We narrowed to 11,082 results for: CHL
-
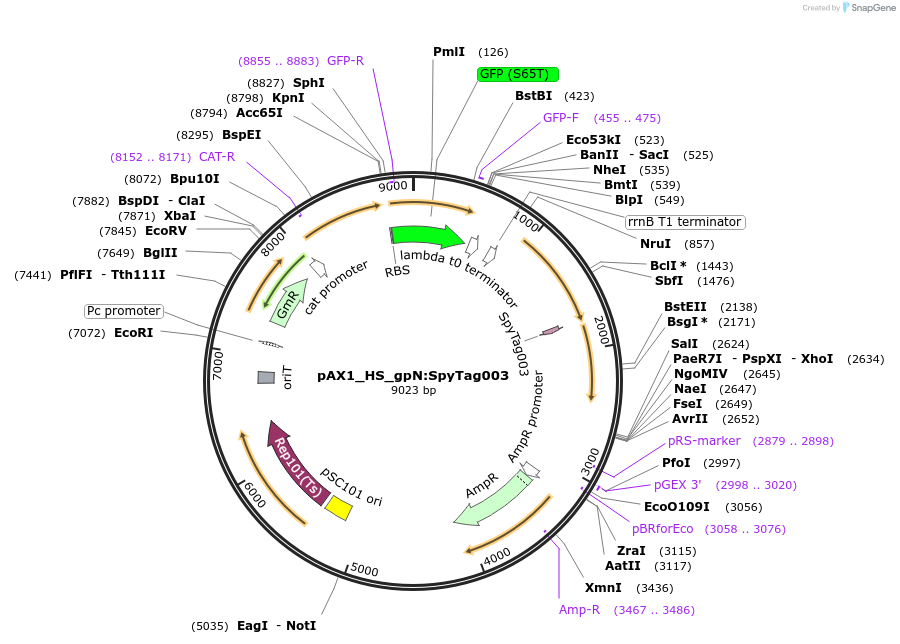
Plasmid#225194PurposeUsed for adding C-terminal SpyTag003 to the E. coli HS P2 phage major capsid proteinDepositorInsertgpN homology arms and SpyTag003
ExpressionBacterialAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
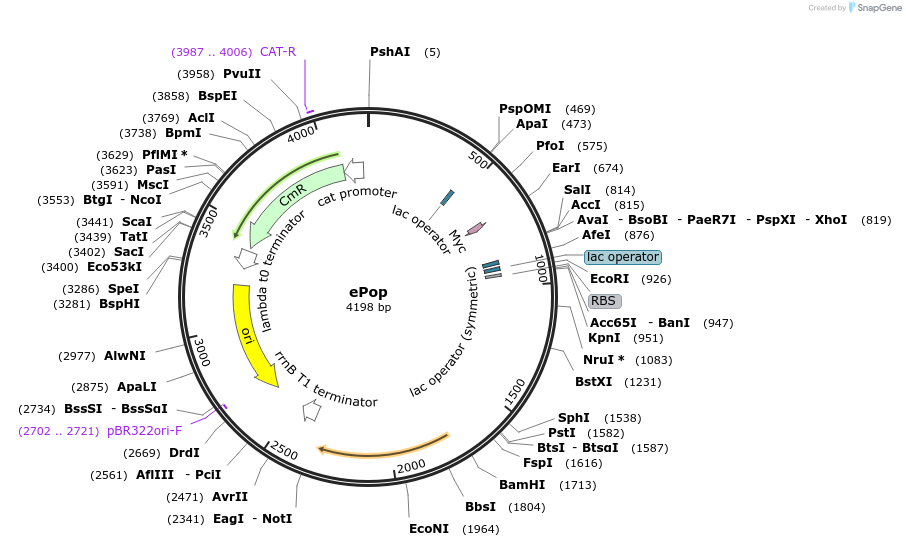
ePop
Plasmid#50953PurposeExpresses E lysis protein that causes in subpopulation death when threshold cell density is reached, resulting in oscillationsDepositorInsertsE lysis protein
LuxI
LuxR
MutationEarly stop codon introduced in mutated luxR resul…Available SinceApril 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
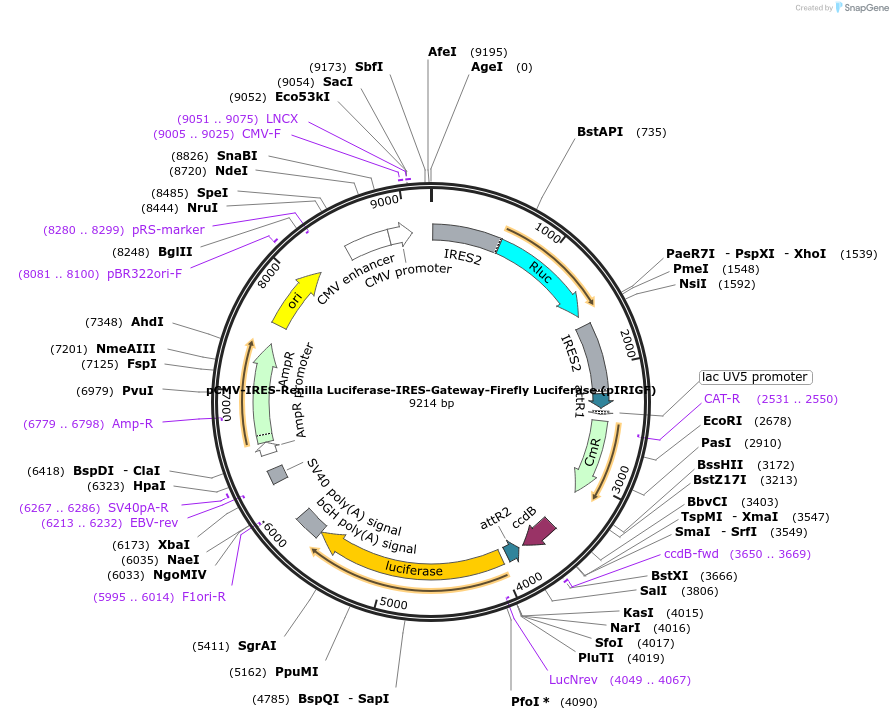
pCMV-IRES-Renilla Luciferase-IRES-Gateway-Firefly Luciferase (pIRIGF)
Plasmid#101139PurposeLuciferase reporter plasmid. Note that both renilla luciferase and ORF-firefly luciferase are encoded by a single mRNA and both are translated independently via internal ribosomal binding sites.DepositorTypeEmpty backboneExpressionMammalianPromoterCMVAvailable SinceOct. 11, 2017AvailabilityAcademic Institutions and Nonprofits only -
scOrange chromoprotein
Plasmid#117840PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses scOrange chromoprotein in E. coliDepositorInsertpromoter, RBS, scOrange
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
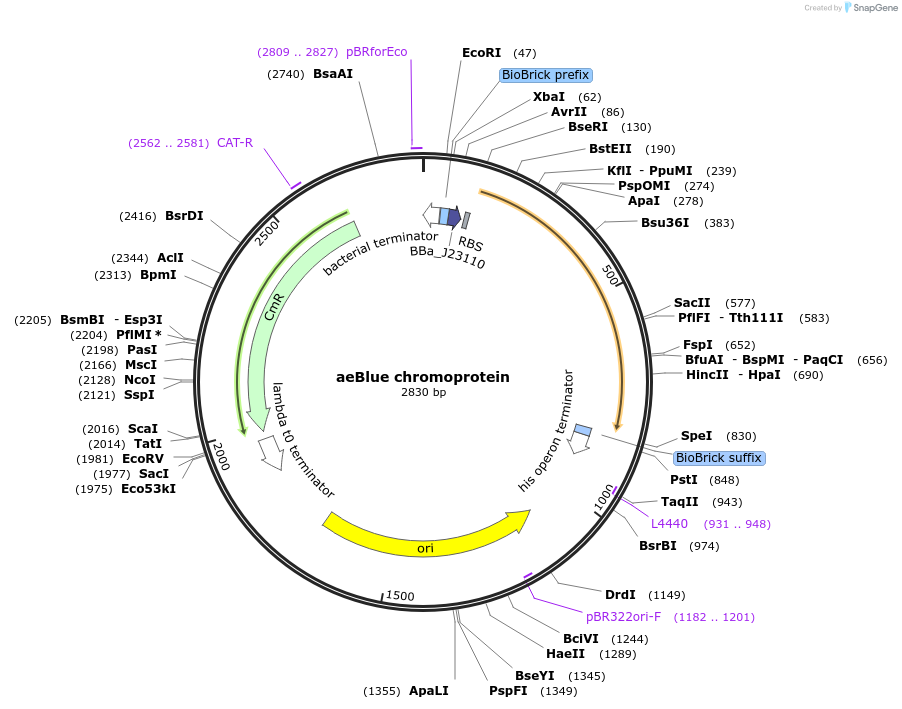
aeBlue chromoprotein
Plasmid#117846PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses aeBlue chromoprotein in E. coliDepositorInsertpromoter, RBS, aeBlue
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
amilGFP chromoprotein
Plasmid#117842PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amilGFP chromoprotein in E. coliDepositorInsertpromoter, RBS, amilGFP
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
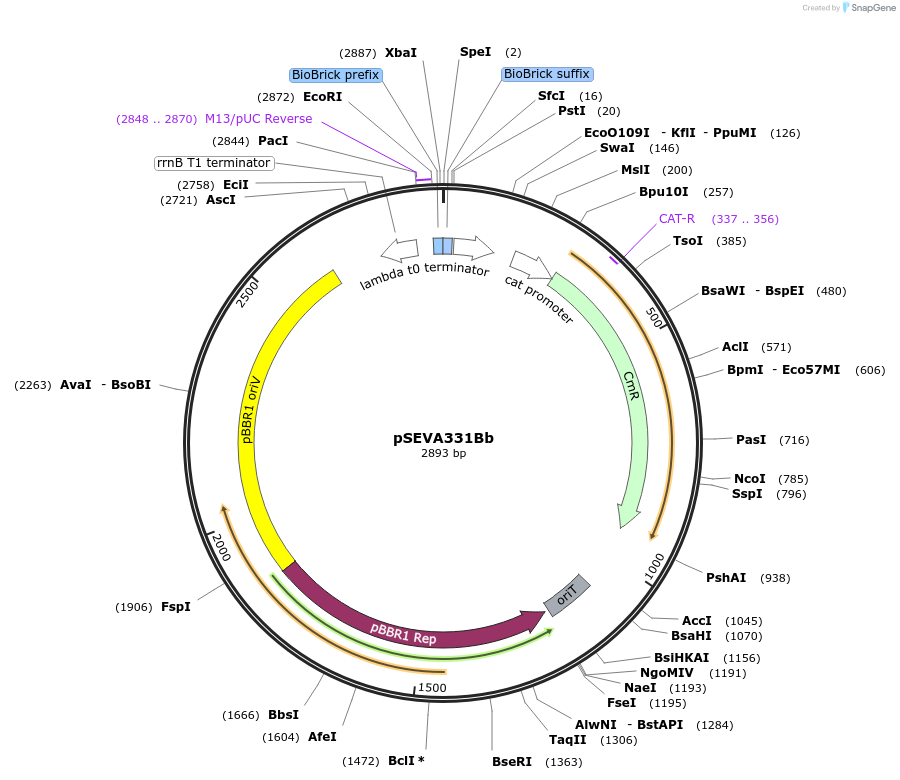
pSEVA331Bb
Plasmid#78269PurposepSEVA331 with Biobrick multiple cloning siteDepositorTypeEmpty backboneAvailable SinceNov. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
pLX303
Plasmid#25897Purpose3rd generation lentiviral Gateway destination vector. Blasticidin selection.DepositorTypeEmpty backboneUseLentiviral; Gateway destination vectorExpressionMammalianAvailable SinceJuly 30, 2010AvailabilityAcademic Institutions and Nonprofits only -
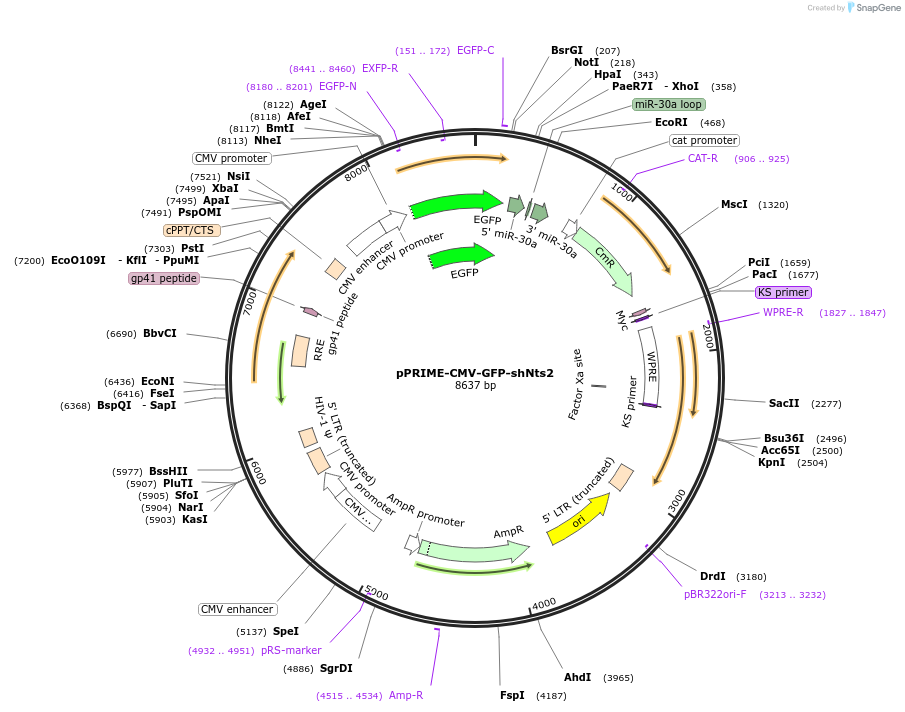
pPRIME-CMV-GFP-shNts2
Plasmid#132714PurposeEncodes short hairpin RNA (shRNA) #2 that targets the 3’-untranslated region of the rat NtsDepositorAvailable SinceOct. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
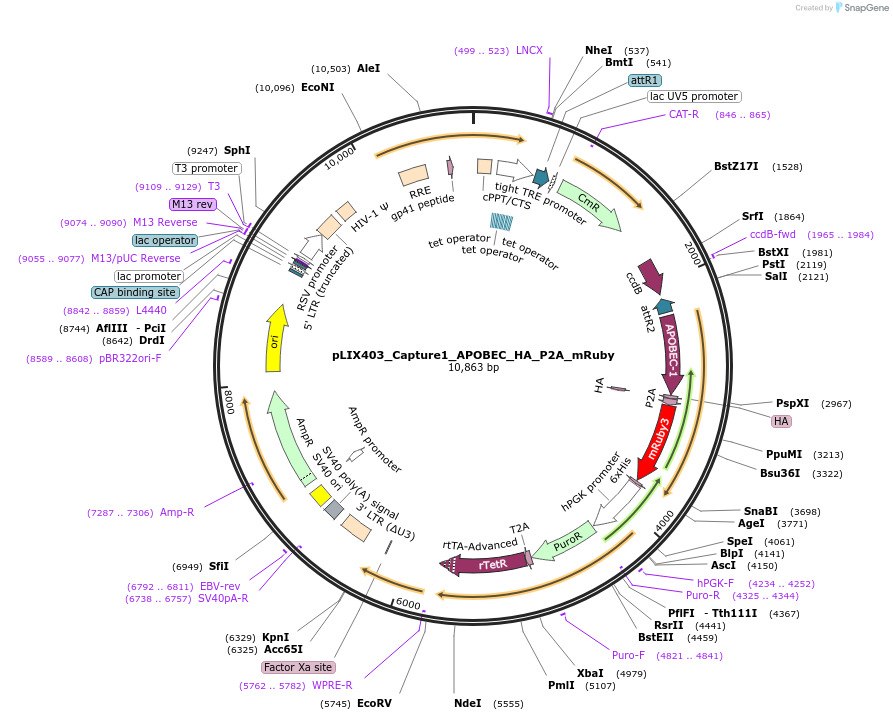
pLIX403_Capture1_APOBEC_HA_P2A_mRuby
Plasmid#183901PurposeInducible lentiviral expression, TRE-gateway-APOBEC-HA-P2A-mRuby; PGK-puro-2A-rtTADepositorTypeEmpty backboneUseLentiviral; Gateway destination vector, doxycycli…TagsCapture1_APOBEC_HA_P2A_mRubyExpressionMammalianPromoterTRE promoter, Tet ONAvailable SinceOct. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
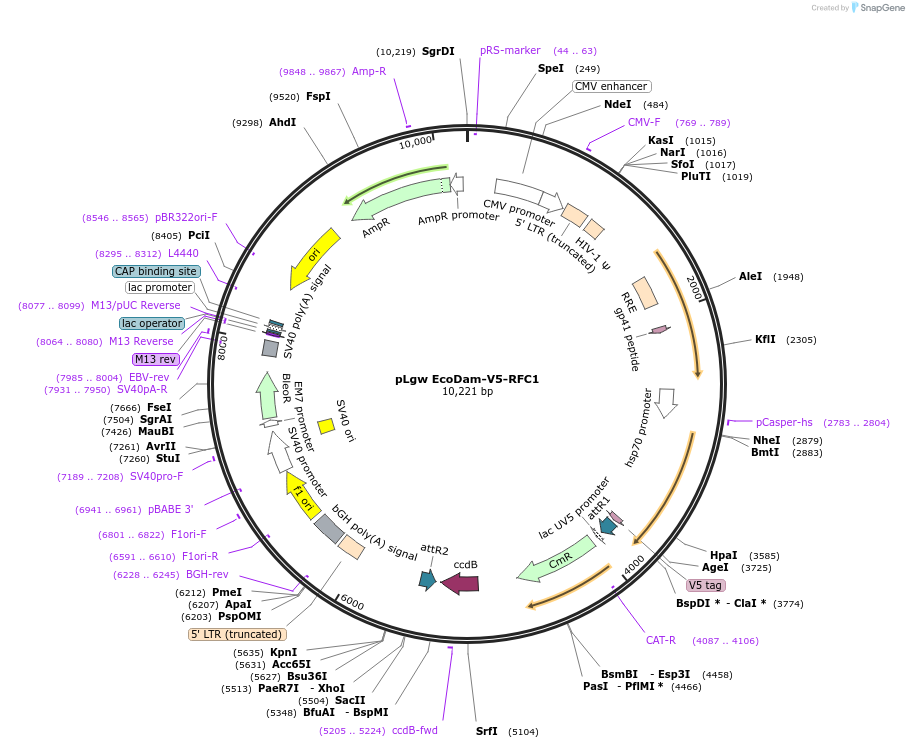
pLgw EcoDam-V5-RFC1
Plasmid#59209PurposeMammalian DamID lentiviral vector for fusing gene of interest with Dam-V5 using Gateway cloningDepositorTypeEmpty backboneUseLentiviral; DamidTagsDam and V5ExpressionMammalianPromoterHeat Shock Minimal PromoterAvailable SinceJune 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
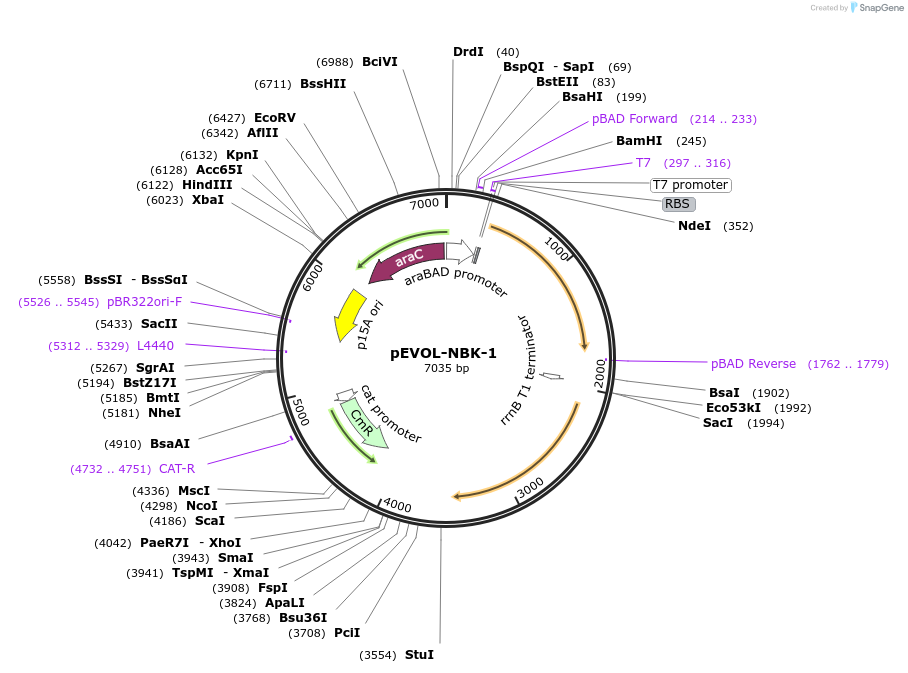
pEVOL-NBK-1
Plasmid#207639PurposeA plasmid for the expression of M. mazei pyrrolysine aminoacyl-tRNA synthetase/tRNA pair (PylRS/tRNAPyl) in E. Coli.DepositorInsertPyl-tRNACUA and 2x pyrrolysyl-tRNA synthetase (MmPylRS)
ExpressionBacterialMutationThe evolved synthetase NBK-1 contains mutations Y…PromoteraraBADAvailable SinceOct. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
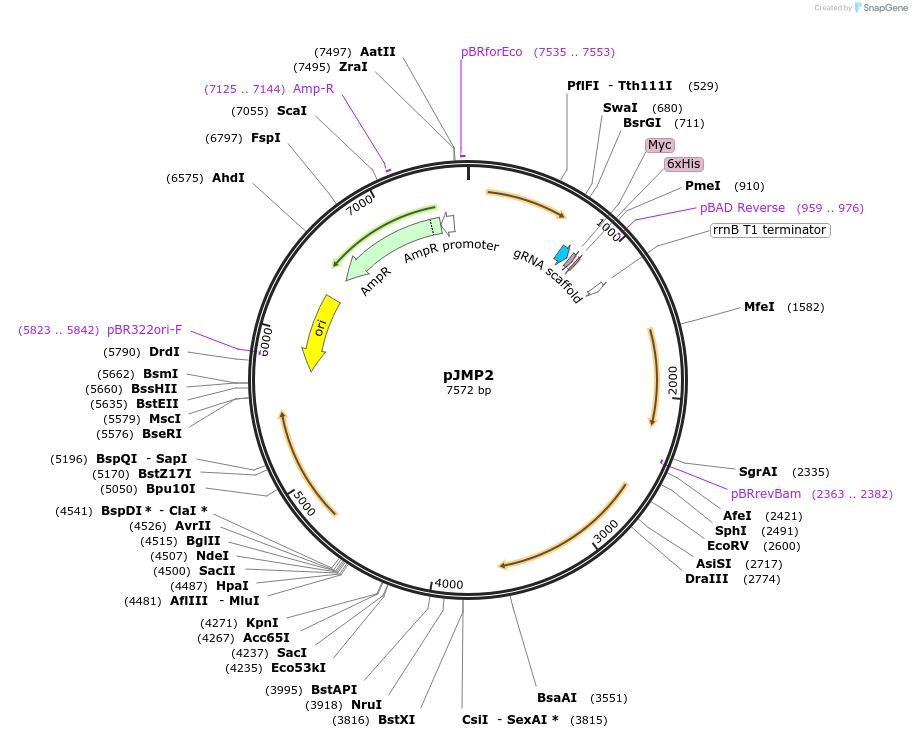
pJMP2
Plasmid#79874PurposeBacillus subtilis sgRNA expression vector; integrates into amyEDepositorInsertsgRNA RR1
UseCRISPRExpressionBacterialPromotervegAvailable SinceSept. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
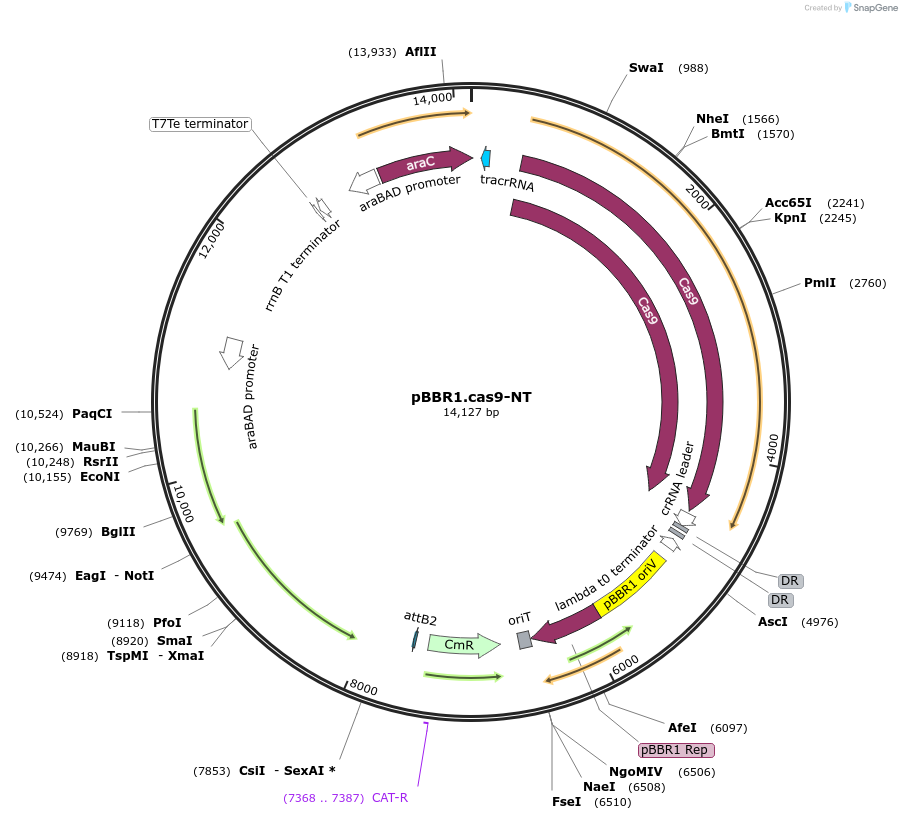
pBBR1.cas9-NT
Plasmid#197871PurposeConstitutive expression of Cas9 endonuclease, trancRNA and crRNA with a randomly generated spacer sequence (non-targeting)DepositorInsertstrancCRNA
Cas9
crRNA (non-chromosomal (E. coli/S. flexneri)-targeting spacer sequence)
ExpressionBacterialAvailable SinceJune 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
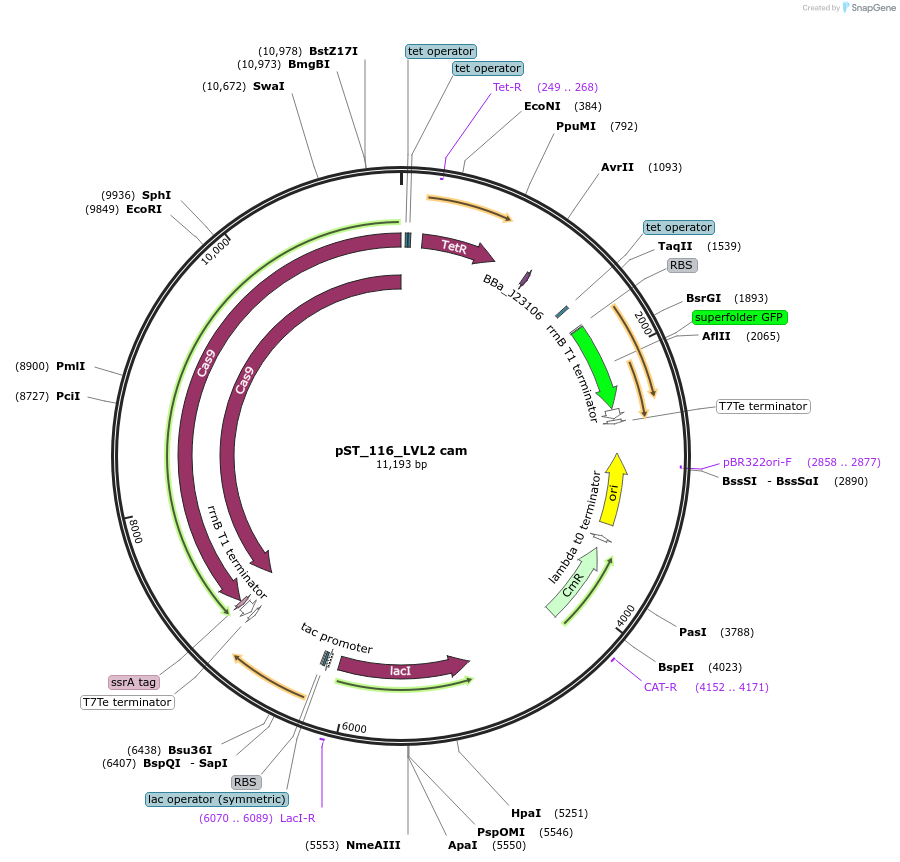
pST_116_LVL2 cam
Plasmid#179332PurposeNT-CRISPR plasmid for a single gRNA.DepositorInsertPtac tfoX, Ptet cas9, PJ23106 acrIIA4, Ptet gRNA with sfGFP dropout
UseCRISPRAvailable SinceMarch 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
tsPurple chromoprotein
Plasmid#117848PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses tsPurple chromoprotein in E. coliDepositorInsertpromoter, RBS, tsPurple
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
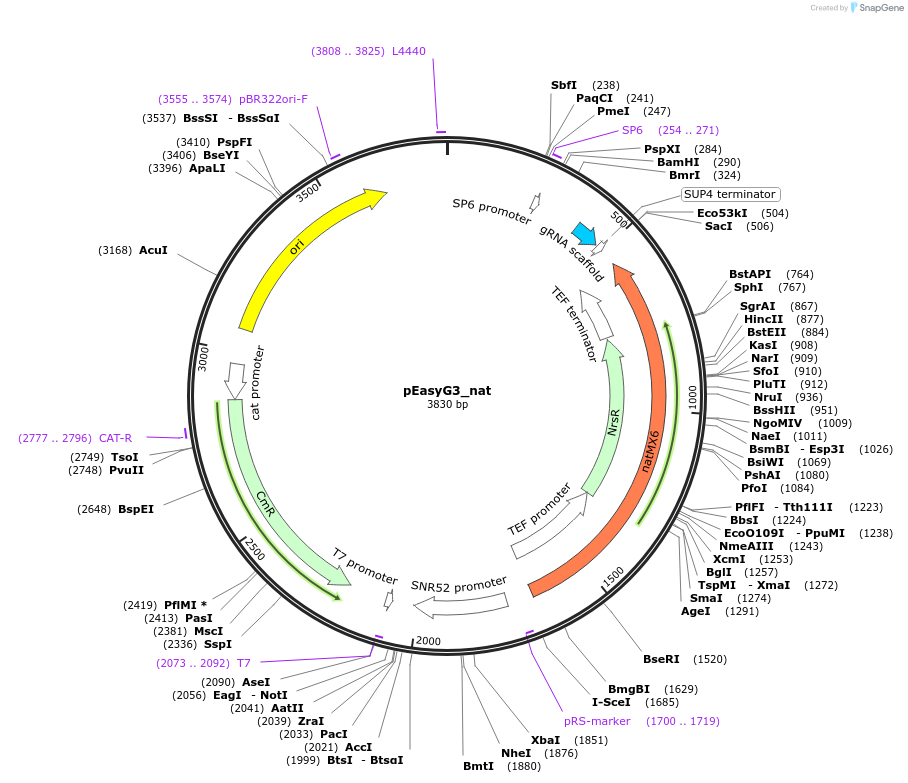
pEasyG3_nat
Plasmid#184918PurposeTemplate to generate via PCR two gRNAs for expression in S. cerevisiae. The PCR product from pEasyG3_nat recombines in vivo with a PCR product from pEasyG3_mic.DepositorInsertgRNA scaffold
UseCRISPRExpressionBacterialAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
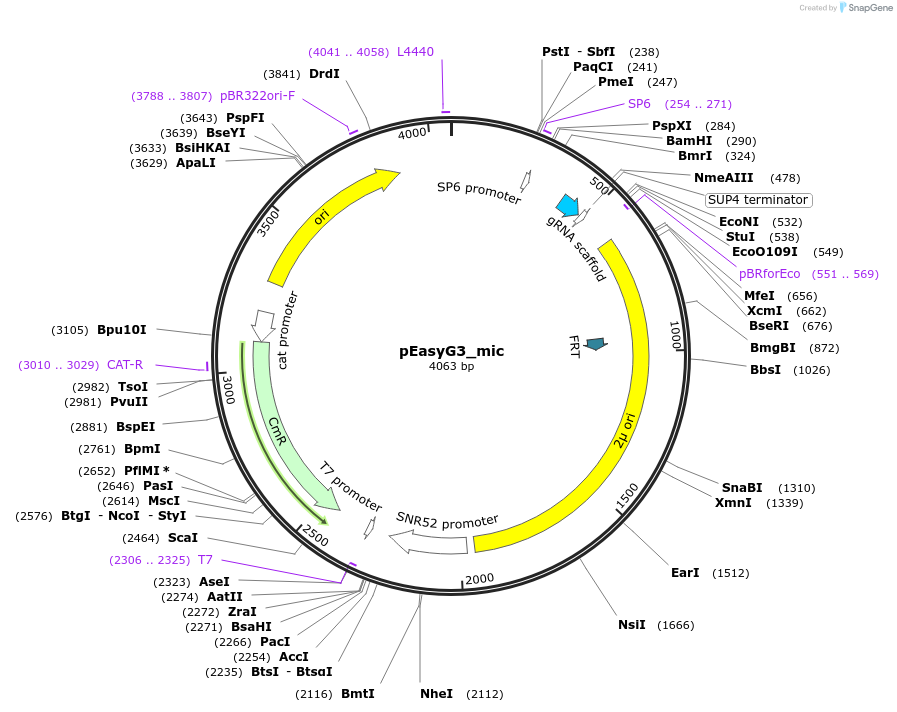
pEasyG3_mic
Plasmid#184920PurposeTemplate to generate via PCR two gRNAs for expression in S. cerevisiae. The PCR product from pEasyG3_mic recombines in vivo with a PCR product from pEasyG3_zeo/nat/hph.DepositorInsertgRNA scaffold
UseCRISPRExpressionBacterialAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
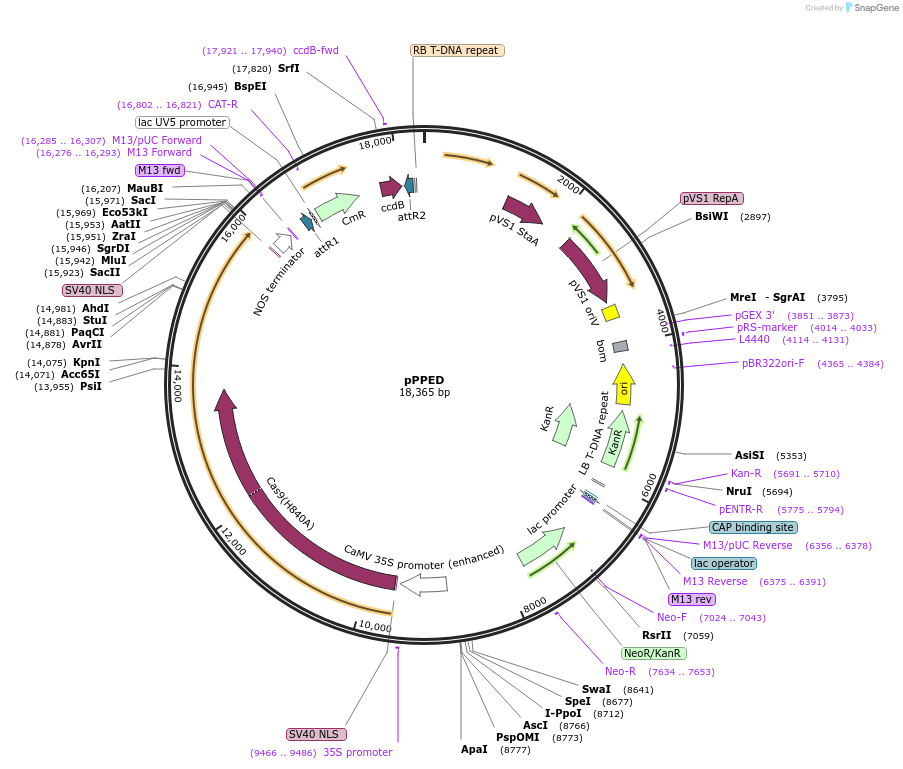
pPPED
Plasmid#162468PurposeDestination Expression vector for dicot plant prime editingDepositorInsertsCas9H840A
attR gateway destination cassette
TagsMLV reverse transcriptaseExpressionPlantMutationChange histidine 840 to Alanine based sequence o…Promoter2x35SAvailable SinceDec. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
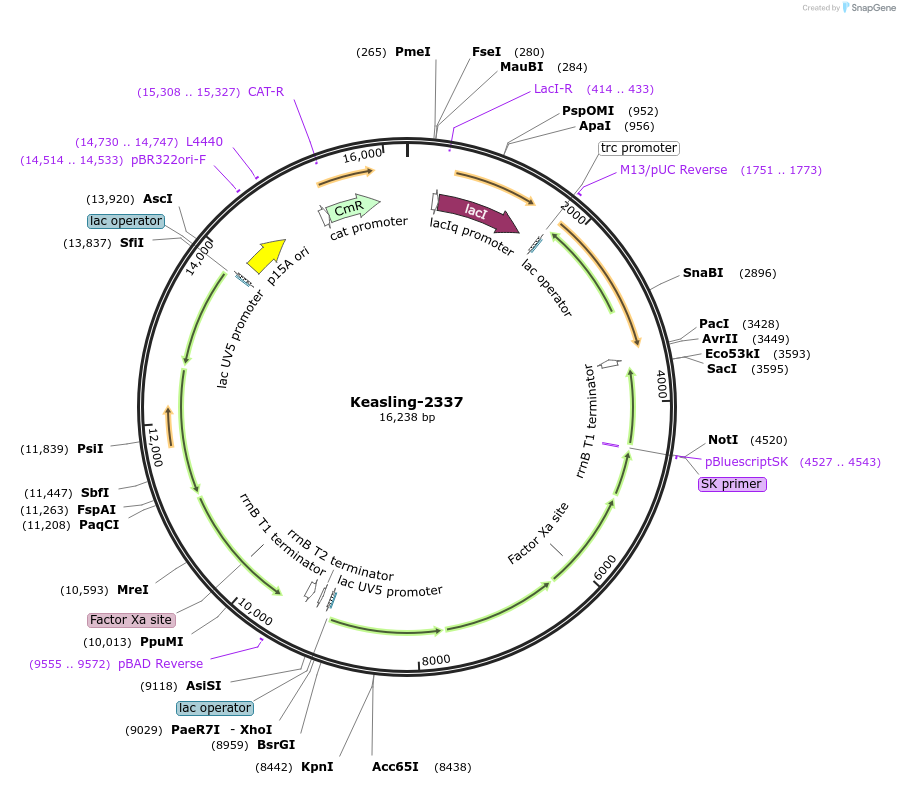
Keasling-2337
Plasmid#100168PurposeUsed to improve production from engineered biosynthetic pathways include optimizing codon usage, enhancing production of rate-limiting enzymes, and eliminating the accumulation of toxic intermediates or byproducts to improve cell growthDepositorInsertlacIq-Ptrc-ADS
ExpressionBacterialPromotertrcAvailable SinceSept. 15, 2017AvailabilityAcademic Institutions and Nonprofits only -
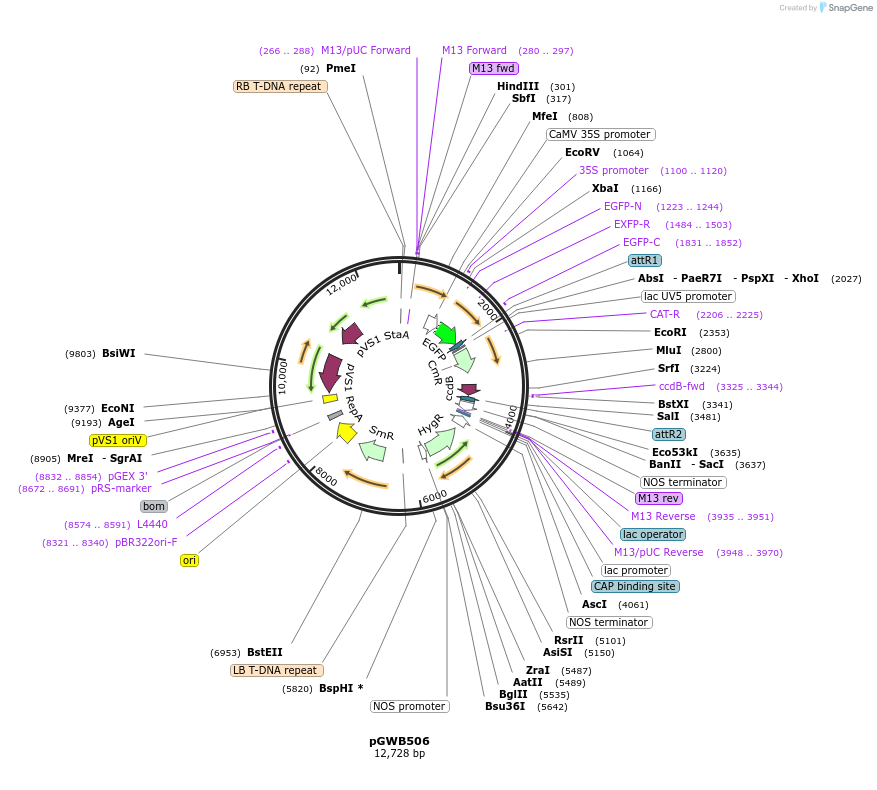
pGWB506
Plasmid#74848PurposeGateway cloning compatible binary vector for N-terminal fusion with sGFP (CaMV35S promoter).DepositorTypeEmpty backboneExpressionPlantPromoterCaMV35SAvailable SinceAug. 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
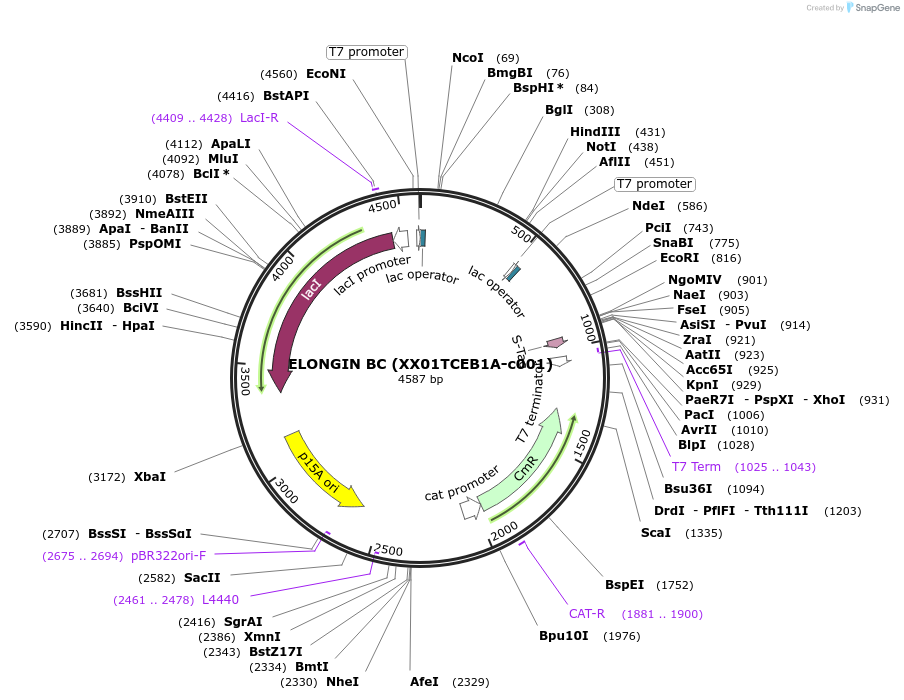
ELONGIN BC (XX01TCEB1A-c001)
Plasmid#110274PurposeElongin B and Elongin C co-expression plasmid for structure determination; Full length Elongin B & amino acids 17-112 of Elogin CDepositorExpressionBacterialMutationElongin B is full length. Elongin C contains a.a…PromoterT7Available SinceMay 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
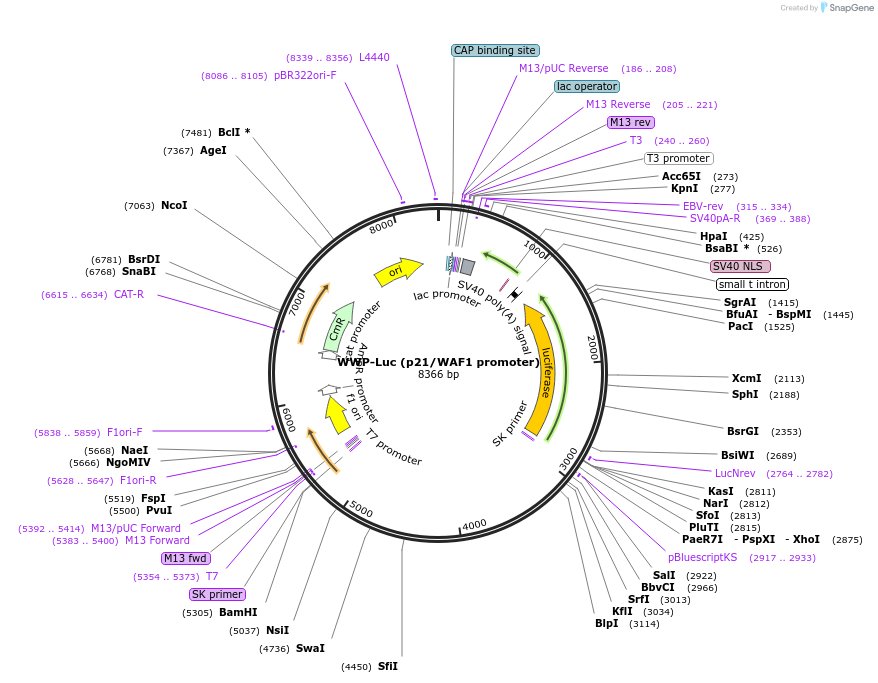
WWP-Luc (p21/WAF1 promoter)
Plasmid#16451DepositorAvailable SinceJune 11, 2008AvailabilityAcademic Institutions and Nonprofits only -
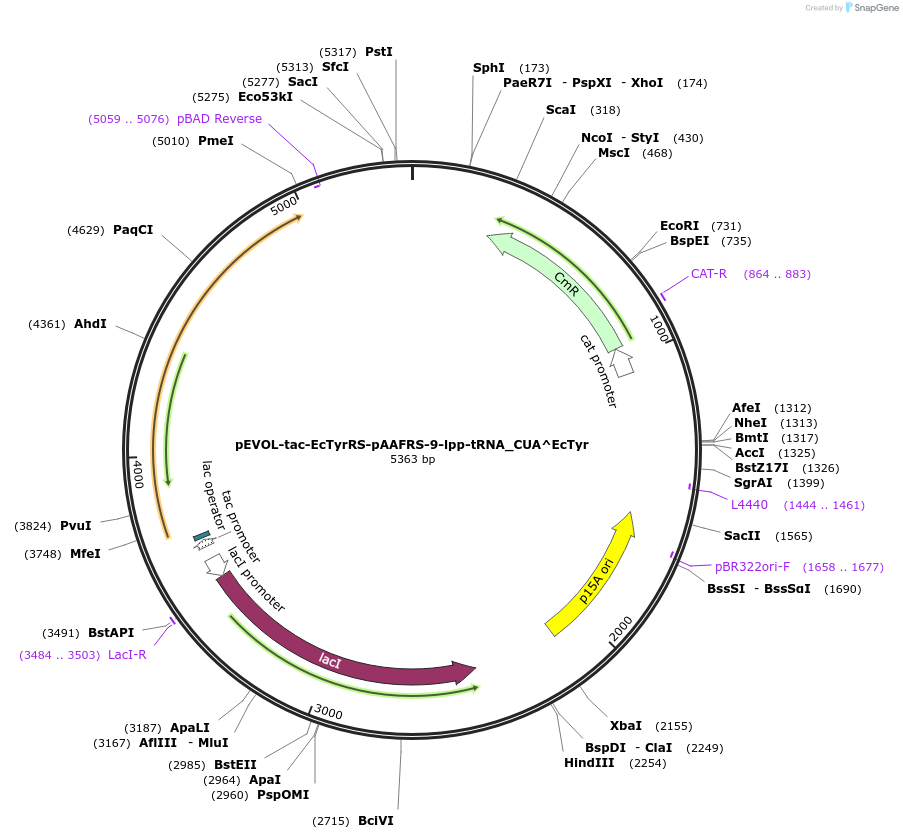
pEVOL-tac-EcTyrRS-pAAFRS-9-lpp-tRNA_CUA^EcTyr
Plasmid#218767Purposeencodes EcTyrRS-pAAFRS-9 and EcTyr-tRNA-TAGDepositorInsertEcTyrRS-pAAFRS-9
ExpressionBacterialMutationY37G, L71V, D182C, F183Y, L186C, D265RPromotertacIAvailable SinceJune 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
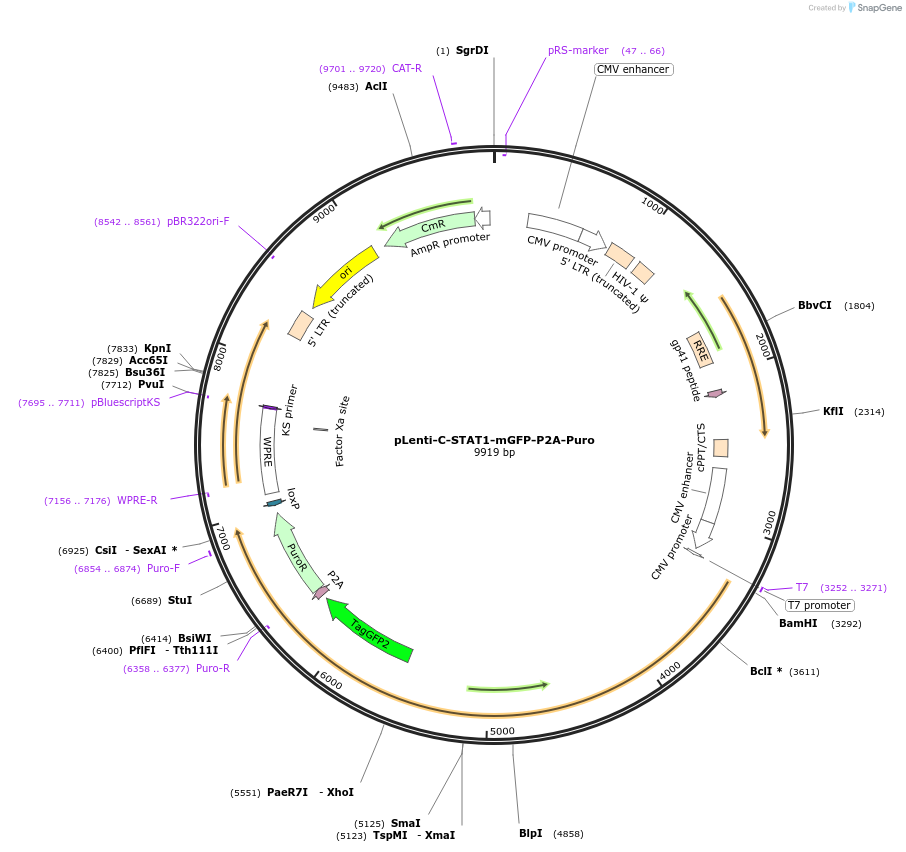
pLenti-C-STAT1-mGFP-P2A-Puro
Plasmid#199345Purposeexpress murine STAT1 protein fused with mGFPDepositorInsertsignal transducer and activator of transcription 1 (Stat1 Mouse)
UseLentiviralTagsmGFPExpressionMammalianPromoterCMVAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
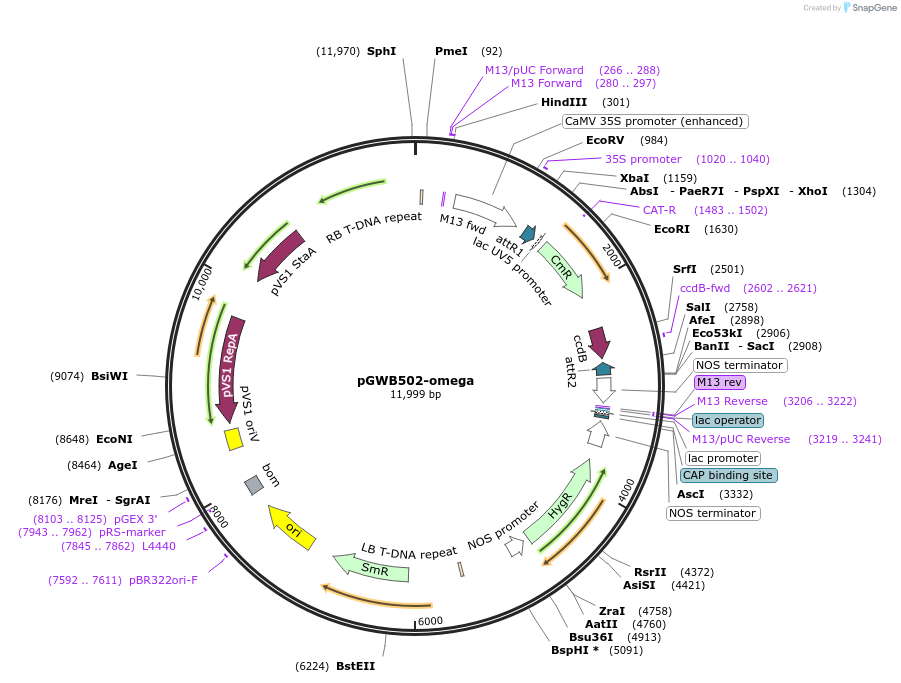
pGWB502-omega
Plasmid#74845PurposeGateway cloning compatible binary vector for expression of gene by 2xCaMV35S enhancer and omega sequence.DepositorTypeEmpty backboneExpressionPlantPromoter2xCaMV35S enhancer and omegaAvailable SinceOct. 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
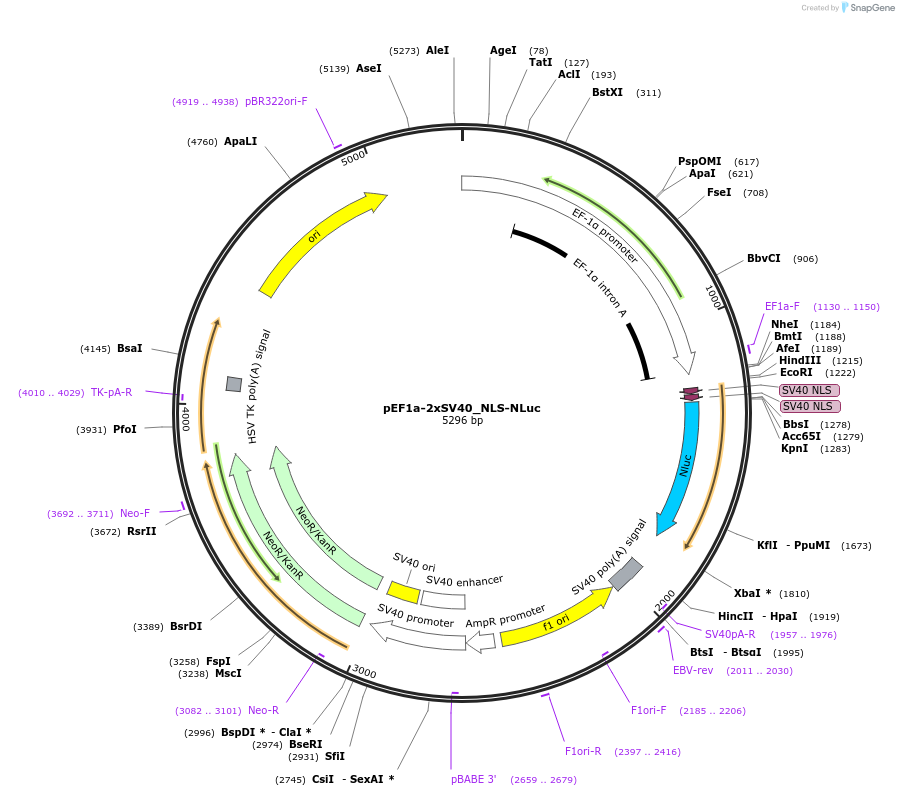
pEF1a-2xSV40_NLS-NLuc
Plasmid#135953PurposeExpresses the NanoLuciferase targeted to the nucleus in mammalian cells.DepositorInsertNanoLuciferase
Tags2xSV40-NLSExpressionMammalianPromoterEF1aAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
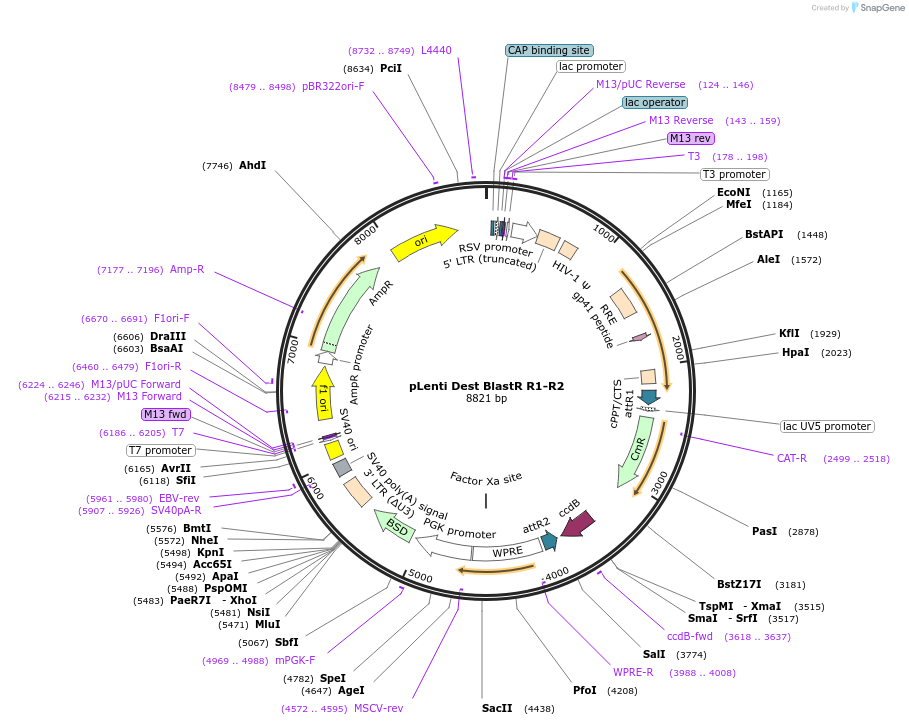
pLenti Dest BlastR R1-R2
Plasmid#84574PurposeGateway destination vector expressing Blasticidin resistance under a PGK promoterDepositorTypeEmpty backboneUseLentiviral; GatewayExpressionMammalianPromoterPGKAvailable SinceJan. 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
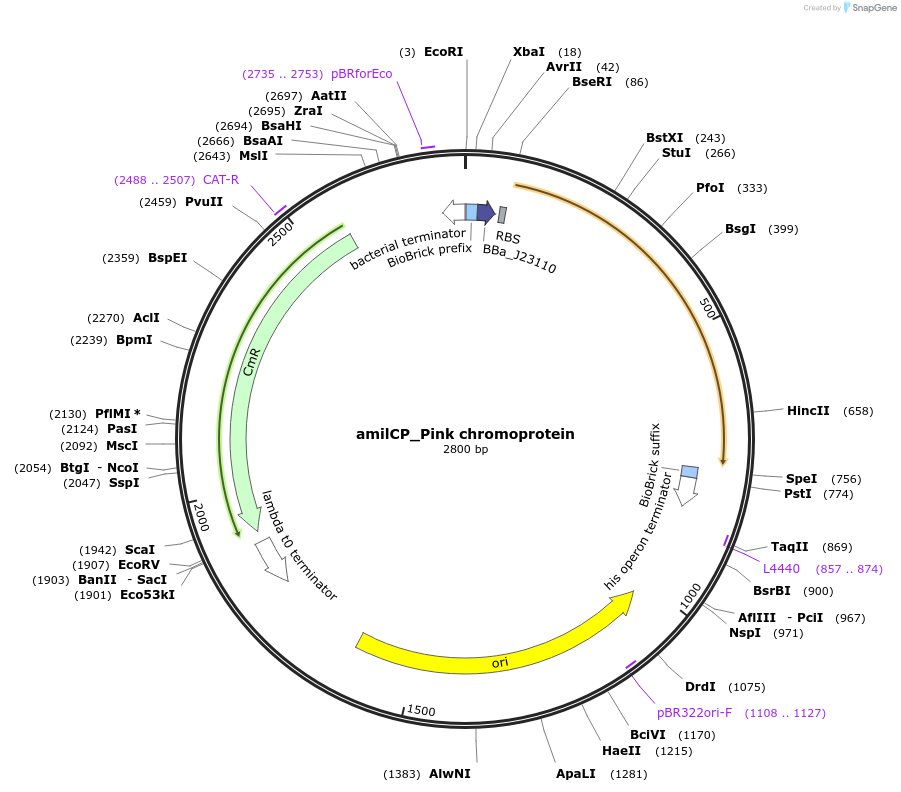
amilCP_Pink chromoprotein
Plasmid#117851PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amilCP_Pink chromoprotein in E. coliDepositorInsertpromoter, RBS, amilCP_Pink
UseSynthetic Biology; Escherichia coliMutation64A,65C, BioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
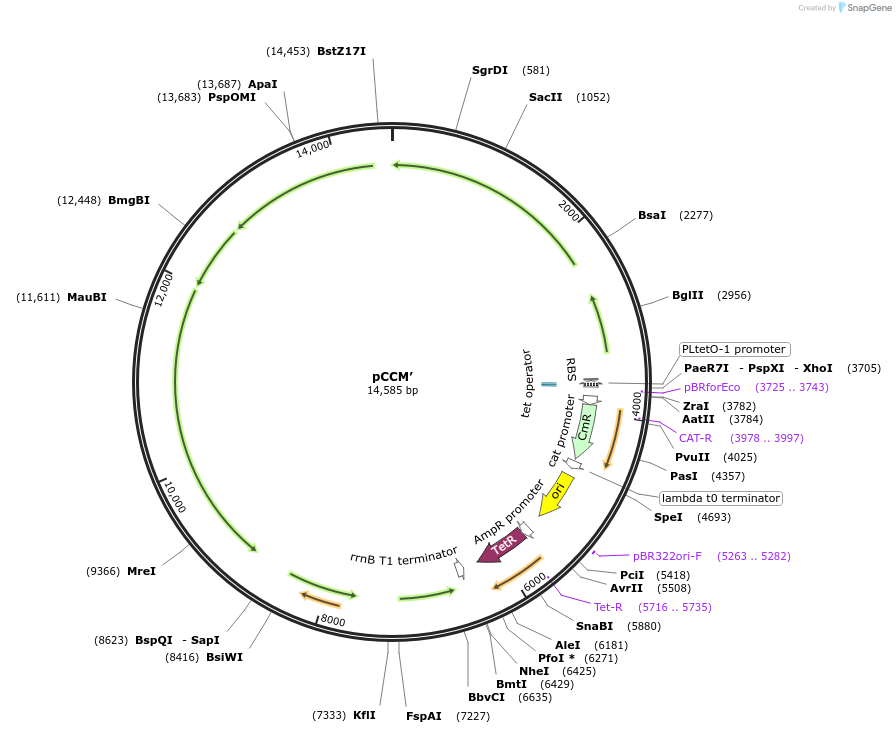
pCCM’
Plasmid#162709PurposepCCM reconstructed after selection for CCMB1 growth on glycerol under ambient airDepositorInsertSecond CCM operon cloned from H. neapolitanus
ExpressionBacterialMutationp15A origin replaced with high-copy colE1 originPromoterPLtet0-1 promoterAvailable SinceJan. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
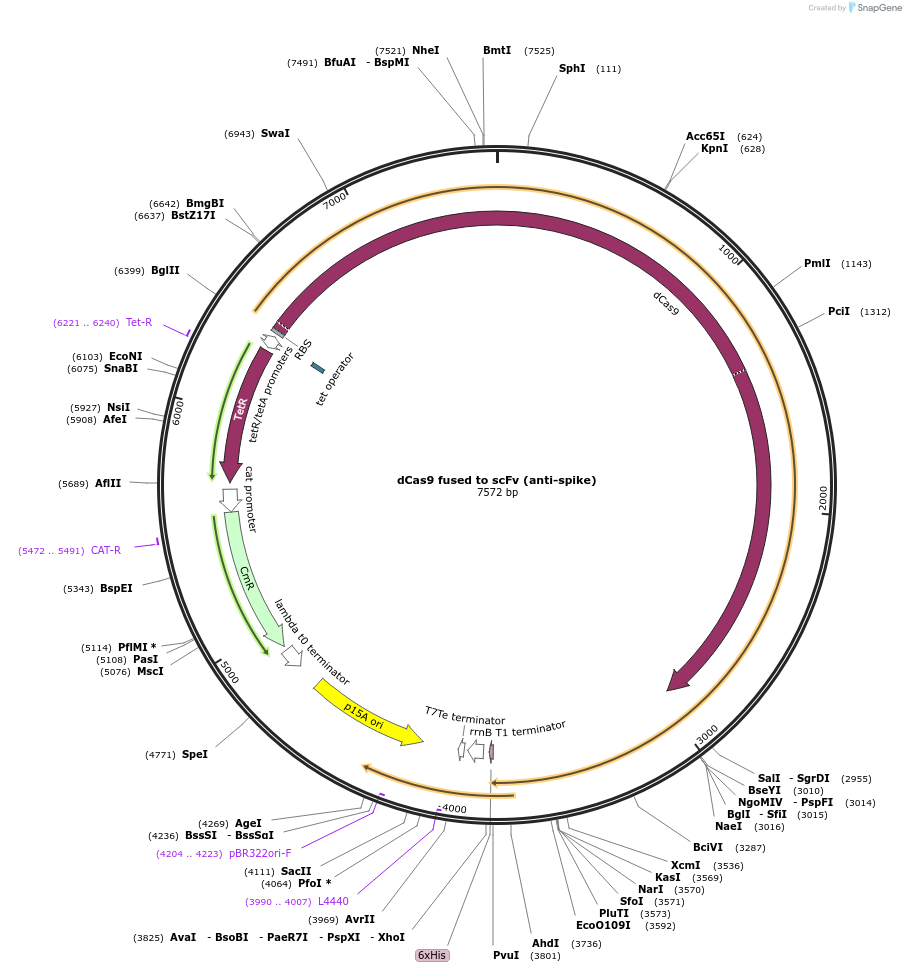
dCas9 fused to scFv (anti-spike)
Plasmid#186421PurposeExpression of dCas9 with C-terminal nanobody fusion recognizing spike protein from SARS-CoV-2DepositorInsertdCas9-scFv fusion (anti-SARS-CoV-2 spike)
UseCRISPRTags6HisAvailable SinceOct. 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
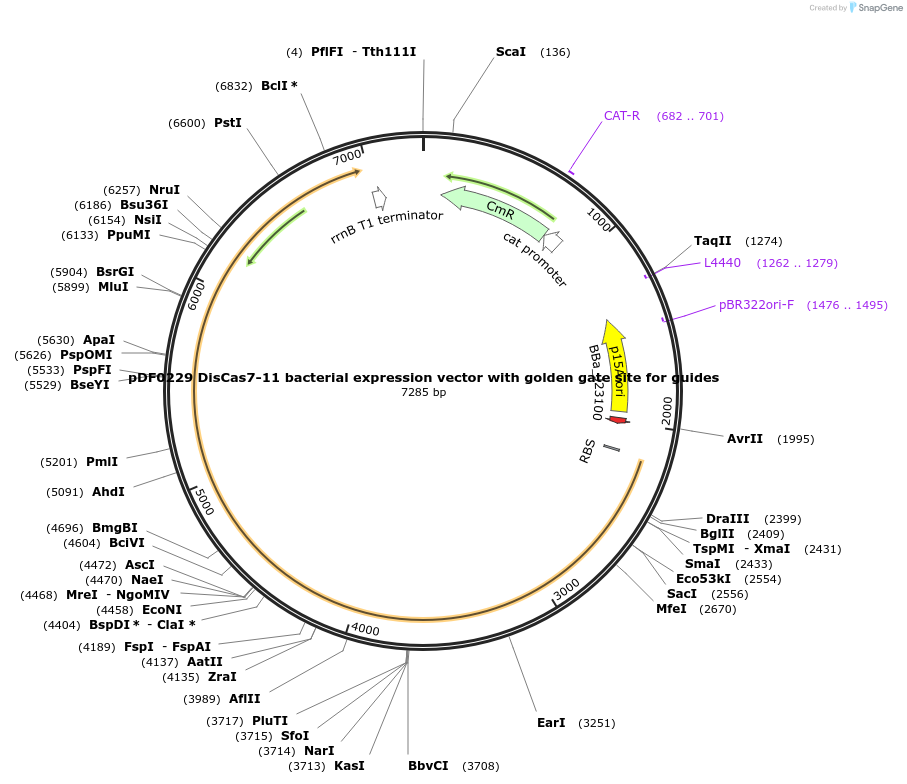
pDF0229 DisCas7-11 bacterial expression vector with golden gate site for guides
Plasmid#172506PurposeEncodes DisCas7-11 protein alone with a golden gate site for spacer cloningDepositorInsertDisCas7-11 bacterial expression vector with golden gate site for guides
ExpressionBacterialAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
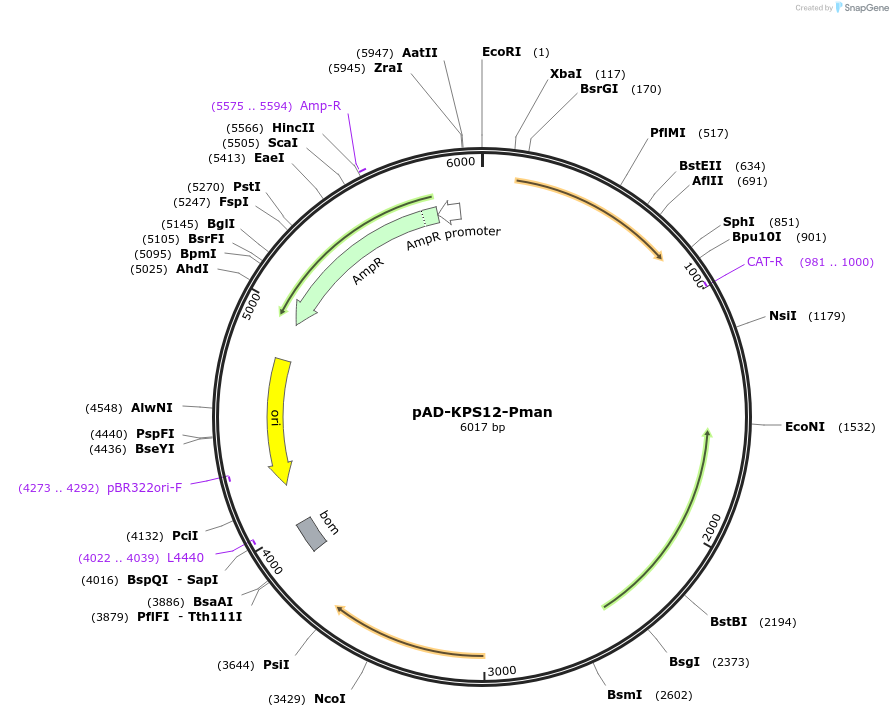
pAD-KPS12-Pman
Plasmid#133832PurposeFluorescent protein mKate(KPS12) variant for optimized expression in Bacillus mycoides; under control of mannose inducible promoter.DepositorInsertmKate(KPS12)
UseE.coli/bacillus spp. shuttle vectorExpressionBacterialPromoterPmanAvailable SinceNov. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
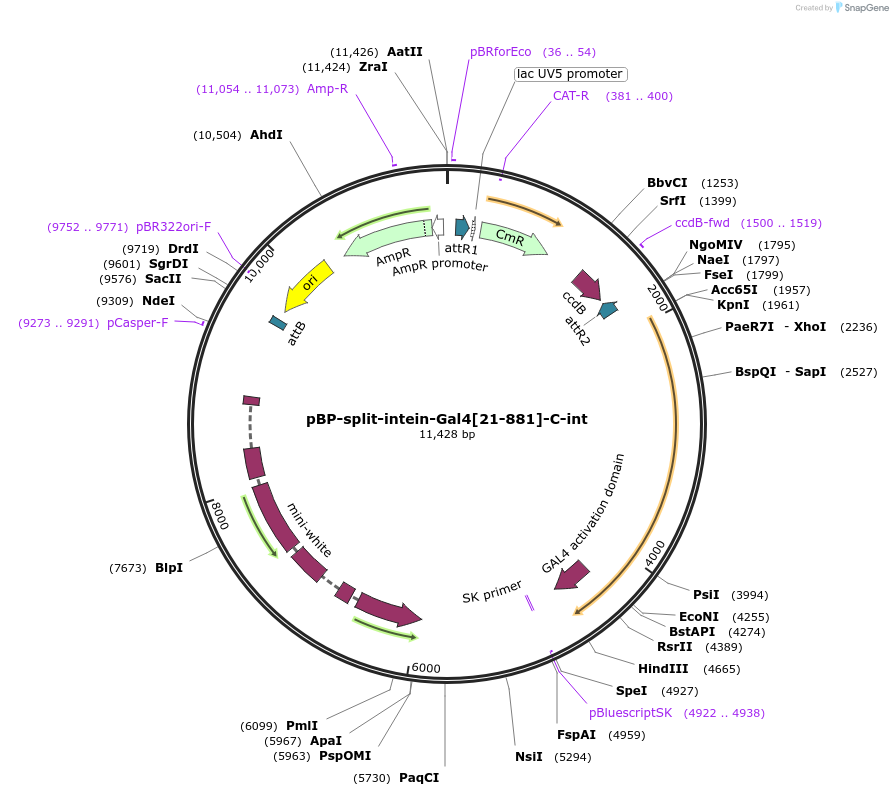
pBP-split-intein-Gal4[21-881]-C-int
Plasmid#199202PurposeGateway destination vector for enhancer-driven split-intein Gal4[21-881]C-int expressionDepositorInsertsplit-intein-Gal4[21-881]-C-int
ExpressionInsectAvailable SinceAug. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
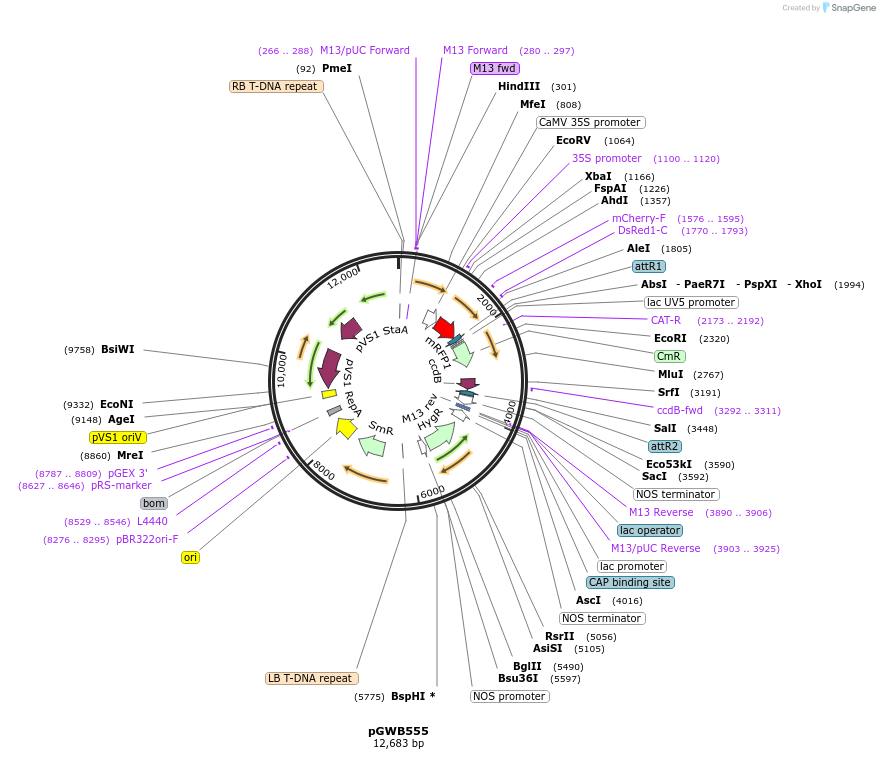
pGWB555
Plasmid#74885PurposeGateway cloning compatible binary vector for N-terminal fusion with mRFP (CaMV35S promoter).DepositorTypeEmpty backboneExpressionPlantPromoterCaMV35SAvailable SinceNov. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
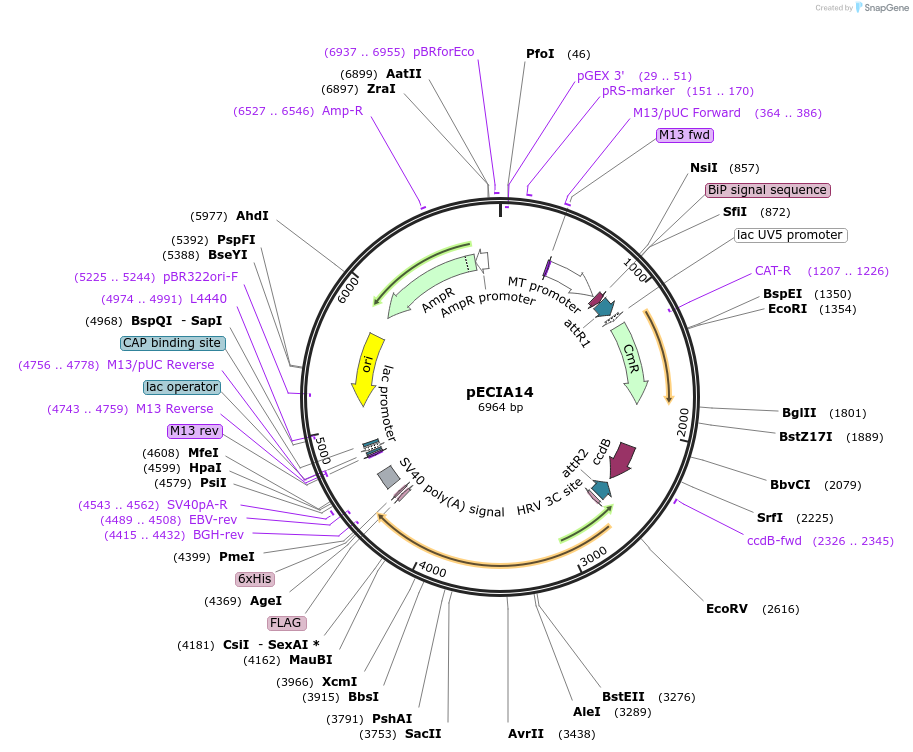
pECIA14
Plasmid#47051PurposeThis is a Gateway destination vector for secreted expression in Drosophila cell culture by CuSO4 induction. Inserts will be pentamerized and tagged with Alkaline phosphatase, FLAG and His tagsDepositorTypeEmpty backboneUseSecreted expression in drosophila cultureTagsAlkaline Phosphatase (human, placental), Flag, HR…ExpressionInsectPromoterMetallothionein (Copper-Inducible)Available SinceAug. 30, 2013AvailabilityAcademic Institutions and Nonprofits only -
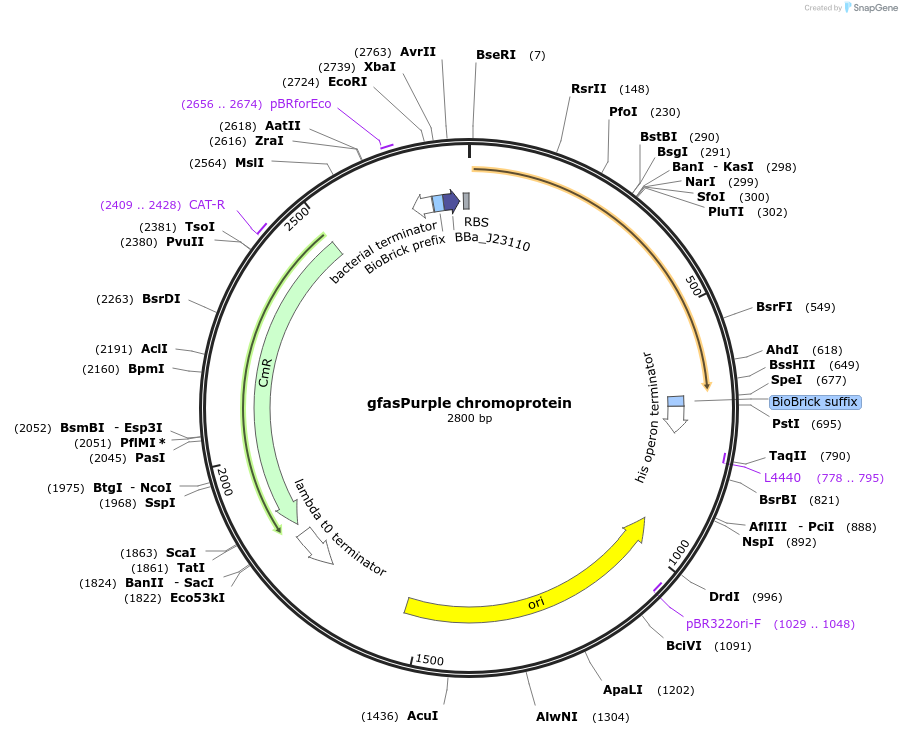
gfasPurple chromoprotein
Plasmid#117849PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses gfasPurple chromoprotein in E. coliDepositorInsertpromoter, RBS, gfasPurple
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
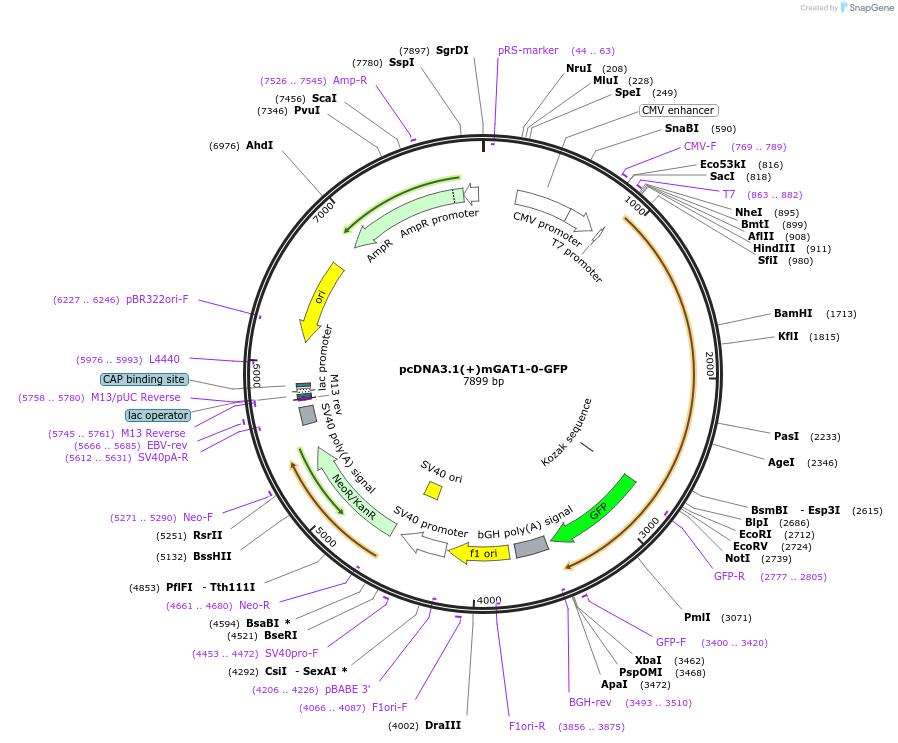
pcDNA3.1(+)mGAT1-0-GFP
Plasmid#41662DepositorInsertMus musculus GABA transporter 1 (Slc6a1 Synthetic, Mouse)
TagsGFPExpressionMammalianMutationC-terminal hydrophobic isoleucine residue added a…PromoterCMVAvailable SinceApril 27, 2016AvailabilityAcademic Institutions and Nonprofits only -
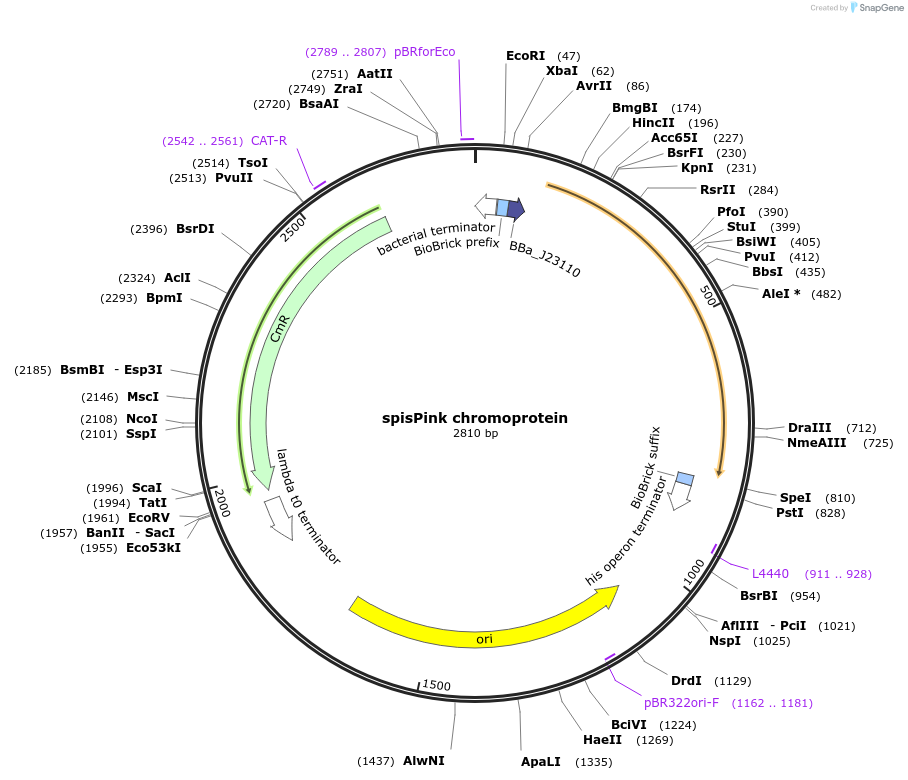
spisPink chromoprotein
Plasmid#117839PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses spisPink chromoprotein in E. coliDepositorInsertpromoter, RBS, spisPink
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceJuly 31, 2019AvailabilityAcademic Institutions and Nonprofits only -
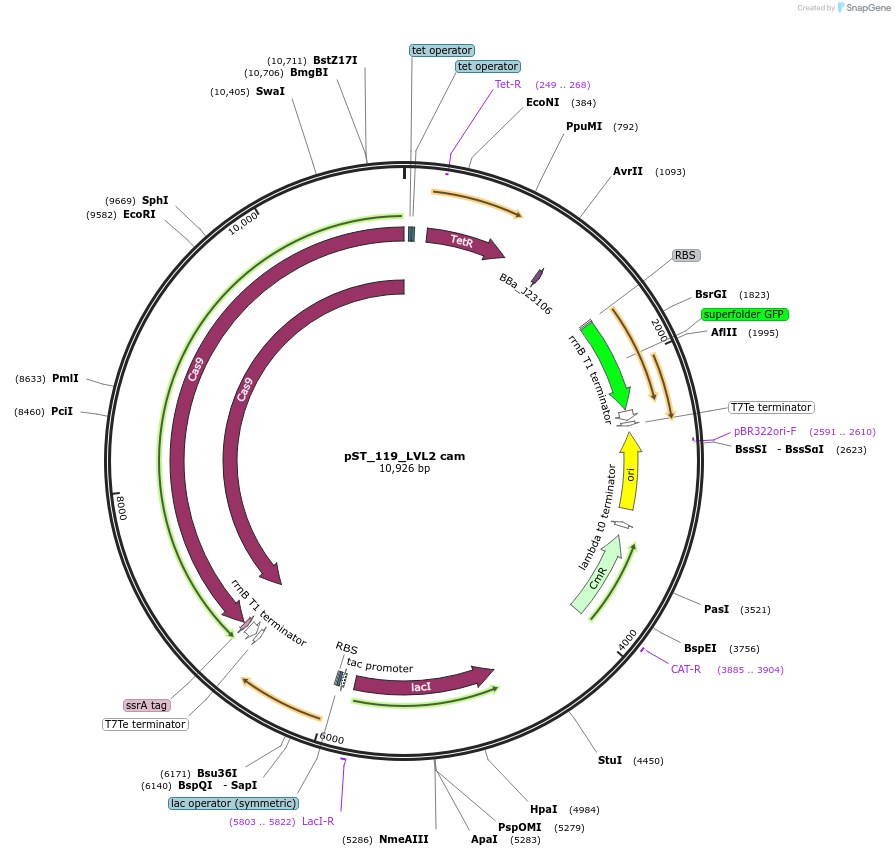
pST_119_LVL2 cam
Plasmid#179333PurposeNT-CRISPR plasmid for integration of multiple gRNAs.DepositorInsertPtac tfoX, Ptet cas9, PJ23106 acrIIA4, sfGFP dropout to be replaced with gRNAs
UseCRISPRAvailable SinceMarch 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
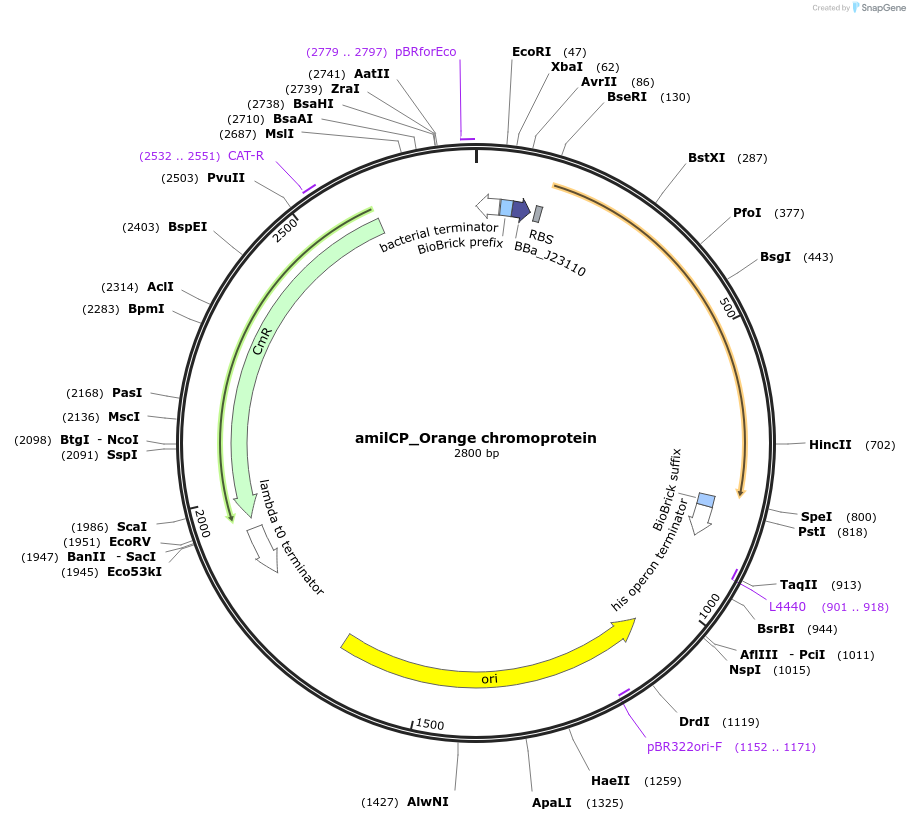
amilCP_Orange chromoprotein
Plasmid#117850PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses amilCP_Orange chromoprotein in E. coliDepositorInsertpromoter, RBS, amilCP_Orange
UseSynthetic Biology; Escherichia coliMutation64V,65G, BioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
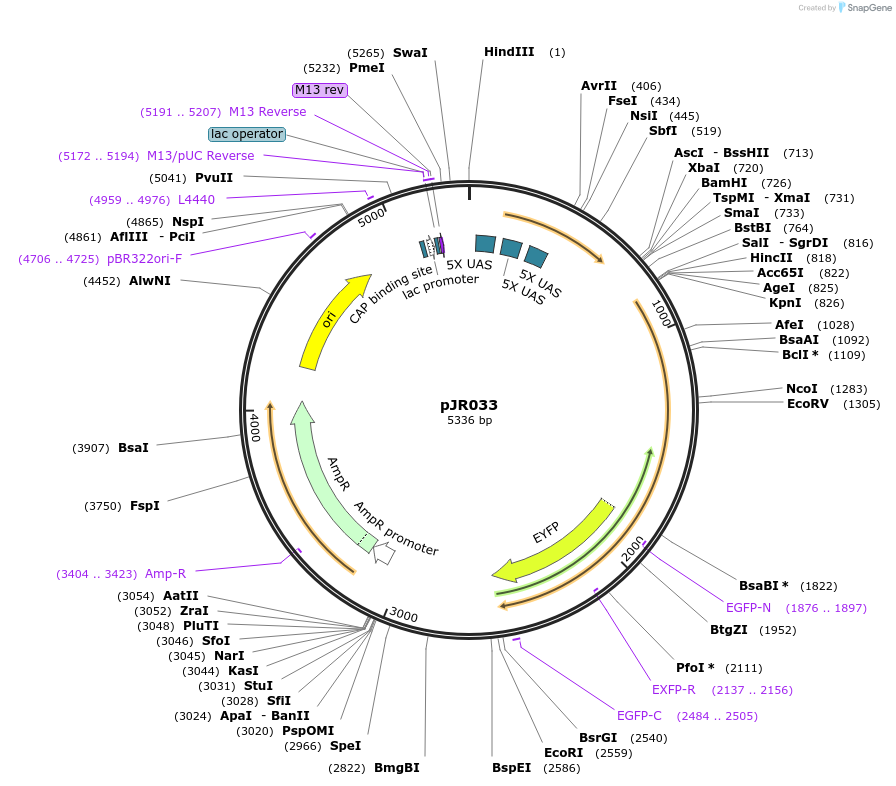
pJR033
Plasmid#198822PurposeChloride-conducting channelrhodopsinDepositorInsert15xUAS::UAS-iC1-c2-TS-EYFP::let-858 3'UTR
ExpressionWormAvailable SinceApril 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
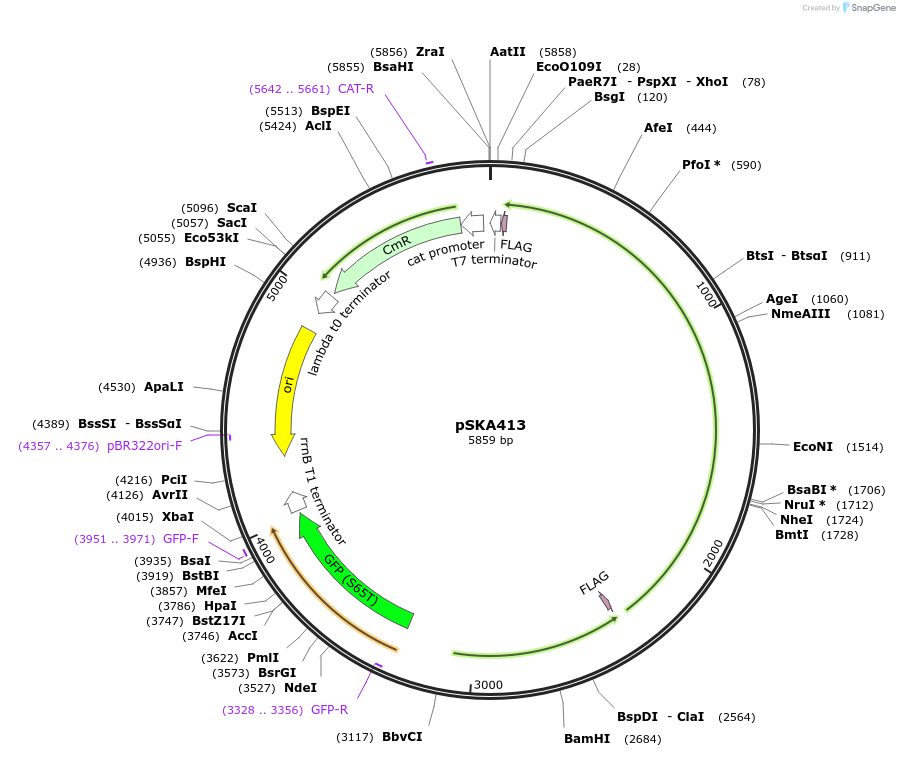
pSKA413
Plasmid#80381PurposeConstitutive CcaS and CcaR with gfpmut3 under the cpcG2Δ59 promoter.DepositorInsertsccaS (complement)
ccaR (complement)
gfpmut3
UseSynthetic Biology; OptogeneticsTagsFLAG tagExpressionBacterialPromoterccaR and cpcG2Δ59Available SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
asPink chromoprotein
Plasmid#117838PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses asPink chromoprotein in E. coliDepositorInsertpromoter, RBS, asPink
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
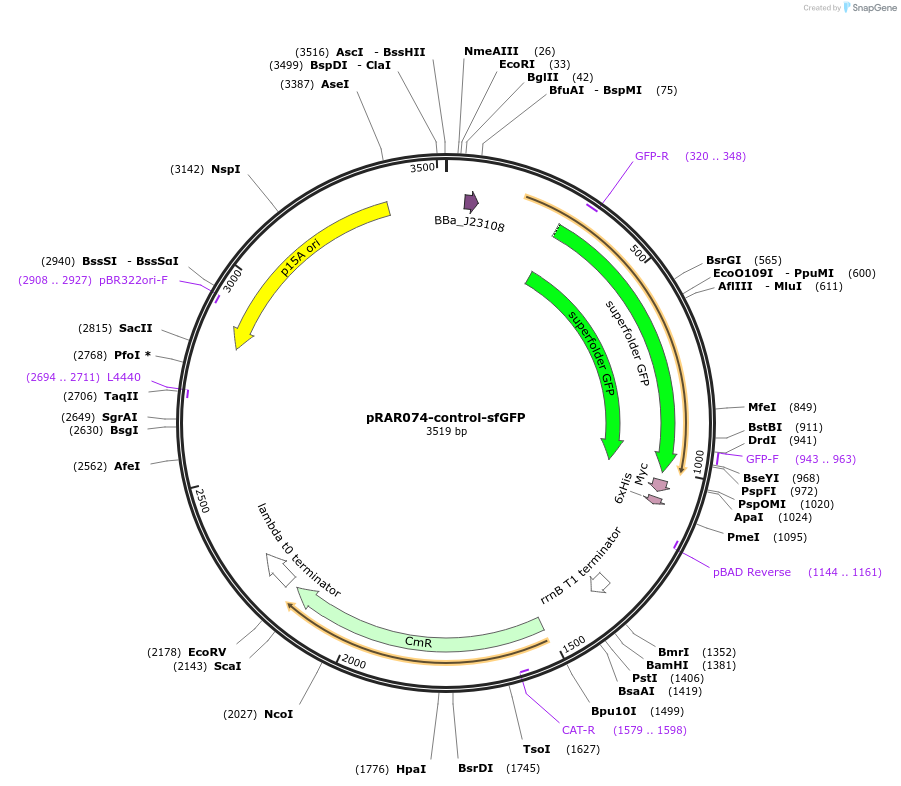
pRAR074-control-sfGFP
Plasmid#185032PurposeJ23108 promoter, scrambled 5'UTR, 90 nt of l31 coding region, glycine linker, sfGFP, trrnBDepositorInsertL31-sfGFP
ExpressionBacterialMutationNucleotides of rpmE 5'UTR scrambled, Only fi…Available SinceJuly 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
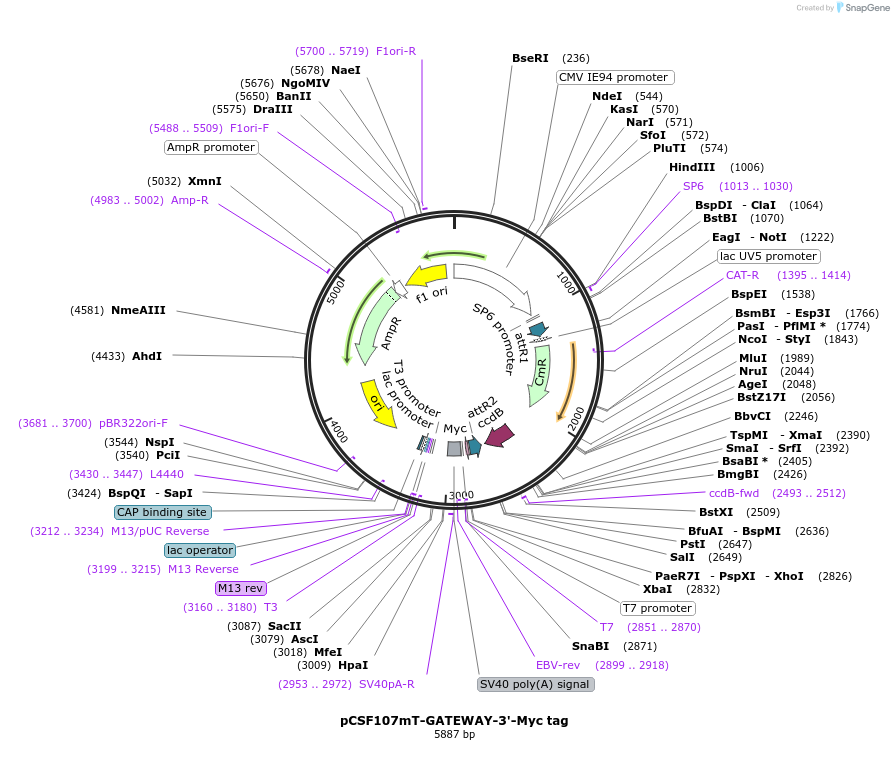
pCSF107mT-GATEWAY-3'-Myc tag
Plasmid#67617PurposeA pCS2 based vector to generate a 3'-Myc tag using GATEWAY cloning technologyDepositorTypeEmpty backboneUseIn vitro transcription or translation off of an s…TagsC-terminal MycExpressionMammalianPromoterCMV and SP6Available SinceAug. 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
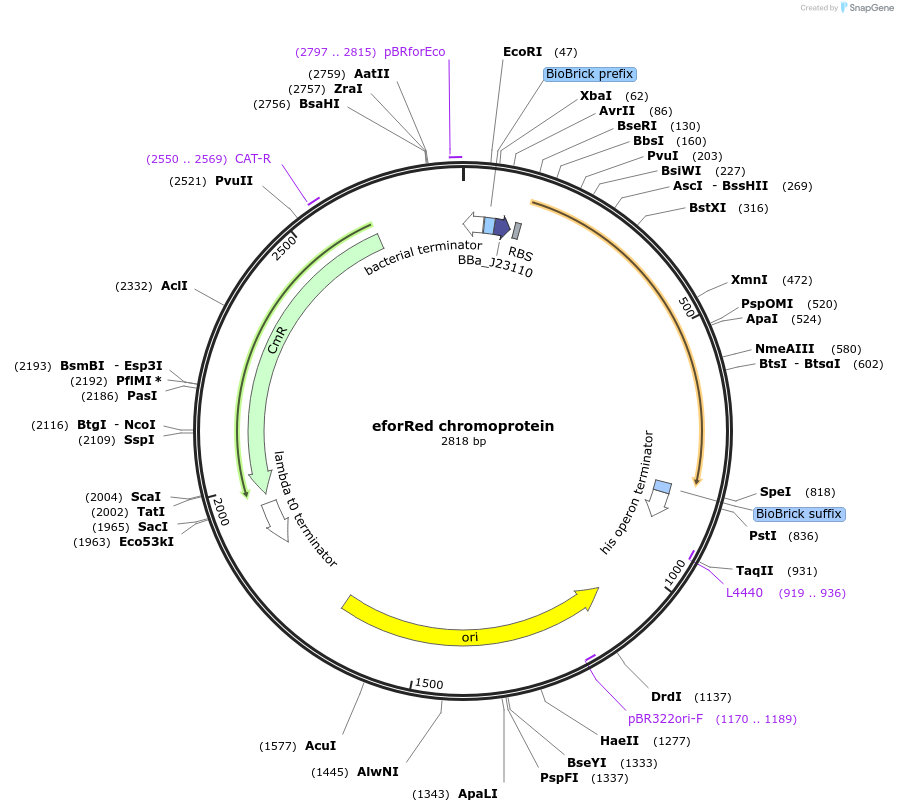
eforRed chromoprotein
Plasmid#117837PurposeBioBrick pSB1C3 plasmid that constitutively overexpresses eforRed chromoprotein in E. coliDepositorInsertpromoter, RBS, eforRed
UseSynthetic Biology; Escherichia coliMutationBioBrick sites removedAvailable SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
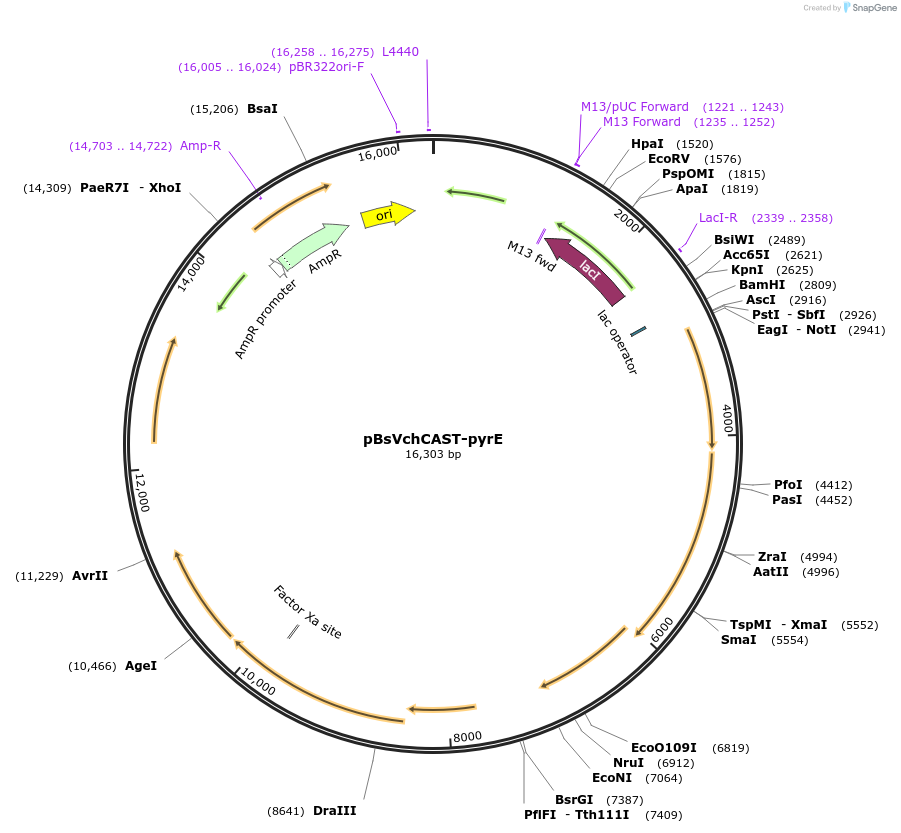
pBsVchCAST-pyrE
Plasmid#203813PurposeExpression of CRISPR-associated transposases, shuttle vector for Bacillus subtilis / E. coliDepositorInsertVchTniQ, VchCas5/8, VchCas7, VchCas6, VchTnsABC, CRISPR(pyrE)
UseCRISPRExpressionBacterialAvailable SinceAug. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
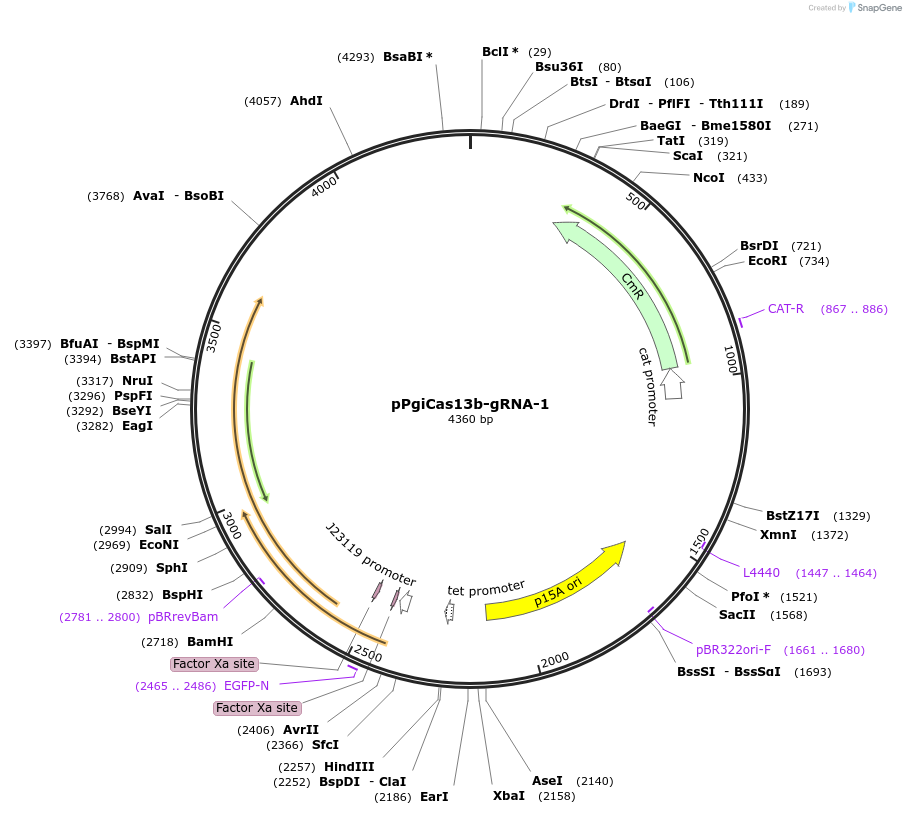
pPgiCas13b-gRNA-1
Plasmid#184834PurposeConstitutive expression of single-spacer CRISPR array with spacer #1 targeting deGFP mRNA for PgiCas13b in bacteria.DepositorInsertConstitutive expression of single-spacer CRISPR array with spacer #1 targeting deGFP mRNA for PgiCas13b in bacteria.
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterJ23119Available SinceNov. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
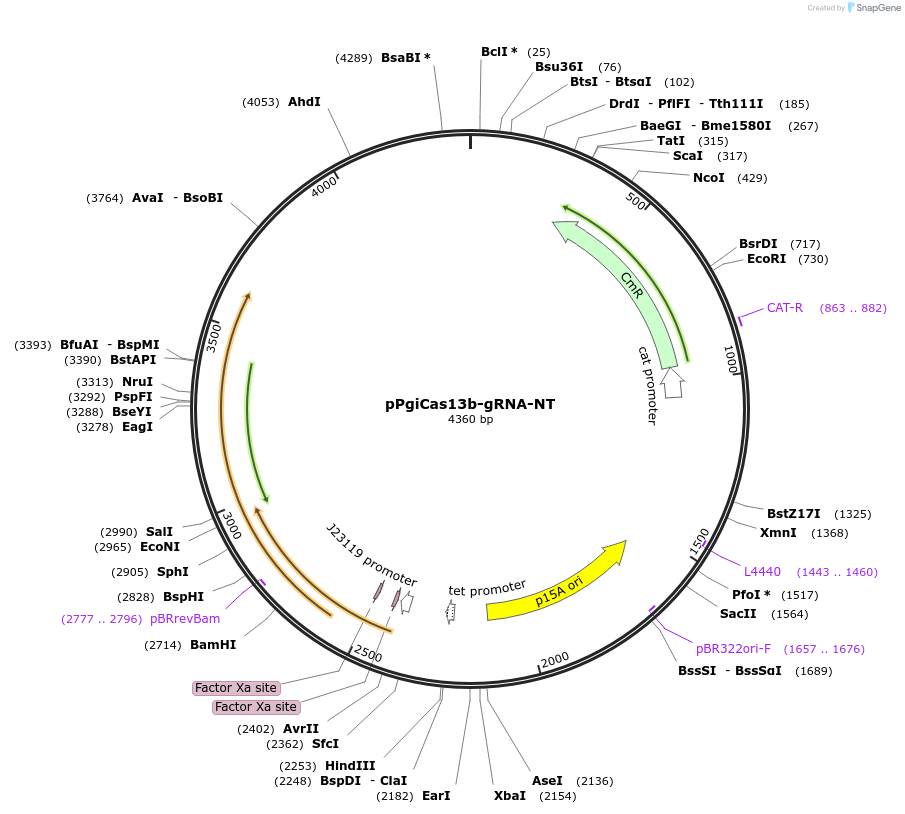
pPgiCas13b-gRNA-NT
Plasmid#184835PurposeConstitutive expression of single-spacer CRISPR array with non-targeting spacer for PgiCas13b in bacteria.DepositorInsertConstitutive expression of single-spacer CRISPR array with a nontargeting spacer for PgiCas13b in bacteria.
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterJ23119Available SinceNov. 9, 2022AvailabilityAcademic Institutions and Nonprofits only