169,603 results
-
Plasmid#16639DepositorAvailable SinceMarch 10, 2008AvailabilityAcademic Institutions and Nonprofits only
-
pLVX-Flag-CDK19-WT-IRES-ZsGreen
Plasmid#68858PurposeExpresses Human CDK19 Wild-TypeDepositorInsertCyclin-Dependent Kinase 19 (CDK19 Human)
UseLentiviralTagsFlagExpressionMammalianPromoterEF1αAvailable SinceMarch 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
pCAX FLAG APP
Plasmid#30154DepositorInsertAmyloid Precursor Protein (APP Human)
TagsFLAGExpressionMammalianMutationwild type splice variant 751Available SinceJuly 29, 2011AvailabilityAcademic Institutions and Nonprofits only -
pJC2722 - pCDNA4TO-CMV-3xFLAG-Halo-AARS1
Plasmid#250254PurposecDNA expression of affinity-tagged AARS1.DepositorAvailable SinceJan. 15, 2026AvailabilityAcademic Institutions and Nonprofits only -
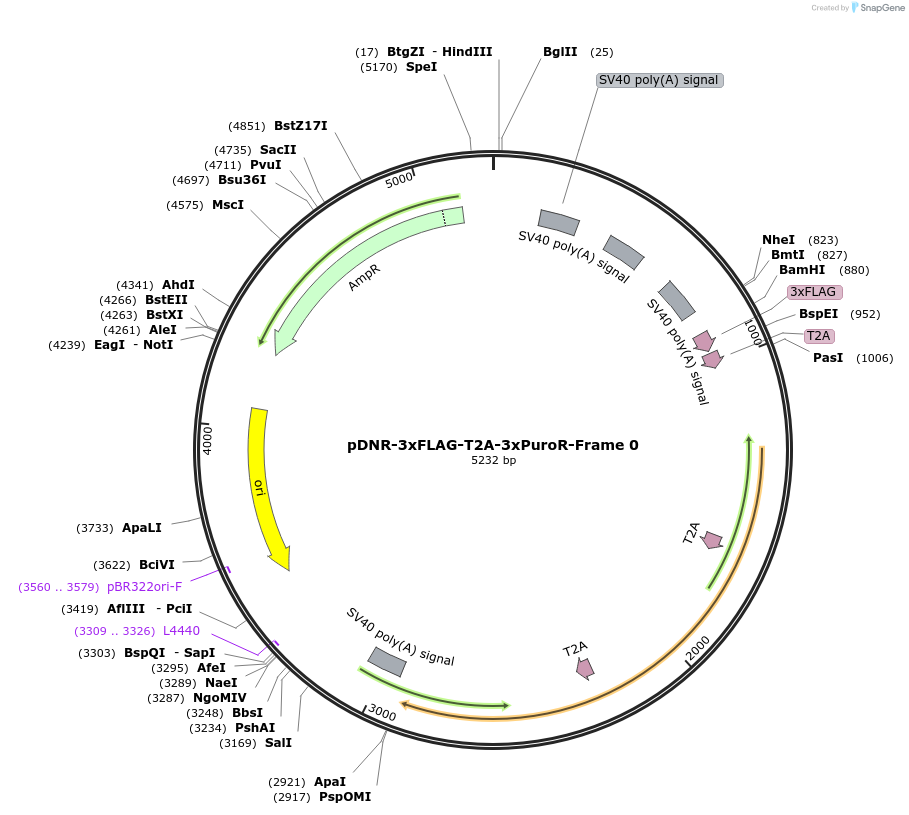
pDNR-3xFLAG-T2A-3xPuroR-Frame 0
Plasmid#245866PurposeHITAG Donor plasmid for Frame 0 with 3xFLAG tag and triple Puromycin resistant marker (3xPuroR)DepositorInsertpuromycin resistant marker
UseCRISPR and Synthetic BiologyExpressionBacterial and MammalianPromoterEndogenous gene promoter after knock-inAvailable SinceJan. 2, 2026AvailabilityAcademic Institutions and Nonprofits only -
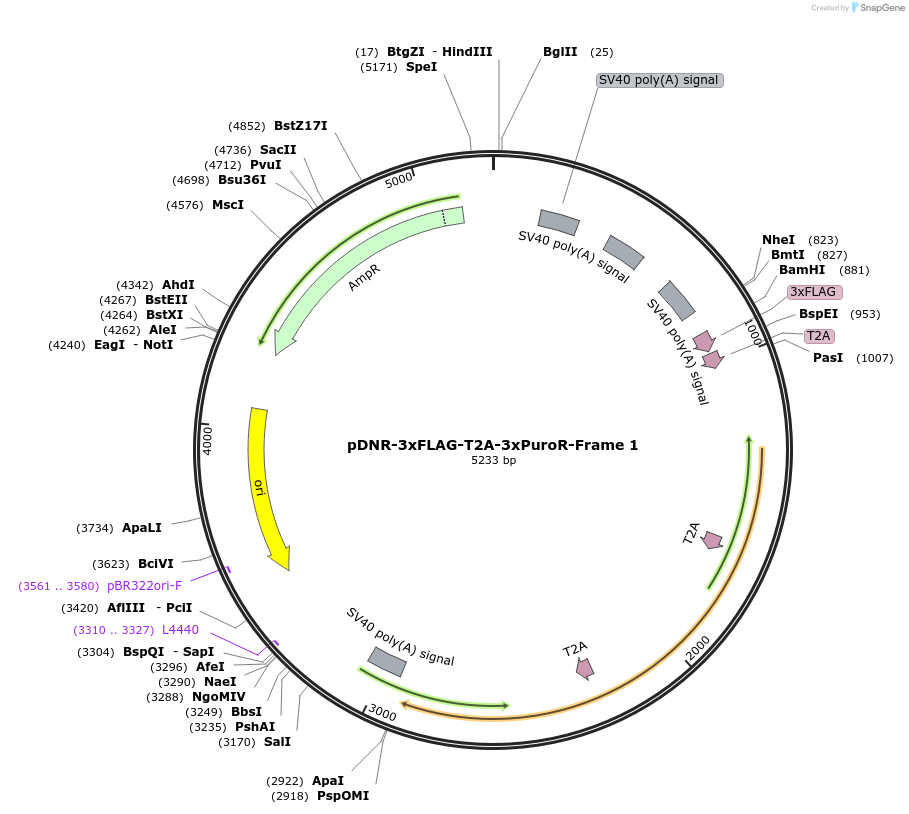
pDNR-3xFLAG-T2A-3xPuroR-Frame 1
Plasmid#245867PurposeHITAG Donor plasmid for Frame 1 with 3xFLAG tag and triple Puromycin resistant marker (3xPuroR)DepositorInsertampicilin resistant marker
UseSynthetic BiologyExpressionBacterial and MammalianAvailable SinceJan. 2, 2026AvailabilityAcademic Institutions and Nonprofits only -
pDNR-3xFLAG-T2A-3xPuroR-Frame 2
Plasmid#245868PurposeHITAG Donor plasmid for Frame 2 with 3xFLAG tag and triple Puromycin resistant marker (3xPuroR)DepositorInsert3xFLAG
UseSynthetic BiologyExpressionBacterial and MammalianAvailable SinceJan. 2, 2026AvailabilityAcademic Institutions and Nonprofits only -
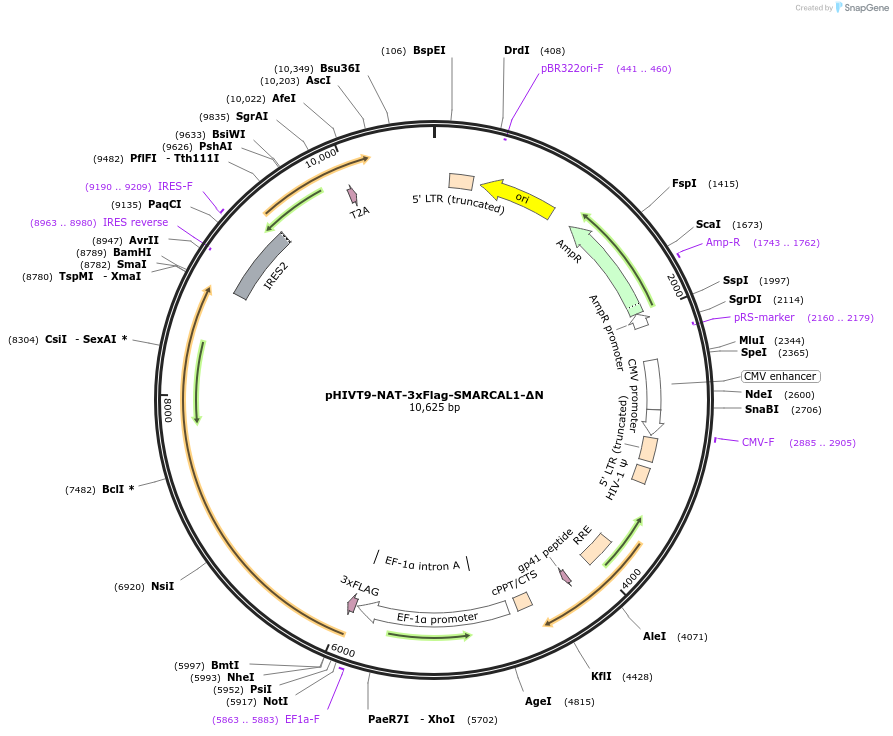
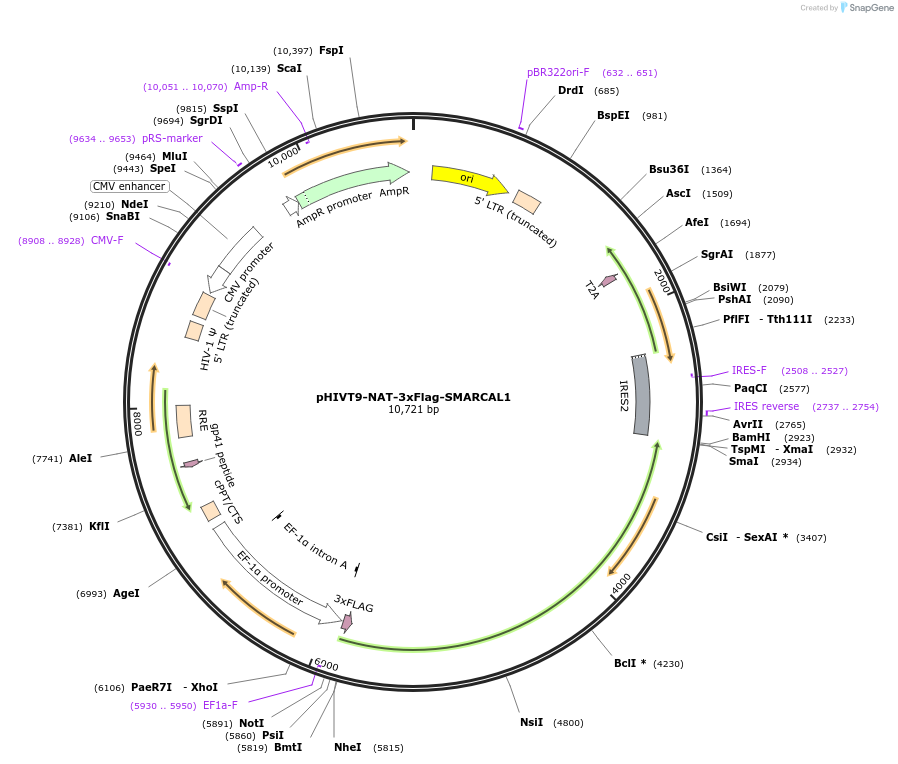
pHIVT9-NAT-3xFlag-SMARCAL1-ΔN
Plasmid#246936PurposeLentiviral vector that expresses Flag-tagged SMARCAL1 with a deletion of the RPA-binding domain located in the N-terminal region (ΔN)DepositorInsertSMARCAL1 (SMARCAL1 Human)
UseLentiviralTags3XFLAGExpressionMammalianPromoterEF-1-alpha promoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
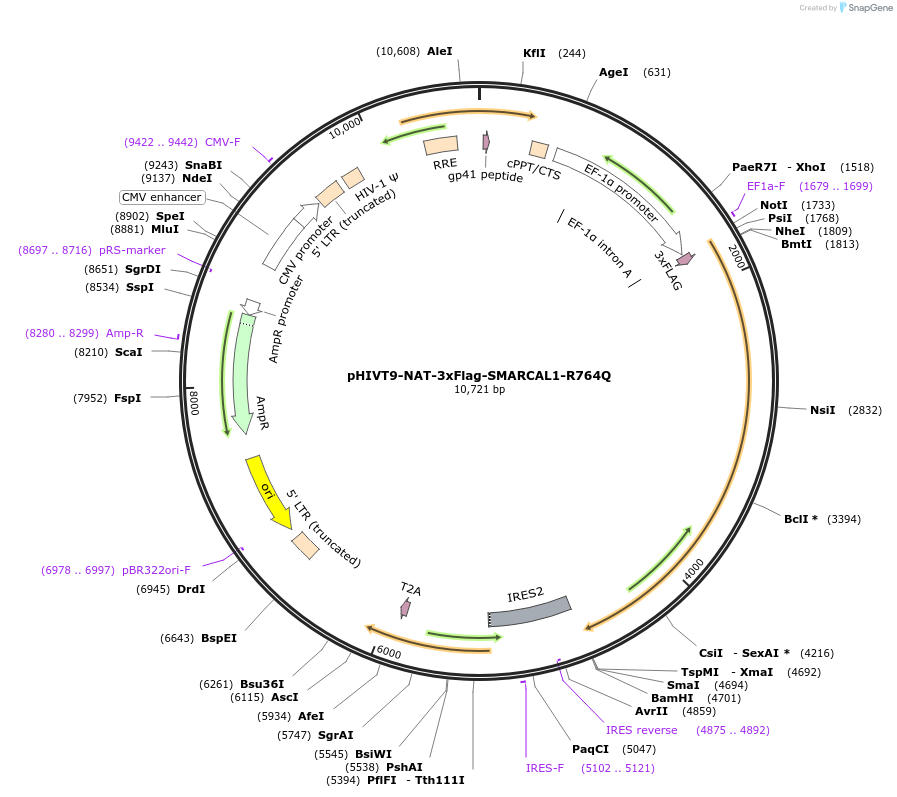
pHIVT9-NAT-3xFlag-SMARCAL1-R764Q
Plasmid#246937PurposeLentiviral vector that expresses Flag-tagged SMARCAL1 with a mutation in the ATP translocase domain (R764Q)DepositorInsertSMARCAL1 (SMARCAL1 Human)
UseLentiviralTags3XFLAGExpressionMammalianPromoterEF-1-alpha promoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
pHIVT9-NAT-3xFlag-SMARCAL1
Plasmid#246935PurposeLentiviral vector that expresses wildtype Flag-tagged SMARCAL1DepositorInsertSMARCAL1 (SMARCAL1 Human)
UseLentiviralTags3XFLAGExpressionMammalianPromoterEF-1-alpha promoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
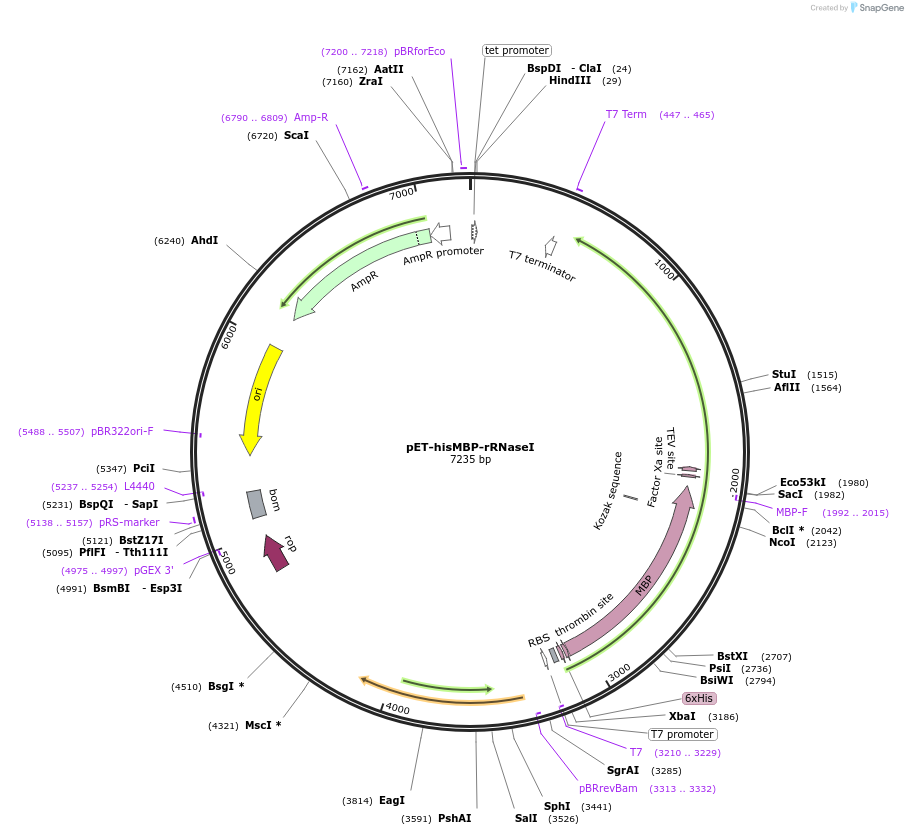
pET-hisMBP-rRNaseI
Plasmid#216673PurposeExpresses hisMBP-tagged rat oxidation-resistant RNase inhibitor in E. coli hostsDepositorAvailable SinceNov. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
pFN1-Aad9
Plasmid#244449PurposeA shuttle plasmid compatible with Fusobacterium nucleatum ATCC10953 or ATCC25586, carrying a spectinomycin resistance marker (aadA gene).DepositorInsertspFn1 Fn ori
Aad9
ExpressionBacterialAvailable SinceOct. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
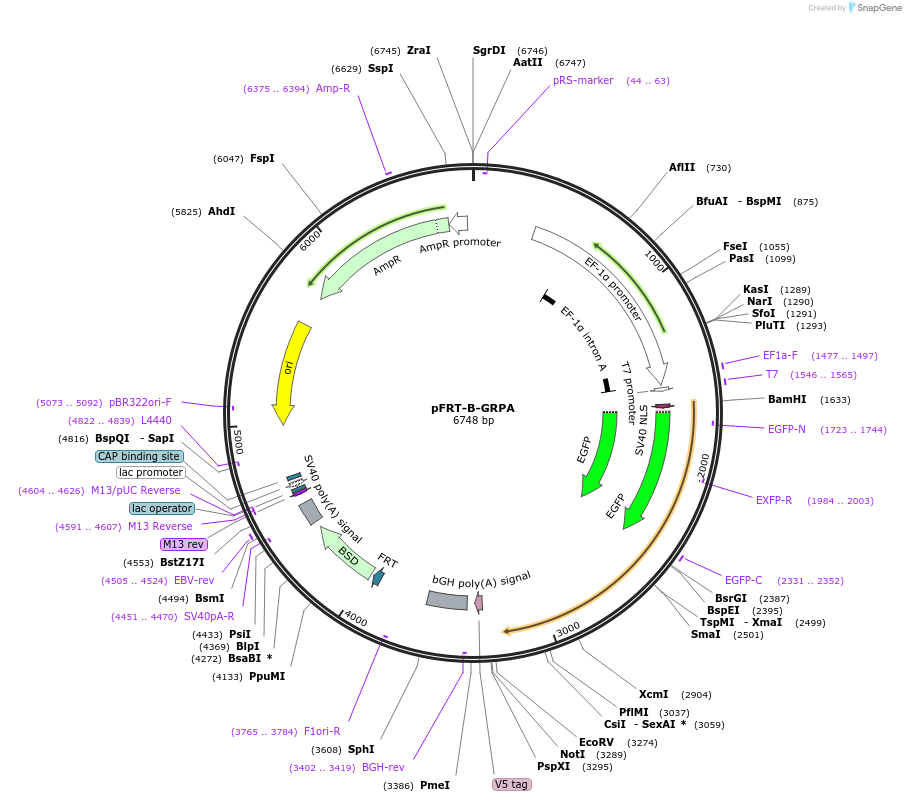
pFRT-B-GRPA
Plasmid#226222PurposeHuman RPA34 tagged to GFP under a EF1alpha promotor for generating stable HeLa Kyoto GFP-RPA34 using the Flp-In recombination systemDepositorAvailable SinceOct. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
pBI121::smRatioGFP:AQ
Plasmid#240453PurposeExpression of indicator protein fusion (soluble modified ratiometric pHluorin & Aequorin) for monitoring calcium concentrations and pH in the cytoplasm of higher plants. See Resource Information.DepositorInsertFusion of soluble modified ratiometric pHluorin and Aequorin
ExpressionBacterial and PlantMutationRatiometric GFP (ratiometric pHluorin) (PMID 9671…PromoterCauliflower mosaic virus 35S promoter (GenBank AJ…Available SinceSept. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
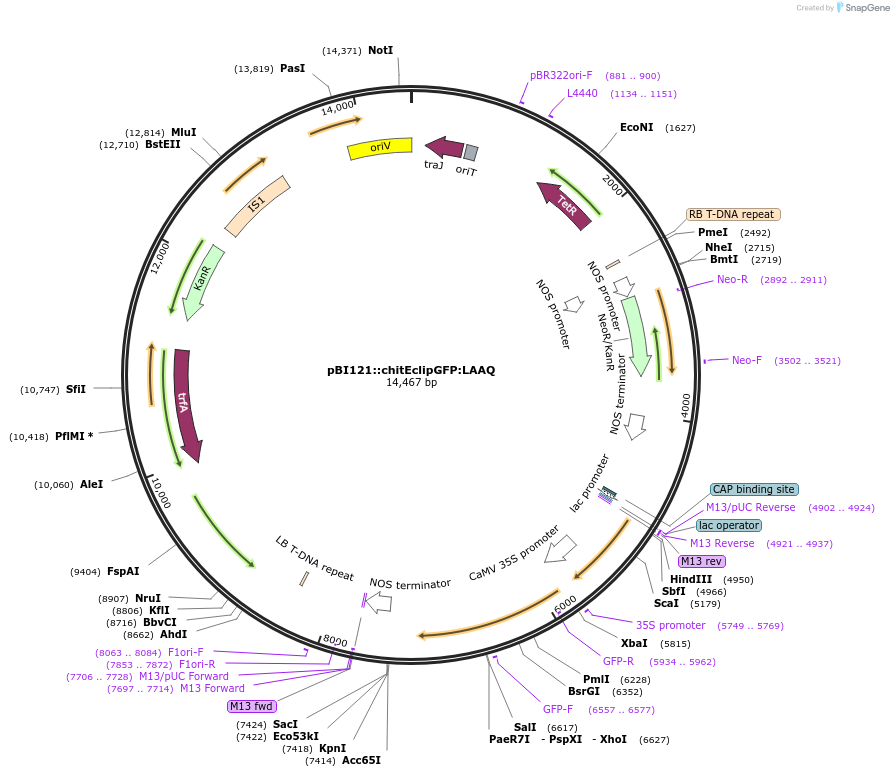
pBI121::chitEclipGFP:LAAQ
Plasmid#240452PurposeExpression of indicator protein fusion (modified ecliptic pHluorin & low affinity Aequorin) for monitoring calcium concentrations and pH in cell walls of higher plants. See Resource Information.DepositorInsertFusion of Chitinase signal, soluble modified ecliptic pHluorin and low affinity Aequorin (D119A)
ExpressionBacterial and PlantMutationEcliptic GFP (ecliptic pHluorin) (PMID 9671304); …PromoterCauliflower mosaic virus 35S promoter (GenBank AJ…Available SinceSept. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
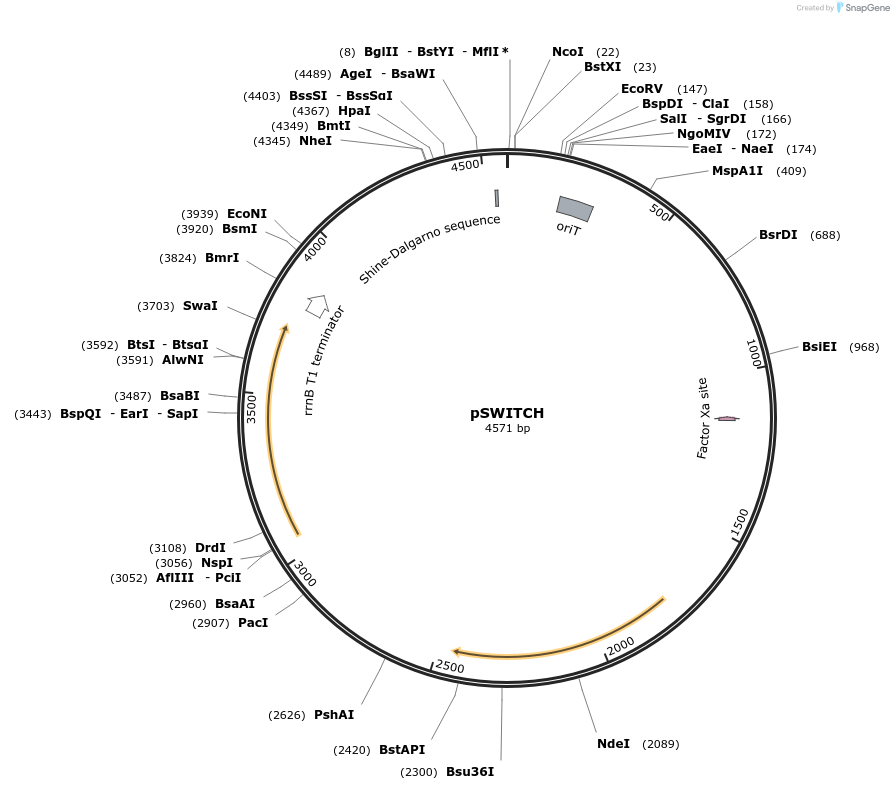
pSWITCH
Plasmid#222402PurposeExpression in Streptococcus pyogenesDepositorTypeEmpty backboneExpressionBacterialAvailable SinceJuly 2, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
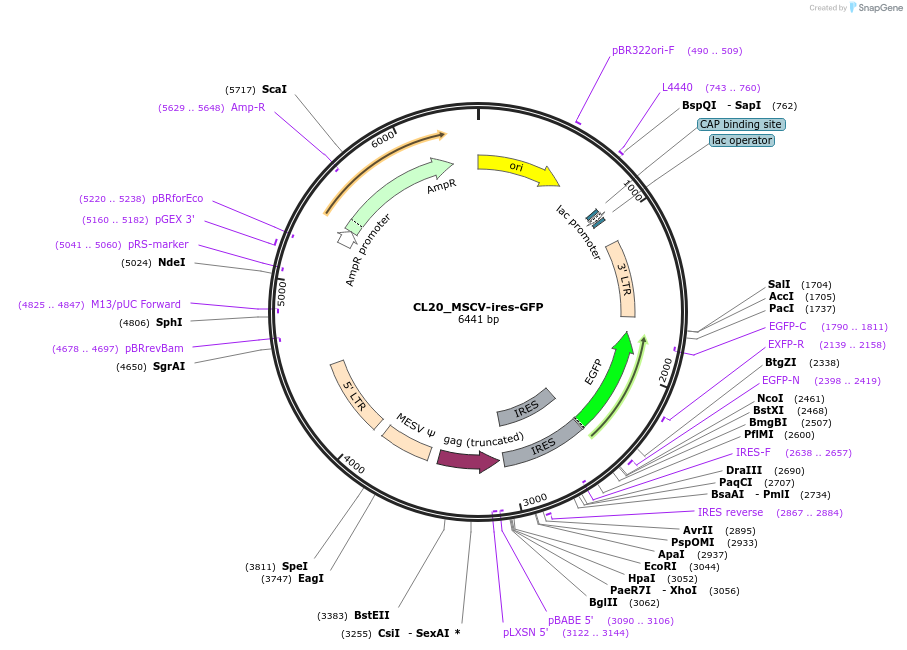
CL20_MSCV-ires-GFP
Plasmid#237496PurposeRetroviral empty vectorDepositorTypeEmpty backboneUseRetroviralExpressionMammalianAvailable SinceJune 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
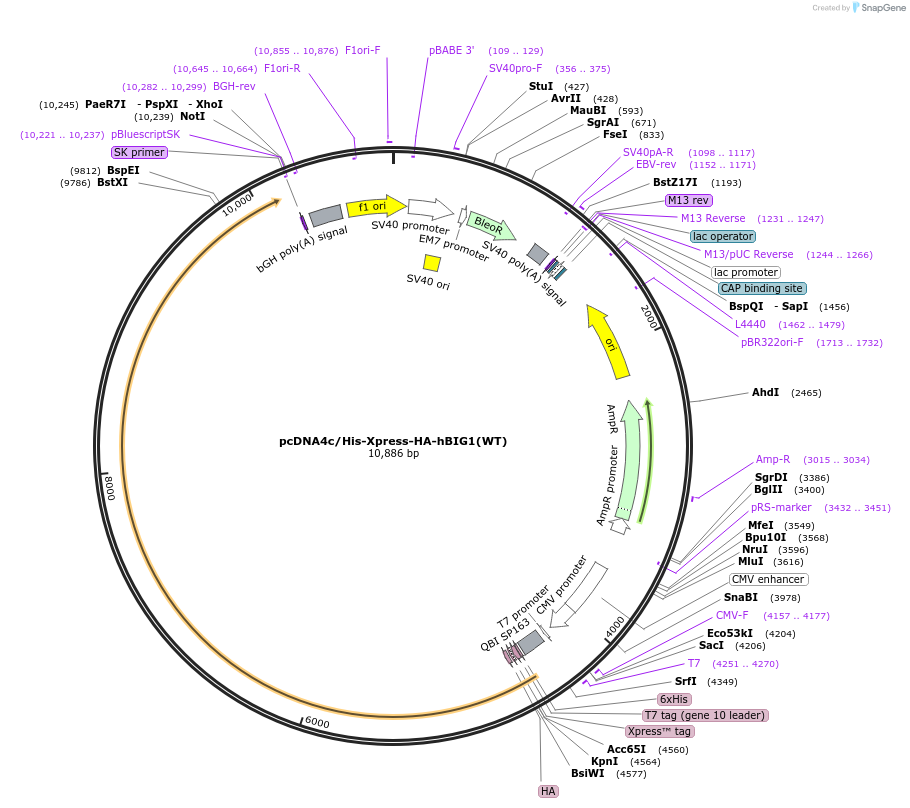
pcDNA4c/His-Xpress-HA-hBIG1(WT)
Plasmid#79431PurposeExpresses N-terminally His6, Xpress and HA-tagged BIG1(WT) in mammalian cellsDepositorAvailable SinceJune 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
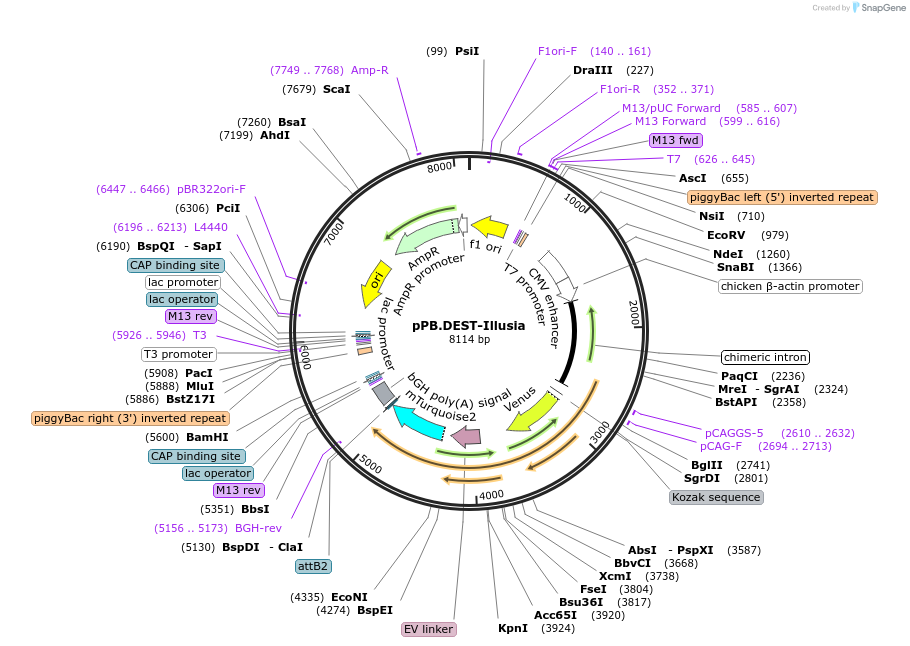
pPB.DEST-Illusia
Plasmid#215444PurposePiggybac expression vector with Illusia FRET reporter for integrin phosphorylationDepositorInsertIllusia
UseTransposon-based stable expressionExpressionMammalianPromoterCAGAvailable SinceMay 29, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
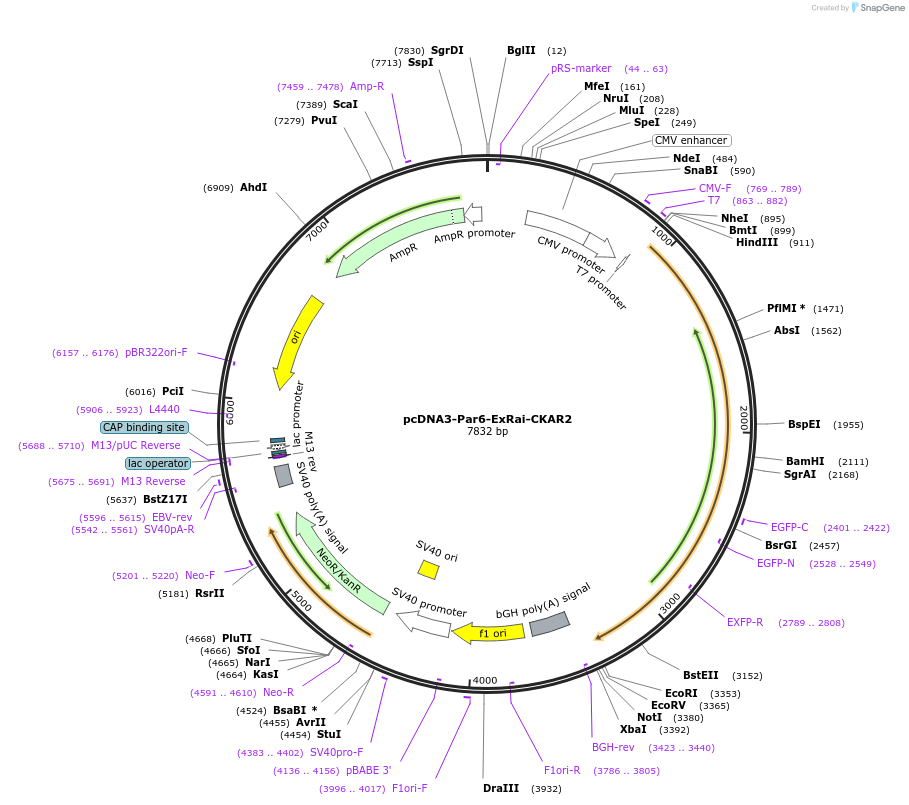
pcDNA3-Par6-ExRai-CKAR2
Plasmid#236101PurposeSubcellularly targeted enhanced excitation-ratiometric biosensor for monitoring Protein Kinase C activity in living cells for selective detection of aPKC activity within the Par complex.DepositorAvailable SinceMay 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
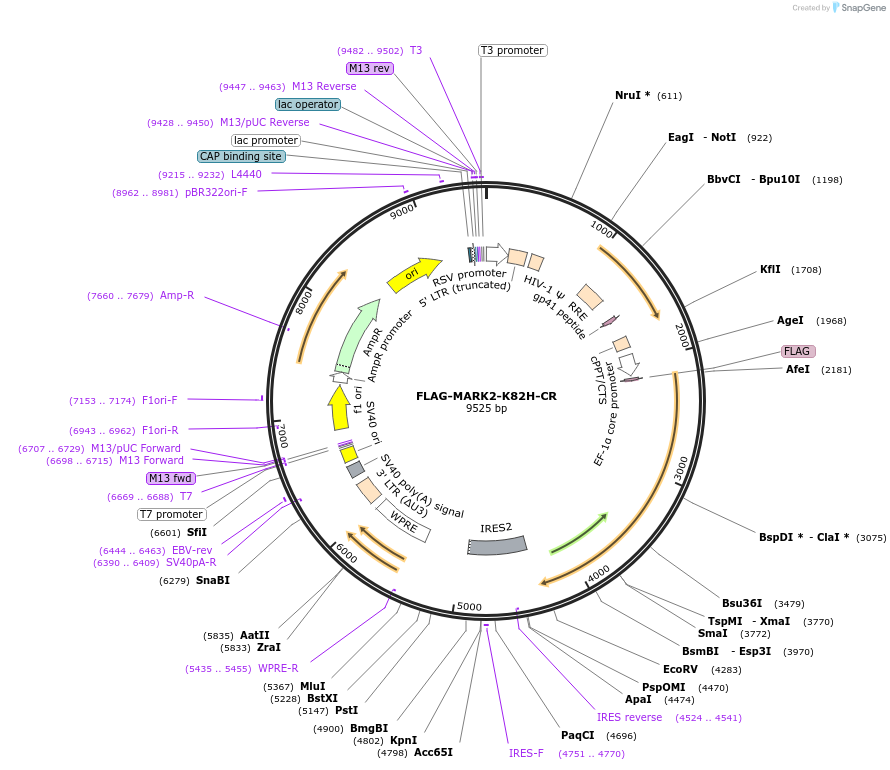
FLAG-MARK2-K82H-CR
Plasmid#229425PurposeLentiviral or overpexression of cDNADepositorInsertMARK2-K82H (MARK2 Human)
UseLentiviralTagsFLAGExpressionMammalianMutationKinase-dead mutant K82H; silent mutation CRISPR r…PromoterEFSAvailable SinceDec. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
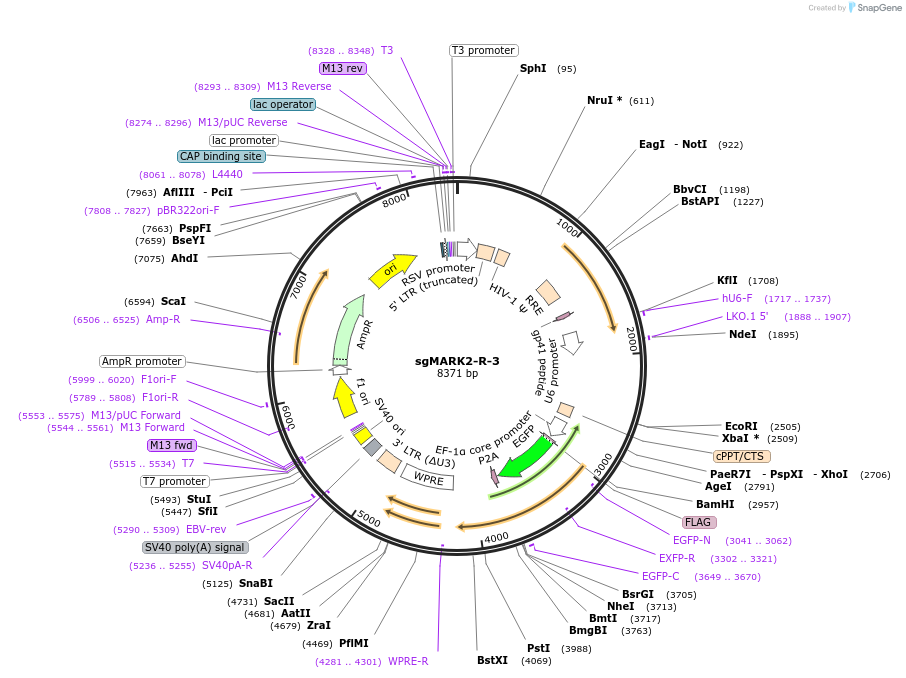
sgMARK2-R-3
Plasmid#229436Purposeknockout of MARK2DepositorInsertsgMARK2-R-3 (MARK2 Human)
UseCRISPR and LentiviralExpressionMammalianPromoterhU6;bU6;EFSAvailable SinceDec. 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
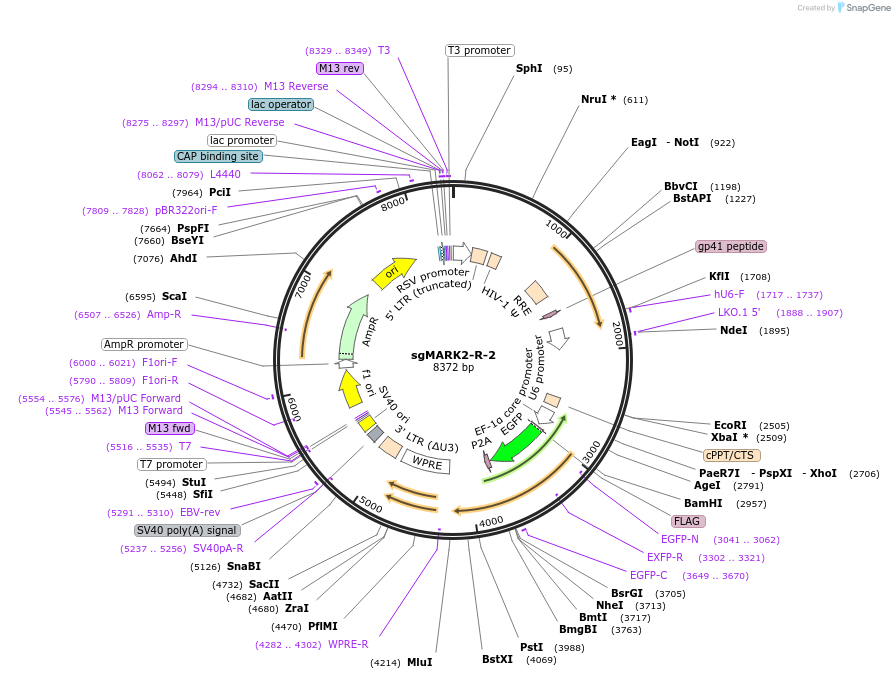
sgMARK2-R-2
Plasmid#229435Purposeknockout of MARK2DepositorInsertsgMARK2-R-2 (MARK2 Human)
UseCRISPR and LentiviralExpressionMammalianPromoterhU6;bU6;EFSAvailable SinceDec. 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
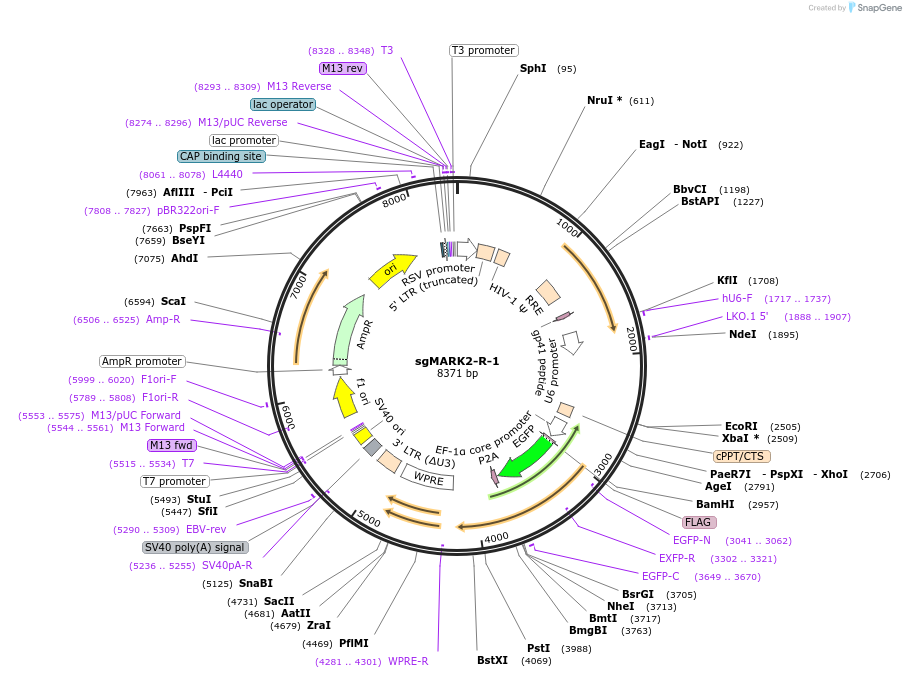
sgMARK2-R-1
Plasmid#229434Purposeknockout of MARK2DepositorInsertsgMARK2-R-1 (MARK2 Human)
UseCRISPR and LentiviralExpressionMammalianPromoterhU6;bU6;EFSAvailable SinceDec. 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
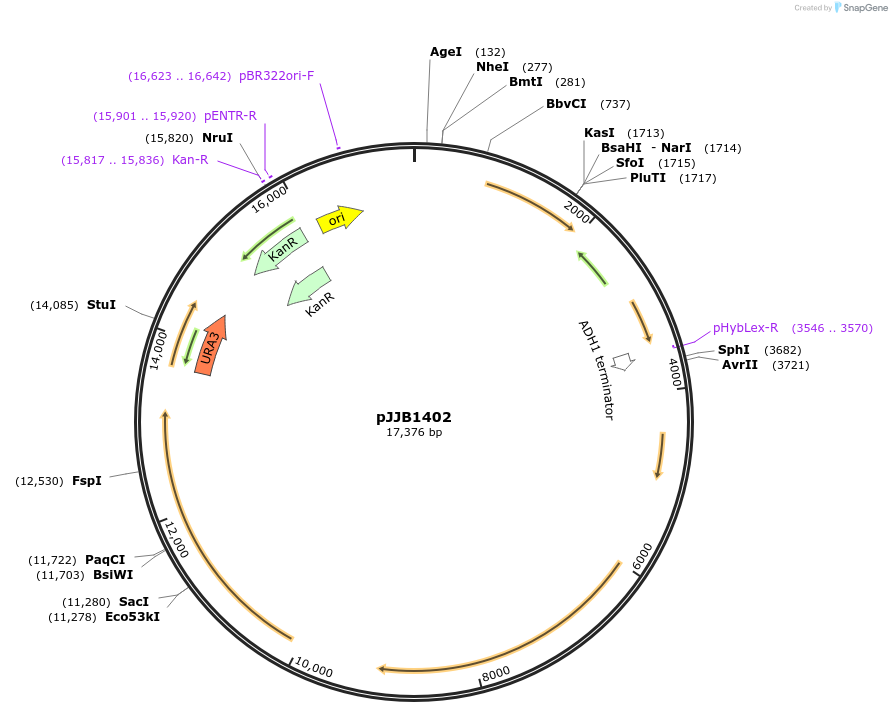
pJJB1402
Plasmid#218616PurposeExpress Pex15, Pex22, Pex4, Pex1, and Pex6DepositorInsertPex15
ExpressionYeastAvailable SinceOct. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
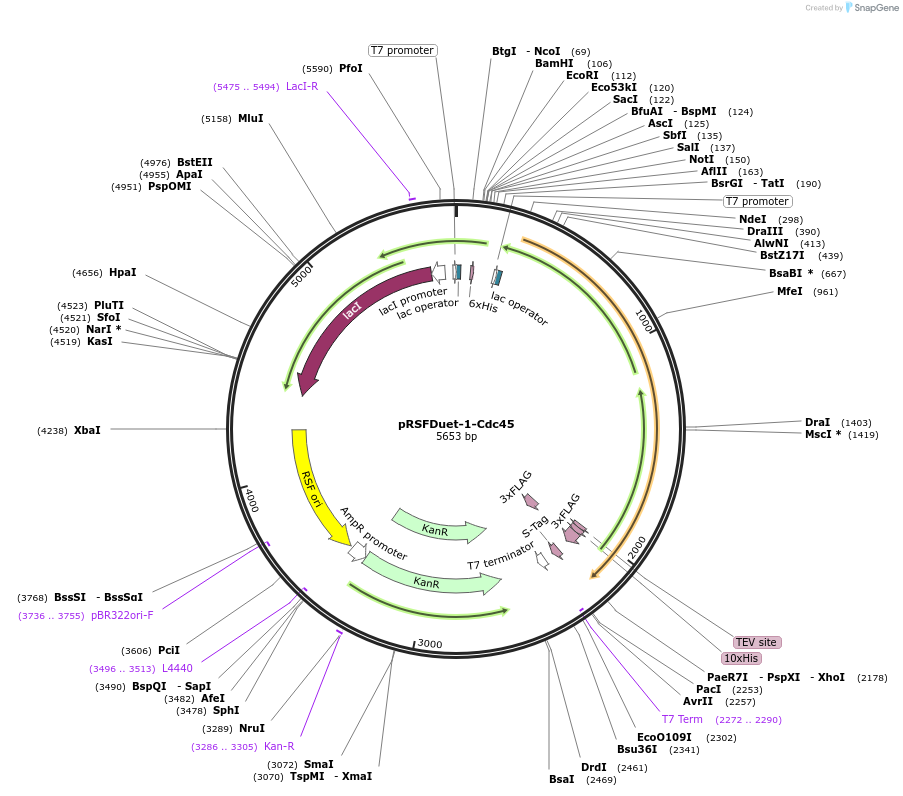
pRSFDuet-1-Cdc45
Plasmid#208796PurposeXenopus laevis FLAG5-His10-TEV-Cdc45DepositorAvailable SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
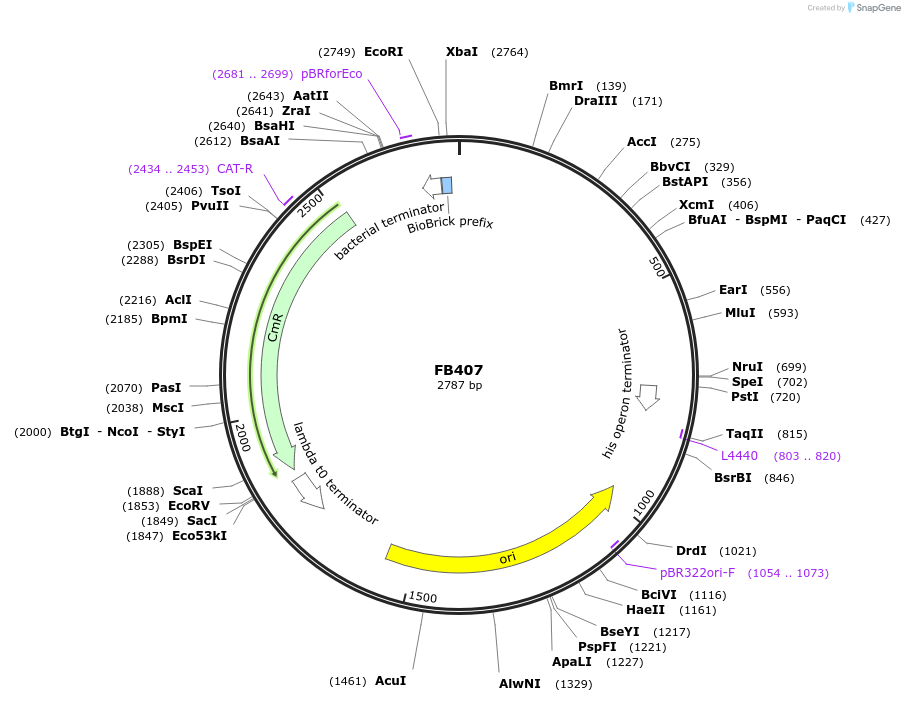
FB407
Plasmid#203628PurposePromoter of the elongation factor 1-alpha gene from P. digitatum (PDIG_59570).DepositorInsertPef1A
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
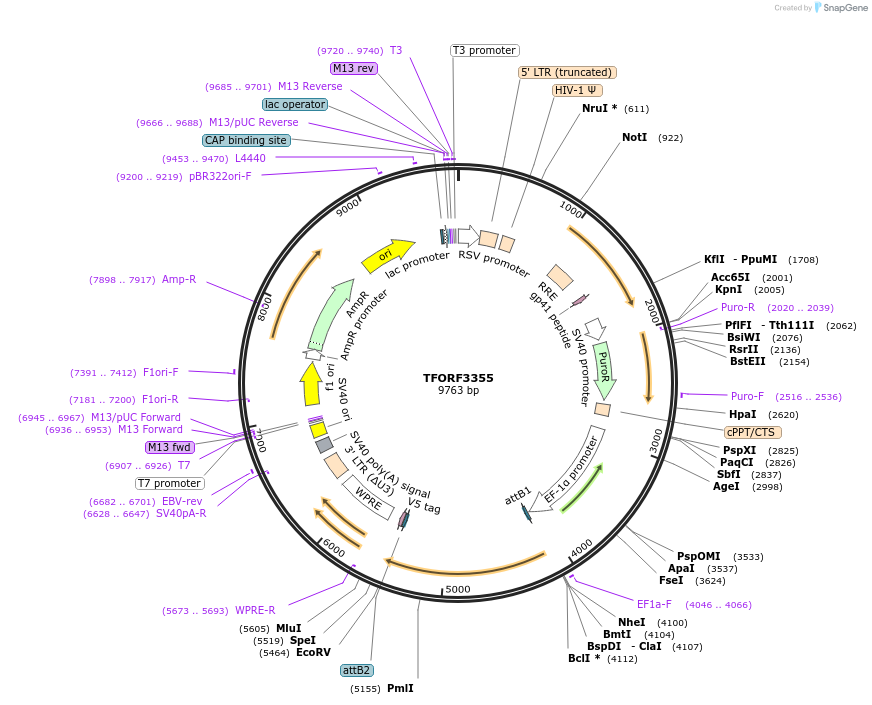
TFORF3355
Plasmid#144831PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceAug. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
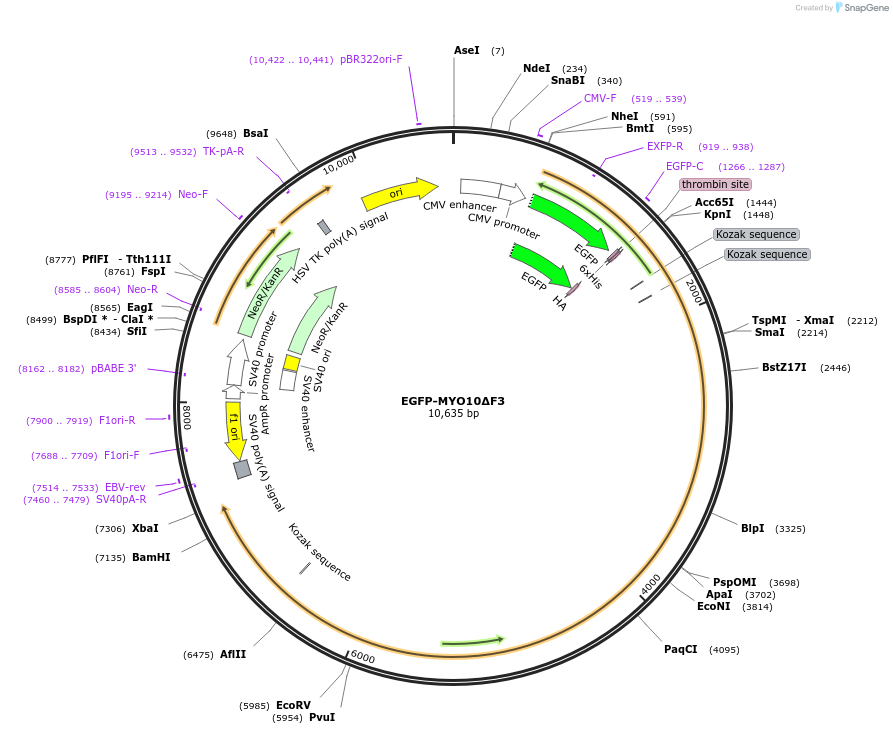
EGFP-MYO10ΔF3
Plasmid#194857PurposeExpress EGFP-MYO10 with the F3 FERM domain lobes deleted (1-1951) in mammalian cells.DepositorInsertMYO10 ΔF3 (MYO10 Human)
TagsEGFPExpressionMammalianMutationTruncated MYO10 construct. MYO10 amino acids 1-1…PromoterCMVAvailable SinceMarch 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
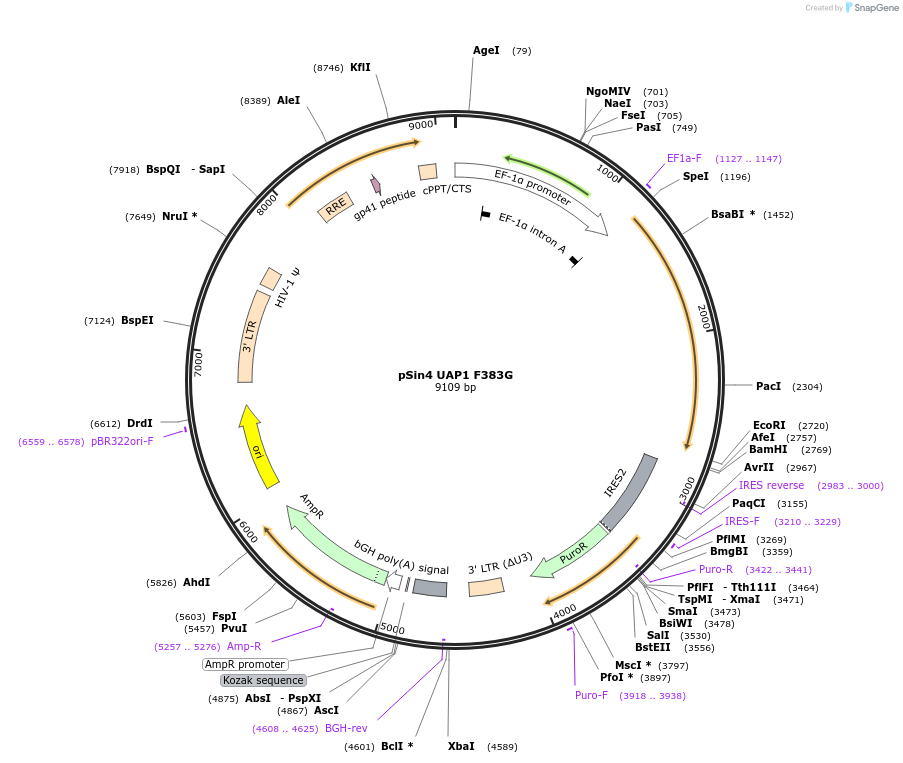
pSin4 UAP1 F383G
Plasmid#169891PurposeUsed for generation of stable mammalian cell lines expressing UAP1 with with F383G mutationDepositorInsertUDP-N-acetylhexosamine pyrophosphorylase (UAP1 Human)
UseLentiviralTagsNoneExpressionMammalianMutationChanged Phenylalanine (F) at position 383 to glyc…Available SinceJune 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
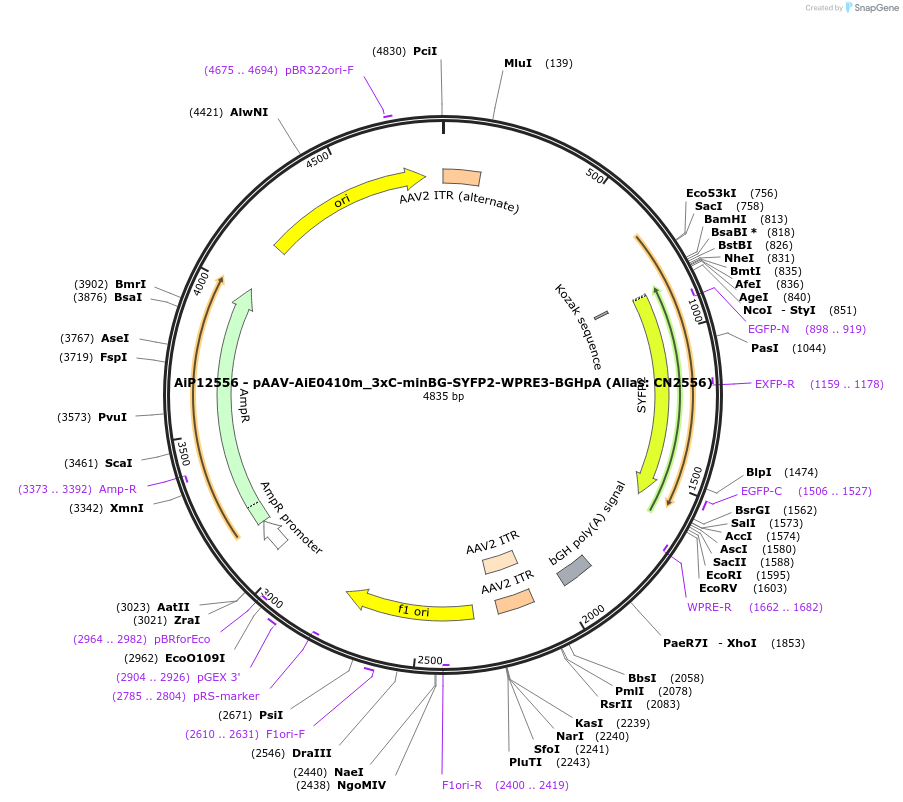
AiP12556 - pAAV-AiE0410m_3xC-minBG-SYFP2-WPRE3-BGHpA (Alias: CN2556)
Plasmid#208144PurposeAiE0410m_3xC2 is an optimized enhancer sequence designed to drive AAV-mediated transgene expression in brain oligodendrocytesDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
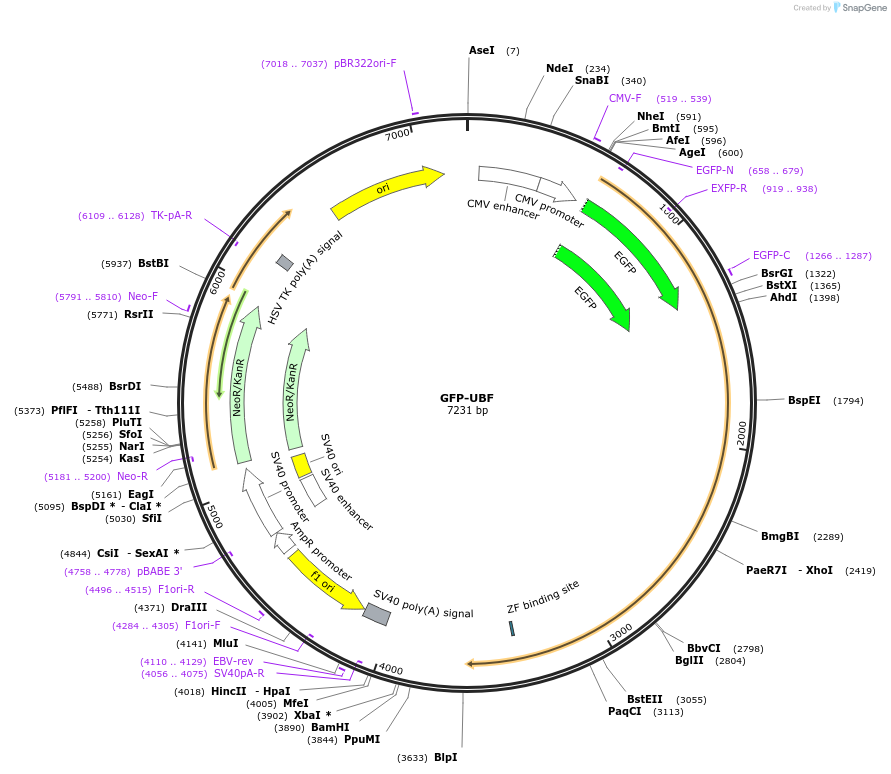
GFP-UBF
Plasmid#17656DepositorAvailable SinceApril 28, 2008AvailabilityAcademic Institutions and Nonprofits only -
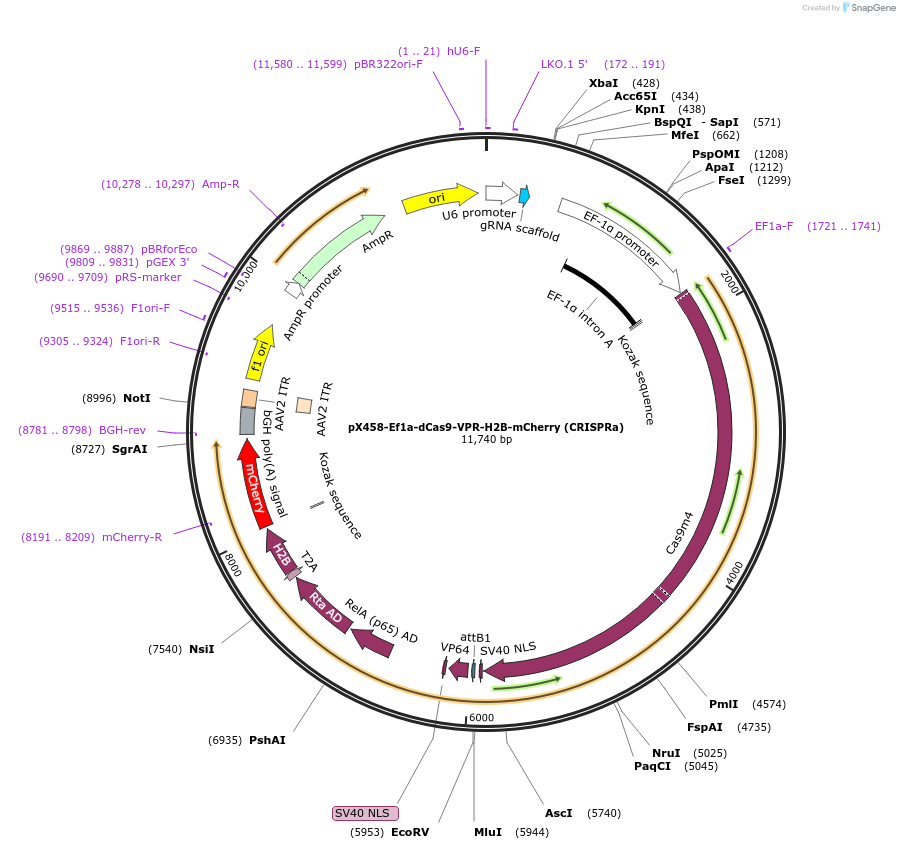
pX458-Ef1a-dCas9-VPR-H2B-mCherry (CRISPRa)
Plasmid#175572PurposeInactivated Cas9 (dCas9) fusion to VPR for the purpose of targeted activationDepositorTypeEmpty backboneUseCRISPRAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
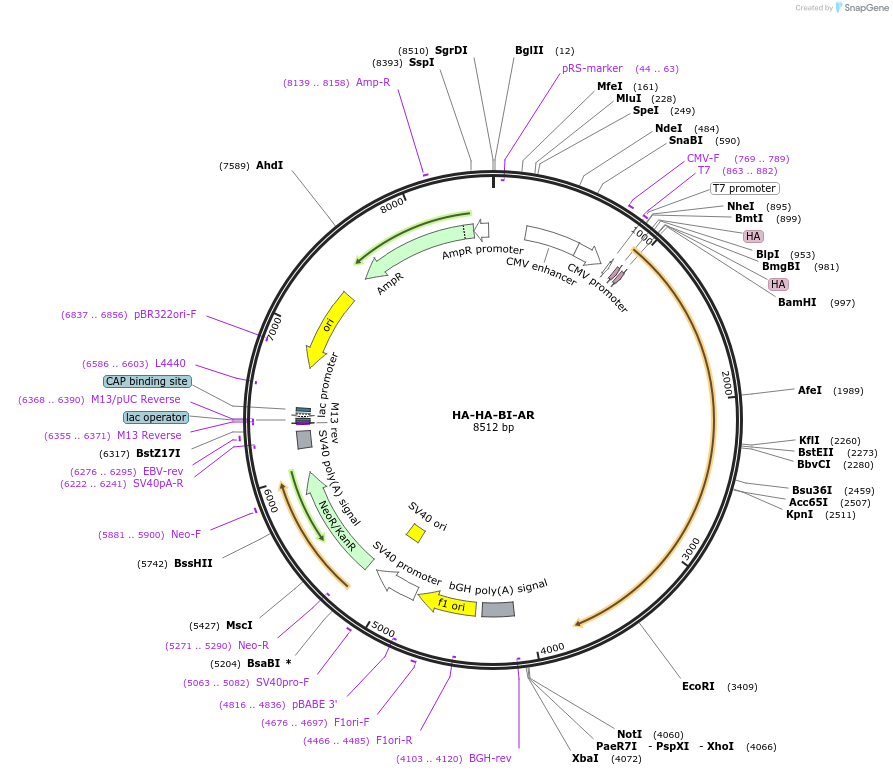
HA-HA-BI-AR
Plasmid#171234PurposeMammalian expression of 2xHA-tagged human androgen receptor (hAR)DepositorInsert2xHA-tagged human androgen receptor (AR Human)
TagsHAExpressionBacterial and MammalianPromoterCMVAvailable SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
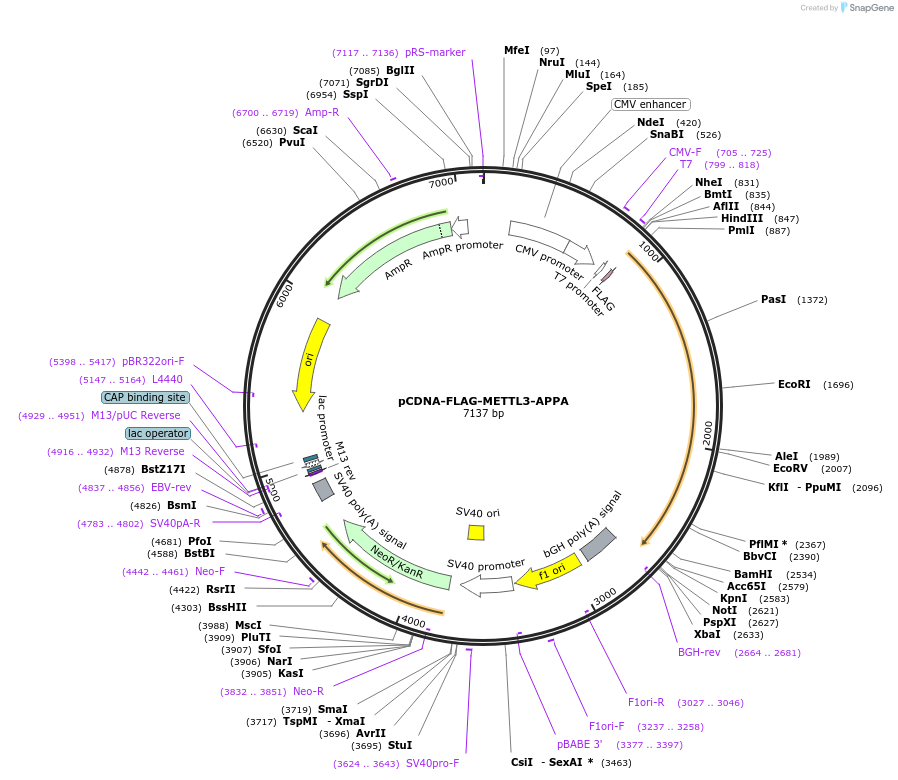
pCDNA-FLAG-METTL3-APPA
Plasmid#160251PurposeExpress catalytic inactive FLAG-METTL3 in human cellsDepositorInsertmethyltransferase like 3 (METTL3 Human)
TagsFLAGExpressionMammalianMutationchanges in aa 395–398, DPPW → APPAPromoterCMVAvailable SinceNov. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
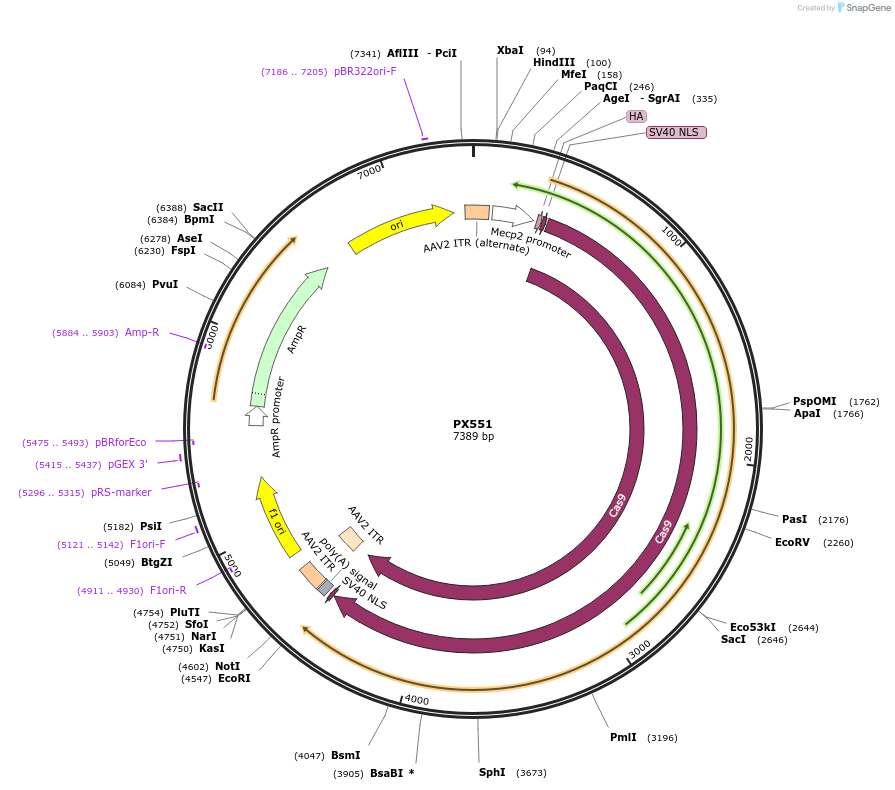
PX551
Plasmid#60957PurposepAAV-pMecp2-SpCas9-spA (AAV-SpCas9). AAV plasmid expressing Cas9 in neurons under control of truncated mecp2 promoter.DepositorInsertSpCas9
UseAAV and CRISPRTagsHAExpressionMammalianPromoterpMecp2Available SinceNov. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
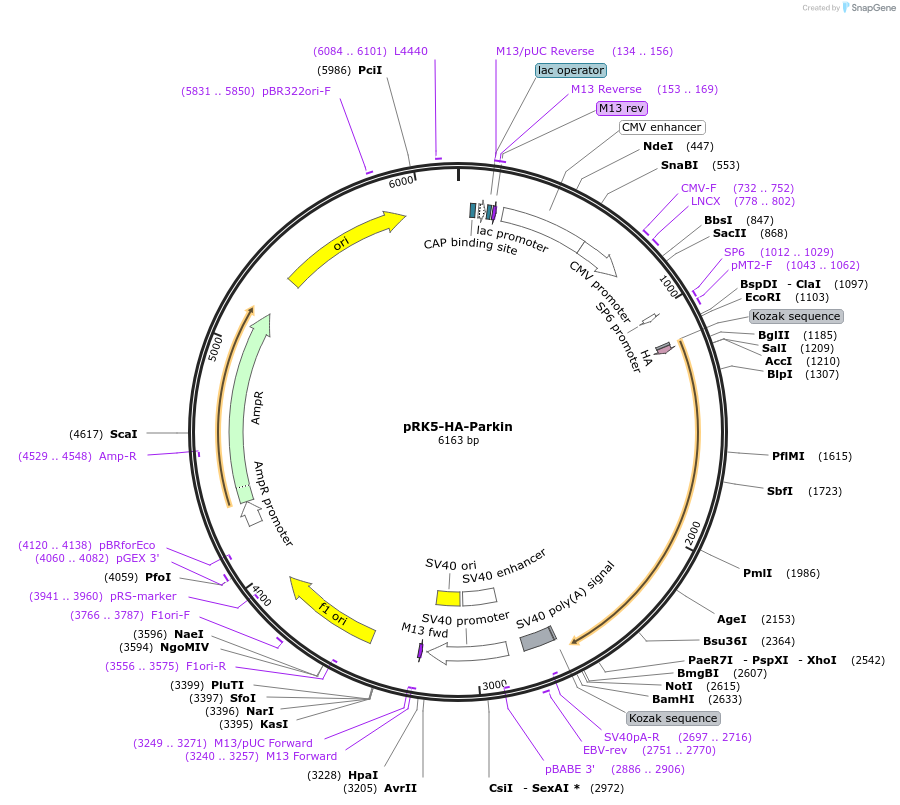
pRK5-HA-Parkin
Plasmid#17613DepositorAvailable SinceApril 3, 2008AvailabilityAcademic Institutions and Nonprofits only -
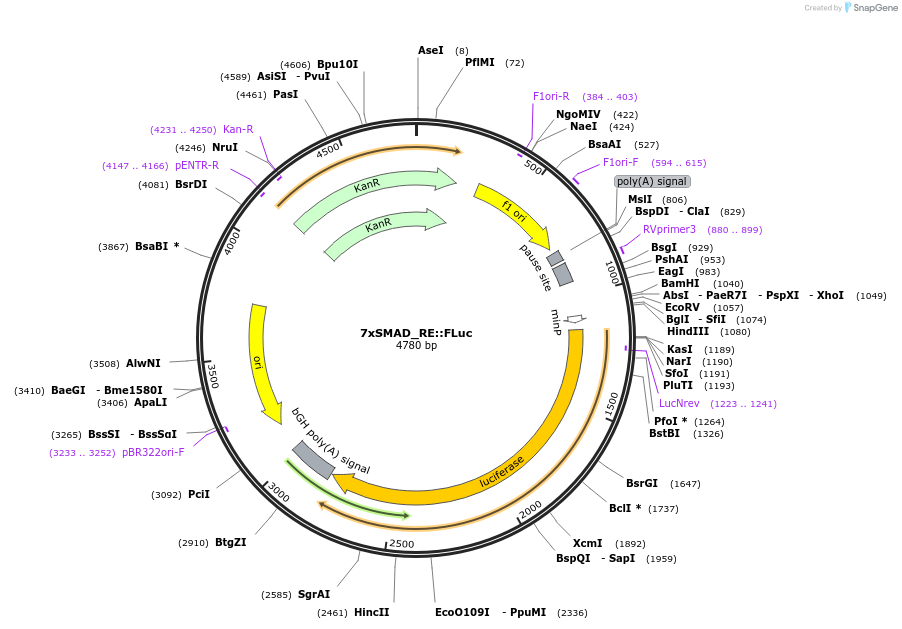
7xSMAD_RE::FLuc
Plasmid#124531PurposeSMAD2 FLuc luciferase reporterDepositorInsertSMAD FLuc reporter
UseLuciferase and Synthetic BiologyExpressionMammalianAvailable SinceJune 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
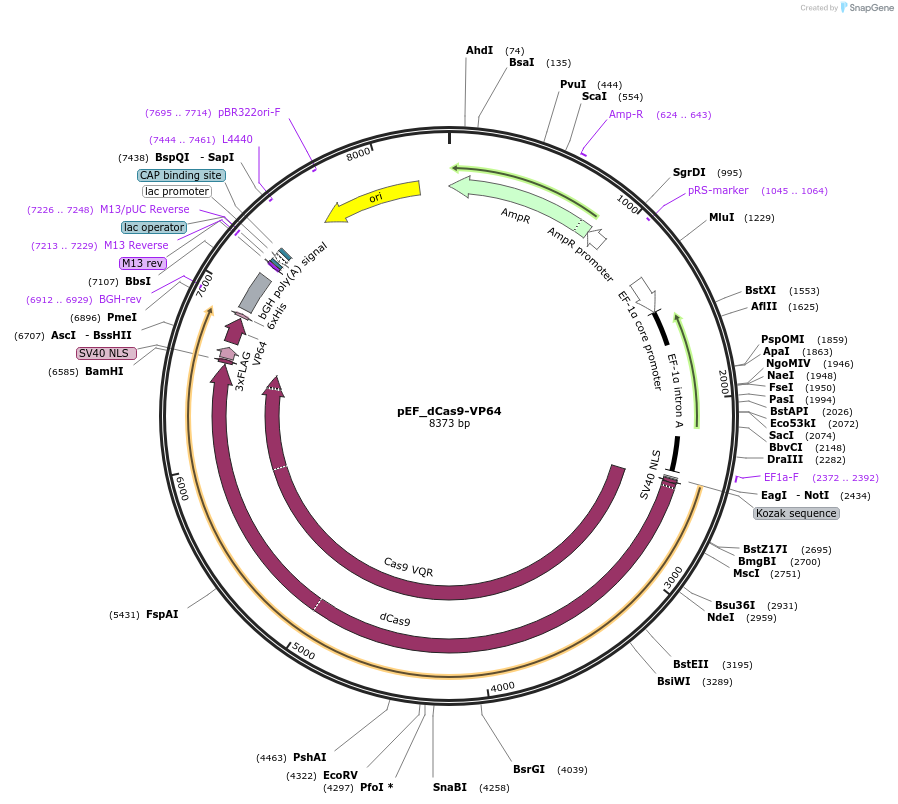
pEF_dCas9-VP64
Plasmid#68417PurposeTransient expression of Sp-dCas9 fused to the VP64 transcription activator, in mammalian cells, under an EF1-alpha promoter.DepositorInsertdCas9
UseCRISPRTags3xFLAG and VP64ExpressionMammalianMutationD10A/H841APromoterhuman EF1[alpha]Available SinceSept. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
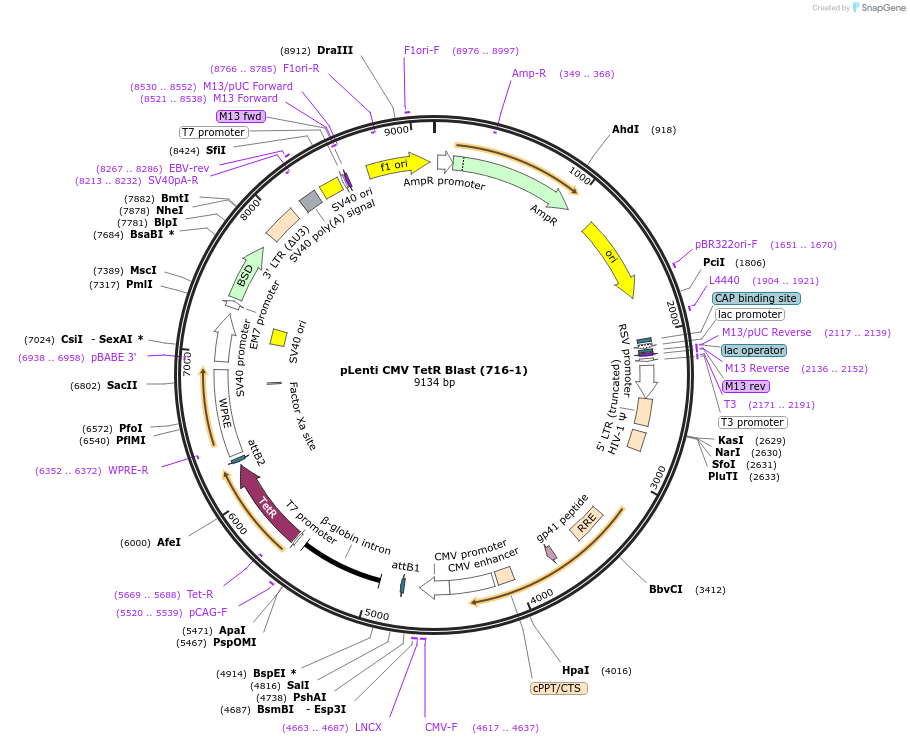
pLenti CMV TetR Blast (716-1)
Plasmid#17492Purpose3rd gen lentiviral Tetracycline repressor expression vector, CMV promoter, BlastDepositorInsertTetracycline repressor
UseLentiviralExpressionMammalianAvailable SinceAug. 12, 2009AvailabilityAcademic Institutions and Nonprofits only -
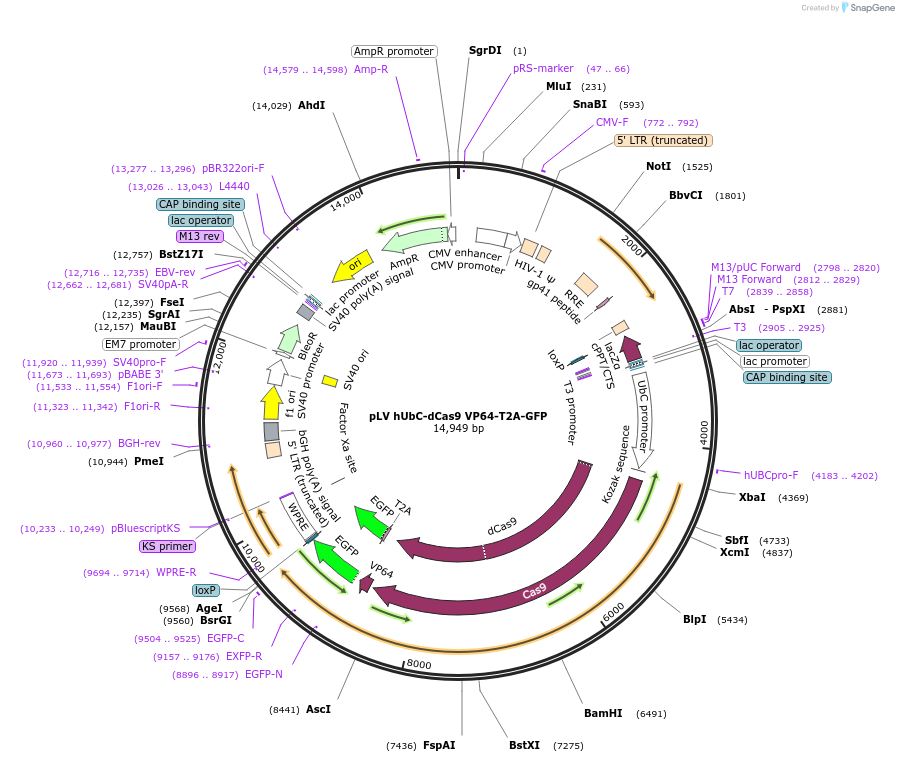
pLV hUbC-dCas9 VP64-T2A-GFP
Plasmid#53192PurposeCo-expresses human optimized S. pyogenes dCas9-VP64 and GFPDepositorInserthumanized dead Cas9 VP64 T2A GFP
UseCRISPR and LentiviralTagsFlagExpressionMammalianMutationD10A and H840APromoterhUbCAvailable SinceJune 25, 2014AvailabilityAcademic Institutions and Nonprofits only -
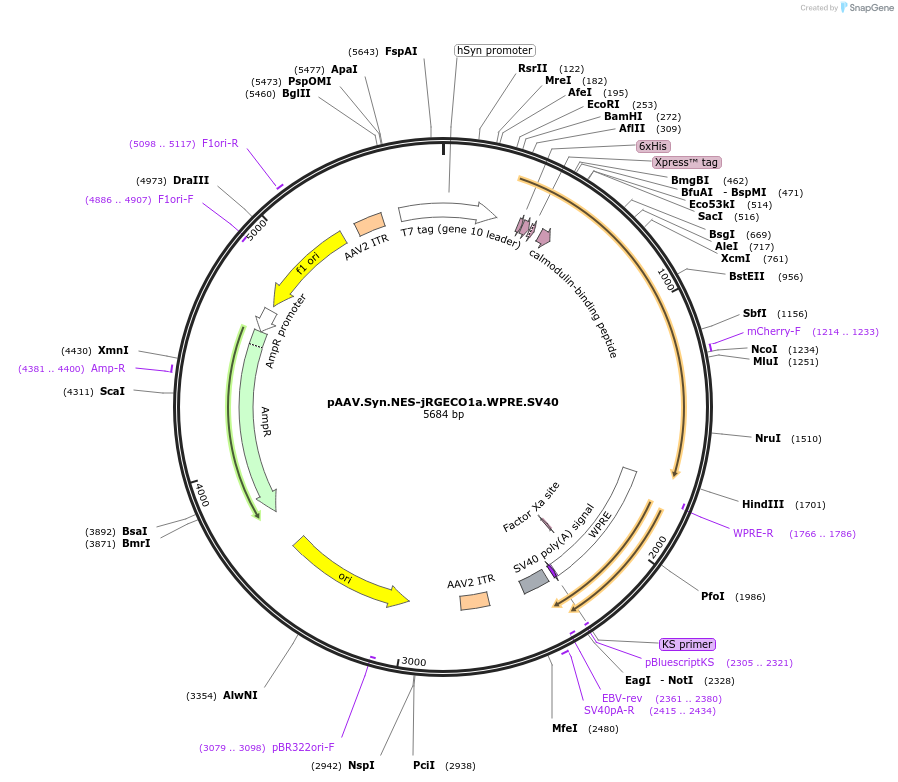
pAAV.Syn.NES-jRGECO1a.WPRE.SV40
Plasmid#100854PurposeAAV expression of jRGECO1a, a red fluorescent calcium sensor protein, from the Synapsin promoterDepositorHas ServiceAAV PHP.eB, AAV Retrograde, AAV1, and AAV9InsertjRGECO1a
UseAAVExpressionMammalianPromoterSynapsinAvailable SinceNov. 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
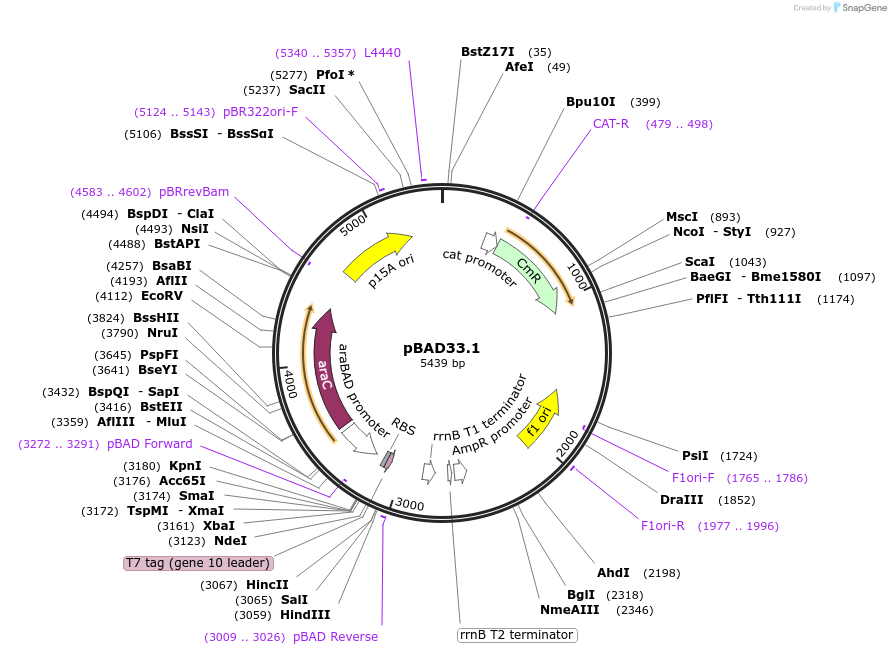
pBAD33.1
Plasmid#36267DepositorTypeEmpty backboneExpressionBacterialPromoterPBADAvailable SinceOct. 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
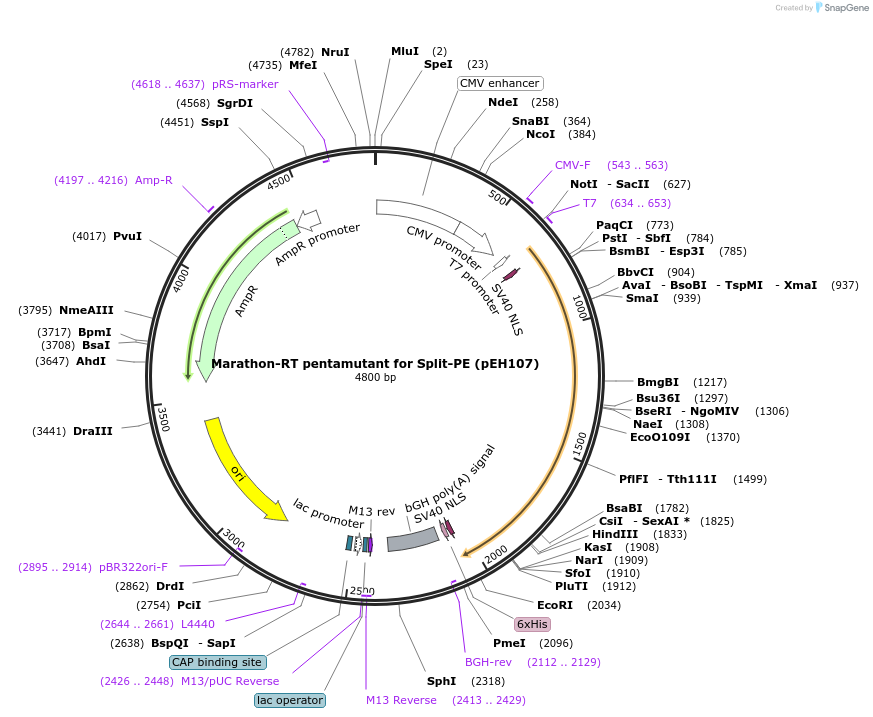
Marathon-RT pentamutant for Split-PE (pEH107)
Plasmid#190110PurposeExpresses Marathon-RT pentamutant (D14R-N26R-D74R-N116K-N197R)DepositorInsertMarathon-RT
TagsbpNLSExpressionMammalianMutationD14R, N26R, D74R, N116K, N197RPromoterCMVAvailable SinceSept. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
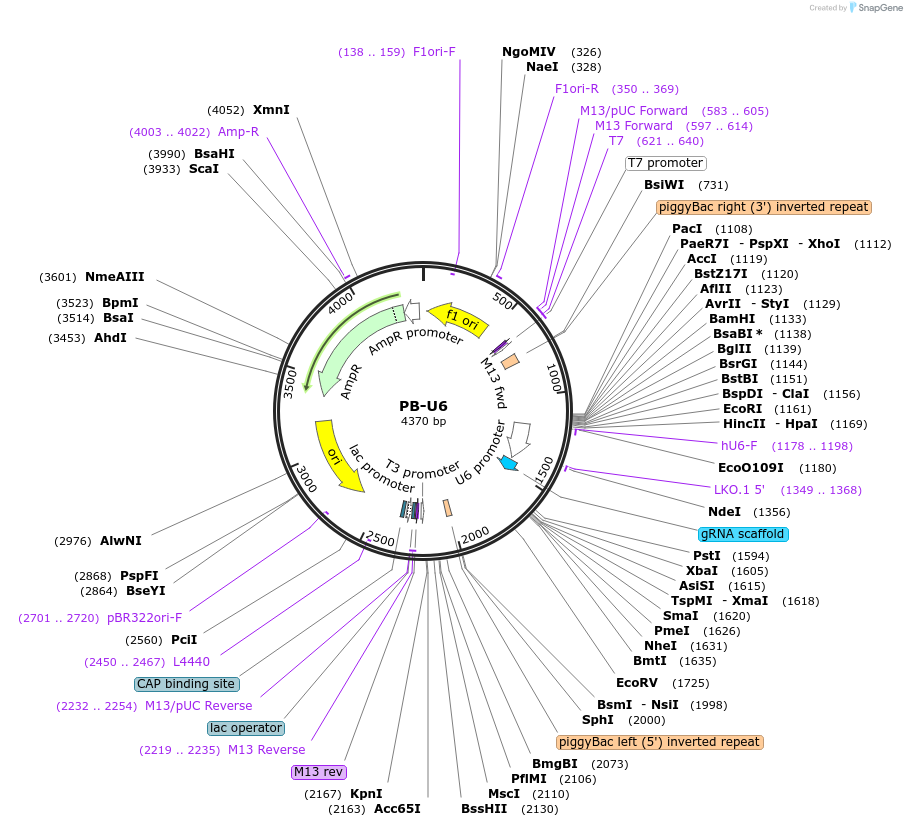
PB-U6
Plasmid#120358PurposepiggyBac transposon vector for sgRNA expression.DepositorTypeEmpty backboneUseCRISPR; Piggybac transposonExpressionMammalianPromoterU6Available SinceApril 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
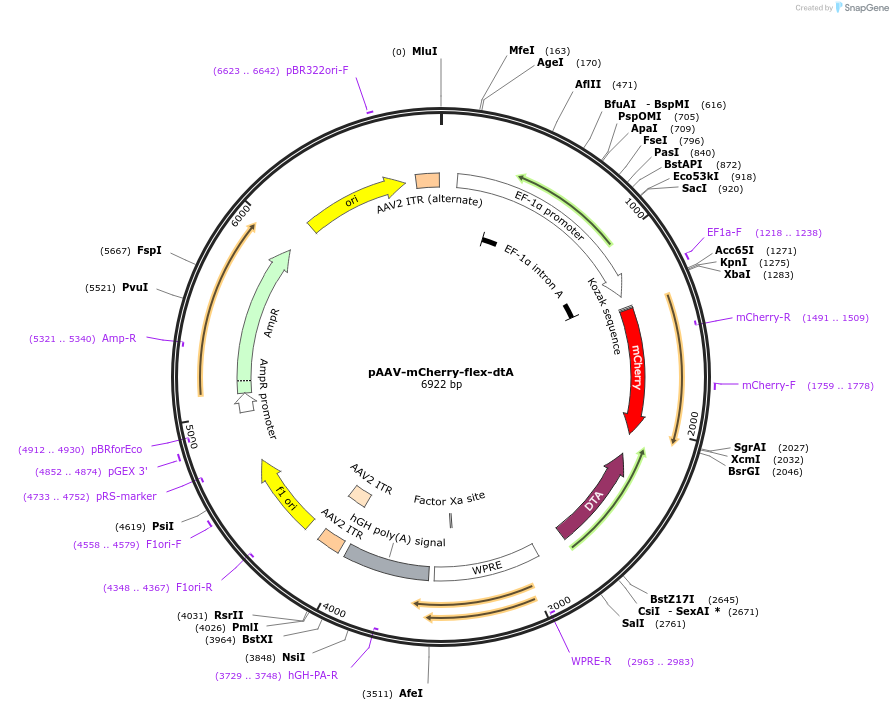
pAAV-mCherry-flex-dtA
Plasmid#58536PurposeExpresses diphtheria toxin A subunit in Cre positive cells, and mCherry in all the infected cellsDepositorAvailable SinceAug. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
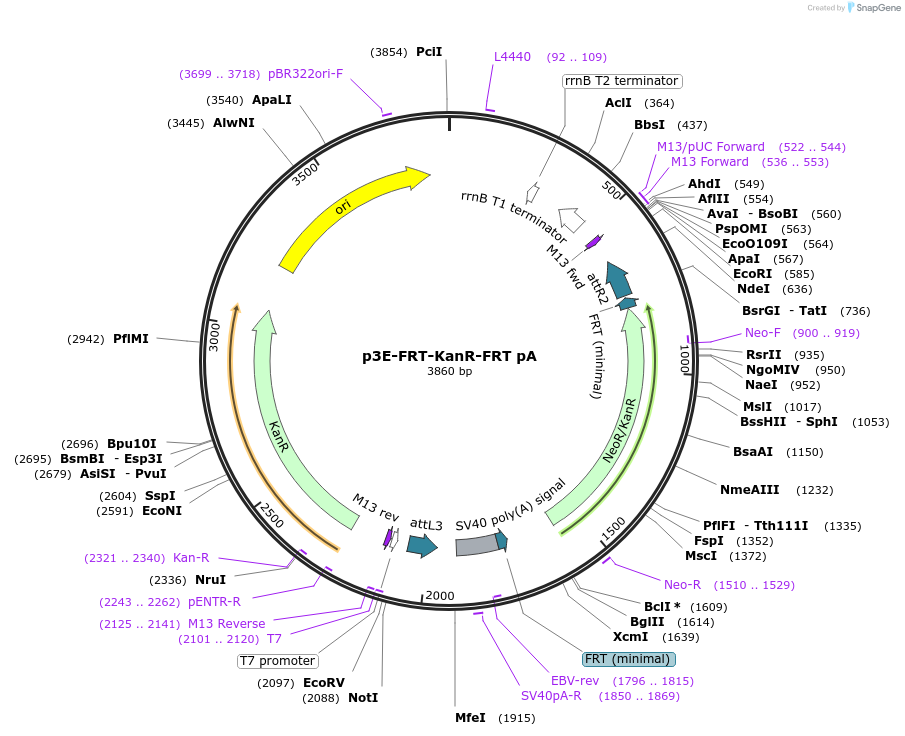
p3E-FRT-KanR-FRT pA
Plasmid#82601Purpose3' entry vector containing FRT-flanked kanamycin resistance cassette (KanR) with pA for FLP-induced kanamycin resistance, drug selectionDepositorInsertFRT-Kan-FRT
PromoterNoneAvailable SinceDec. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
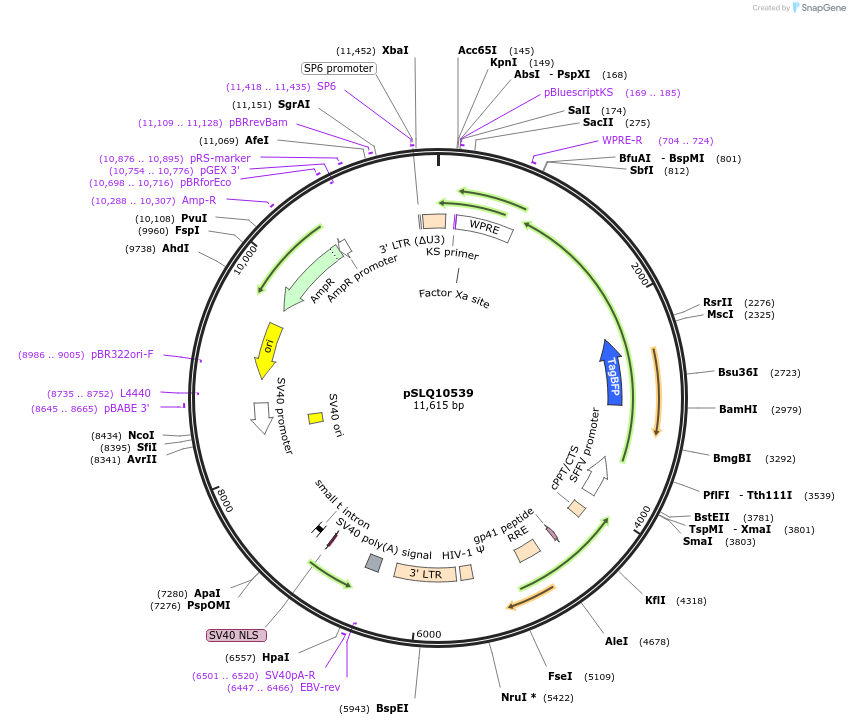
pSLQ10539
Plasmid#239270PurposeExpresses PYL1-DDX6 for recruiting RNA to the p-body via CRISPR-TODepositorAvailable SinceSept. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
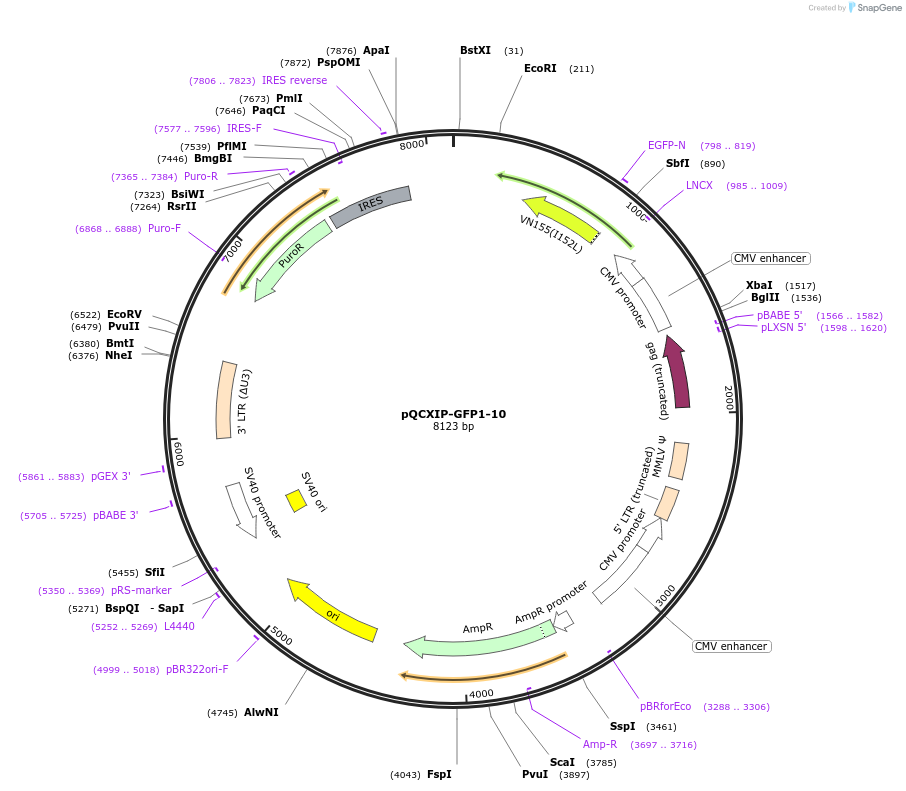
pQCXIP-GFP1-10
Plasmid#68715PurposeSplit GFP assayDepositorInsertGFP1-10
UseRetroviralExpressionMammalianPromoterCMVAvailable SinceFeb. 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
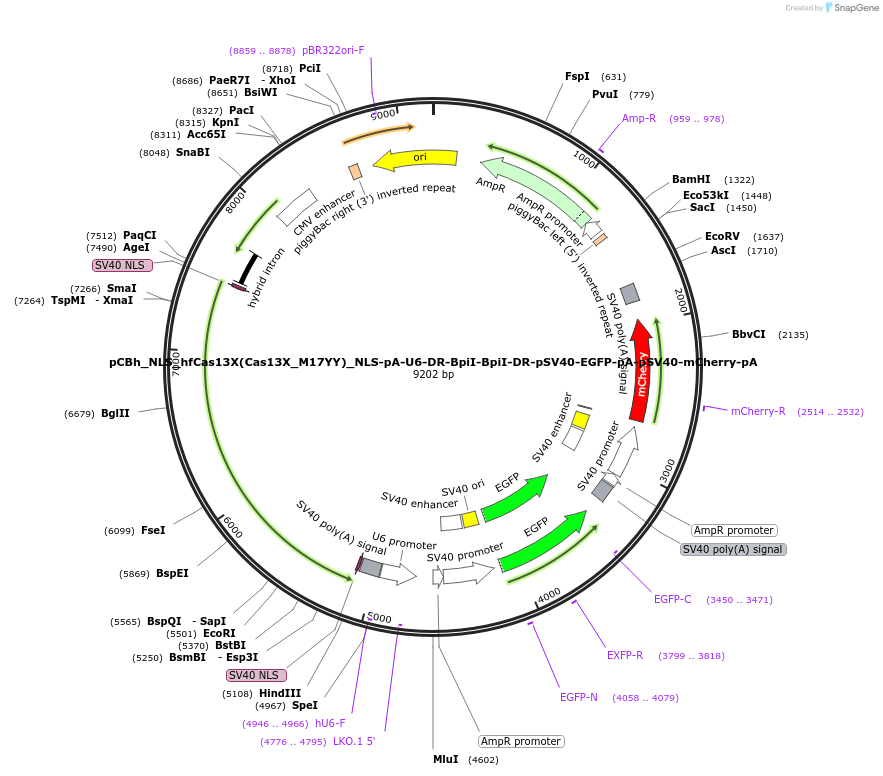
pCBh_NLS_hfCas13X(Cas13X_M17YY)_NLS-pA-U6-DR-BpiI-BpiI-DR-pSV40-EGFP-pA-pSV40-mCherry-pA
Plasmid#190033Purposevector for encoding a human codon-optimized High-fidelity Cas13X (hfCas13X) driven by CBh promoter, guide RNAs compatible with Cas13X driven by hU6, EGFP and mCherry driven by SV40 promotersDepositorInserthumanized hfCas13X
ExpressionMammalianMutationY672A,Y676APromoterCBh, SV40, hU6Available SinceSept. 12, 2022AvailabilityAcademic Institutions and Nonprofits only