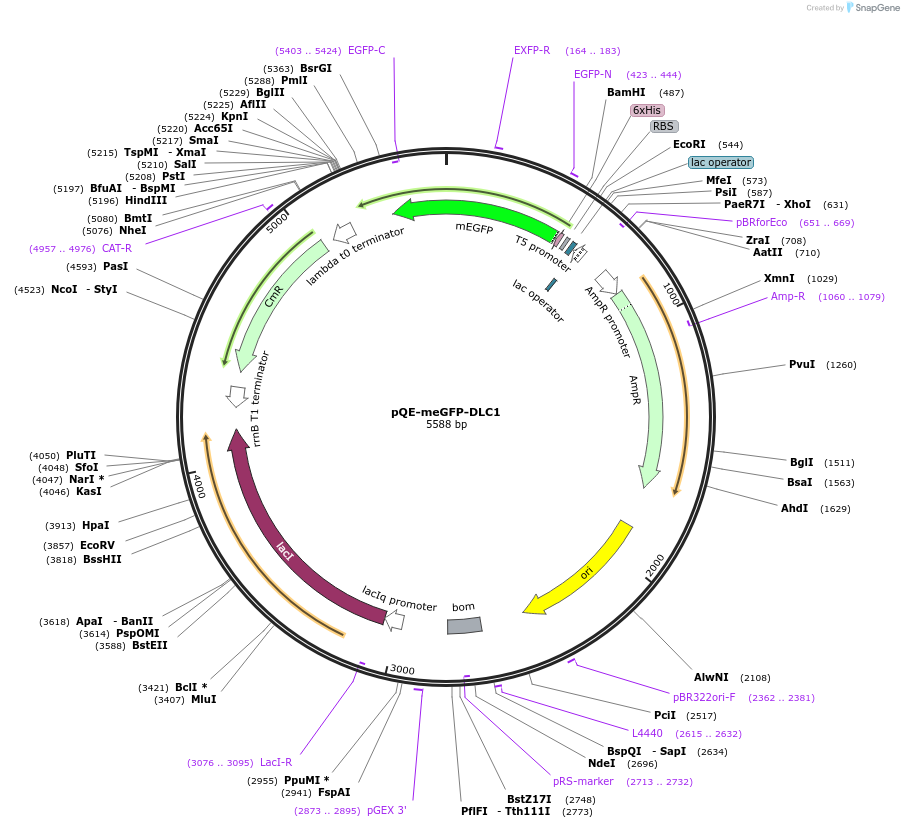
pQE-meGFP-DLC1
Plasmid#192048PurposePlasmid expresses DLC1 binding peptide to Talin R8 domain in chimera with meGFPDepositorInsertDLC1 (DLC1 Human)
ExpressionBacterialAvailable SinceFeb. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
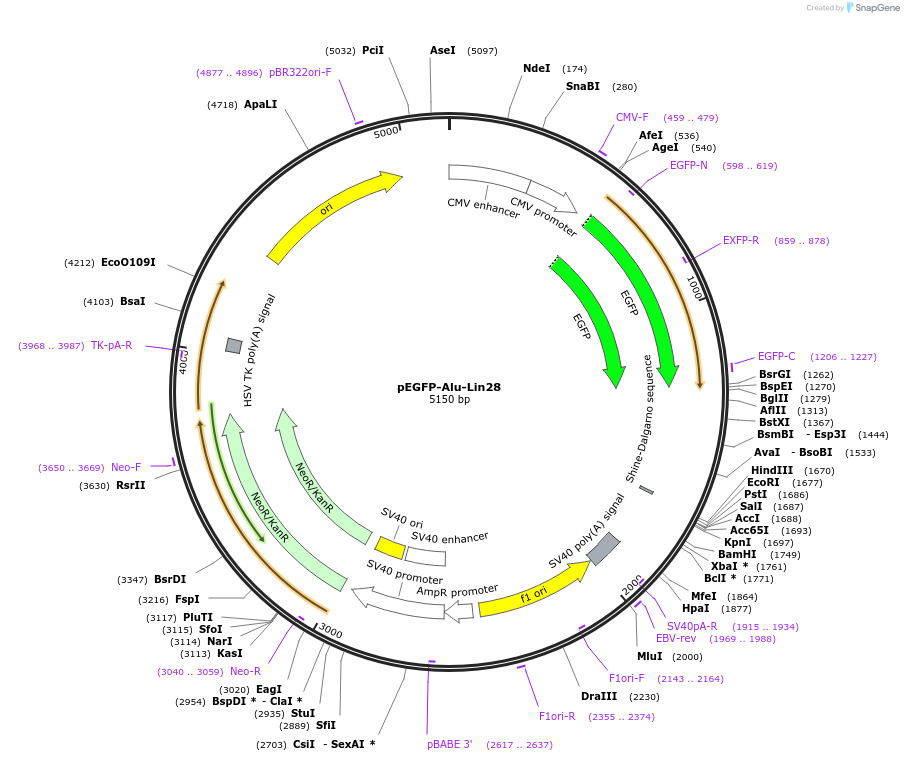
pEGFP-Alu-Lin28
Plasmid#92346PurposePlasmid for expression of GFP gene with a Lin28 gene derived Alu element containing 3'UTR.DepositorInsertLin28 gene derived Alu element containing 3'UTR (LIN28A Human)
ExpressionMammalianPromoterCMVAvailable SinceJune 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
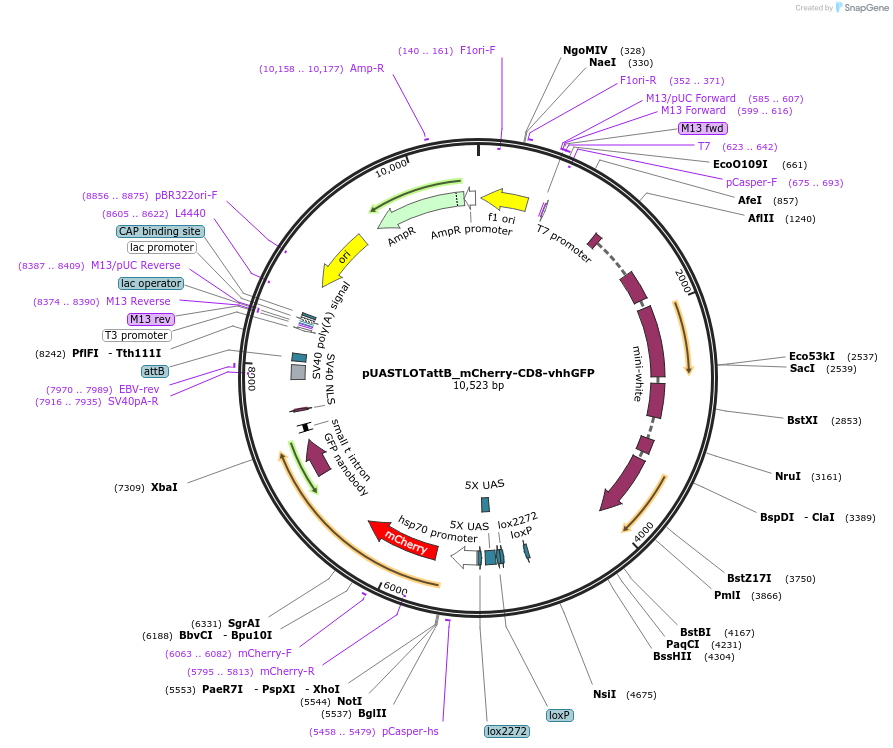
pUASTLOTattB_mCherry-CD8-vhhGFP
Plasmid#163930PurposeExpression of Morphotrap-intracellular (Grab-FP-intracellular) under control of UAS or LOP enhancerDepositorInsertFusion of vhhGPF4 nanobody (intracellular) with mouse CD8 transmembrane protein and mCherry fluorophor
UseCre/LoxTagsmCherryExpressionInsectAvailable SinceJan. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
bdSUMO-R9-GFP-HiBit
Plasmid#170990PurposeFor bacterial expression of bdSUMO tagged muGFP with R9 peptide and NanoLuc complementation peptide variant #86DepositorInsert14x-His-bdSUMO-R9-muGFP-HiBiT
UseLuciferaseExpressionBacterialPromoterT7Available SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
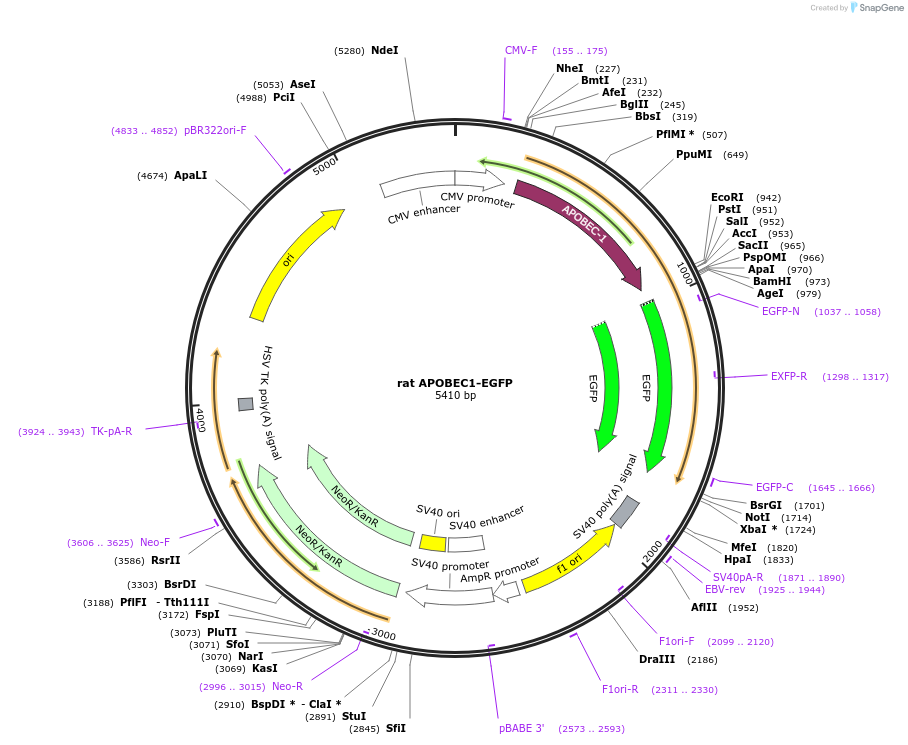
rat APOBEC1-EGFP
Plasmid#112857Purposeplasmid for the expression of a rat APOBEC1-EGFP C-terminal fusion proteinDepositorInsertapolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (Apobec1 Rat)
ExpressionMammalianPromoterCMV promoterAvailable SinceAug. 16, 2018AvailabilityAcademic Institutions and Nonprofits only -
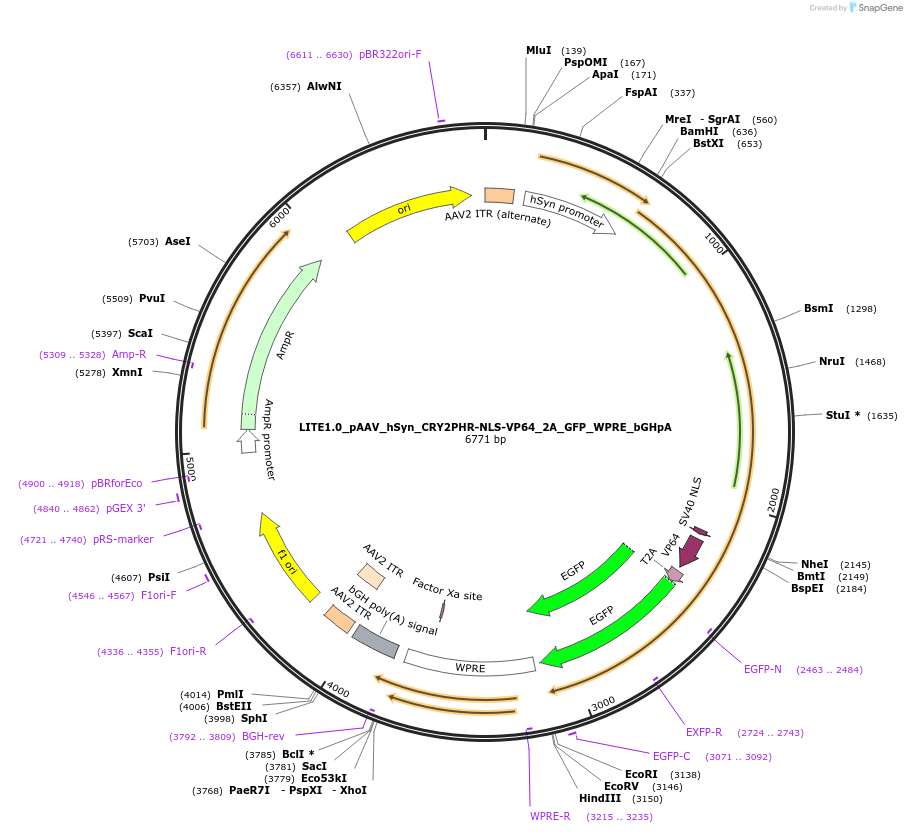
LITE1.0_pAAV_hSyn_CRY2PHR-NLS-VP64_2A_GFP_WPRE_bGHpA
Plasmid#48253PurposeLITE1.0 CRY2PHR fused to VP64 transcriptional activator domain. Binds to CIB1 upon blue light stimulation. Synapsin promoter for neuronal expression. See LITE2.0 for optimized LITE activators.DepositorInsertCRY2PHR-NLS-VP64
UseAAV; TaleTags2A_GFPExpressionMammalianPromoterEF1-alphaAvailable SinceOct. 15, 2013AvailabilityAcademic Institutions and Nonprofits only -
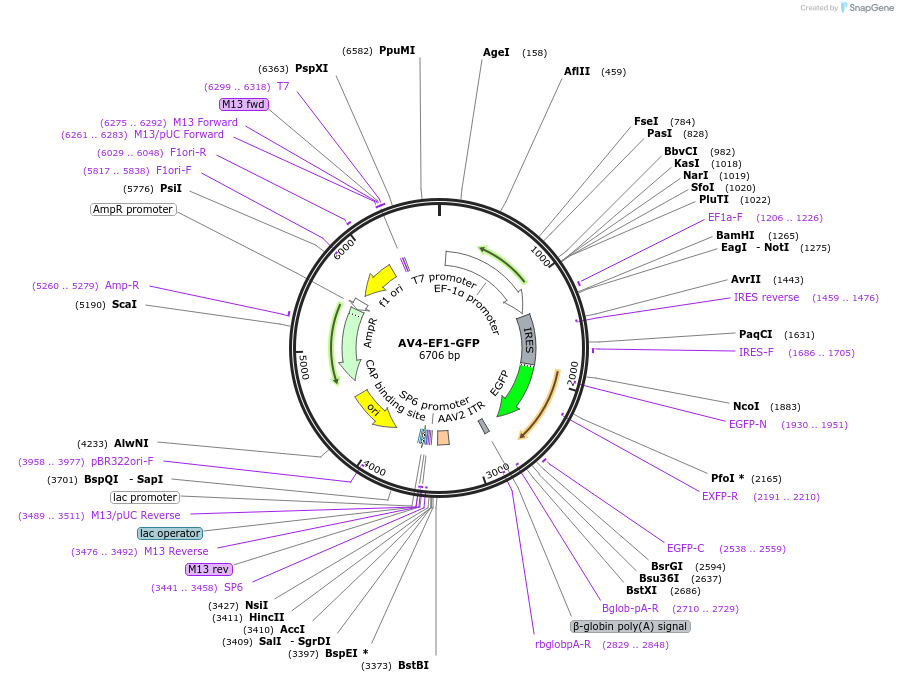
AV4-EF1-GFP
Plasmid#124971PurposeControl for AV4-EF1-NMNAT2DepositorInsertGFP
UseAAVPromoterEF1-alphaAvailable SinceMay 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
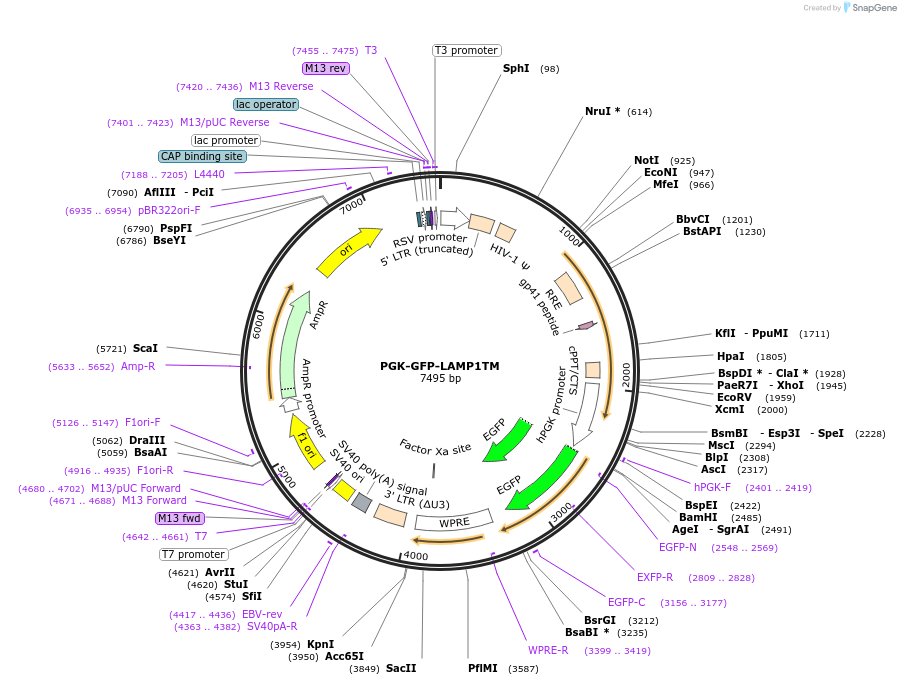
PGK-GFP-LAMP1TM
Plasmid#234874PurposeLentiviral plasmid expressing GFP fused to the transmembrane domain and cytosolic tail of LAMP-1. Designed as a control vector for PGK-HA-PGRN-LAMP1TM.DepositorInsertEGFP
UseLentiviralTagsLAMP-1 transmembrane domain and cytosolic tailExpressionMammalianPromoterPGKAvailable SinceApril 15, 2025AvailabilityAcademic Institutions and Nonprofits only -
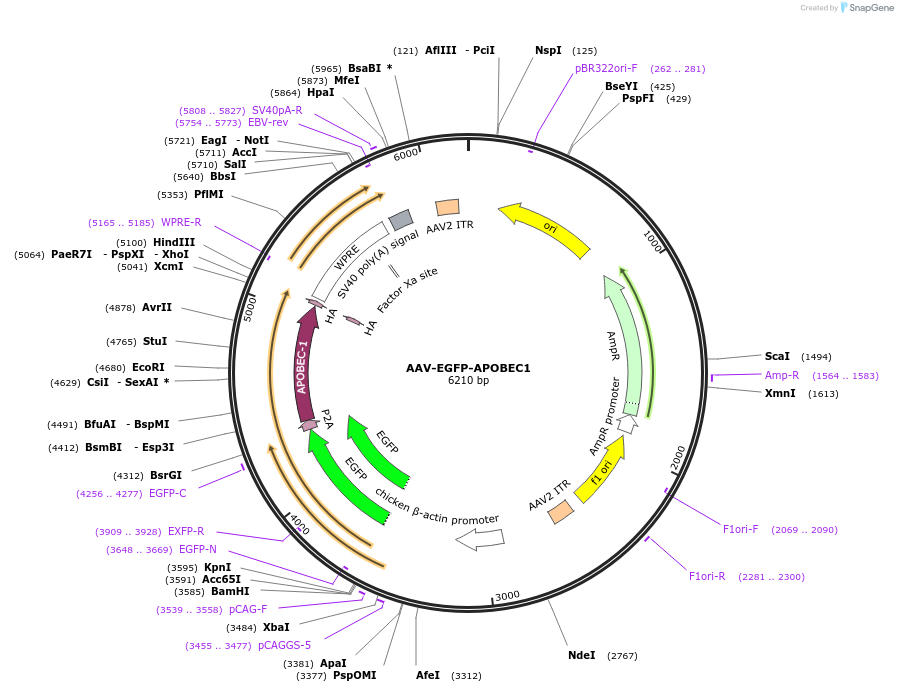
AAV-EGFP-APOBEC1
Plasmid#209321PurposeAAV packaging vector containing a EGFP-P2A expression cassette, APOBEC1 expression.DepositorInsertAPOBEC1-HA
UseAAVTagsHAAvailable SinceMay 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
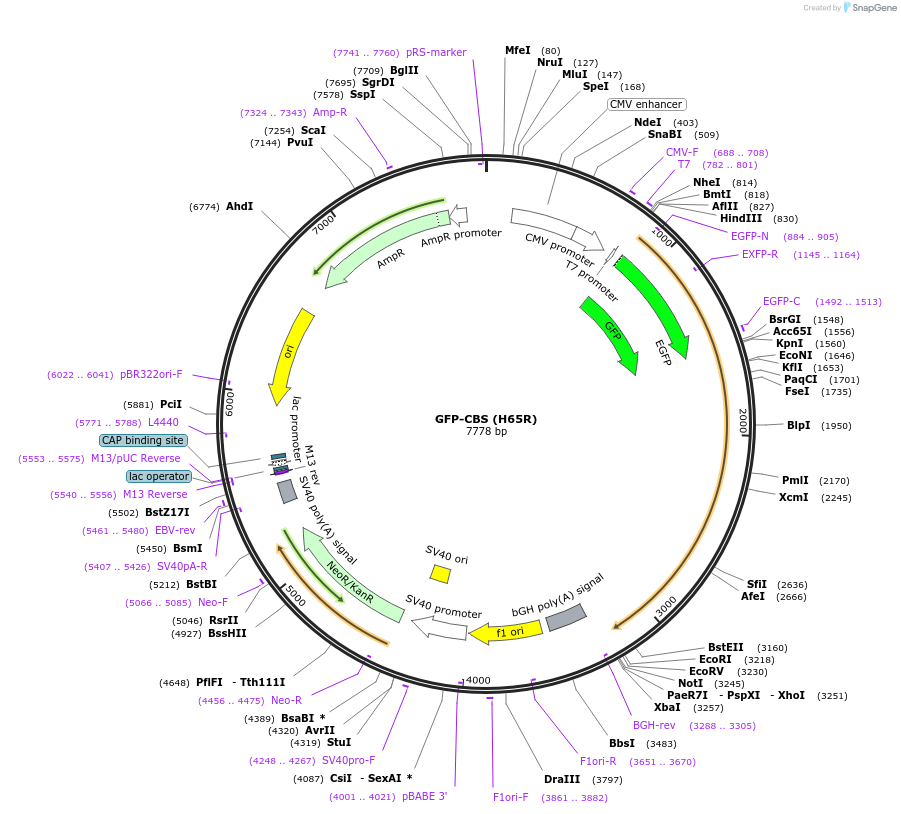
GFP-CBS (H65R)
Plasmid#182924PurposeMammalian expression plasmid for human CBS mutant H65RDepositorInsertCystathionine Beta-Synthase (CBS Human)
TagseGFPExpressionMammalianMutationchanged histidine 65 to argininePromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
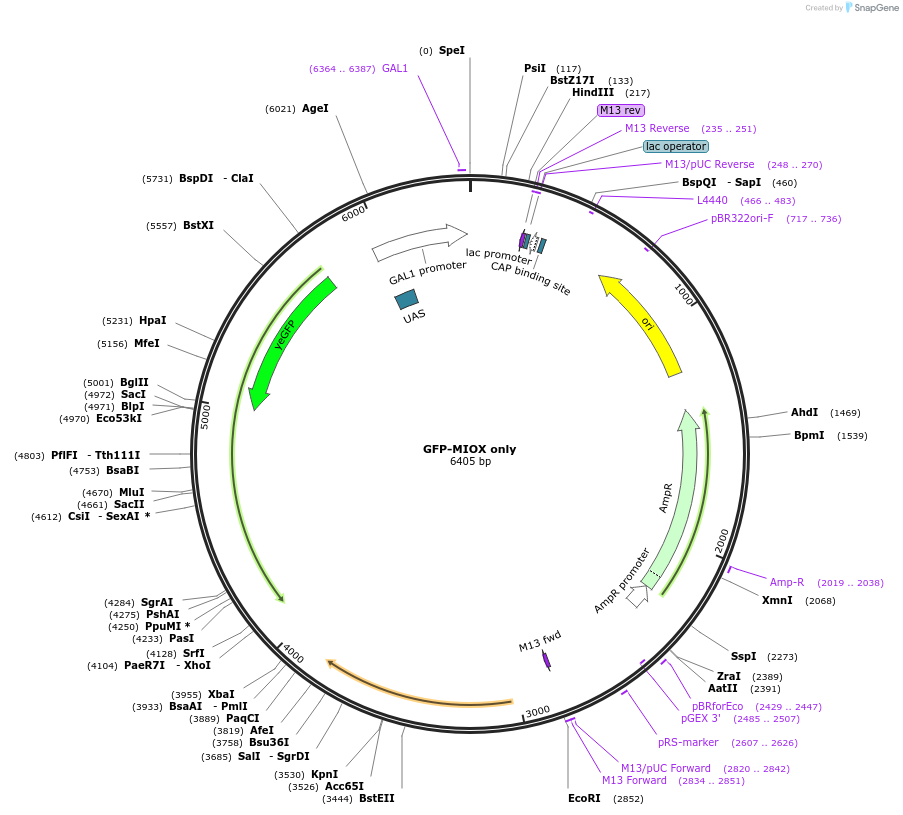
GFP-MIOX only
Plasmid#166683PurposeYeast integrative plasmid for expressing mouse myo-inositol oxygenase fused to GFP (GAL10 promoter). Contains uracil auxotrophic marker (K. lactis URA3).DepositorInsertGFP-MIOX
UseSynthetic BiologyExpressionYeastPromoterGAL10Available SinceApril 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
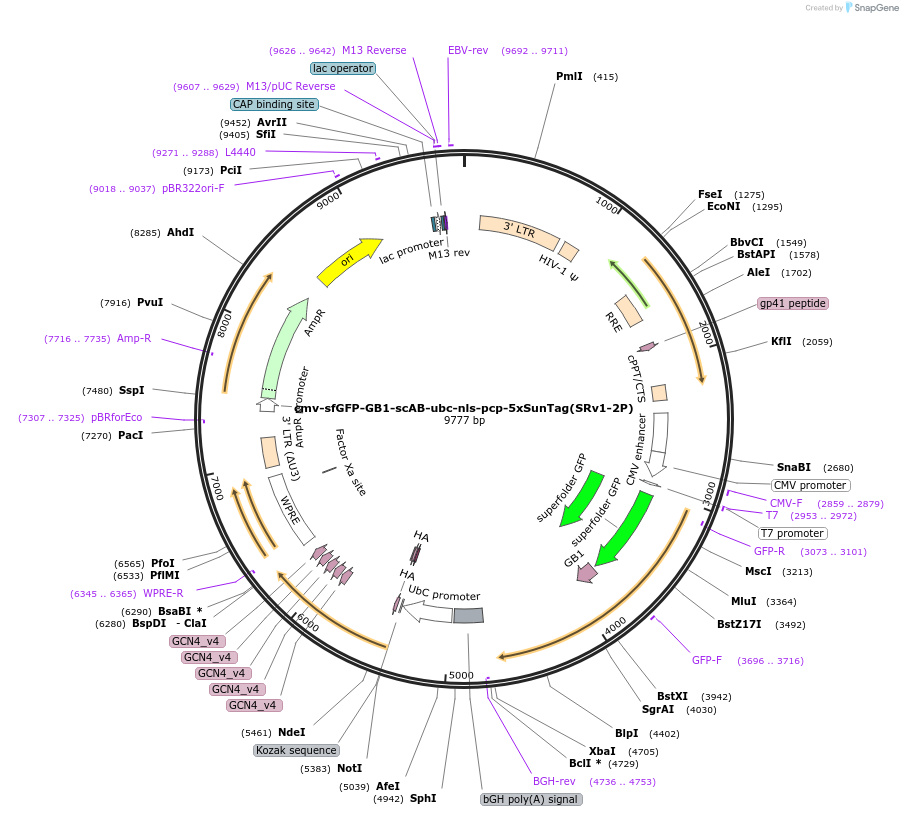
cmv-sfGFP-GB1-scAB-ubc-nls-pcp-5xSunTag(SRv1-2P)
Plasmid#185800Purposeone plasmid encoding scFv GFP and pcp-5xSunTag components for SunRISER SRv1DepositorInsertcmv-sfGFP-GB1-scAB--ubc-nls-pcp-5xSunTag
ExpressionMammalianAvailable SinceJuly 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
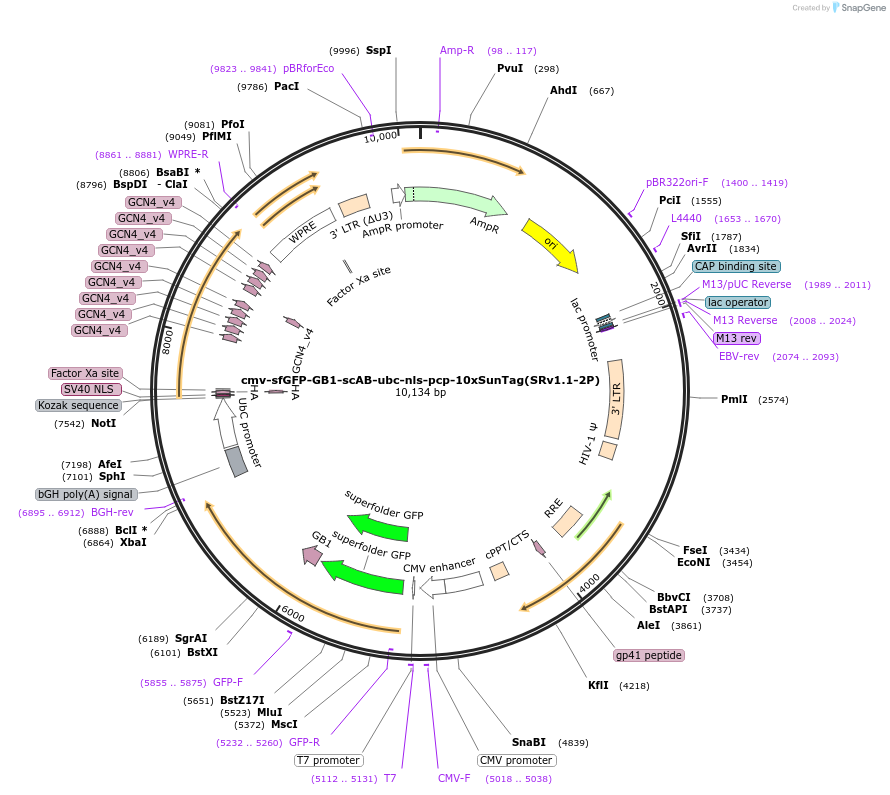
cmv-sfGFP-GB1-scAB-ubc-nls-pcp-10xSunTag(SRv1.1-2P)
Plasmid#185801Purposeone plasmid encoding scFv GFP and pcp-10xSunTag components for SunRISER SRv1.1DepositorInsertcmv-sfGFP-GB1-scAB--ubc-nls-pcp-10xSunTag
ExpressionMammalianAvailable SinceJuly 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
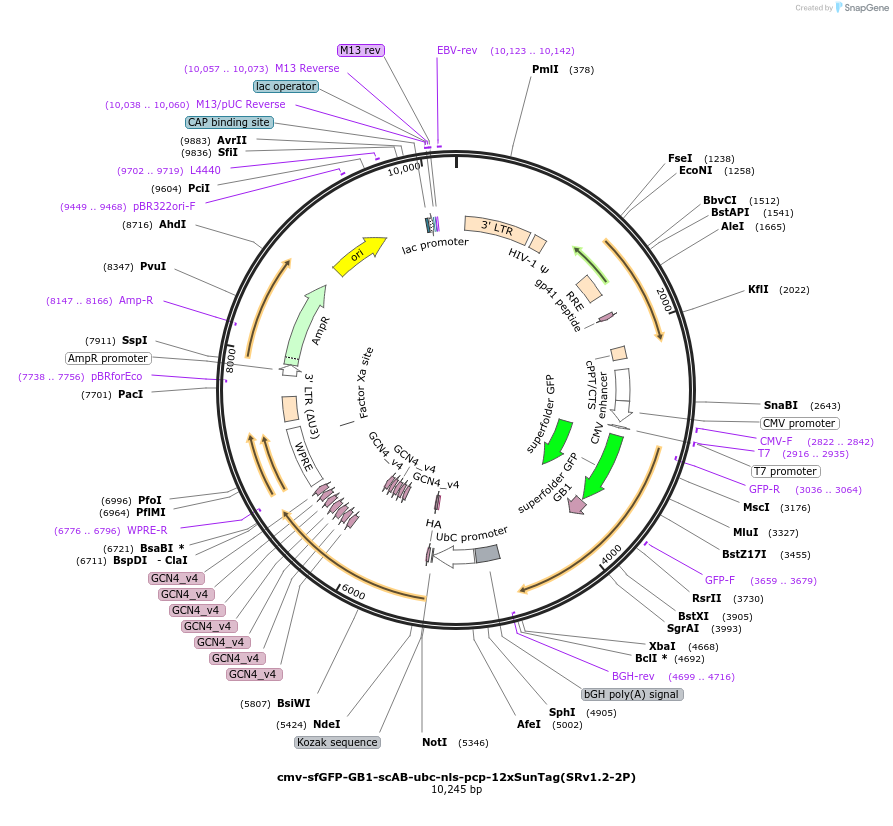
cmv-sfGFP-GB1-scAB-ubc-nls-pcp-12xSunTag(SRv1.2-2P)
Plasmid#185802Purposeone plasmid encoding scFv GFP and pcp-10xSunTag components for SunRISER SRv1.2DepositorInsertcmv-sfGFP-GB1-scAB--ubc-nls-pcp-12xSunTag
ExpressionMammalianAvailable SinceJuly 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
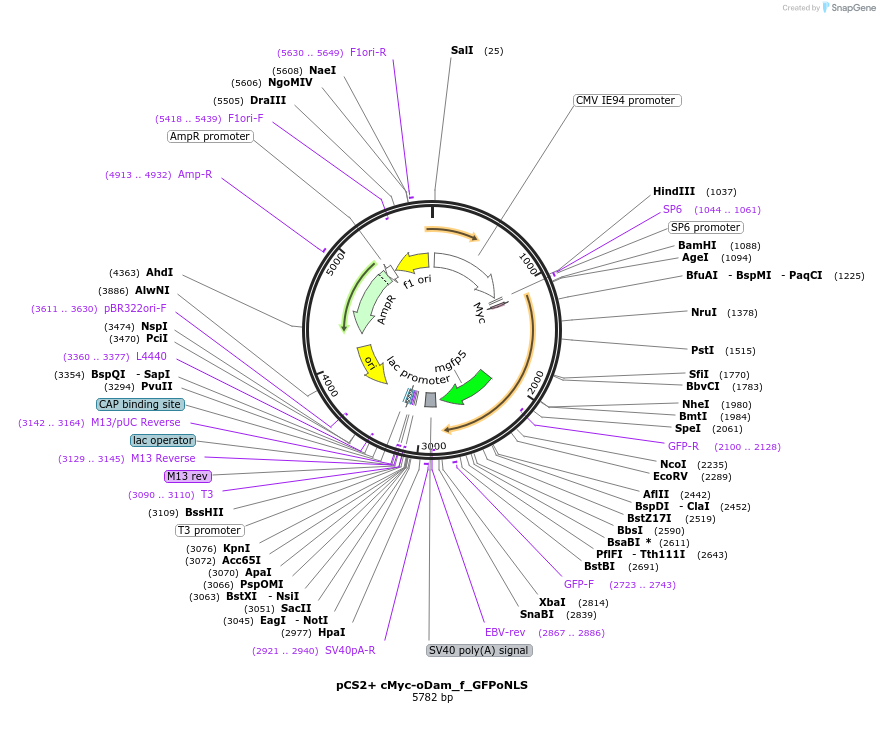
pCS2+ cMyc-oDam_f_GFPoNLS
Plasmid#85818PurposeiDamID plasmid. To express transiently the c-Myc-tagged and optimized E. coli Dam adenine methyltransferase fused to the nuclear-localized mmGFP via flexylinker.DepositorInsertcMyc oDam_f_GFPoNLS
TagscMyc and oNLS (optimized nuclear localization sig…ExpressionBacterial, Insect, Mammalia…MutationL122A. Codon optimization compatible to work in M…Available SinceJan. 4, 2017AvailabilityAcademic Institutions and Nonprofits only -
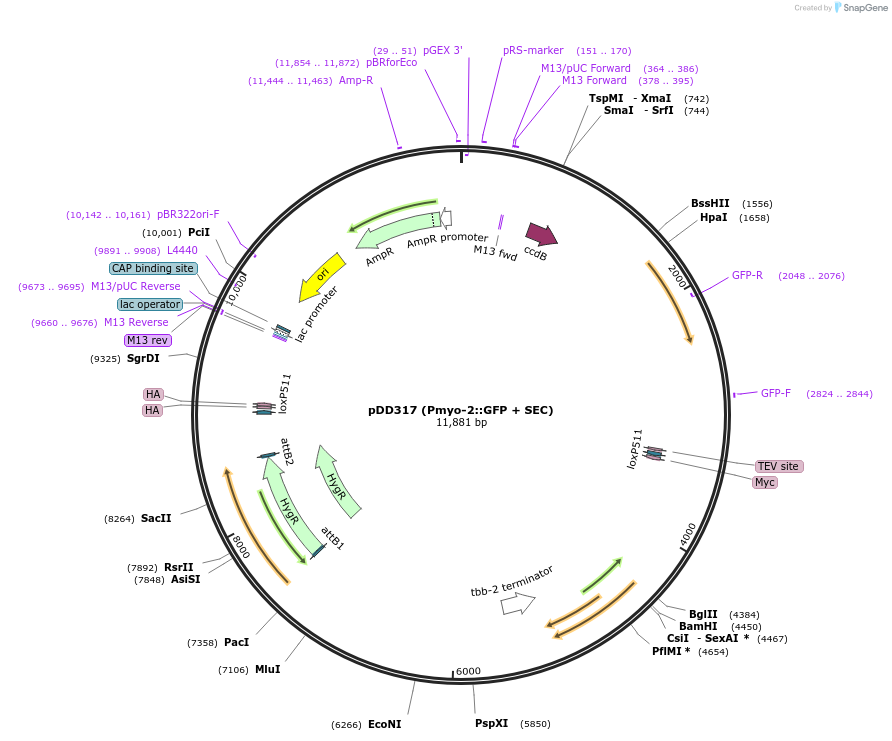
pDD317 (Pmyo-2::GFP + SEC)
Plasmid#73344PurposeSEC vector carry Pmyo-2::GFP and ccdB sites for cloning homology armsDepositorInsertPmyo-2::GFP + SEC
UseCRISPR and Cre/LoxTagsGFPExpressionWormPromotermyo-2Available SinceMarch 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
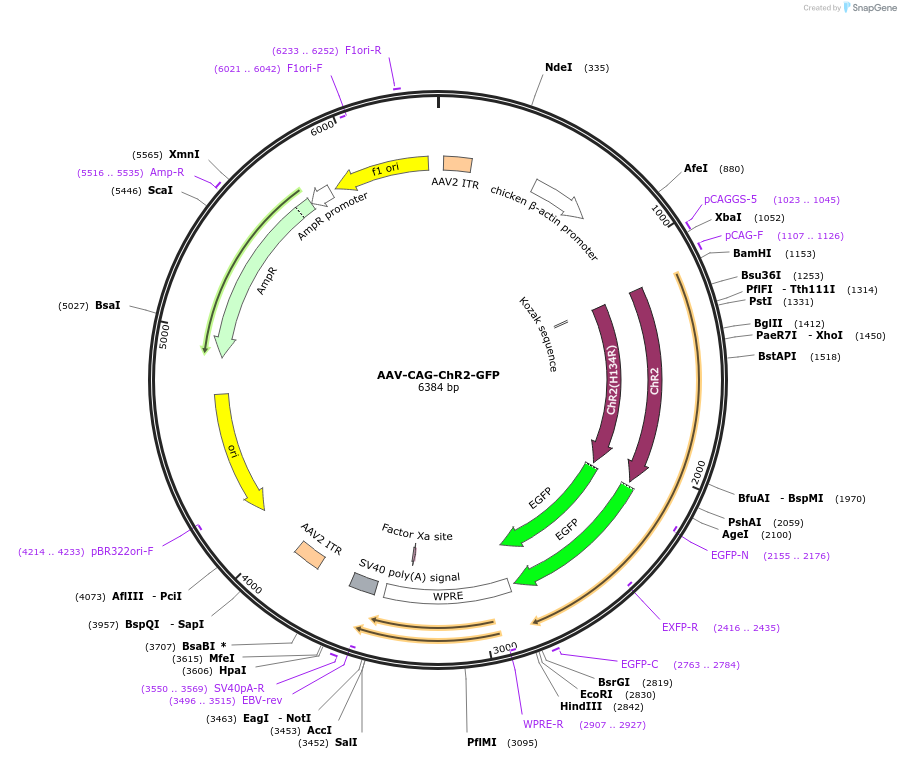
AAV-CAG-ChR2-GFP
Plasmid#26929PurposeAAV-mediated expression of ChR2-GFP for neuronal excitationDepositorInsertChR2
UseAAV; Adeno-associated virusTagsGFPExpressionMammalianPromoterCAGAvailable SinceJan. 11, 2011AvailabilityAcademic Institutions and Nonprofits only -
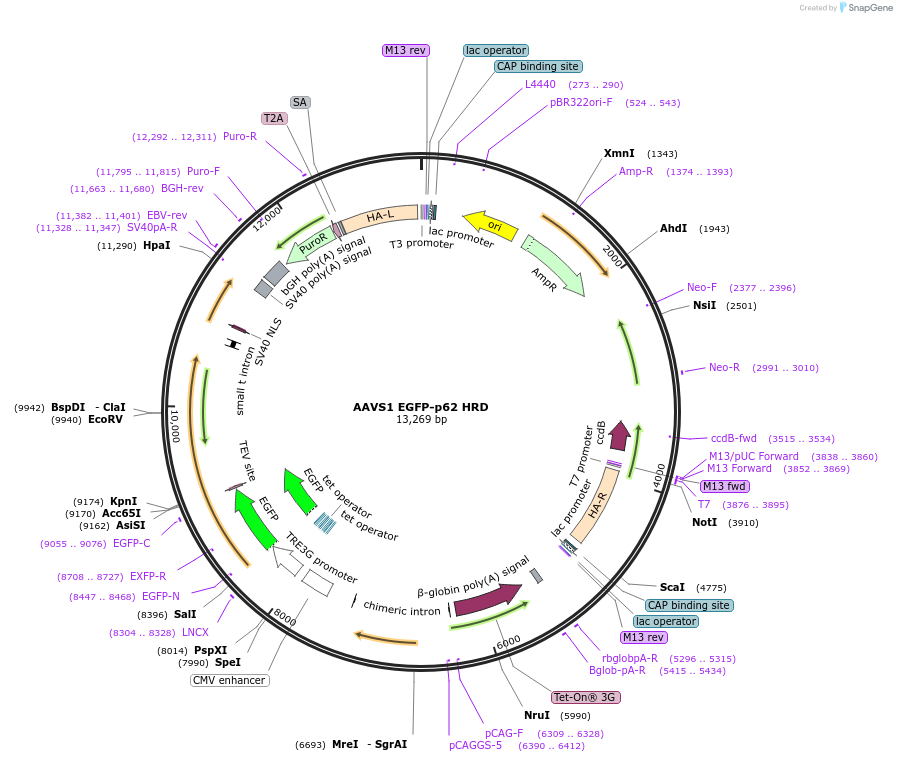
AAVS1 EGFP-p62 HRD
Plasmid#207550PurposeHomologous recombination donor to integrate and expression cassette for eGFP-p62 in the AAVS1 locus of human cellsDepositorInsertTET inducible EGFP-P62
TagsEGFPExpressionMammalianPromoterEndogenousAvailable SinceApril 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
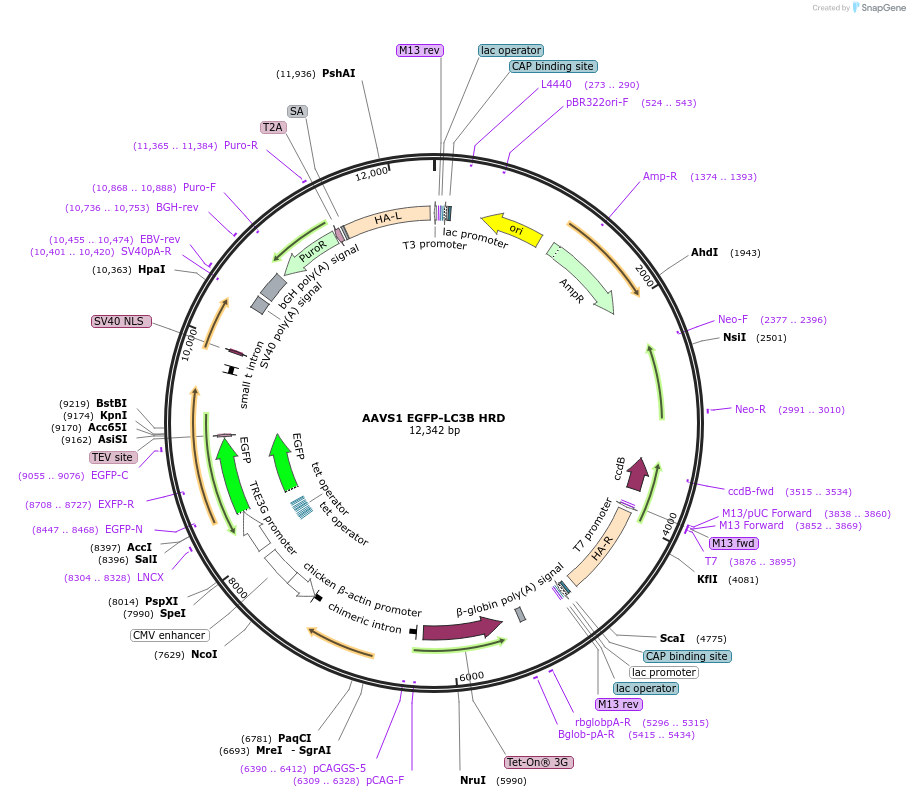
AAVS1 EGFP-LC3B HRD
Plasmid#207551PurposeHomologous recombination donor to integrate and expression cassette for eGFP-LC3B in the AAVS1 locus of human cellsDepositorInsertTET inducible EGFP-LC3B
TagsEGFPExpressionMammalianPromoterEndogenousAvailable SinceApril 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
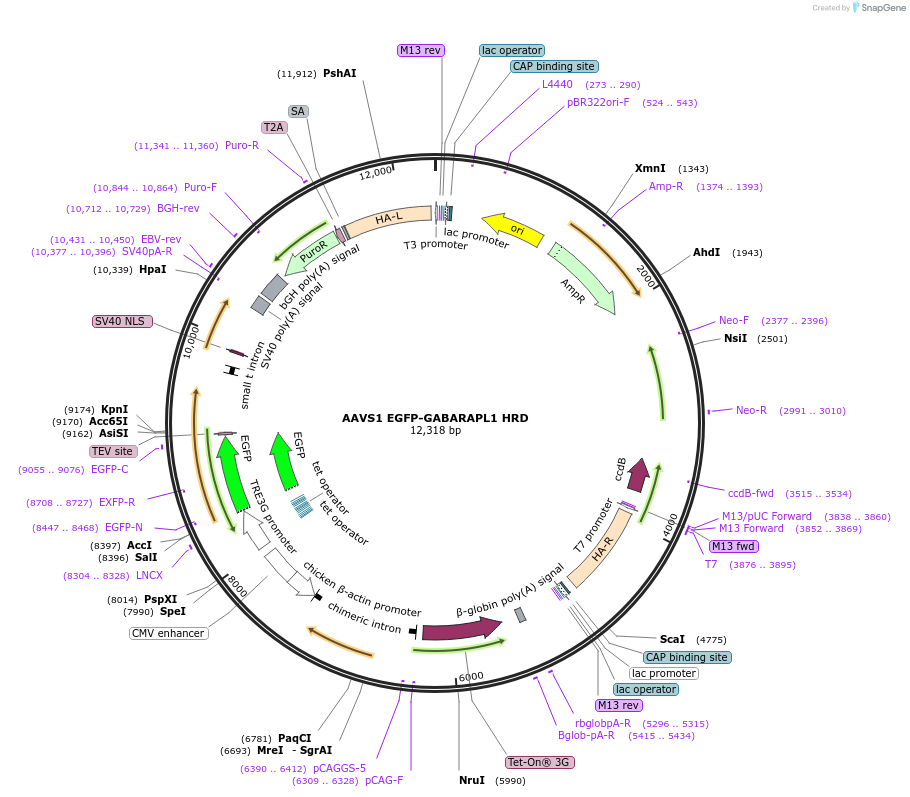
AAVS1 EGFP-GABARAPL1 HRD
Plasmid#207552PurposeHomologous recombination donor to integrate and expression cassette for EGFP-GABRAPL1 in the AAVS1 locus of human cellsDepositorInsertTET inducible EGFP-GABARAPL1
TagsEGFPExpressionMammalianPromoterEndogenousAvailable SinceApril 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
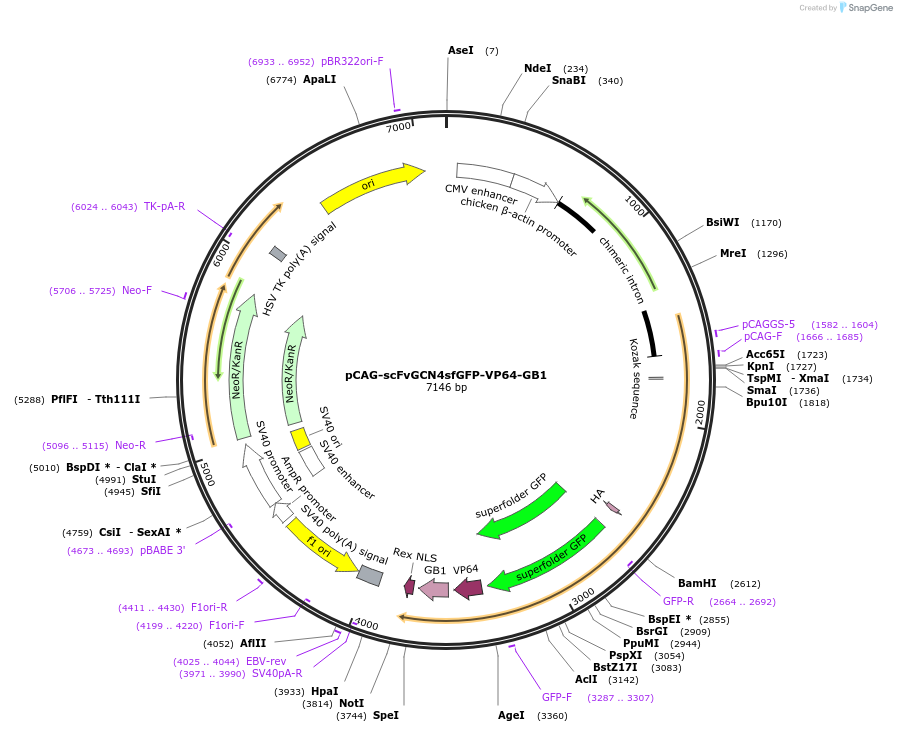
pCAG-scFvGCN4sfGFP-VP64-GB1
Plasmid#141417PurposeExpress scFvGCN4-sfGFP-VP64 fusion protein for dCas9-SunTag mediated targeted gene activationDepositorInsertscFvGCN4sfGFP-VP64-GB1
TagsHA and sfGFPExpressionMammalianPromoterCAGAvailable SinceJune 22, 2020AvailabilityAcademic Institutions and Nonprofits only