We narrowed to 9,586 results for: Coli
-
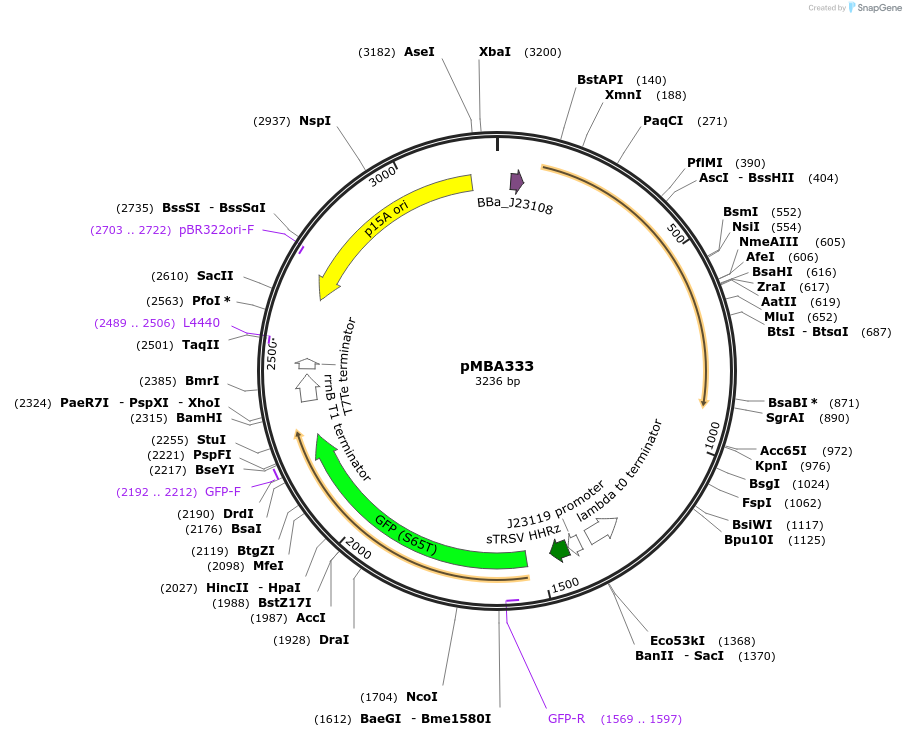
Plasmid#214745PurposeMedium copy thyA-infA plasmid containing gfp and no antibiotic resistance gene (ARG)DepositorInsertBBa_J23119-RiboJ-RBS(21992)-gfpmut3
ExpressionBacterialAvailable SinceMay 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
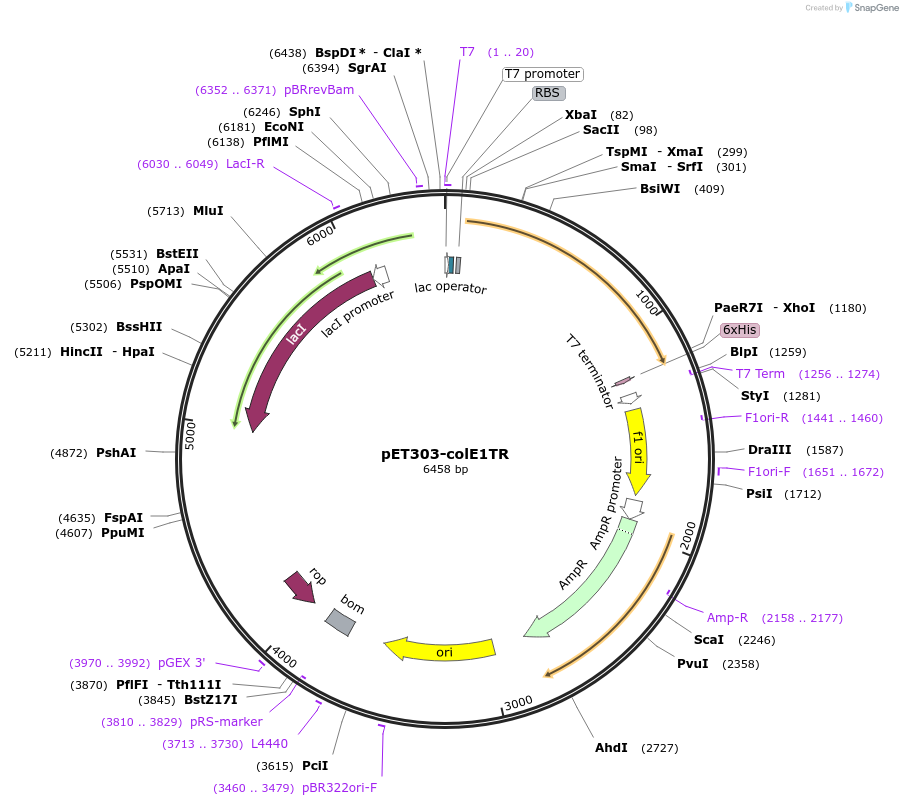
pET303-colE1TR
Plasmid#155181PurposeExpresses colicin E1 residues 1-364DepositorInsertcolicin E1 residues 1-364
TagsC-terminal 6x Histidine tagExpressionBacterialMutationtruncation of colicin E1 including 1-364PromoterT7Available SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
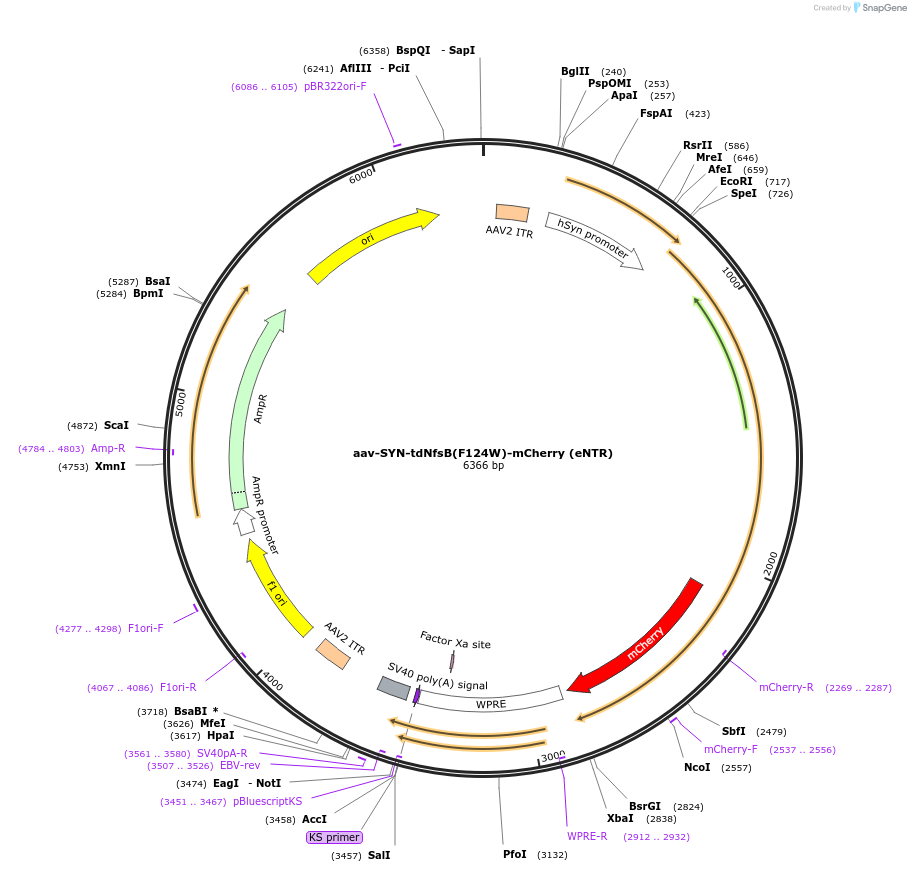
aav-SYN-tdNfsB(F124W)-mCherry (eNTR)
Plasmid#113760PurposeAAV-mediated expression of bacterial tdNfsB-mCherry (eNTR) under the SYN promoterDepositorInserttdNfsB(F124W)-mCherry
UseAAVTagsmCherryExpressionMammalianMutationF124WPromoterSynapsinAvailable SinceOct. 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
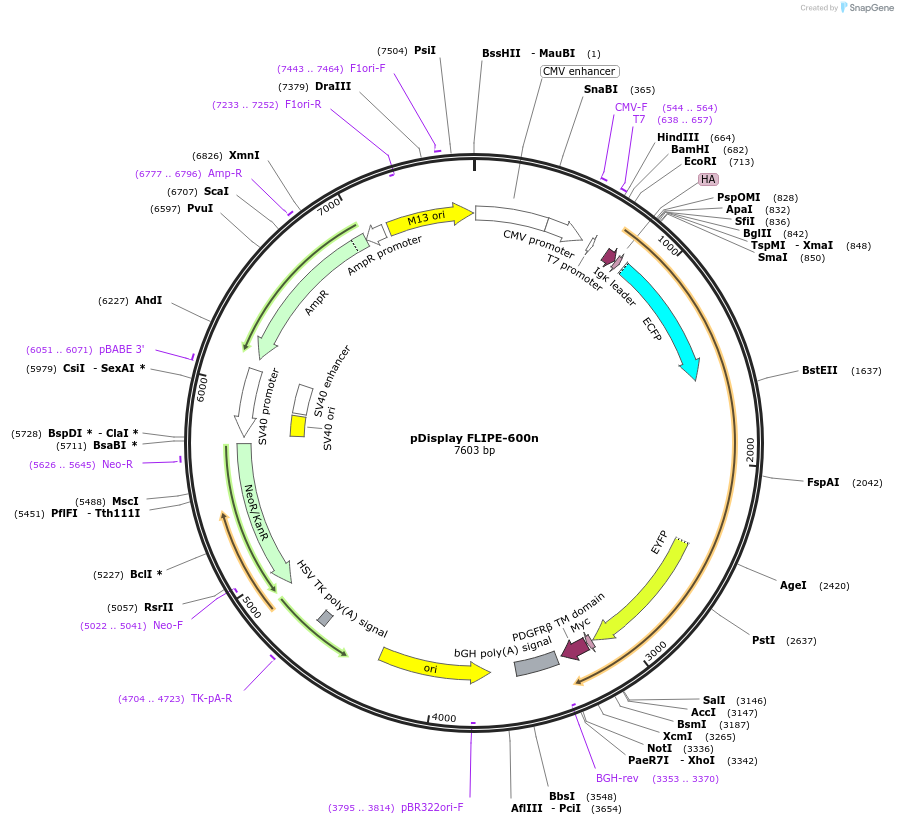
pDisplay FLIPE-600n
Plasmid#13545DepositorInsertFLIP-E WT
TagsECFP and VenusExpressionMammalianAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
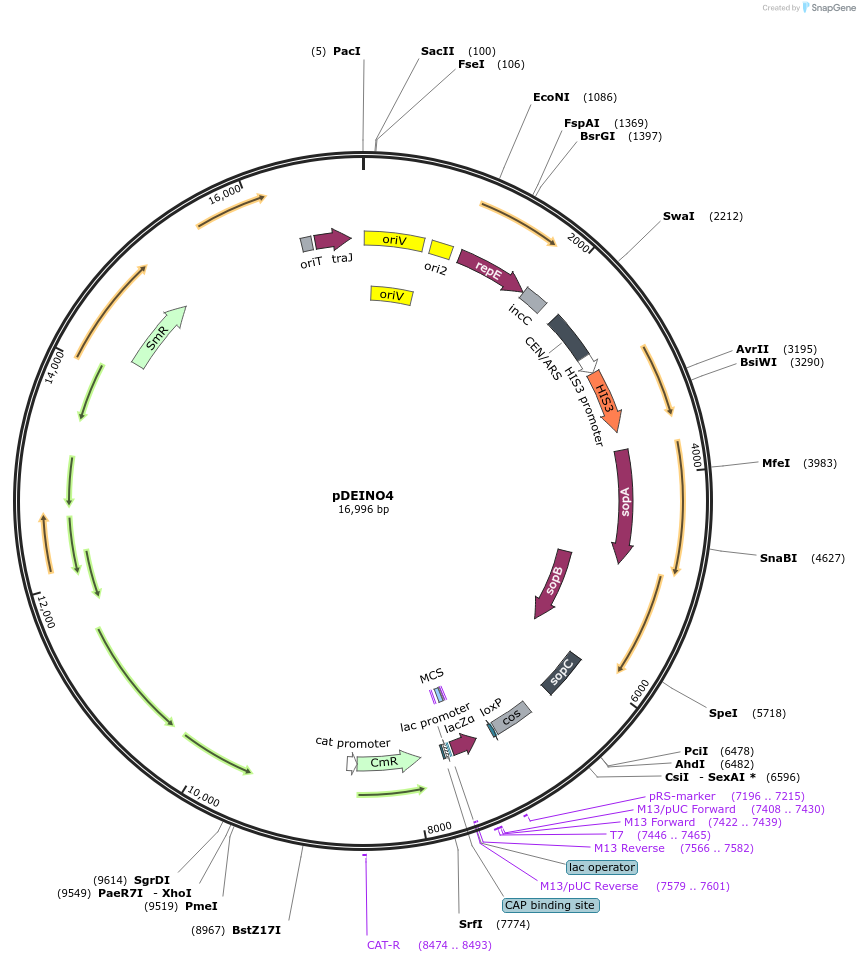
pDEINO4
Plasmid#179488PurposeMulti-host shuttle plasmid for E. coli, D. radiodurans, and S. cerevisiae; contains an oriT for transfer via conjugationDepositorInsertaadA1
UseSynthetic Biology; Bacterial and yeast cloningAvailable SinceMay 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
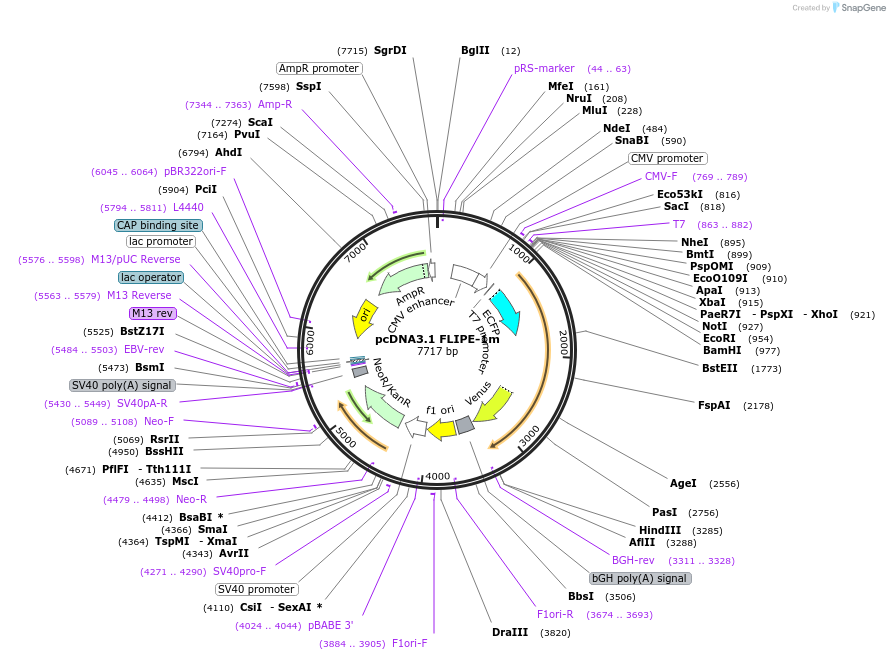
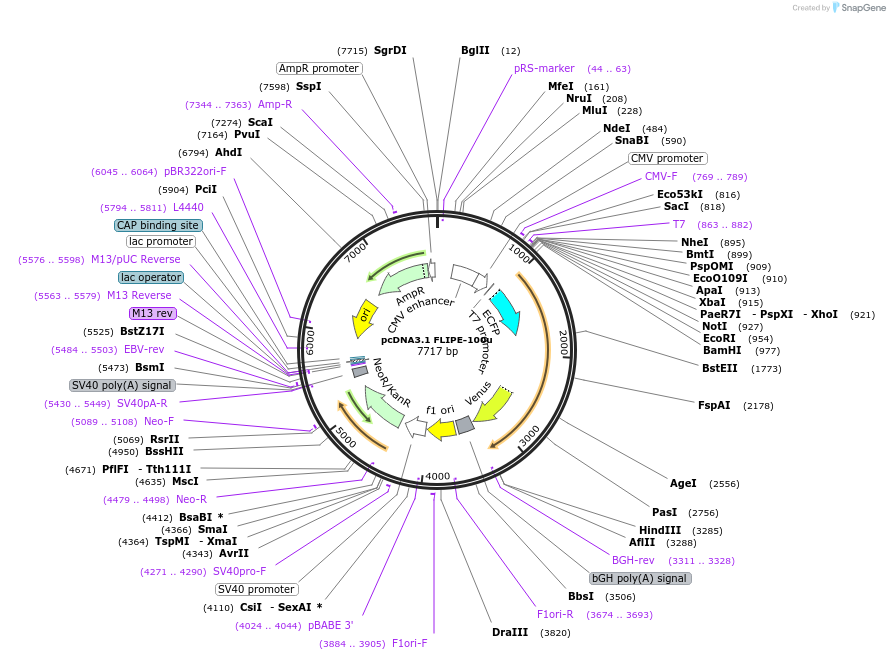
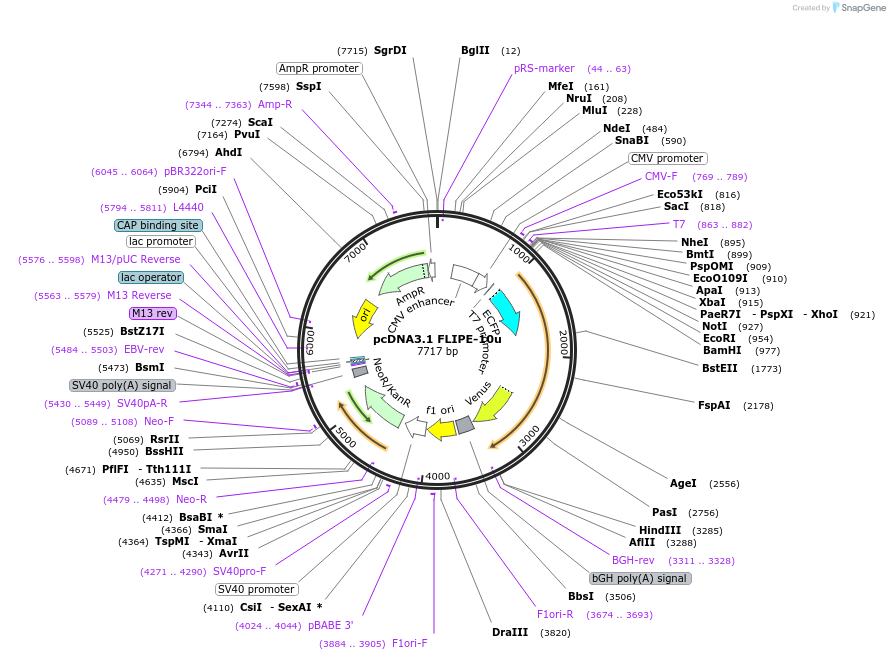
pcDNA3.1 FLIPE-1m
Plasmid#13544DepositorInsertFLIP-E A207W
TagsECFP and VenusExpressionMammalianMutationA207W in binding proteinAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
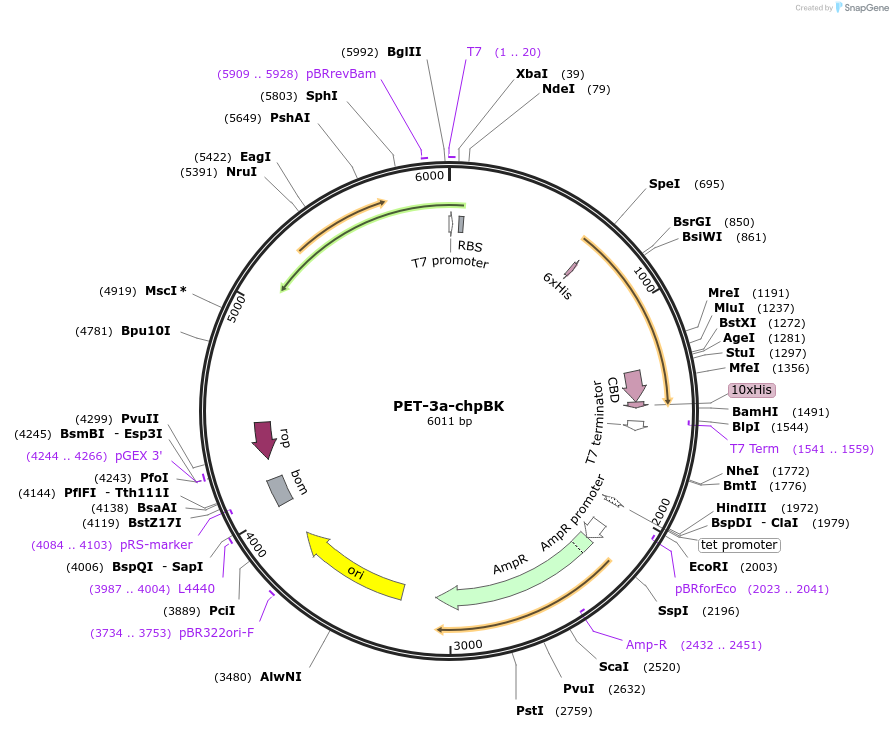
PET-3a-chpBK
Plasmid#155201PurposeTo express recombinant ChpBK proteinDepositorInsertschpS-chpBK-6×His-Tag
GyrA-CBD-10xHis-Tag
Tags10xHis and 6xHisExpressionBacterialAvailable SinceDec. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
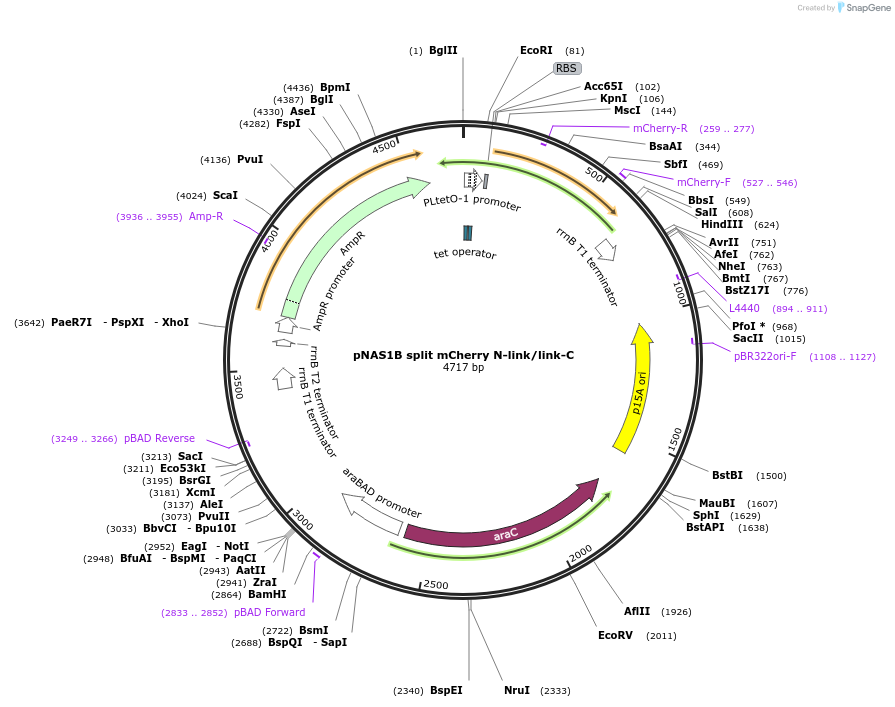
pNAS1B split mCherry N-link/link-C
Plasmid#61965PurposeNegative control duet plasmid for split mCherry assembly assay (i.e., neither half of mCherry is fused to another peptide or protein)DepositorInsertsN-mCherry-link
link-C-mCherry
ExpressionBacterialPromoterPBAD and PLtetOAvailable SinceFeb. 13, 2015AvailabilityAcademic Institutions and Nonprofits only -
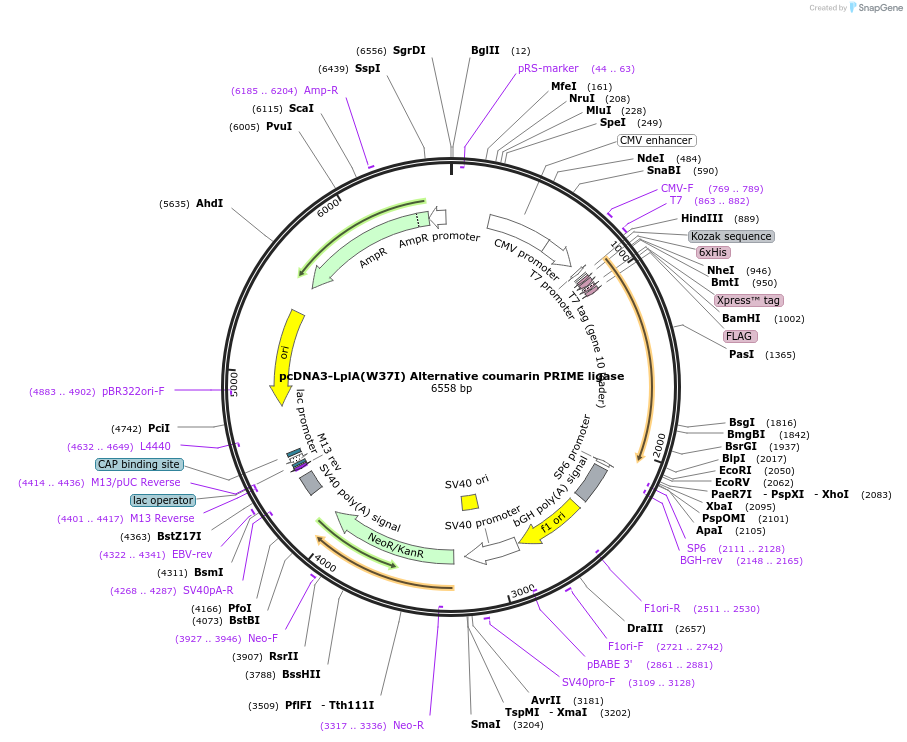
pcDNA3-LplA(W37I) Alternative coumarin PRIME ligase
Plasmid#25846DepositorInsertLplA-W37I
TagsFlag and HisExpressionMammalianMutationW37IAvailable SinceJuly 27, 2010AvailabilityAcademic Institutions and Nonprofits only -
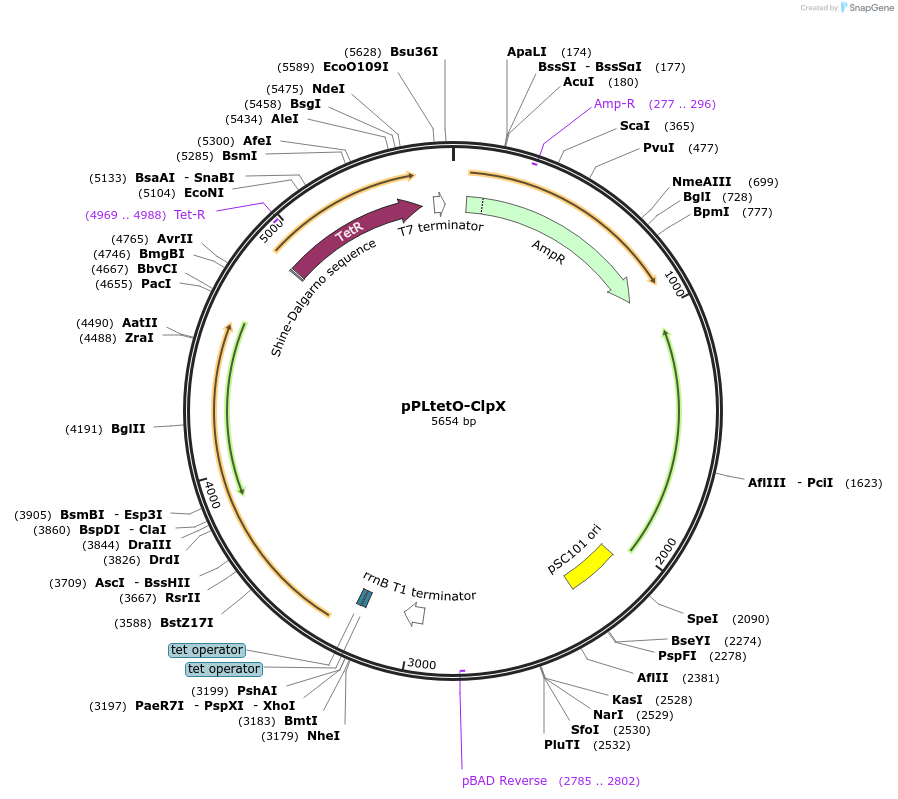
pPLtetO-ClpX
Plasmid#89657PurposePlasmid for titrating synthesis of ClpX with doxycyclineDepositorInsertclpX
UseSynthetic BiologyAvailable SinceMay 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
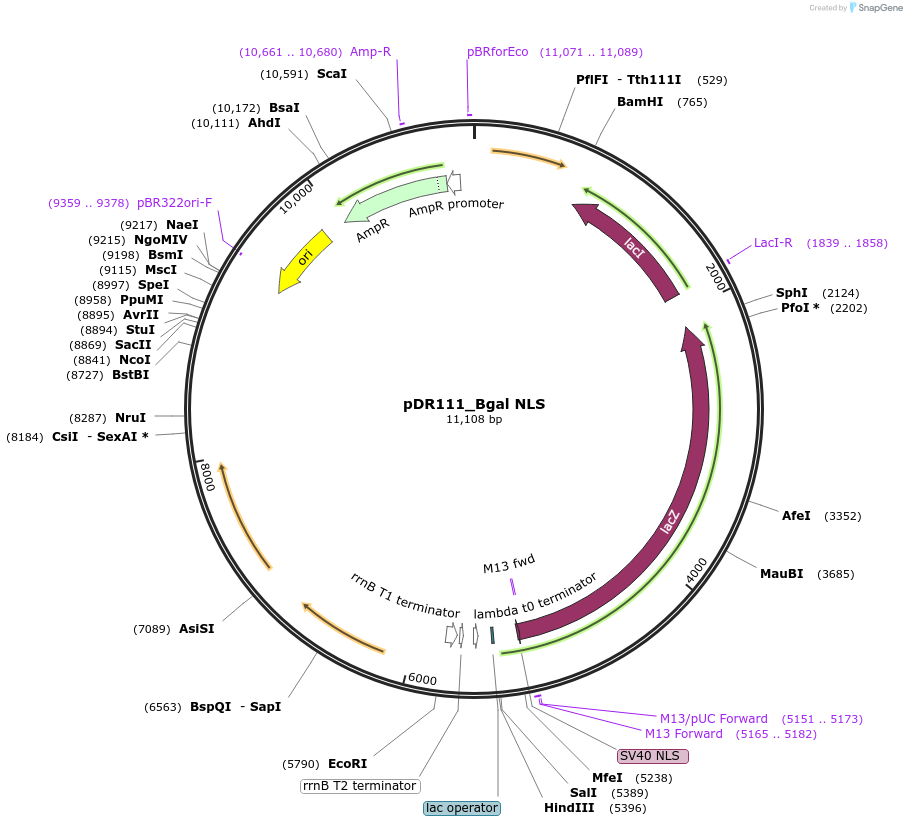
pDR111_Bgal NLS
Plasmid#188396PurposeExpresses beta-galactosidase via Phyper-spank which is optimized for Bacillus subtilis. There is a secretion peptide (PhoD) and nuclear localization signal (SV40) that is fused to beta-galactosidase.DepositorInsertlacZ
TagsPhoD secretion peptide and SV40 NLSExpressionBacterialPromoterPhyper spankAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
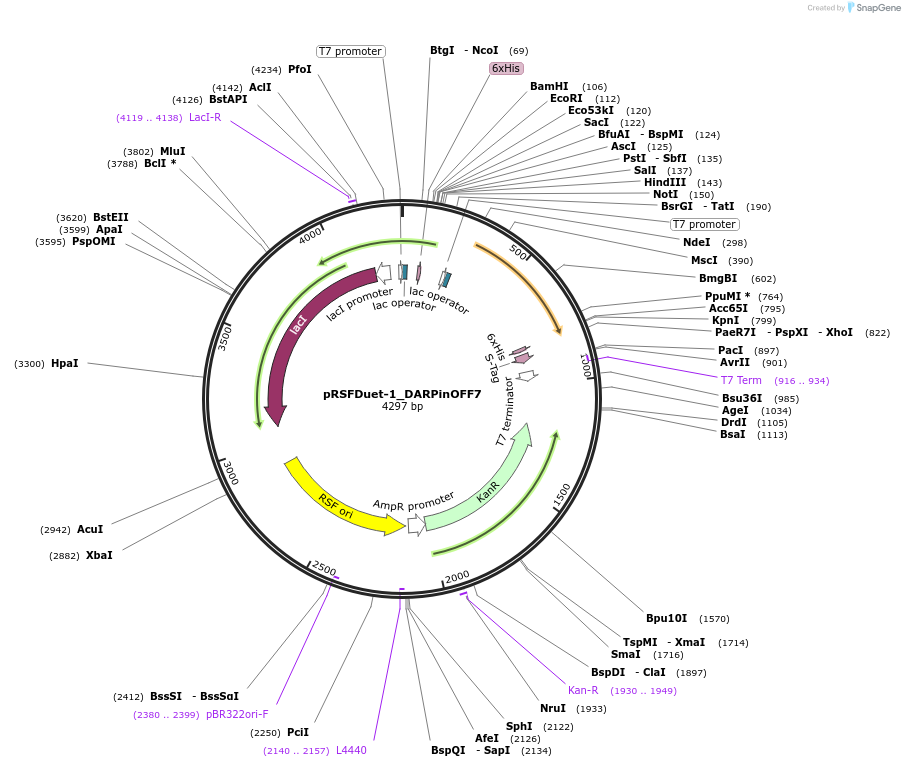
pRSFDuet-1_DARPinOFF7
Plasmid#182396PurposeBacterial expression of DARPinOFF7DepositorInsertDARPinOFF7
ExpressionBacterialPromoterT7Available SinceFeb. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
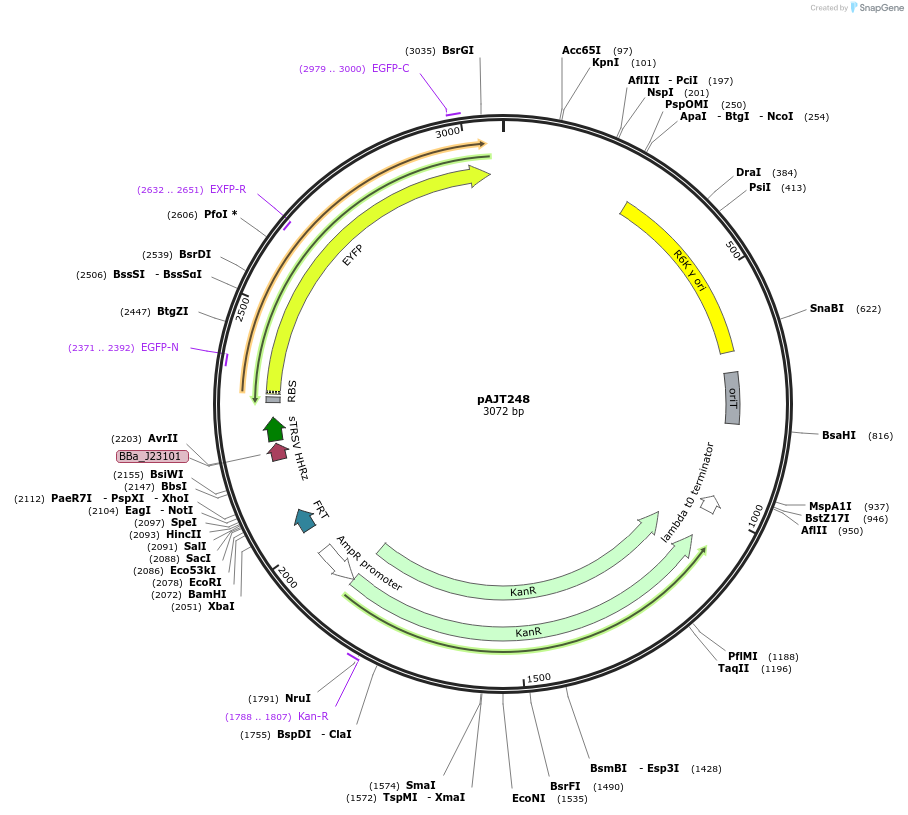
pAJT248
Plasmid#202289PurposeRPU integration plasmid (LPattB2)DepositorInsertJ23101-RiboJ00-B0034-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
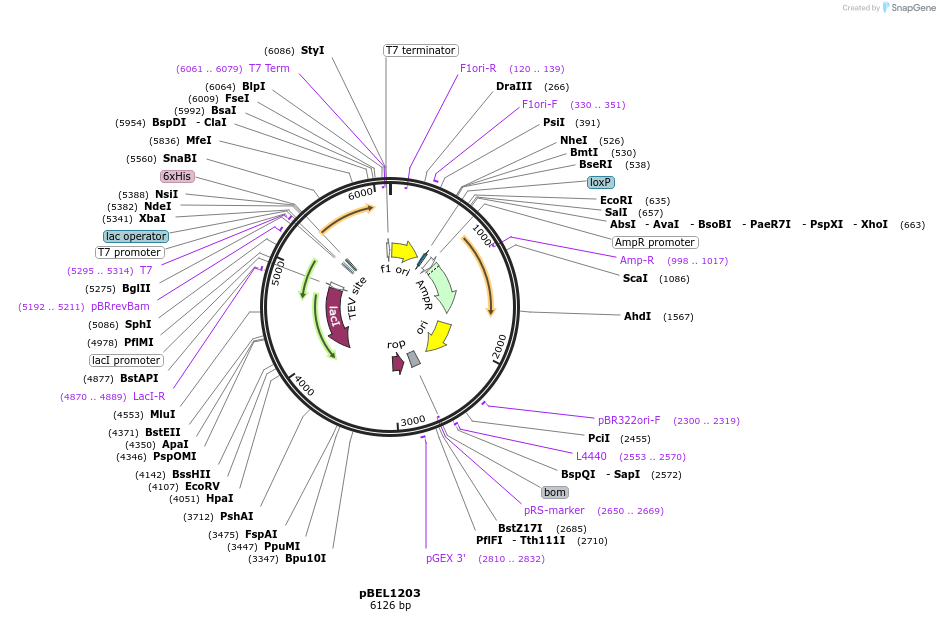
pBEL1203
Plasmid#196024PurposeExpression/purification of 6xHis-TEV-MlaCDepositorInsertMlaC
ExpressionBacterialAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
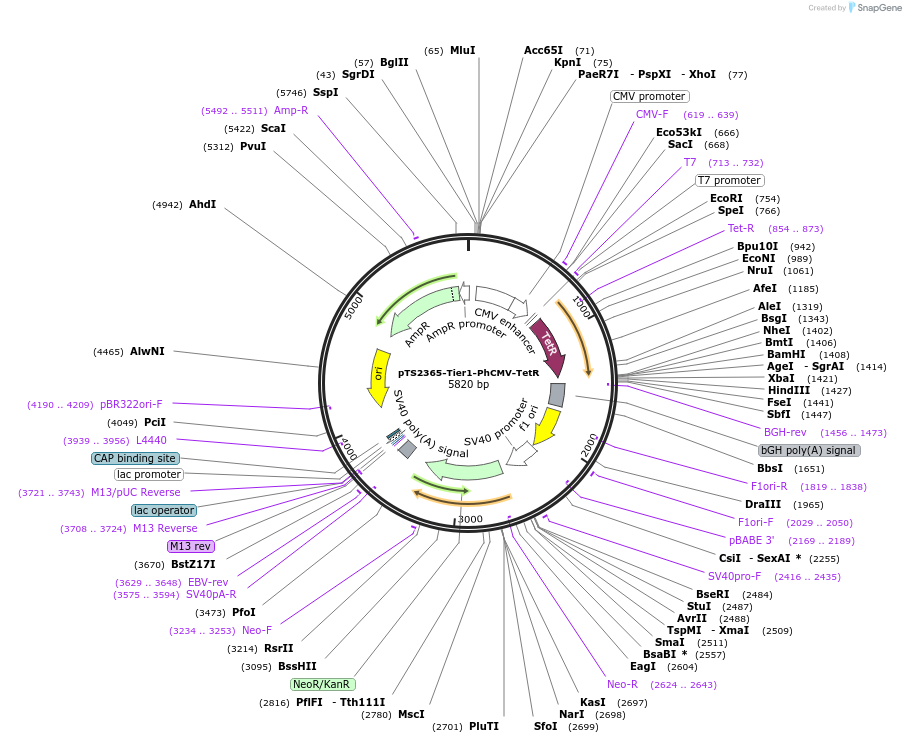
pTS2365-Tier1-PhCMV-TetR
Plasmid#169566PurposeTier-1 vector encoding PhCMV-driven tetracycline dependent regulator protein TetR (PhCMV-TetR-pA).DepositorInsertPCMV-driven tetracycline repressor protein class B from transposon Tn10
ExpressionMammalianPromoterPhCMVAvailable SinceJune 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
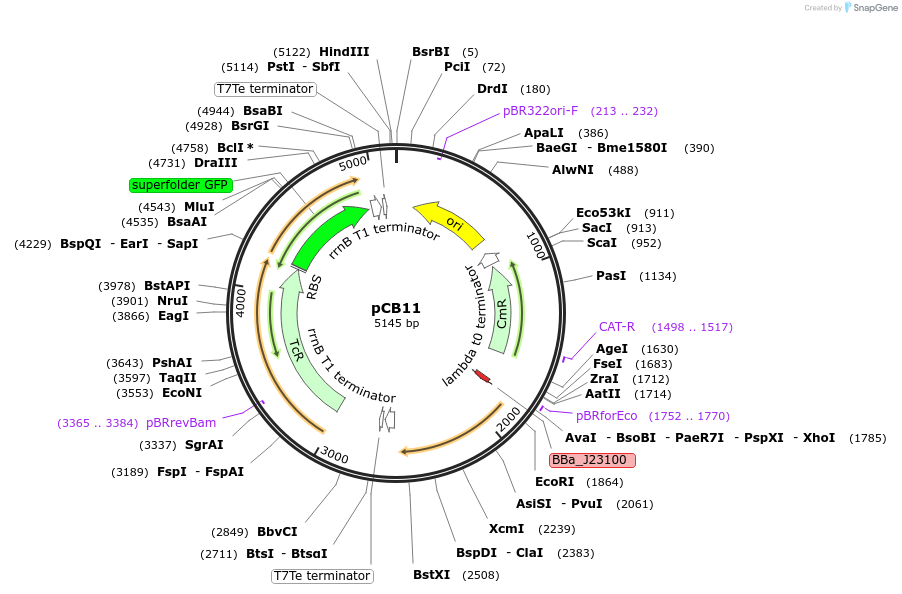
pCB11
Plasmid#201111PurposepSR58.6 (Addgene plasmid #63176) modified to include the tetA gene upstream of sfGFP.DepositorInsertsCcaR
tetA(C)
sfGFP
ExpressionBacterialMutationC-to-T transition at pos. no. 1049 of tetA(C) lea…PromoterJ23100 and PcpcG2-172Available SinceMarch 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
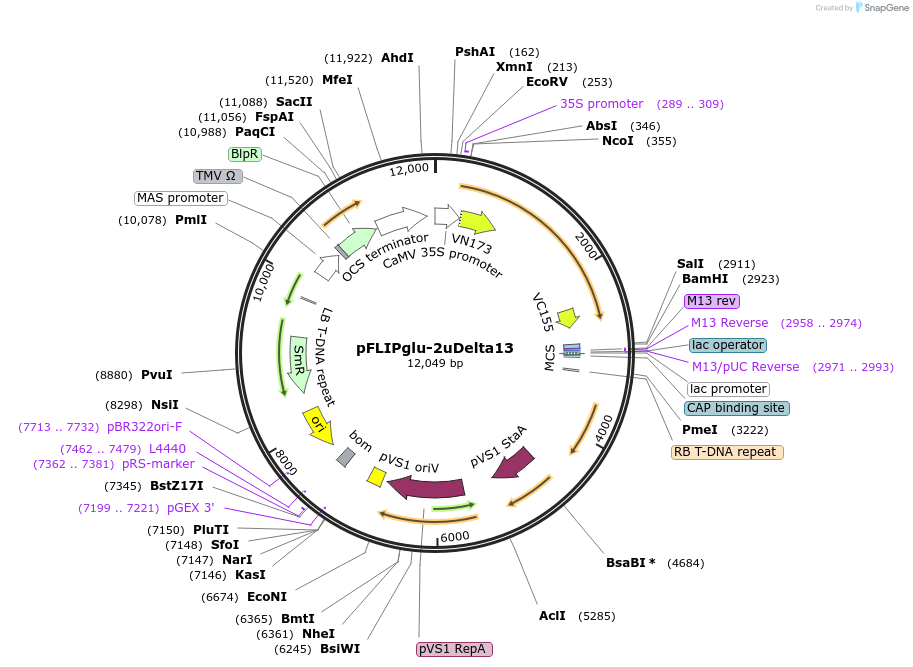
pFLIPglu-2uDelta13
Plasmid#12990DepositorInsertFLIPglu F16F D152A Delta13
UsePlant nanosensorTagsECFP and EYFPExpressionPlantMutationD152A mutation in binding proteinAvailable SinceJan. 5, 2007AvailabilityAcademic Institutions and Nonprofits only -
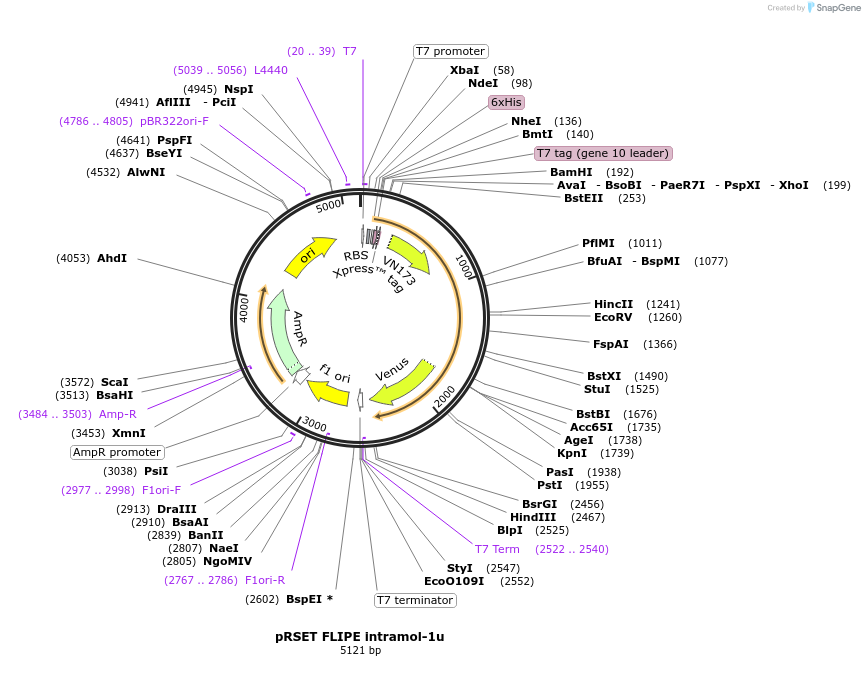
pRSET FLIPE intramol-1u
Plasmid#13549DepositorInsertFLIP-E intramol WT (gltI E. coli)
TagsEYFPExpressionBacterialMutationECFP insert in binding proteinAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
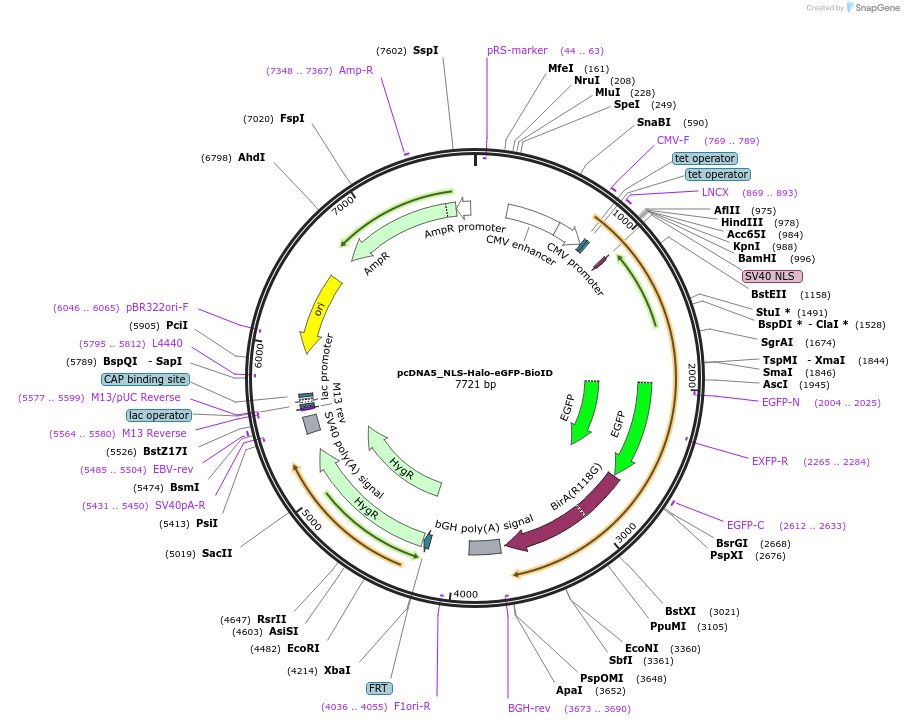
pcDNA5_NLS-Halo-eGFP-BioID
Plasmid#218448Purposeexpresses the NLS-Halo-eGFP-BioID fusion protein via stable Flp-In integrationDepositorInsertNLS-Halo-eGFP-BirA
ExpressionMammalianPromoterCMVAvailable SinceJune 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
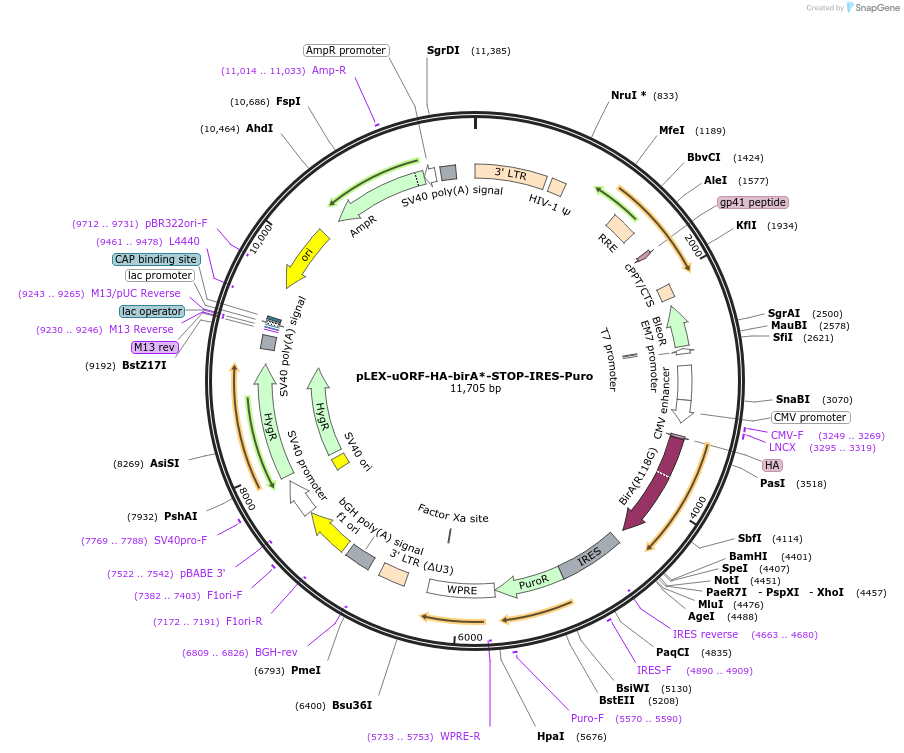
pLEX-uORF-HA-birA*-STOP-IRES-Puro
Plasmid#120558PurposeExpresses control HA-birA* in mammalian cells & for virus productionDepositorInsertbirA
UseLentiviralTagsHAExpressionMammalianMutationR118GPromoterCMVAvailable SinceJan. 30, 2019AvailabilityAcademic Institutions and Nonprofits only -
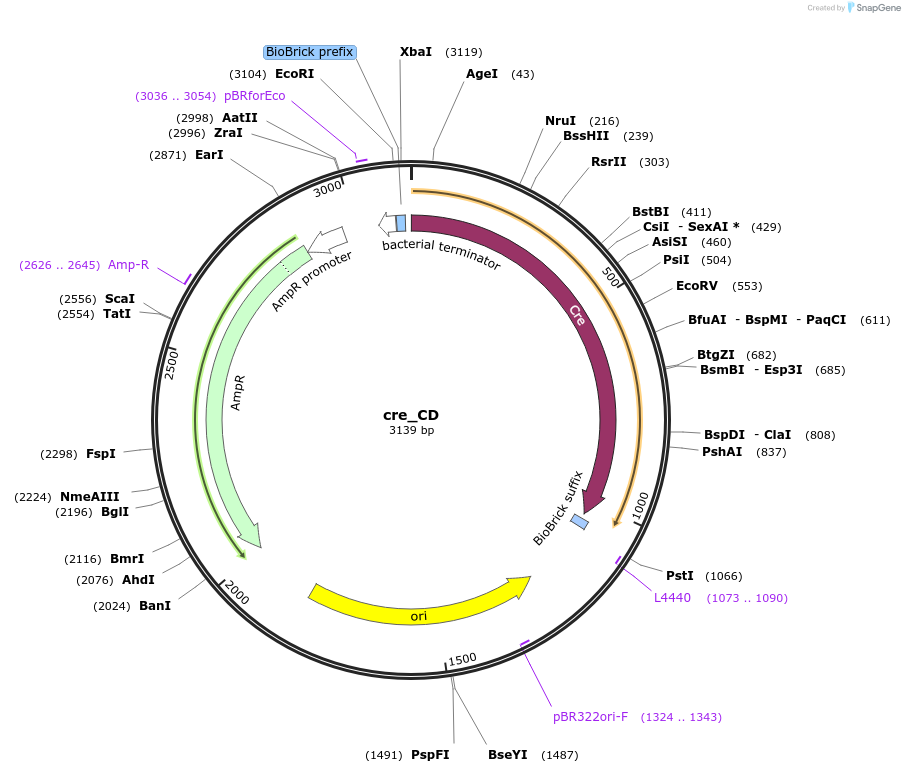
cre_CD
Plasmid#66030PurposeMoClo Basic Part: CDS - cre recombinase (recombines DNA at loxP sites) [C:cre:D]DepositorInsertRecombinase
UseSynthetic BiologyAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
pFLIPglu-600uDelta13
Plasmid#12991DepositorInsertFLIPglu F16A Delta13
UsePlant nanosensorTagsECFP and EYFPExpressionPlantMutationF16A in binding proteinAvailable SinceJan. 5, 2007AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1 FLIPE-100u
Plasmid#13543DepositorInsertFLIP-E S118W
TagsECFP and VenusExpressionMammalianMutationS118W in binding proteinAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
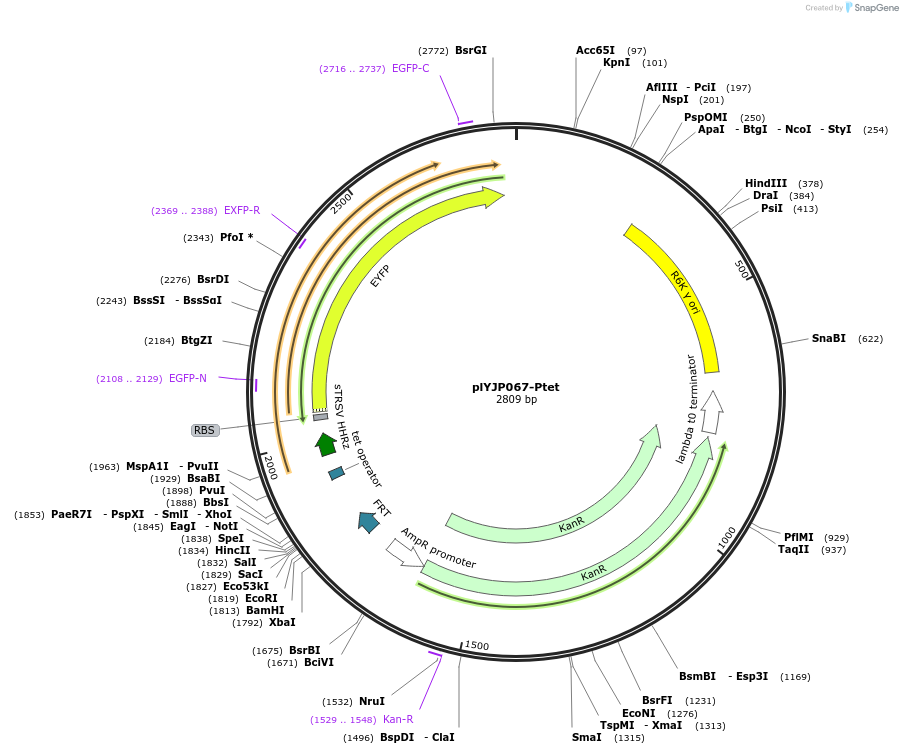
plYJP067-Ptet
Plasmid#202299PurposePTet-YFP plasmid for LPattB2 integrationDepositorInsertPtet-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pET22b-BirA-CPDSalI
Plasmid#38258DepositorInsertbirA (birA E. coli)
Tags6xHis and Vibrio cholerae MARTX toxin cysteine pr…ExpressionBacterialMutationcloned from pGEX4T1-BirAPromoterT7Available SinceJuly 20, 2012AvailabilityAcademic Institutions and Nonprofits only -
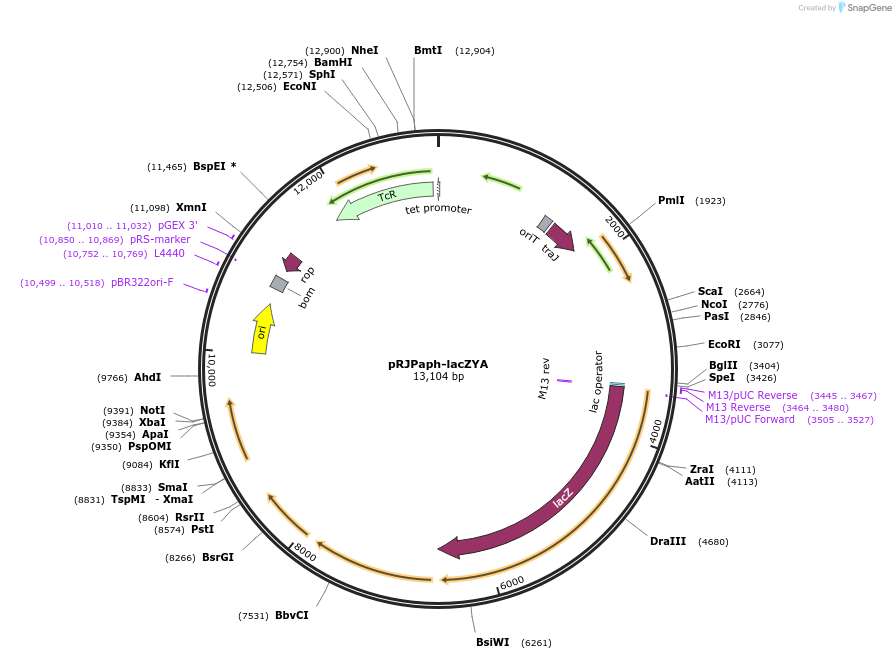
pRJPaph-lacZYA
Plasmid#67037PurposePlasmid for Paph-lacZYA integration downstream of B. diazoefficiens (B. japonicum) scoIDepositorInsertsB. diazoefficiens DNA from scoI downstream region (comprising blr1132')
lacZYA
UseConjugative bacterial vectorAvailable SinceOct. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
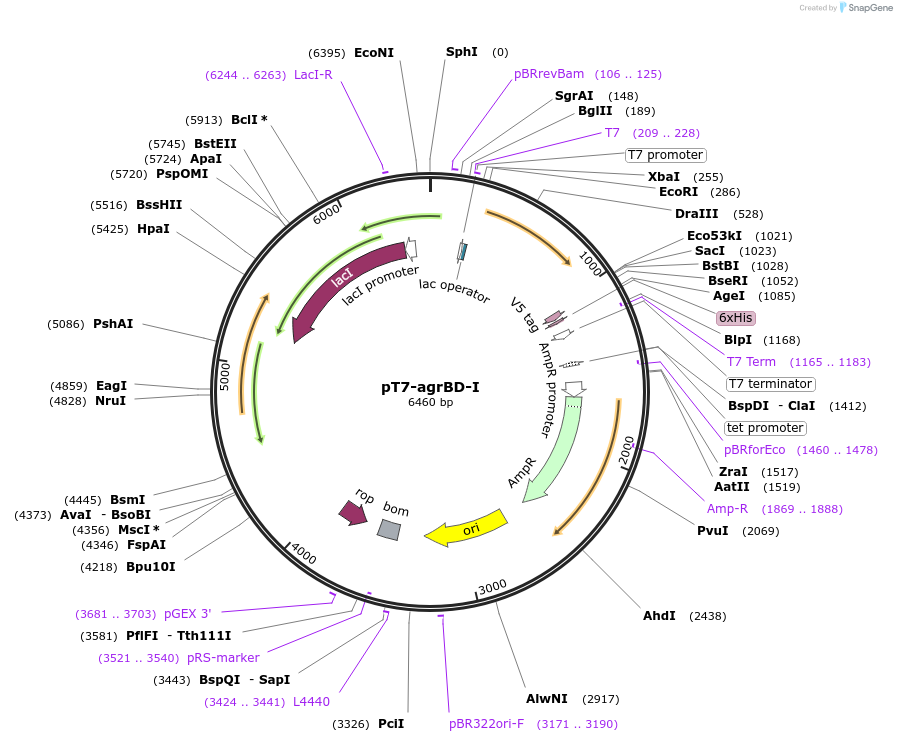
pT7-agrBD-I
Plasmid#53439PurposeT7-dependent expression of the type-I agrB and agrD genes of S. aureus RN4220DepositorInsertagrBD locus
UseSynthetic BiologyTagsV5 epitope, 6x HIS (not expressed on agrB or agrD…ExpressionBacterialMutationnonePromoterT7-lacAvailable SinceJune 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
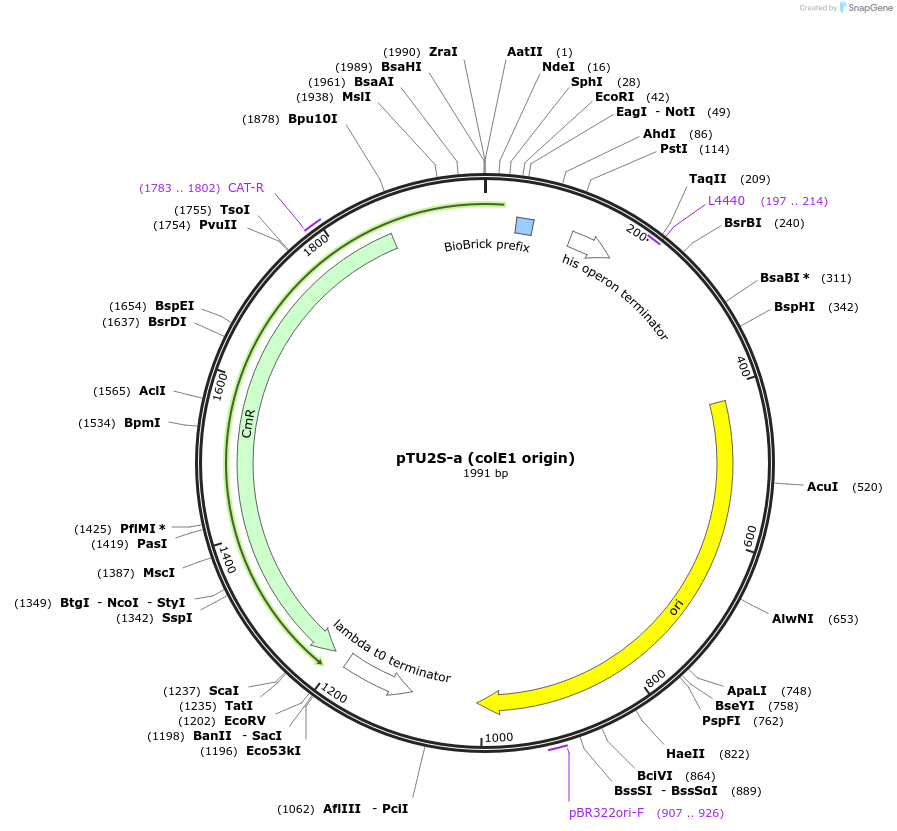
pTU2S-a (colE1 origin)
Plasmid#74091PurposeSecondary module (BpiI) site in pTU2-a backbone with colE1 (medium-copy) origin of replicationDepositorInsertGolden Gate destination vector - Level 2, 2 TU's with secondary module and colE1 origin
UseSynthetic BiologyExpressionBacterialPromoterNoneAvailable SinceApril 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
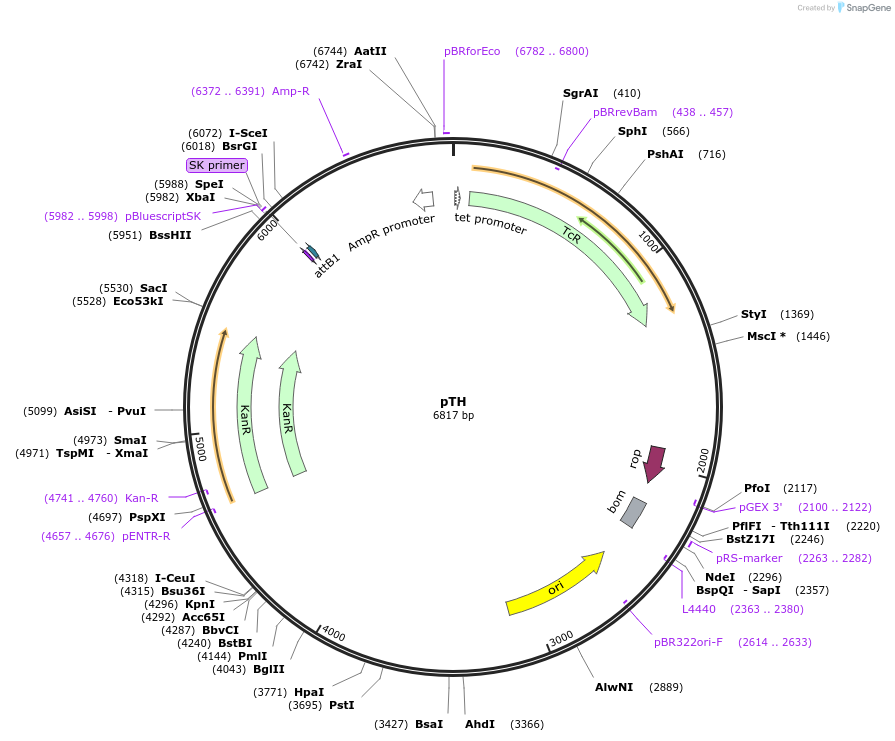
pTH
Plasmid#56659PurposeDual-expression vector for P. tricornutumDepositorInsertKanR
UseSynthetic Biology; Diatom expressionAvailable SinceOct. 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
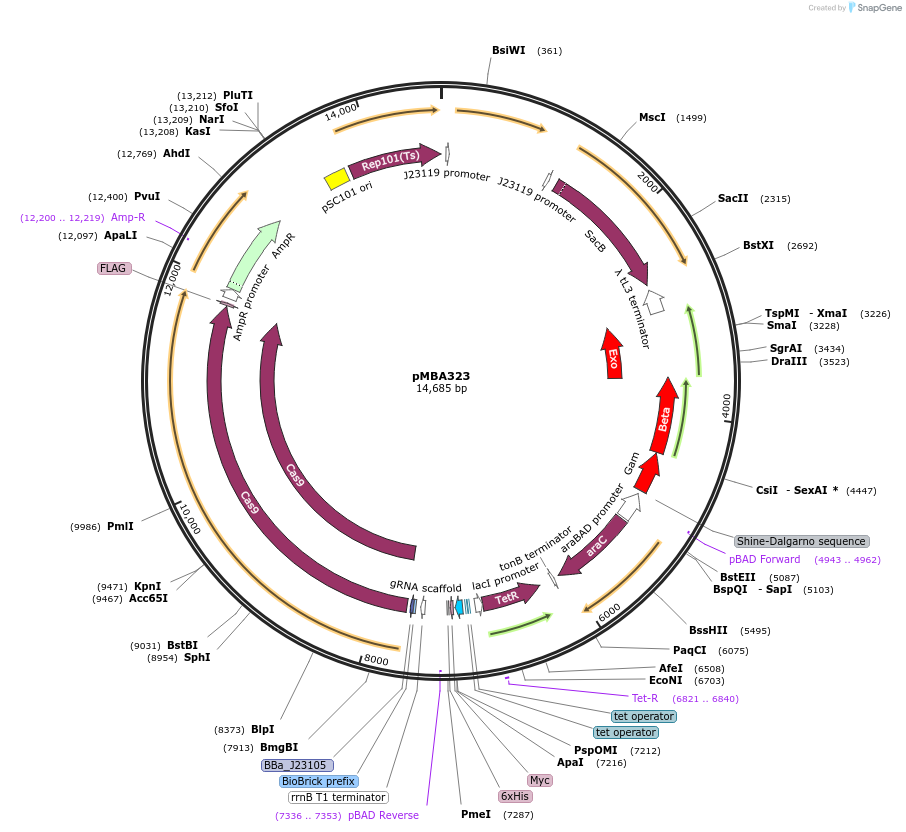
pMBA323
Plasmid#214751PurposeTo create delta(glmS,nadE) working strains and maintaining their viabilityDepositorInsertBBa_J23105-cas9 + pBAD-λ Red genes + ptet-gRNA (pBR322ori)
UseCRISPRExpressionBacterialAvailable SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
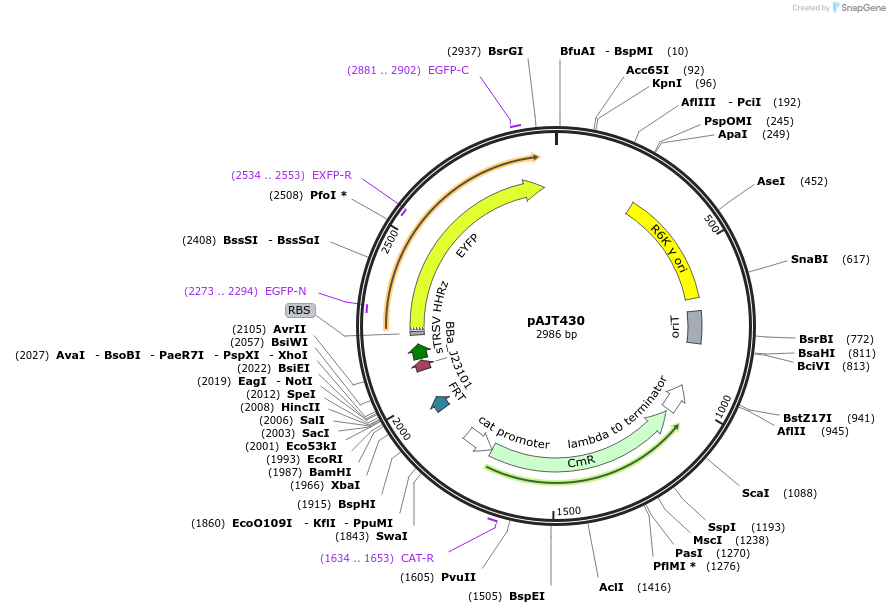
pAJT430
Plasmid#202291PurposeRPU integration plasmid (LPattB7)DepositorInsertJ23101-RiboJ00-B0034-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
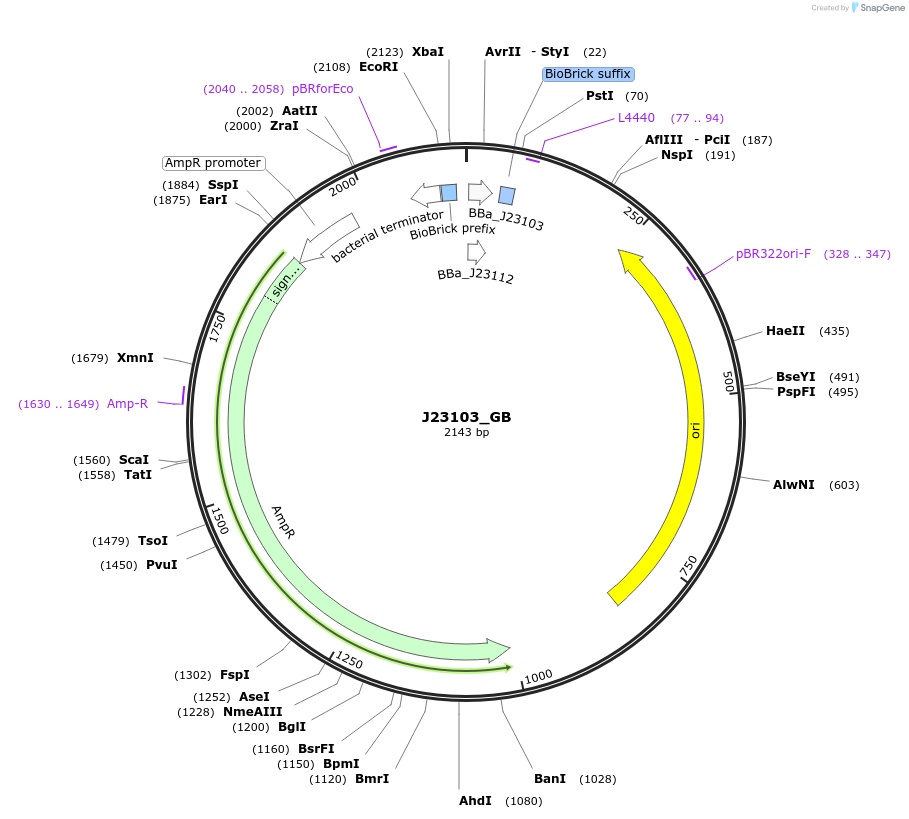
J23103_GB
Plasmid#65991PurposeMoClo Basic Part: Constitutive promoter - Anderson series - low strength [G:J23103:B]DepositorInsertConstitutive promoter
UseSynthetic BiologyAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
J23107_GB
Plasmid#65999PurposeMoClo Basic Part: Constitutive promoter - Anderson series - medium strength [G:J23107:B]DepositorInsertConstitutive promoter
UseSynthetic BiologyAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
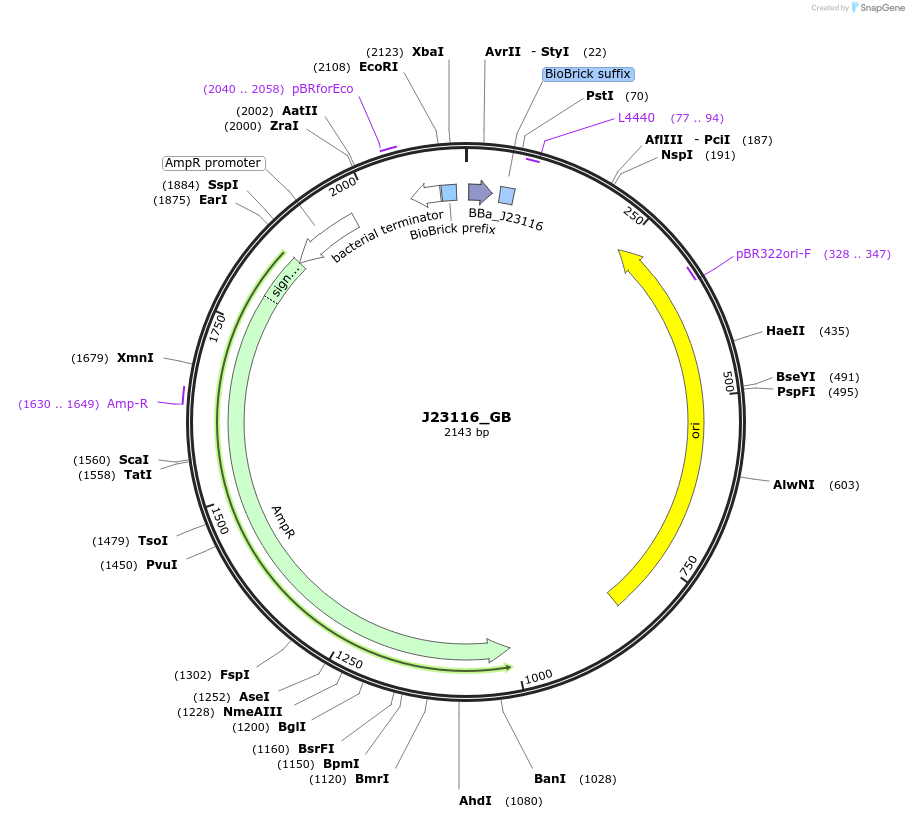
J23116_GB
Plasmid#66003PurposeMoClo Basic Part: Constitutive promoter - Anderson series - low strength [G:J23116:B]DepositorInsertConstitutive promoter
UseSynthetic BiologyAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
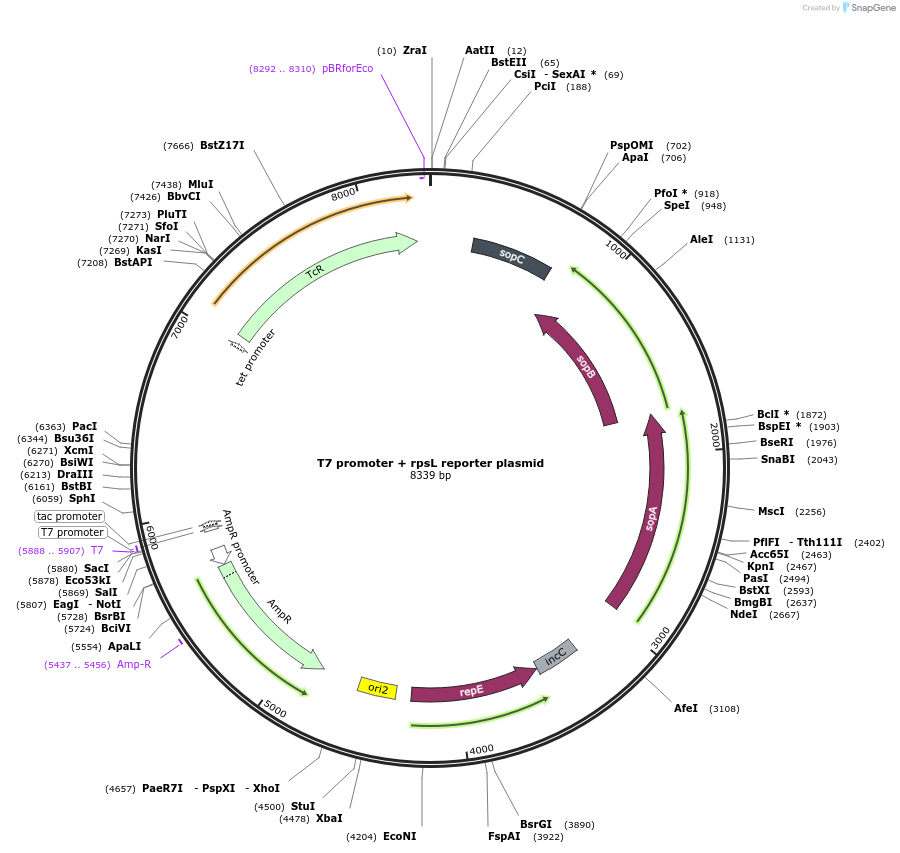
T7 promoter + rpsL reporter plasmid
Plasmid#156455PurposeA mutational reporter plasmid that can gain Streptomycin and/or Tetracycline resistance upon mutation. Contains T7 promoter.DepositorInsertsrpsL
TcR
ExpressionBacterialMutationATG start codon mutated to ACGPromotertac and tetAvailable SinceOct. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
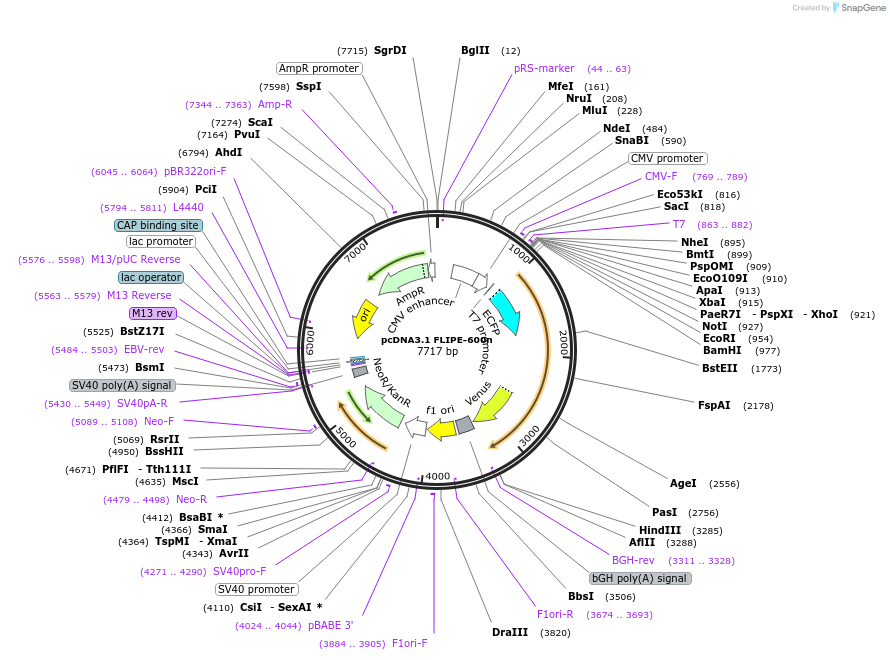
pcDNA3.1 FLIPE-600n
Plasmid#13541DepositorInsertFLIP-E WT
TagsECFP and VenusExpressionMammalianAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
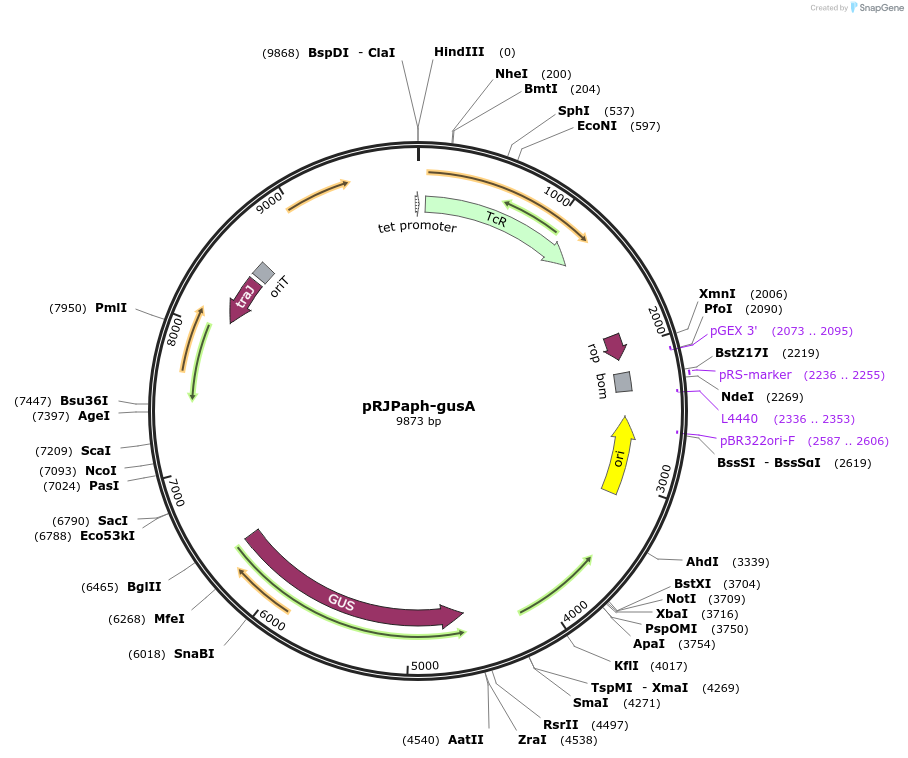
pRJPaph-gusA
Plasmid#67036PurposePlasmid for Paph-gusA integration downstream of B. diazoefficiens (B. japonicum) scoIDepositorInsertsB. diazoefficiens DNA from scoI downstream region (comprising blr1132')
gusA
UseConjugative bacterial vectorAvailable SinceOct. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
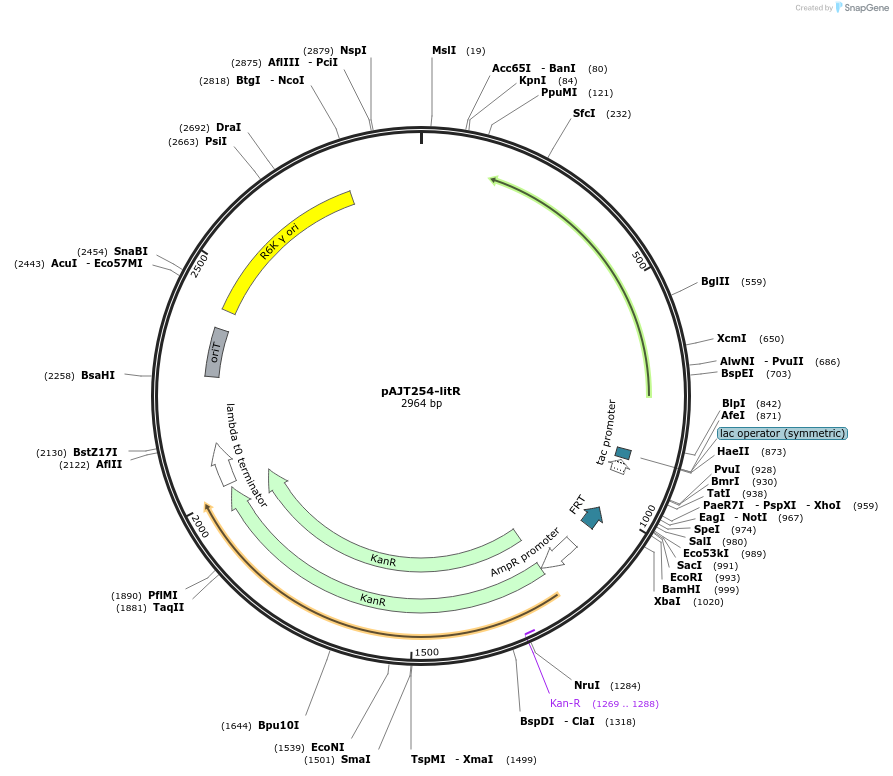
pAJT254-litR
Plasmid#202308PurposelitR repressor plasmid for LPattB2 integrationDepositorInsertlitR
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
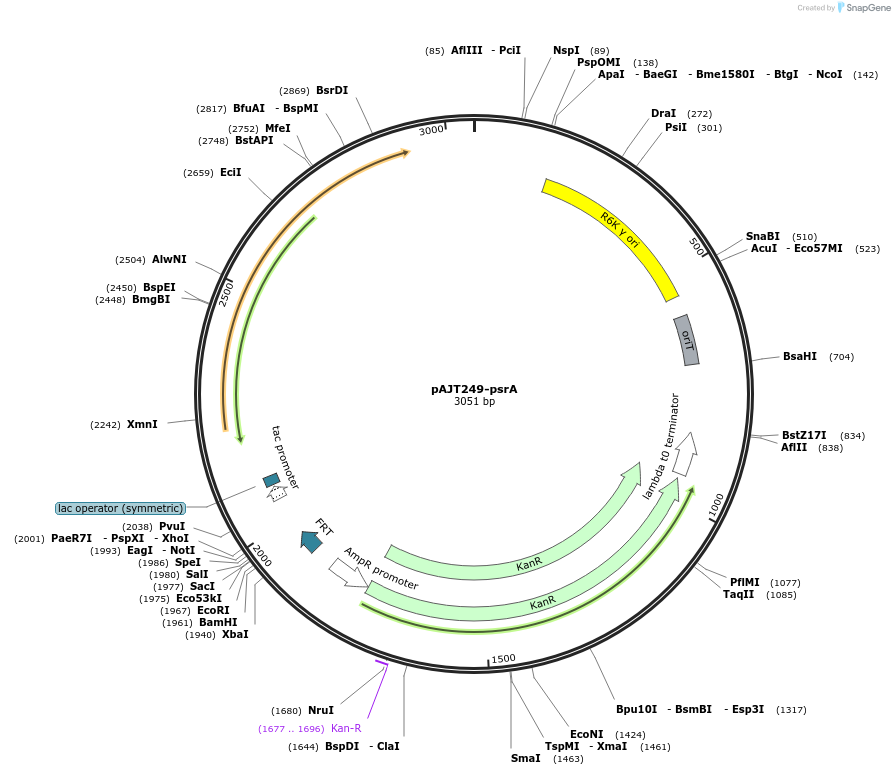
pAJT249-psrA
Plasmid#202304PurposepsrA repressor plasmid for LPattB2 integrationDepositorInsertpsrA
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
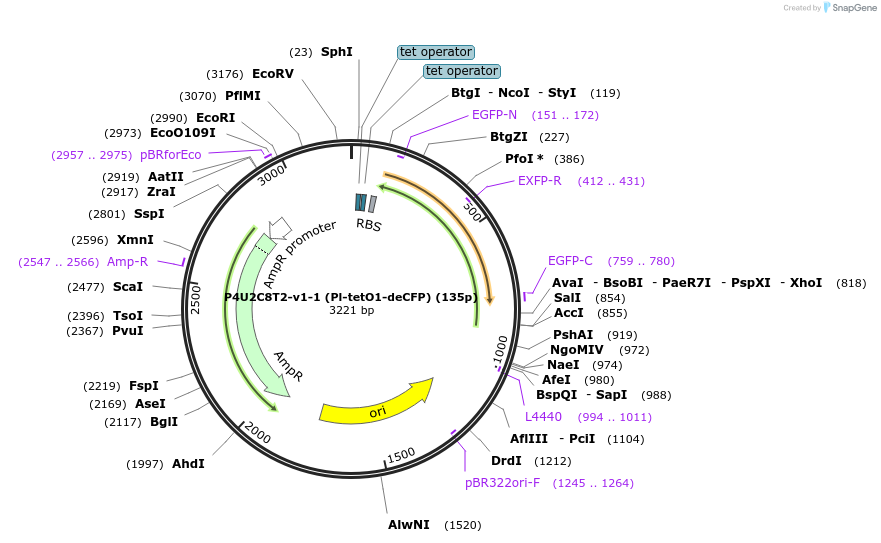
P4U2C8T2-v1-1 (Pl-tetO1-deCFP) (135p)
Plasmid#70211Purpose1 Part of 4-part switchDepositorInsertdeCFP
Available SinceMay 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pCAG-AILR-LplA-ER
Plasmid#42574DepositorInsertLplA(AILR) with ER-retention sequence KDEL
TagsFLAGMutationCompared to wild-type LplA: tryptophan 37 to alan…PromoterCAGAvailable SinceApril 11, 2013AvailabilityAcademic Institutions and Nonprofits only -
pETM41-EcLonA
Plasmid#38330DepositorInsertLon protease fragment (lon Escherichia coli)
TagsHis, MBP, and TEVExpressionBacterialMutationPolyphosphate binding fragment of Lon protease. …PromoterT7Available SinceAug. 10, 2012AvailabilityAcademic Institutions and Nonprofits only -
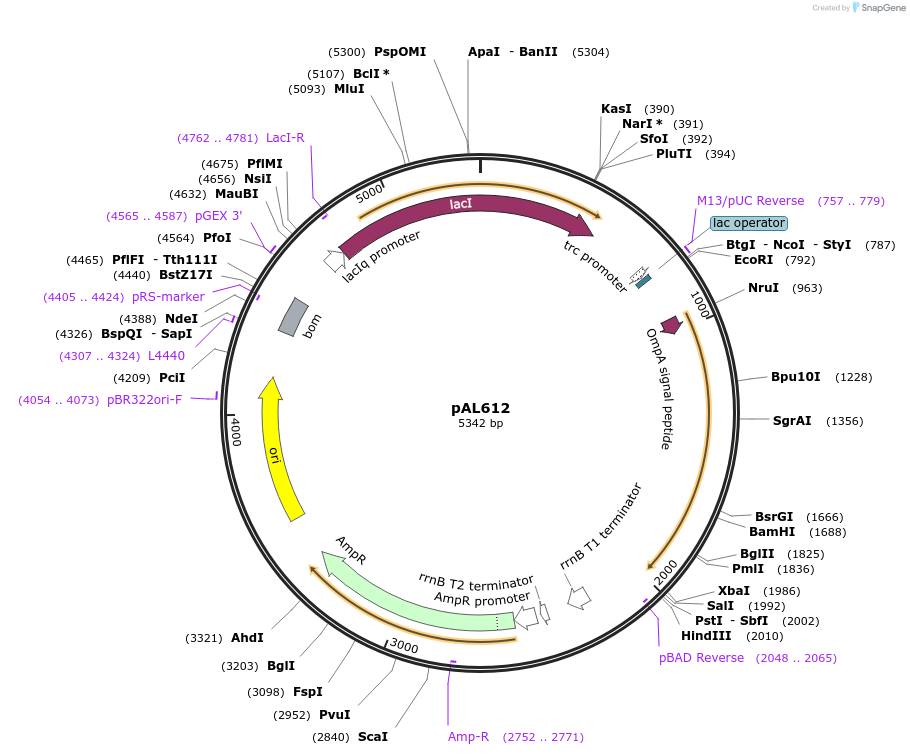
pAL612
Plasmid#119860Purposeprecusor outer membrane protein A, ompA, with mutations Cysteine 290 Serine, Glycine 244 CysteineDepositorInsertouter membrane protein A, ompA, with mutations Cysterine 290 to Serine and Glycine 244 to Cysteine
TagsnoneExpressionBacterialMutationCysterine 290 to Serine and Glycine 244 to Cystei…PromoterlacAvailable SinceDec. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1 FLIPE-10u
Plasmid#13542DepositorInsertFLIP-E A207R
TagsECFP and VenusExpressionMammalianMutationA207R in binding proteinAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
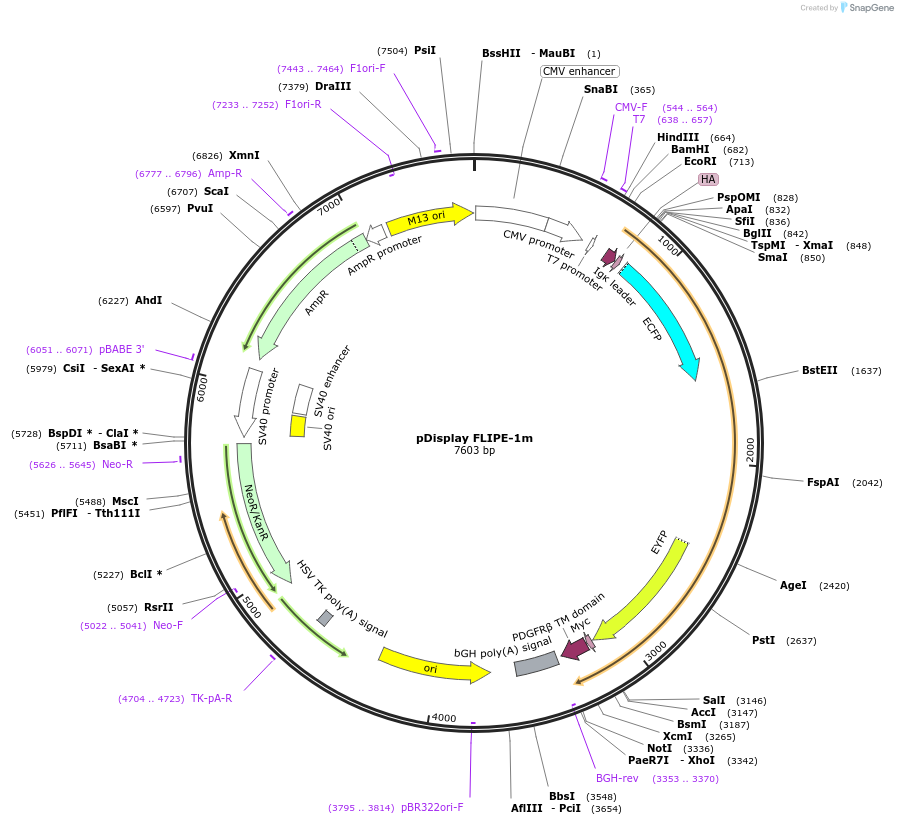
pDisplay FLIPE-1m
Plasmid#13548DepositorInsertFLIP-E A207W
TagsECFP and VenusExpressionMammalianMutationA207W in binding proteinAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
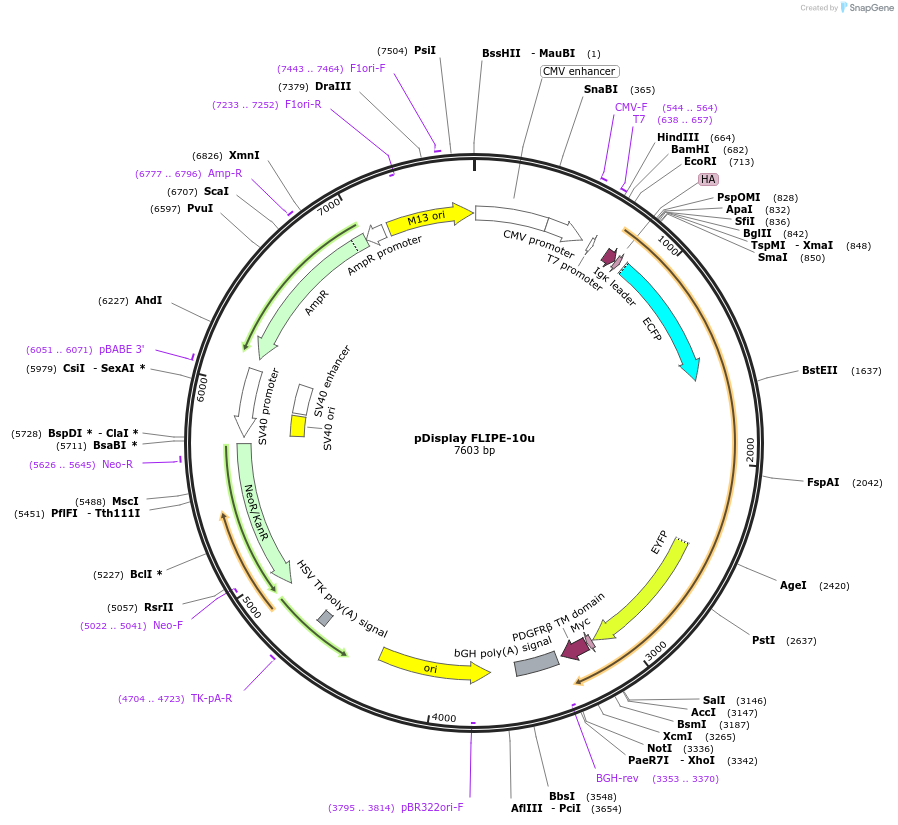
pDisplay FLIPE-10u
Plasmid#13546DepositorInsertFLIP-E A207R
TagsECFP and VenusExpressionMammalianMutationA207R in binding proteinAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
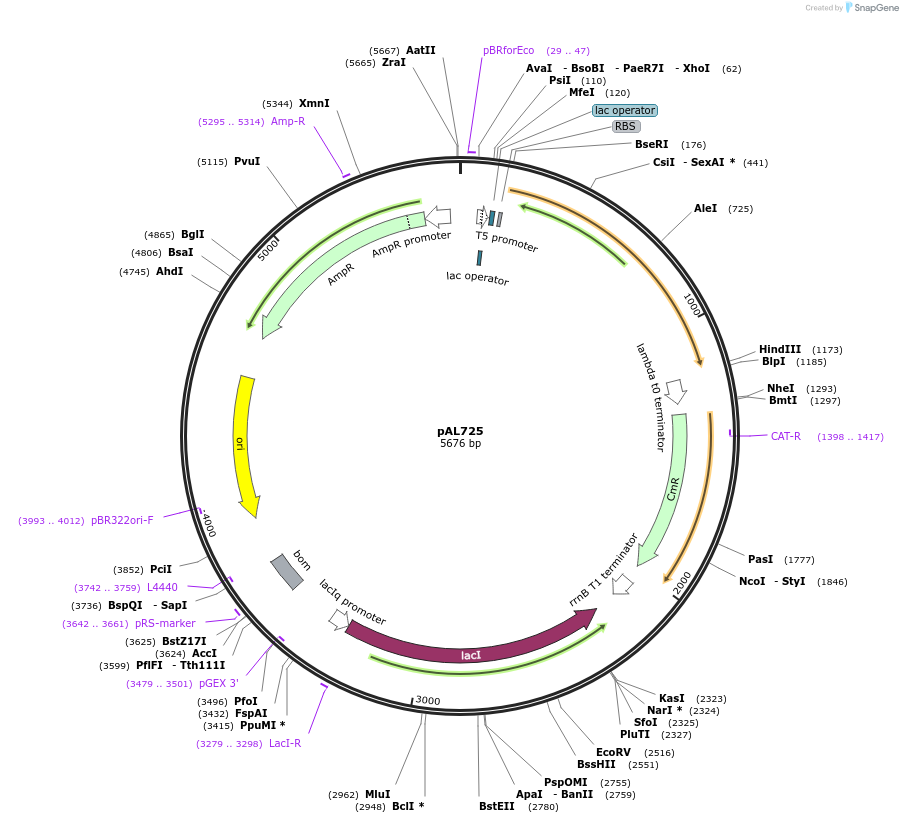
pAL725
Plasmid#119862Purposeprecusor D-galactose/methyl-galactoside ABC transporter periplasmic binding protein, mglB, L244C, D287CDepositorInsertD-galactose/methyl-galactoside ABC transporter periplasmic binding protein, mglB, L244C, D287C (mglB Escherichia coli str. K-12)
TagsnoneExpressionBacterialMutationLeucine 244 to Cysteine, Aspartate 287 to CysteinePromoterT5Available SinceDec. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
pET28a-HsSENP5
Plasmid#71468PurposeEncodes human full length SENP5 optimised for E. coli expression. Suitable for in vitro transcription/translation (T7).DepositorInsertSUMO1/sentrin specific peptidase 5 (SENP5 Human)
TagsHexahistidineExpressionBacterialMutationGene optimised for E. coli expression (Genscript)PromoterT7Available SinceDec. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
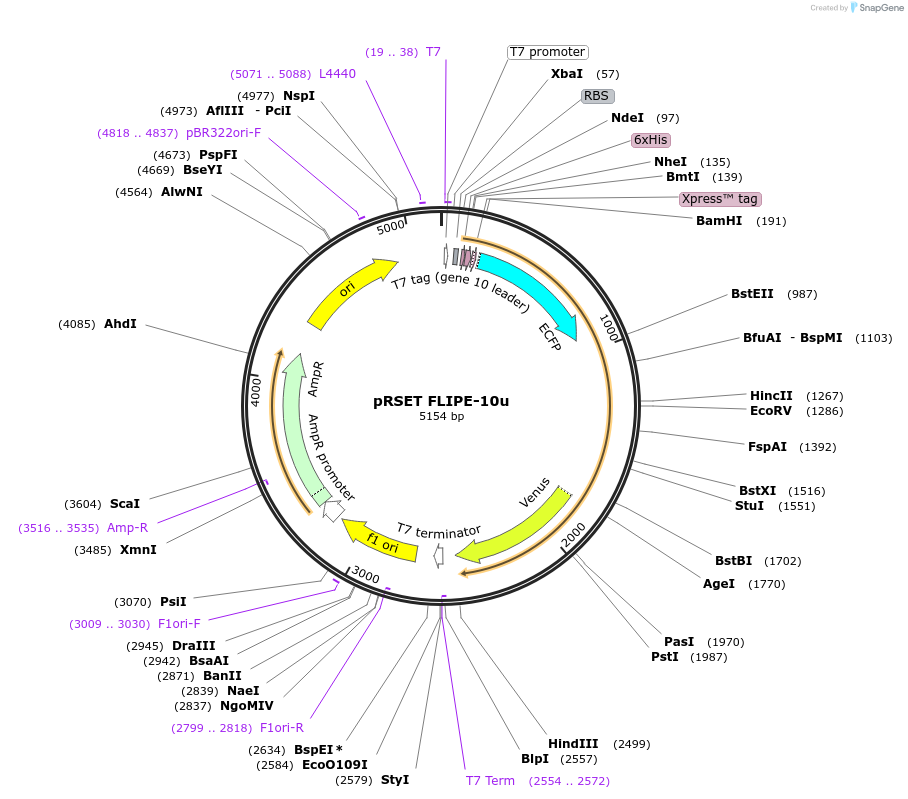
pRSET FLIPE-10u
Plasmid#13538DepositorInsertFLIP-E A207R
TagsECFP and VenusExpressionBacterialMutationA207R in binding proteinAvailable SinceDec. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
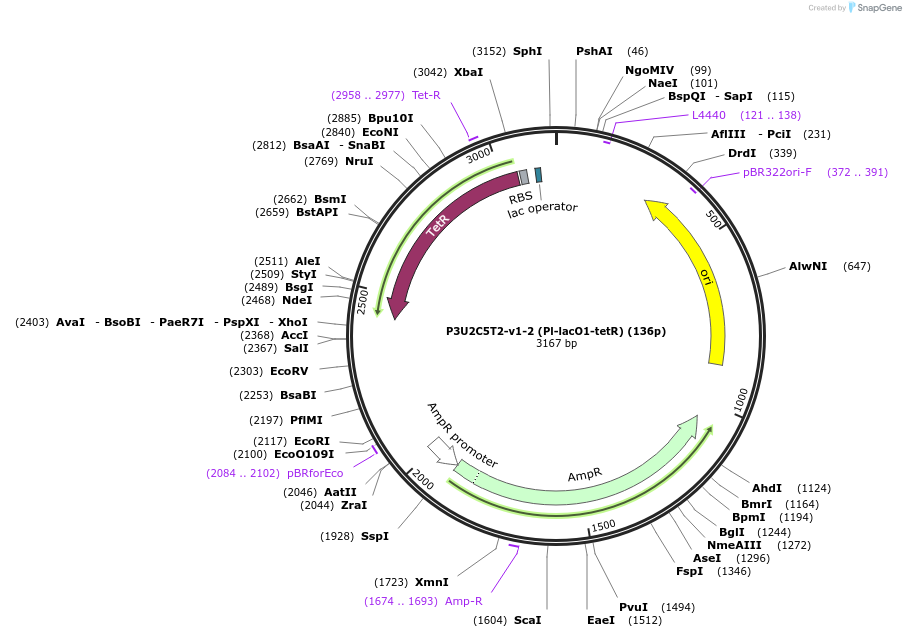
P3U2C5T2-v1-2 (Pl-lacO1-tetR) (136p)
Plasmid#70212Purpose1 Part of 4-part switchDepositorInserttetR
Available SinceMay 19, 2016AvailabilityAcademic Institutions and Nonprofits only