169,450 results
-
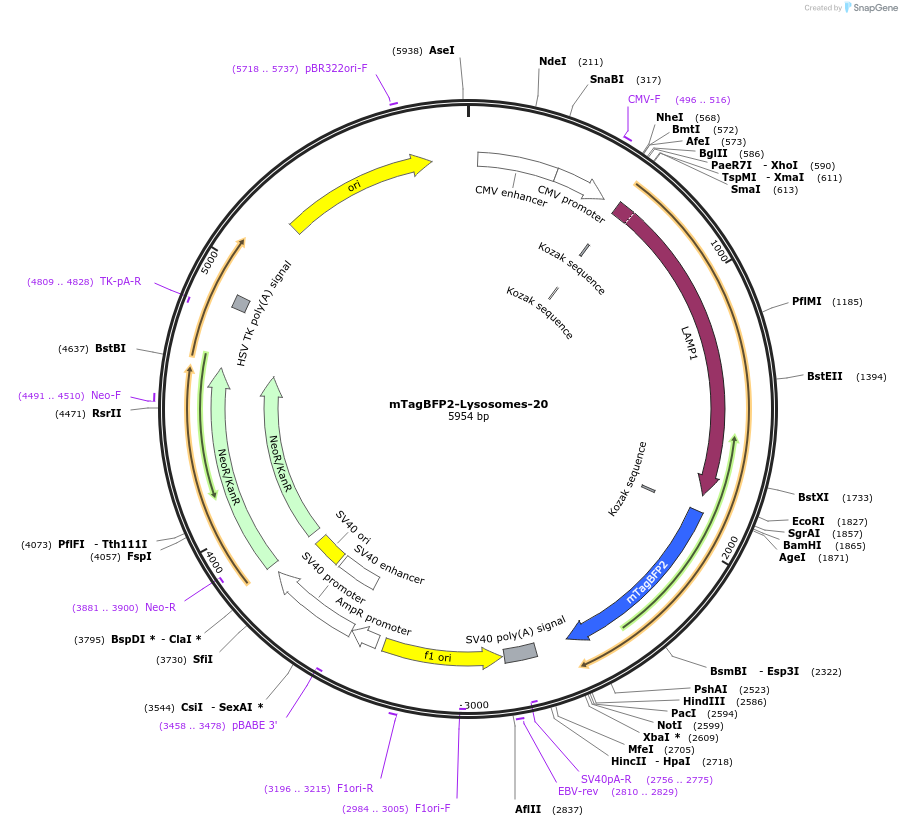
Plasmid#55308PurposeLocalization: Lysosome Membrane, Excitation: 399, Emission: 456DepositorInsertLAMP1
TagsmTagBFP2ExpressionMammalianPromoterCMVAvailable SinceOct. 10, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
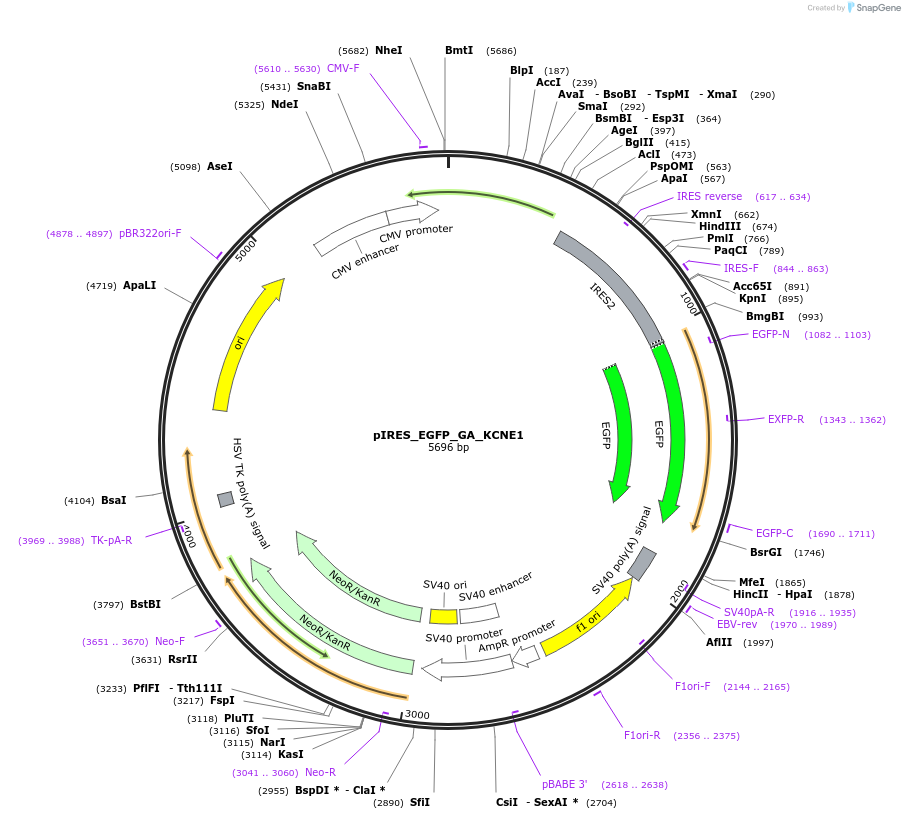
pIRES_EGFP_GA_KCNE1
Plasmid#173160PurposeFor mammalian expression of KCNE1 and creating variantsDepositorAvailable SinceSept. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
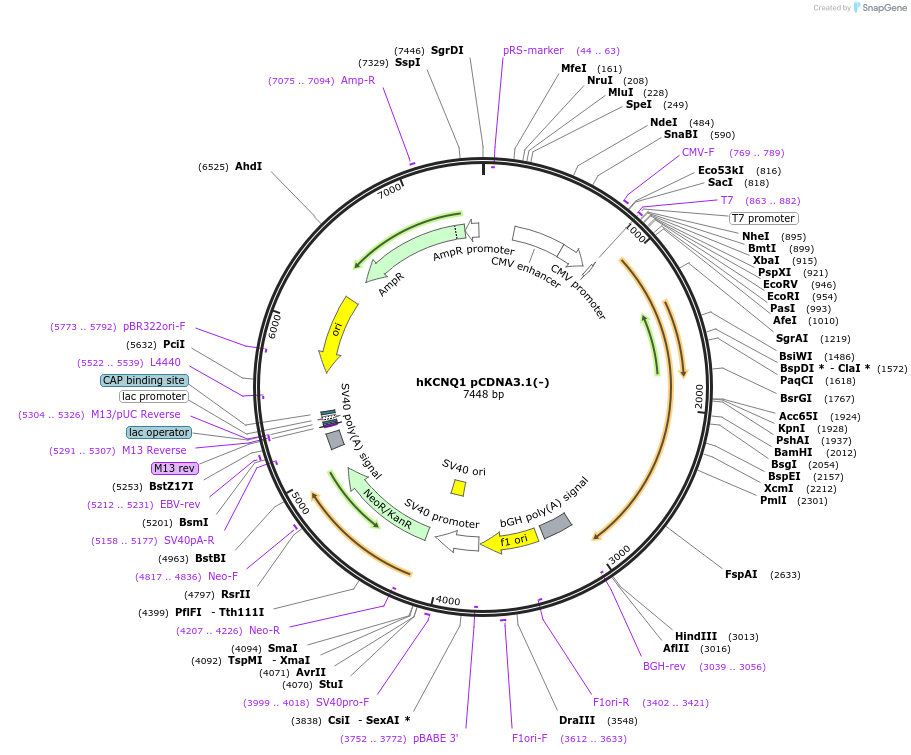
hKCNQ1 pCDNA3.1(-)
Plasmid#219953PurposeExpresses human KCNQ1 in mammalian cellsDepositorInsertKCNQ1 (KCNQ1 Human)
ExpressionMammalianMutationcontains silent sequence variationsPromoterCMVAvailable SinceAug. 13, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
pENN.AAV.CamKII.HI.GFP-Cre.WPRE.SV40 (AAV9)
Viral Prep#105551-AAV9PurposeReady-to-use AAV9 particles produced from pENN.AAV.CamKII.HI.GFP-Cre.WPRE.SV40 (#105551). In addition to the viral particles, you will also receive purified pENN.AAV.CamKII.HI.GFP-Cre.WPRE.SV40 plasmid DNA. Expression of GFP-Cre from CamKII promoter. These AAV preparations are suitable purity for injection into animals.DepositorPromoterCaMKIITagsGFPAvailable SinceAug. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
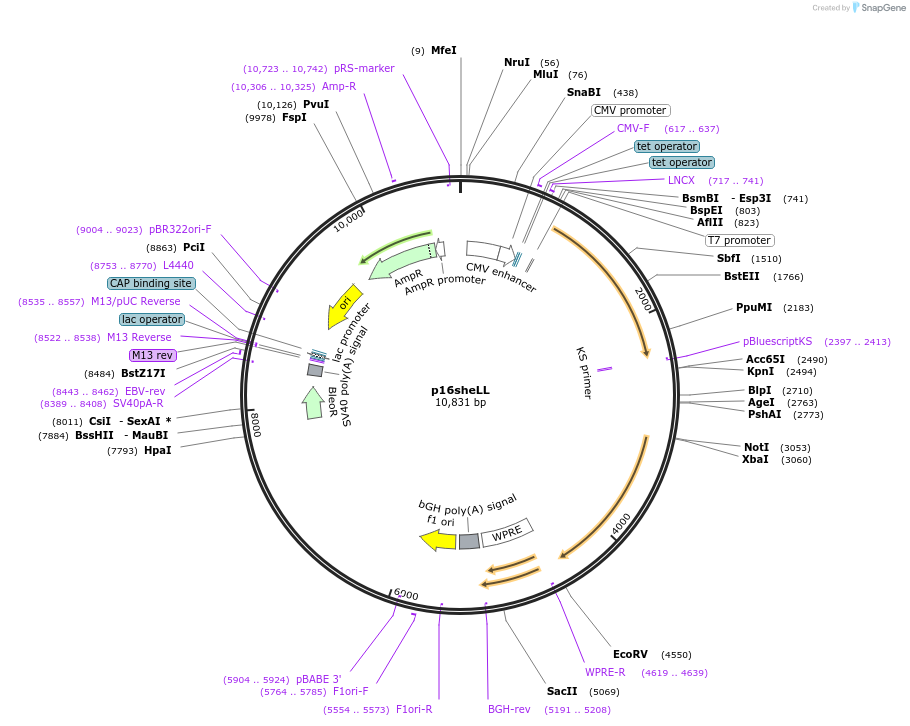
p16sheLL
Plasmid#37320DepositorInsertHPV16L1+L2
ExpressionMammalianPromoterCMVAvailable SinceJuly 8, 2013AvailabilityAcademic Institutions and Nonprofits only -
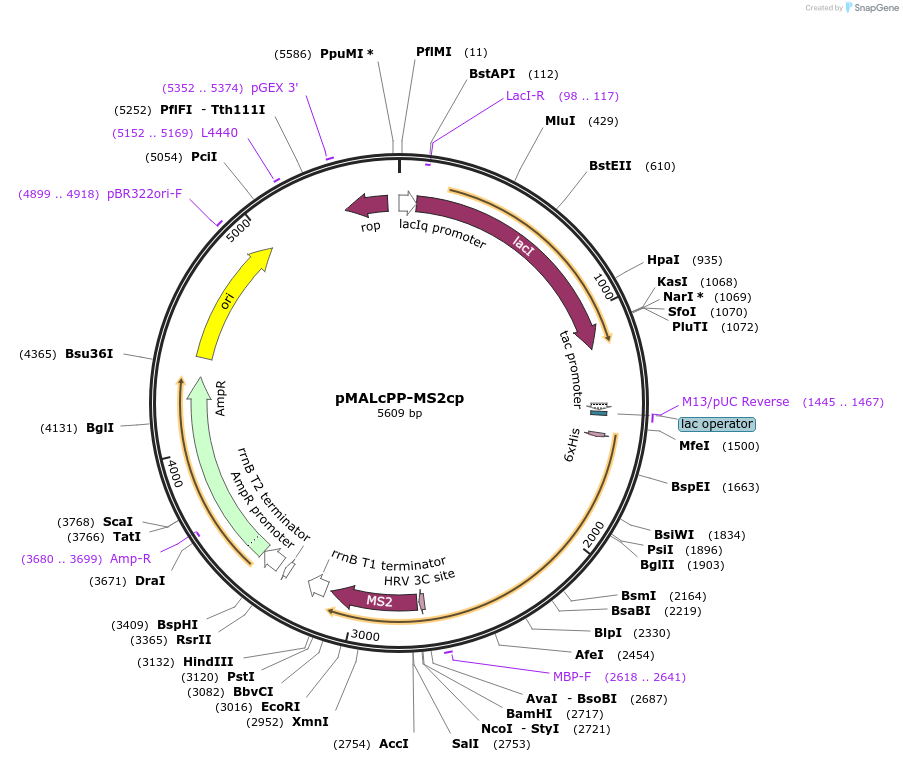
pMALcPP-MS2cp
Plasmid#203351PurposeExpresses His/MBP-tagged MS2-CP V75E/A81G variant in bacteriaDepositorInsertMS2 coat protein
TagsHx6-MBPExpressionBacterialMutationV75E and A81G, to prevent capsid assemblyAvailable SinceJuly 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
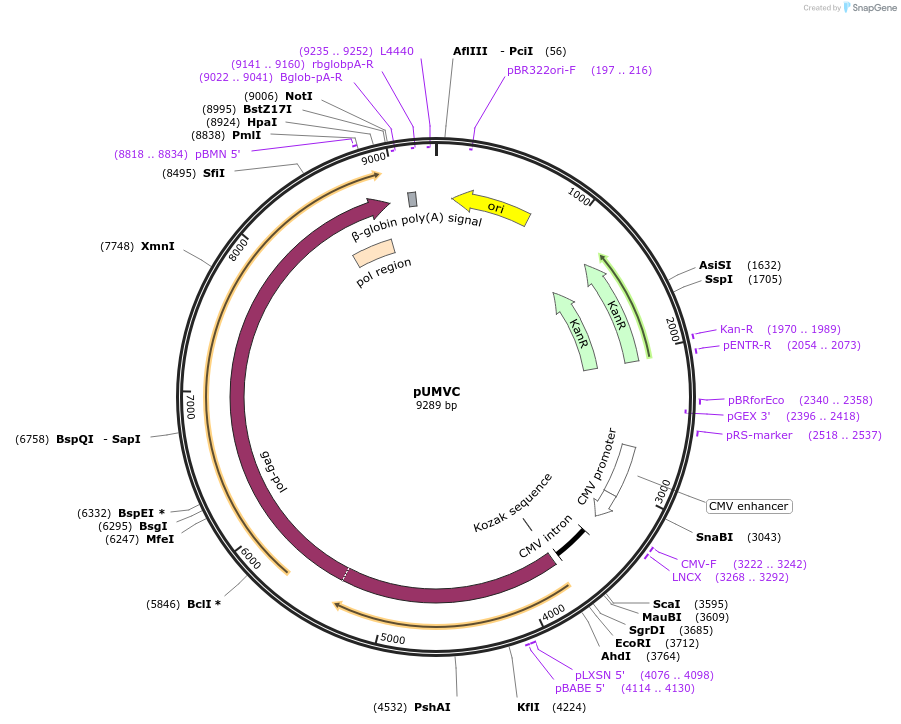
pUMVC
Plasmid#8449PurposePackaging plasmid for producing MuLV retroviral particles. Use in conjunction with an envelope plasmid such as pCMV-VSV-G.DepositorHas ServiceCloning Grade DNATypeEmpty backboneUseRetroviralExpressionMammalianAvailable SinceOct. 4, 2005AvailabilityAcademic Institutions and Nonprofits only -
pAAV.Syn.NES-jRCaMP1b.WPRE.SV40 (AAV9)
Viral Prep#100851-AAV9PurposeReady-to-use AAV9 particles produced from pAAV.Syn.NES-jRCaMP1b.WPRE.SV40 (#100851). In addition to the viral particles, you will also receive purified pAAV.Syn.NES-jRCaMP1b.WPRE.SV40 plasmid DNA. Syn-driven jRCaMP1b calcium sensor. These AAV preparations are suitable purity for injection into animals.DepositorPromoterSynAvailable SinceApril 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
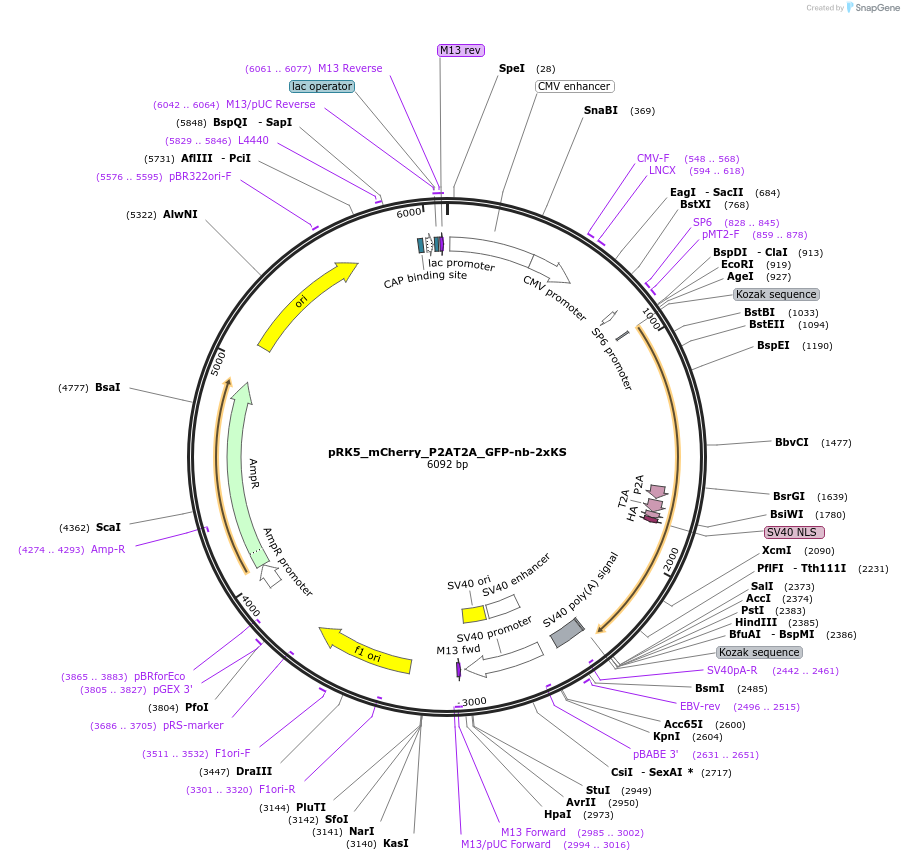
pRK5_mCherry_P2AT2A_GFP-nb-2xKS
Plasmid#238236PurposeFor overexpression of mCherry_P2AT2A_GFP-nb-2xKSDepositorInsertmCherry_P2AT2A_GFP-nb-2xKS
ExpressionMammalianAvailable SinceJune 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
pAAV-Syn-ChrimsonR-tdT (AAV1)
Viral Prep#59171-AAV1PurposeReady-to-use AAV1 particles produced from pAAV-Syn-ChrimsonR-tdT (#59171). In addition to the viral particles, you will also receive purified pAAV-Syn-ChrimsonR-tdT plasmid DNA. Syn-driven ChrimsonR-tdTomato expression for optogenetic neural activation. These AAV preparations are suitable purity for injection into animals.DepositorPromoterSynTagstdTomatoAvailable SinceFeb. 7, 2018AvailabilityAcademic Institutions and Nonprofits only -
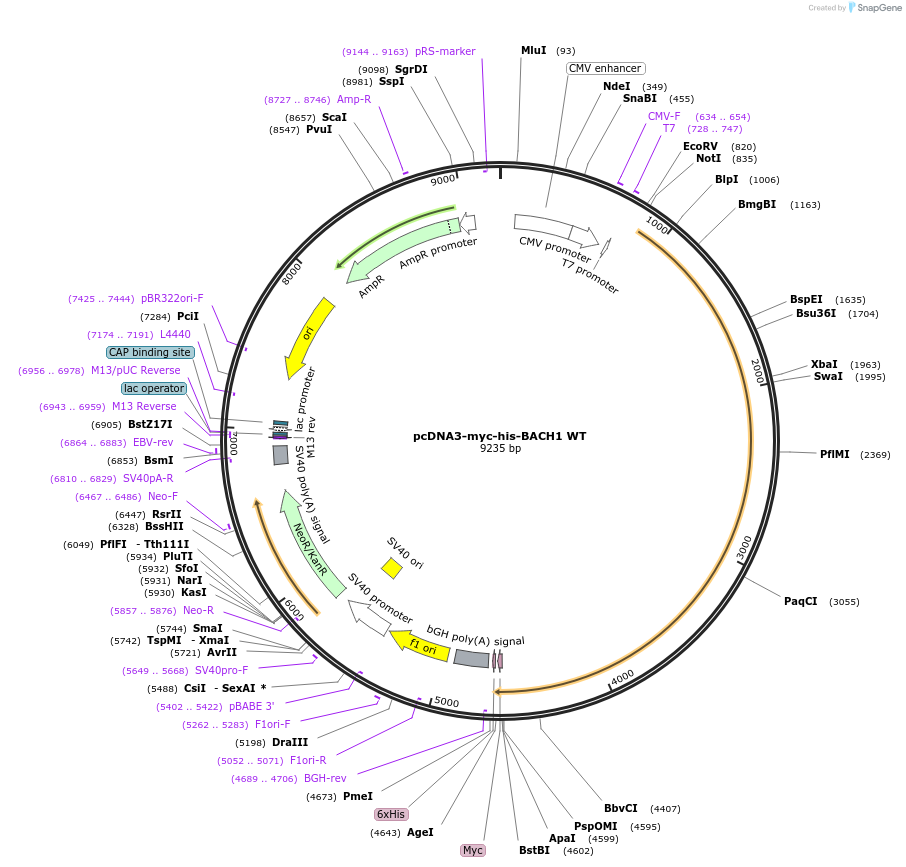
pcDNA3-myc-his-BACH1 WT
Plasmid#17642DepositorAvailable SinceApril 4, 2008AvailabilityAcademic Institutions and Nonprofits only -
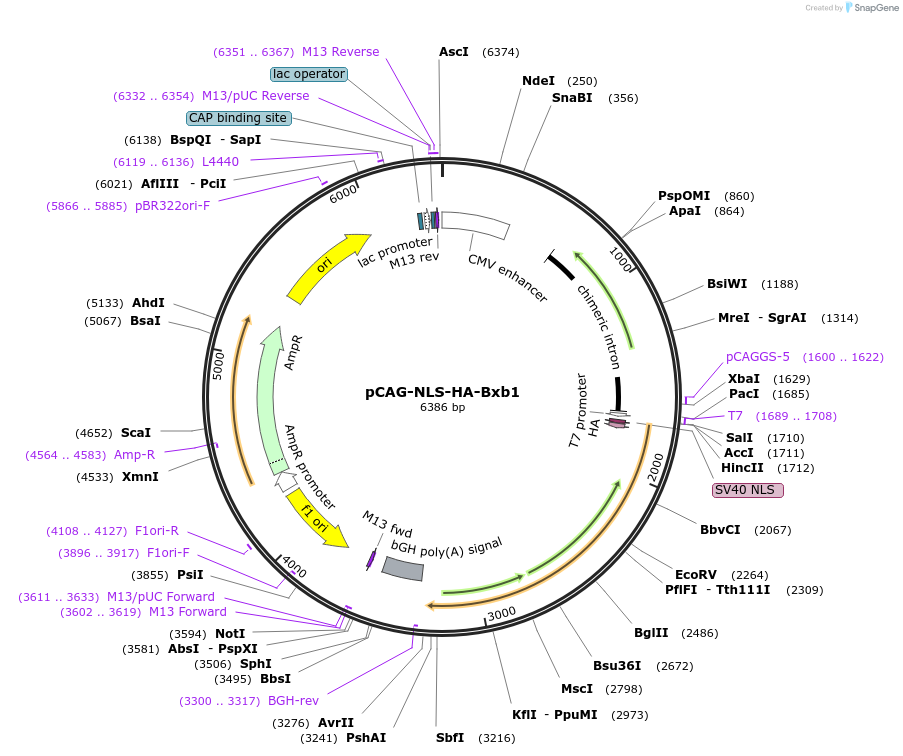
pCAG-NLS-HA-Bxb1
Plasmid#51271PurposeComponent of the Bxb1-rcCre binary site-specific recombinase system, expression of the site-specific recombinase Bxb1 (mouse codon-optimized)DepositorInsertNLS-HA-Bxb1
ExpressionMammalianPromoterCAGAvailable SinceApril 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
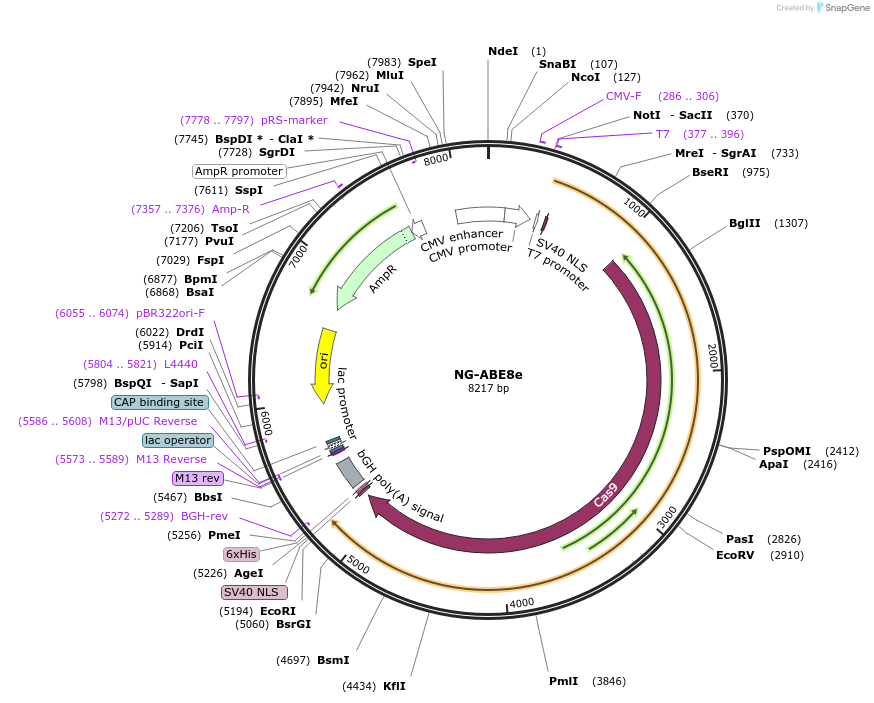
NG-ABE8e
Plasmid#138491PurposeA-to-G base editorDepositorInsertecTadA(8e)-nSpCas9 (NG)
ExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
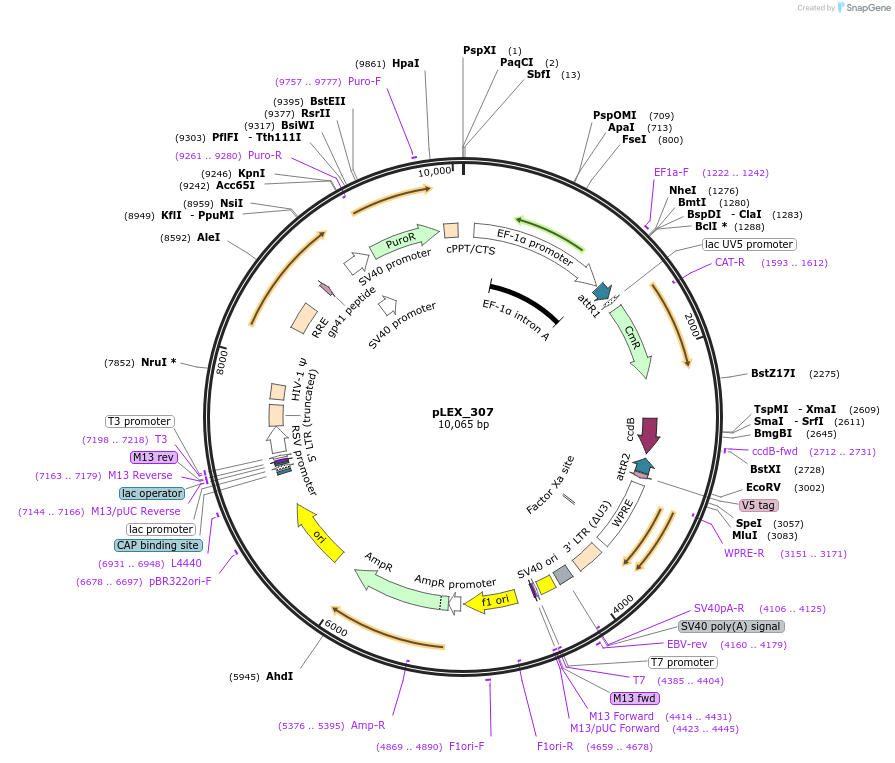
pLEX_307
Plasmid#41392PurposeConstitutive lentiviral expression, SV40-puro; EF1a-gateway-V5 tagDepositorHas ServiceCloning Grade DNATypeEmpty backboneUseLentiviral; Gateway destination vectorTagsV5ExpressionMammalianPromoterEF1aAvailable SinceNov. 30, 2012AvailabilityAcademic Institutions and Nonprofits only -
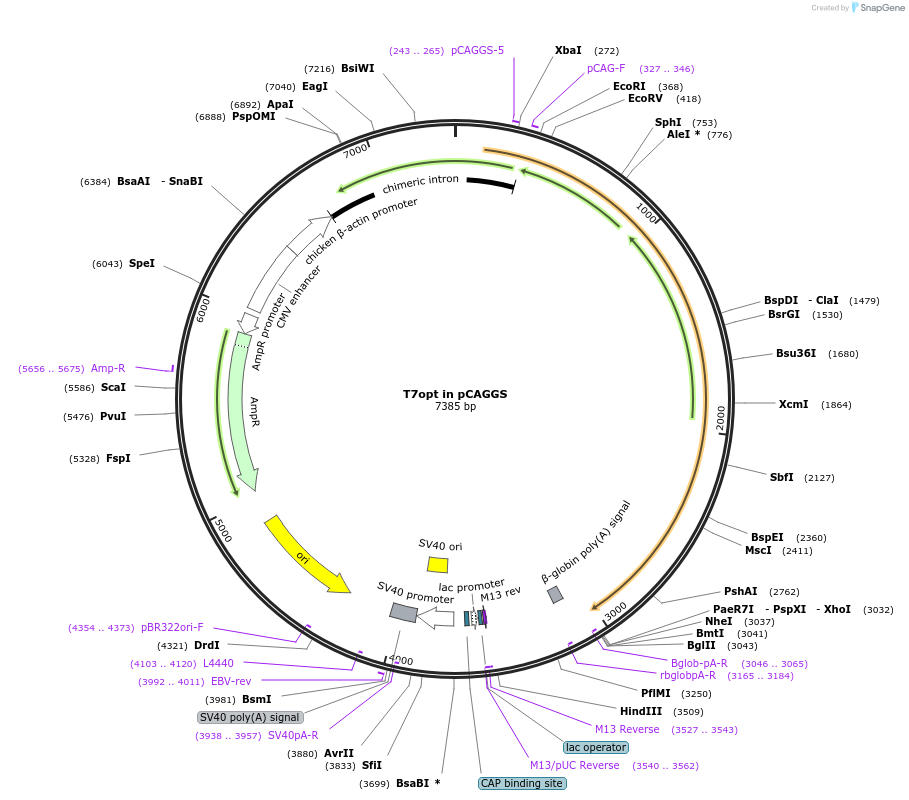
T7opt in pCAGGS
Plasmid#65974PurposeHuman codon-optimized T7 polymeraseDepositorInsertT7-opt
ExpressionMammalianAvailable SinceJuly 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
AiP13781 - pAAV-AiE0452h-minBG-iCre(R297T)-BGHpA (Alias: CN3781) (AAV PHP.eB)
Viral Prep#199777-PHPeBPurposeReady-to-use AAV PHP.eB particles produced from AiP13781 - pAAV-AiE0452h-minBG-iCre(R297T)-BGHpA (Alias: CN3781) (#199777). In addition to the viral particles, you will also receive purified AiP13781 - pAAV-AiE0452h-minBG-iCre(R297T)-BGHpA (Alias: CN3781) plasmid DNA. Cre recombinase expression in striatal indirect pathway medium spiny neurons (D2-MSNs). These AAV were produced with the PHPeB serotype, which permits efficient transduction of the central nervous system. These AAV preparations are suitable purity for injection into animals.DepositorAvailable SinceJuly 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
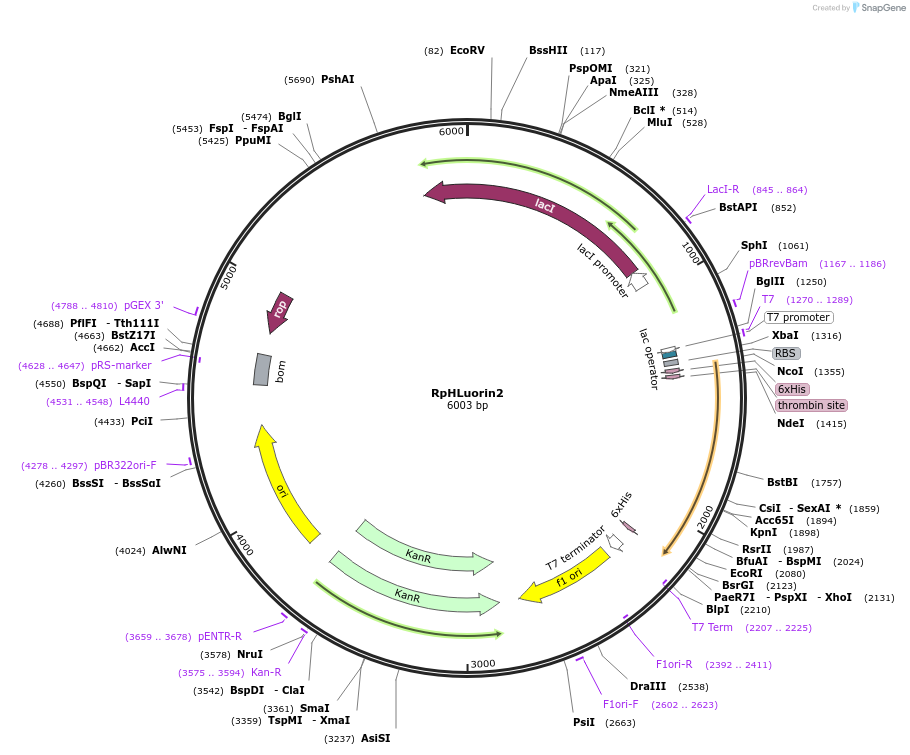
RpHLuorin2
Plasmid#180227Purposefor bacterial expression of recombinant ratiometric pHluorin2DepositorInsertRatiometric pHLuorin2
TagsHis-tag followed by thrombin cleavage siteExpressionBacterialPromoterT7Available SinceMarch 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
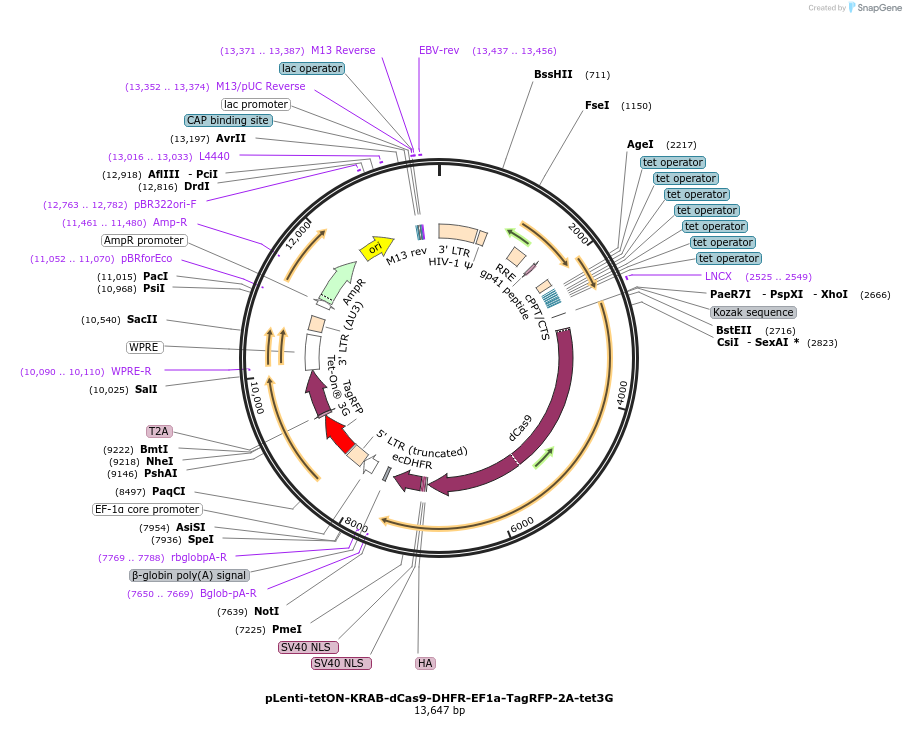
pLenti-tetON-KRAB-dCas9-DHFR-EF1a-TagRFP-2A-tet3G
Plasmid#167935PurposeInducible CRISPRi expressionDepositorInsertKRAB-dCas9-DHFR
UseLentiviralPromotertetONAvailable SinceJune 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
AAV-CamKIIa-jGCaMP8f-WPRE (AAV5)
Viral Prep#176750-AAV5PurposeReady-to-use AAV5 particles produced from AAV-CamKIIa-jGCaMP8f-WPRE (#176750). In addition to the viral particles, you will also receive purified AAV-CamKIIa-jGCaMP8f-WPRE plasmid DNA. CamKIIa-driven expression of calcium sensor GCaMP8f. These AAV preparations are suitable purity for injection into animals.DepositorPromoterCamKIIalphaAvailable SinceMarch 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
pENN.AAV.hSyn.HI.eGFP-Cre.WPRE.SV40 (AAV9)
Viral Prep#105540-AAV9PurposeReady-to-use AAV9 particles produced from pENN.AAV.hSyn.HI.eGFP-Cre.WPRE.SV40 (#105540). In addition to the viral particles, you will also receive purified pENN.AAV.hSyn.HI.eGFP-Cre.WPRE.SV40 plasmid DNA. Synapsin-driven EGFP-Cre expression. These AAV preparations are suitable purity for injection into animals.DepositorPromoterSynTagsEGFPAvailable SinceMarch 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
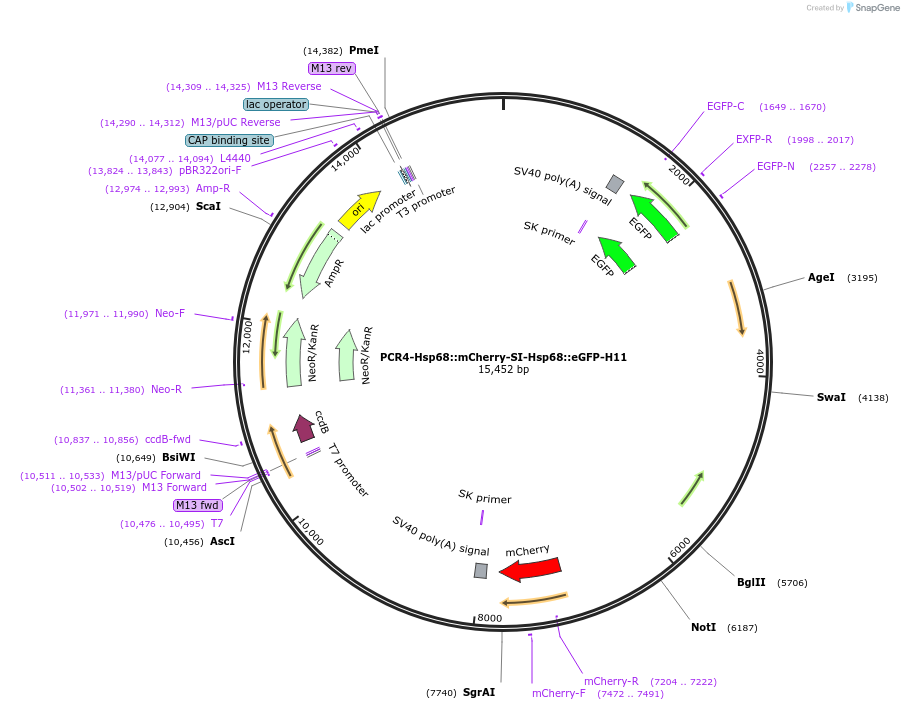
PCR4-Hsp68::mCherry-SI-Hsp68::eGFP-H11
Plasmid#211942Purposedual-enSERT-2.2 vector for site-specific integration of.a two-color enhancer-reporter construct into the H11 locus (separated by a synthetic insulator)DepositorTypeEmpty backboneUseCRISPR and Mouse TargetingExpressionMammalianPromoterHSP68Available SinceMarch 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
pENN.AAV.CamKII.GCaMP6f.WPRE.SV40 (AAV5)
Viral Prep#100834-AAV5PurposeReady-to-use AAV5 particles produced from pENN.AAV.CamKII.GCaMP6f.WPRE.SV40 (#100834). In addition to the viral particles, you will also receive purified pENN.AAV.CamKII.GCaMP6f.WPRE.SV40 plasmid DNA. CamKII-driven GCaMP6f calcium sensor. These AAV preparations are suitable purity for injection into animals.DepositorPromoterCaMKIIAvailable SinceApril 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
pENN.AAV.CamKII 0.4.Cre.SV40 (AAV1)
Viral Prep#105558-AAV1PurposeReady-to-use AAV1 particles produced from pENN.AAV.CamKII 0.4.Cre.SV40 (#105558). In addition to the viral particles, you will also receive purified pENN.AAV.CamKII 0.4.Cre.SV40 plasmid DNA. CamKII-driven Cre expression. These AAV preparations are suitable purity for injection into animals.DepositorPromoterCaMKIIAvailable SinceMay 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
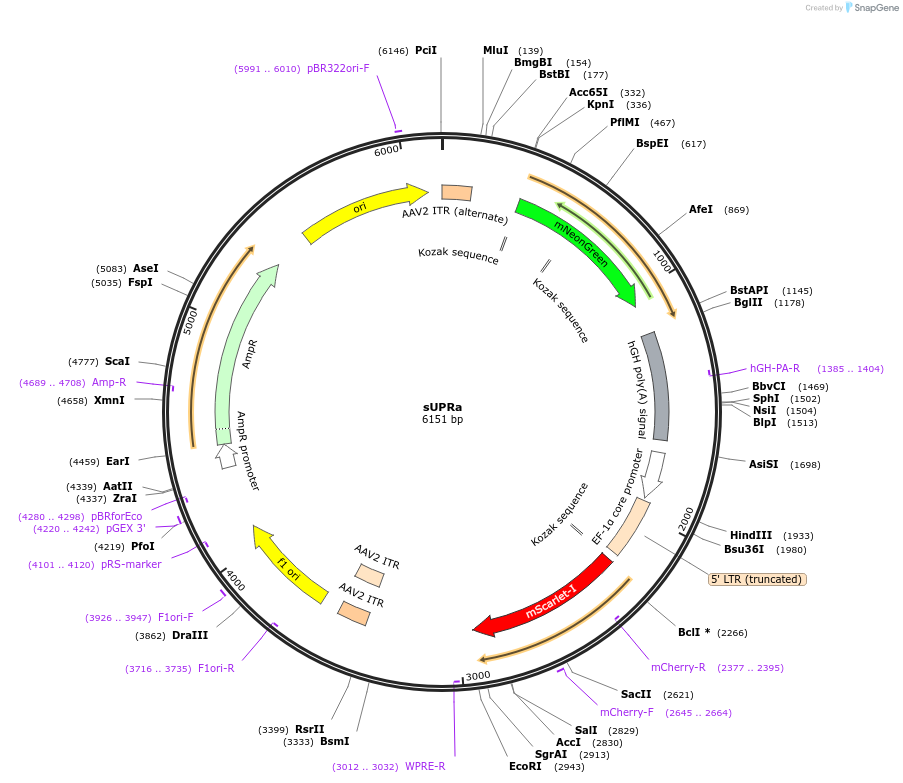
sUPRa
Plasmid#223242PurposesUPRa is a dual promoter and two-color construct, which is designed to report the global unfolded protein response (UPR) and affords unbiased cell detection irrespective of UPR levelsDepositorInsertsUPRa
UseAAVPromoterCustomAvailable SinceOct. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
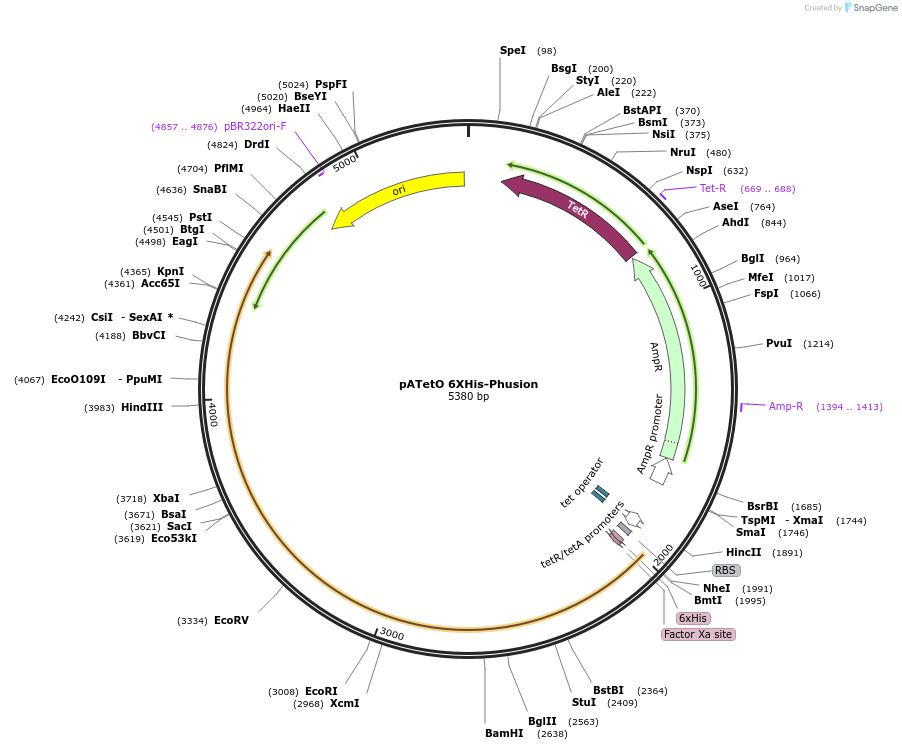
pATetO 6XHis-Phusion
Plasmid#206210PurposeInducible expression of DNA polymeraseDepositorInsertPhusion DNA polymerase
ExpressionBacterialAvailable SinceDec. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
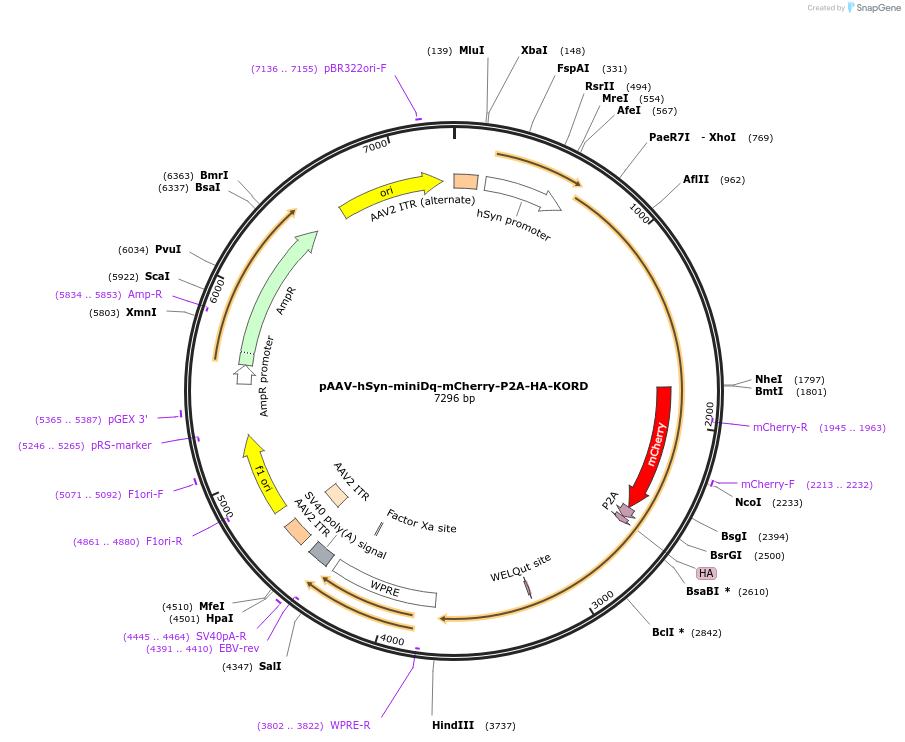
pAAV-hSyn-miniDq-mCherry-P2A-HA-KORD
Plasmid#204359PurposeAAV vector for coexpression of miniDq and KORD under the control of human synapsin promoterDepositorInsertminiDq-mCherry-P2A-HA-KORD
UseAAVTagsHA (for KORD) and mCherry (for miniDq)MutationFor miniDq, the third intracellular loop (ICL3) o…PromoterhSynAvailable SinceOct. 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
pAAV-nEF-Con/Fon DREADD Gi-mCherry (AAV8)
Viral Prep#177672-AAV8PurposeReady-to-use AAV8 particles produced from pAAV-nEF-Con/Fon DREADD Gi-mCherry (#177672). In addition to the viral particles, you will also receive purified pAAV-nEF-Con/Fon DREADD Gi-mCherry plasmid DNA. nEF-driven, Cre- and Flp-dependent hM4D(Gi) receptor with an mCherry reporter for CNO-induced neuronal inhibition. These AAV preparations are suitable purity for injection into animals.DepositorPromoternEFTagsmCherryAvailable SinceFeb. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
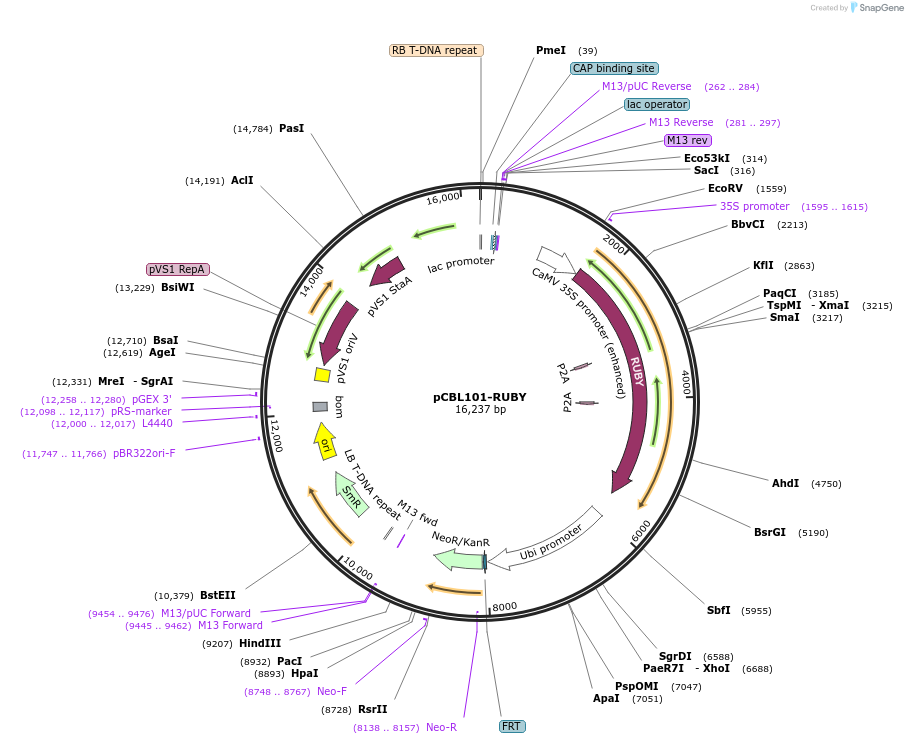
pCBL101-RUBY
Plasmid#199723PurposeExpresses betalain biosynthesis genes under CaMV 35S promoterDepositorInsert35S-RUBY
ExpressionPlantAvailable SinceApril 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
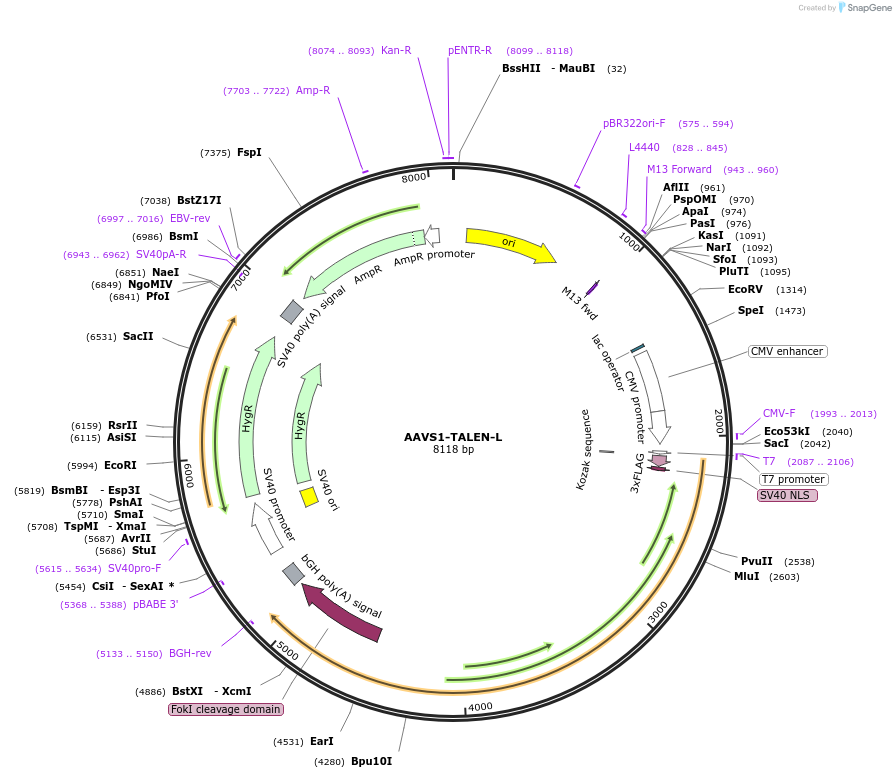
AAVS1-TALEN-L
Plasmid#59025PurposeTALEN designed to target the human AAVS1/PPP1R12C locusDepositorInsertAAVS1-TALEN-L
UseAAV and TALENTags3xFlag and NLSExpressionMammalianPromoterCMVAvailable SinceAug. 12, 2014AvailabilityAcademic Institutions and Nonprofits only -
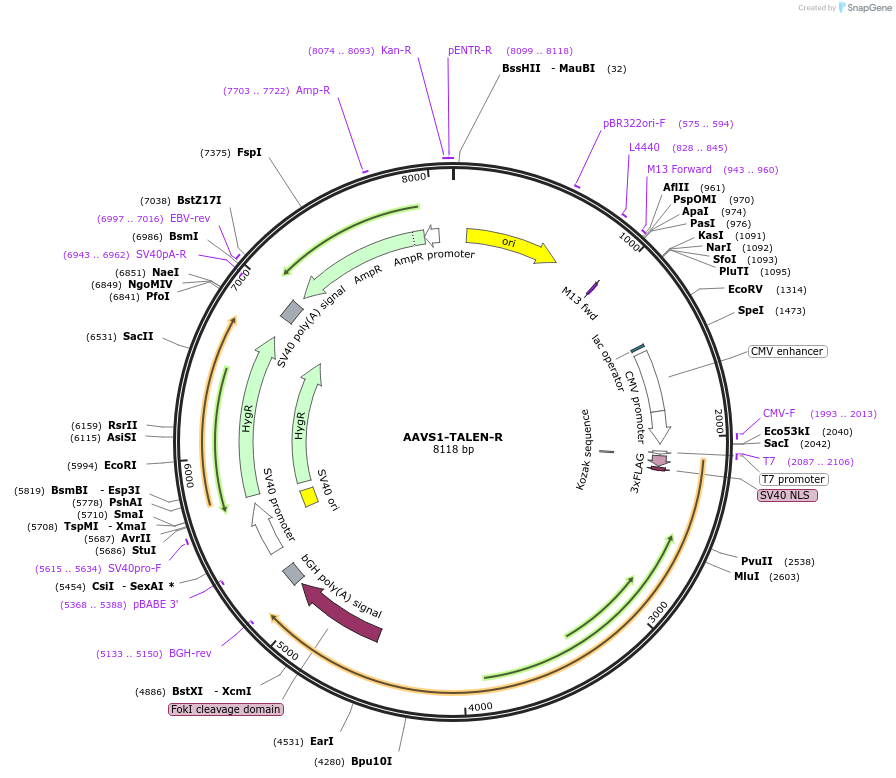
AAVS1-TALEN-R
Plasmid#59026PurposeTALEN designed to target the human AAVS1/PPP1R12C locusDepositorInsertAAVS1-TALEN-R
UseAAV and TALENTags3xFlag and NLSExpressionMammalianPromoterCMVAvailable SinceAug. 12, 2014AvailabilityAcademic Institutions and Nonprofits only -
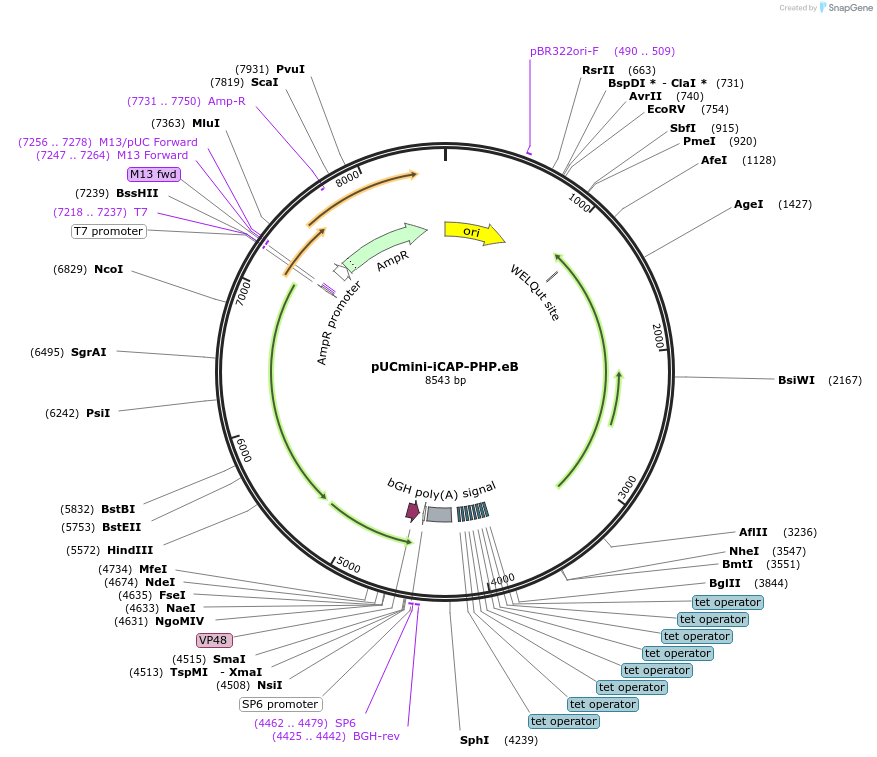
pUCmini-iCAP-PHP.eB
Plasmid#103005Purposenon-standard AAV2 rep-AAV-PHP.eB cap plasmid with AAV cap expression controlled by a tTA-TRE amplification systemDepositorInsertSynthetic construct isolate AAV-PHP.eB VP1 gene
UseAAVExpressionMammalianPromoterp41Available SinceMarch 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
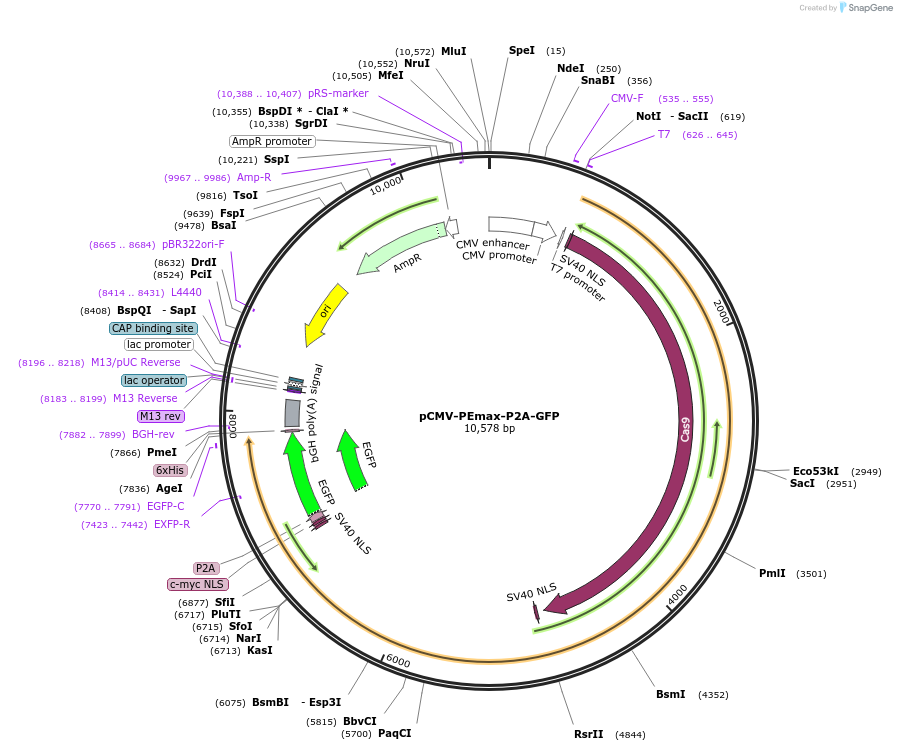
pCMV-PEmax-P2A-GFP
Plasmid#180020PurposeMammalian expression of SpCas9 PEmax prime editor with P2A-EGFP markerDepositorInsertPEmax-P2A-EGFP
TagsP2A-EGFP, SV40 bpNLS, and c-Myc NLSExpressionMammalianMutationDetailed in manuscriptPromoterCMVAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
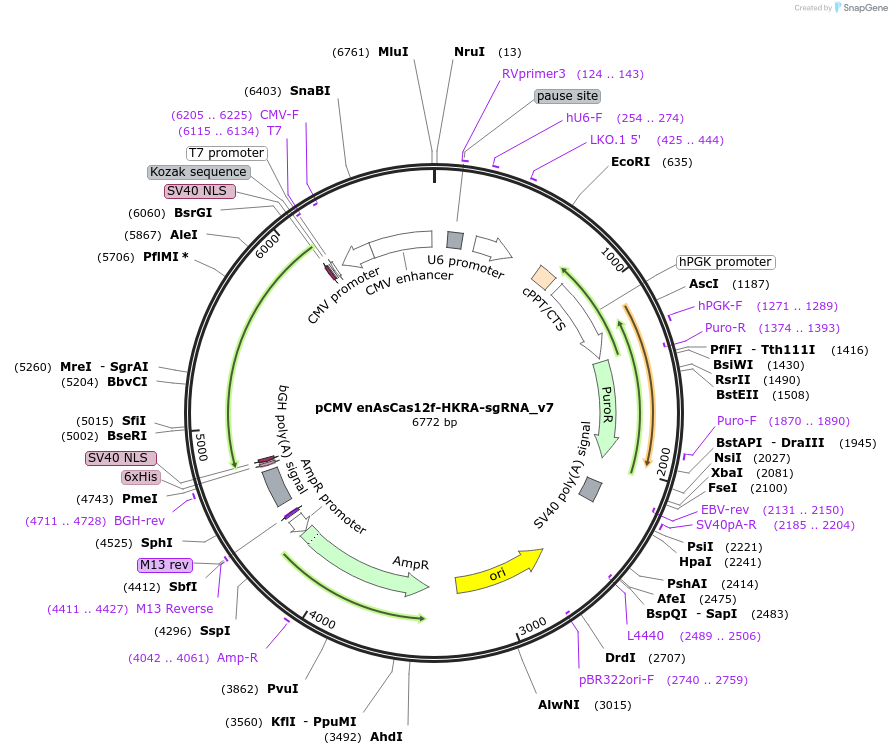
pCMV enAsCas12f-HKRA-sgRNA_v7
Plasmid#208357PurposeExpression of enAsCas12f-HKRA and sgRNA_v7 in mammalian cellsDepositorInsertenAsCas12f-HKRA
ExpressionMammalianMutationI123H/D195K/D208R/V232APromoterCMVAvailable SinceOct. 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
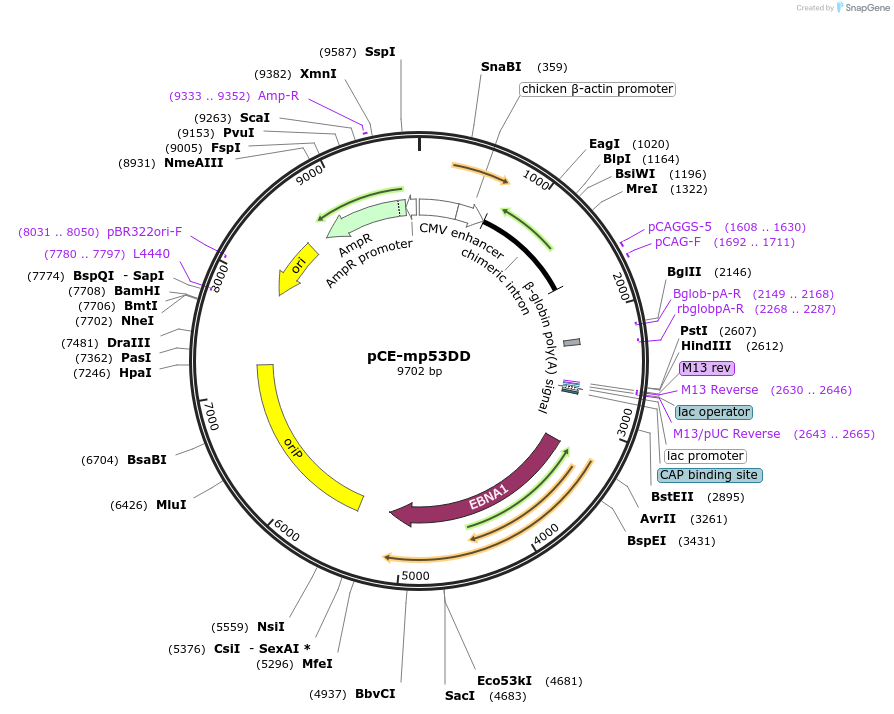
pCE-mp53DD
Plasmid#41856PurposeNon-integrating (episomal) expression of mouse p53DD - p53 carboxy-terminal dominant-negative fragmentDepositorAvailable SinceJuly 18, 2013AvailabilityAcademic Institutions and Nonprofits only -
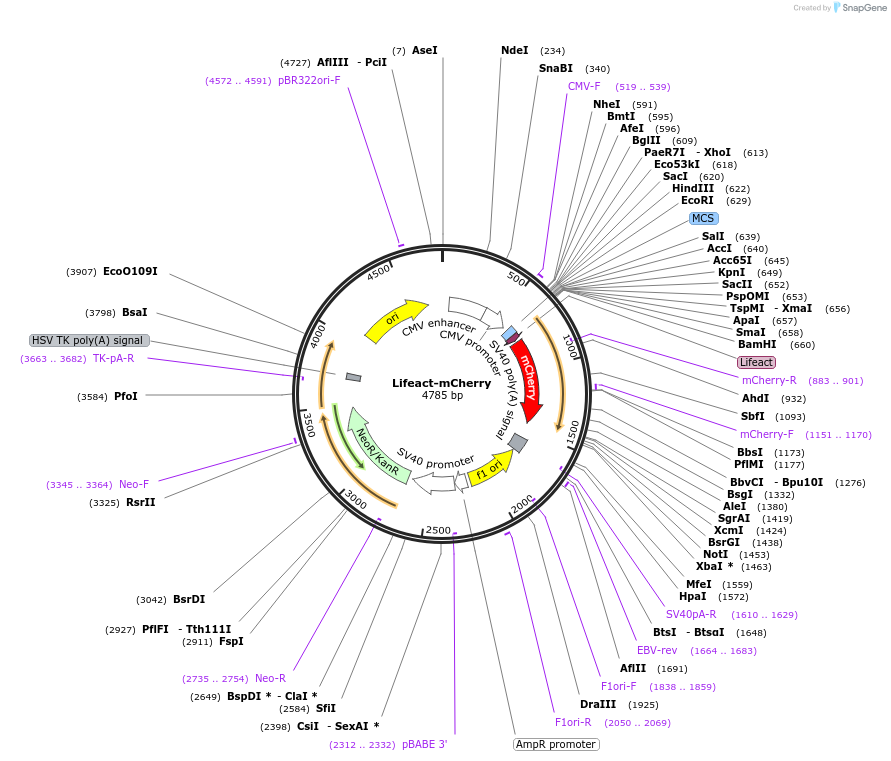
Lifeact-mCherry
Plasmid#193300Purposemammalian transient expression - expression of Lifeact-mCherryDepositorInsertLifeact
TagsmCherryExpressionMammalianAvailable SinceDec. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAAV.Syn.NES-jRGECO1a.WPRE.SV40 (AAV1)
Viral Prep#100854-AAV1PurposeReady-to-use AAV1 particles produced from pAAV.Syn.NES-jRGECO1a.WPRE.SV40 (#100854). In addition to the viral particles, you will also receive purified pAAV.Syn.NES-jRGECO1a.WPRE.SV40 plasmid DNA. Synapsin-driven GECO1a calcium sensor. These AAV preparations are suitable purity for injection into animals.DepositorPromoterSynAvailable SinceApril 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
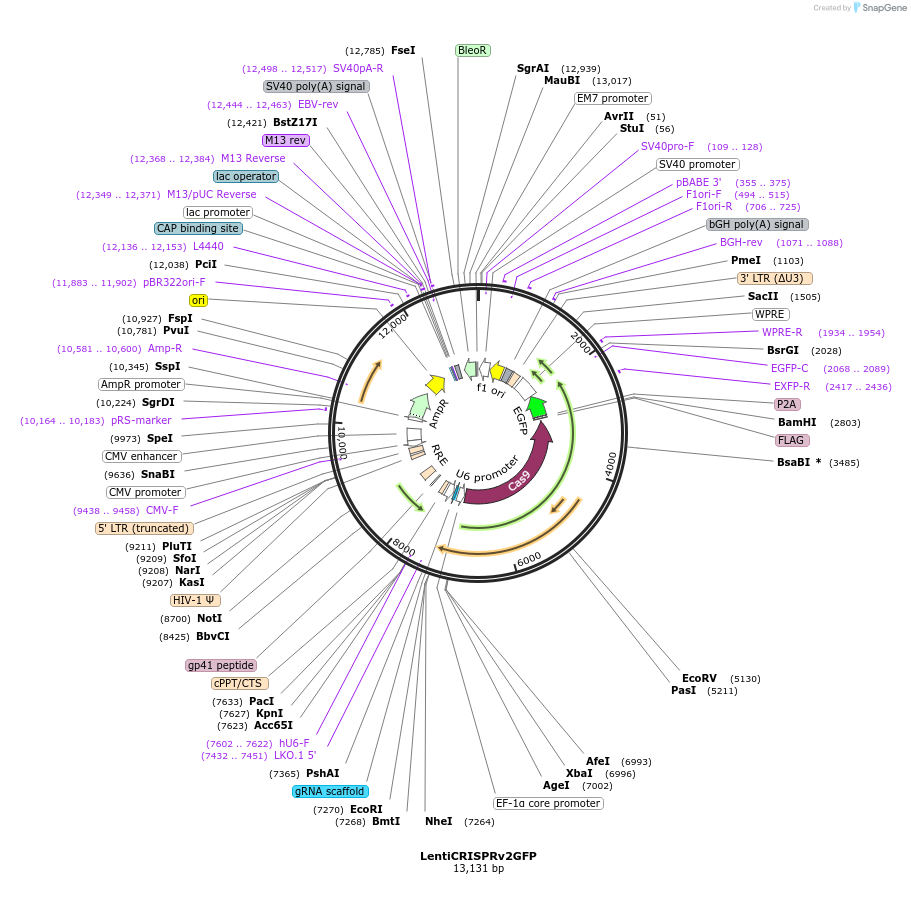
LentiCRISPRv2GFP
Plasmid#82416Purpose3rd generation lentiviral vector expressing GFP alongside Cas9 and an sgRNA cloning siteDepositorInsertGFP
UseCRISPR and LentiviralPromoterEFS (P2A)Available SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
AAV-CamKIIa-jGCaMP8m-WPRE (AAV1)
Viral Prep#176751-AAV1PurposeReady-to-use AAV1 particles produced from AAV-CamKIIa-jGCaMP8m-WPRE (#176751). In addition to the viral particles, you will also receive purified AAV-CamKIIa-jGCaMP8m-WPRE plasmid DNA. CamKIIa-driven expression of calcium sensor GCaMP8m. These AAV preparations are suitable purity for injection into animals.DepositorPromoterCamKIIalphaAvailable SinceFeb. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
hSyn-DIO-somBiPOLES-mCerulean (AAV9)
Viral Prep#154951-AAV9PurposeReady-to-use AAV9 particles produced from hSyn-DIO-somBiPOLES-mCerulean (#154951). In addition to the viral particles, you will also receive purified hSyn-DIO-somBiPOLES-mCerulean plasmid DNA. Synapsin-driven, Cre-dependent expression of soma-targeted BiPOLES for optogenetic inhibition (blue light) and activation (red light). These AAV preparations are suitable purity for injection into animals.DepositorPromoterSynTagsmCerulean (Cre-dependent)Available SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
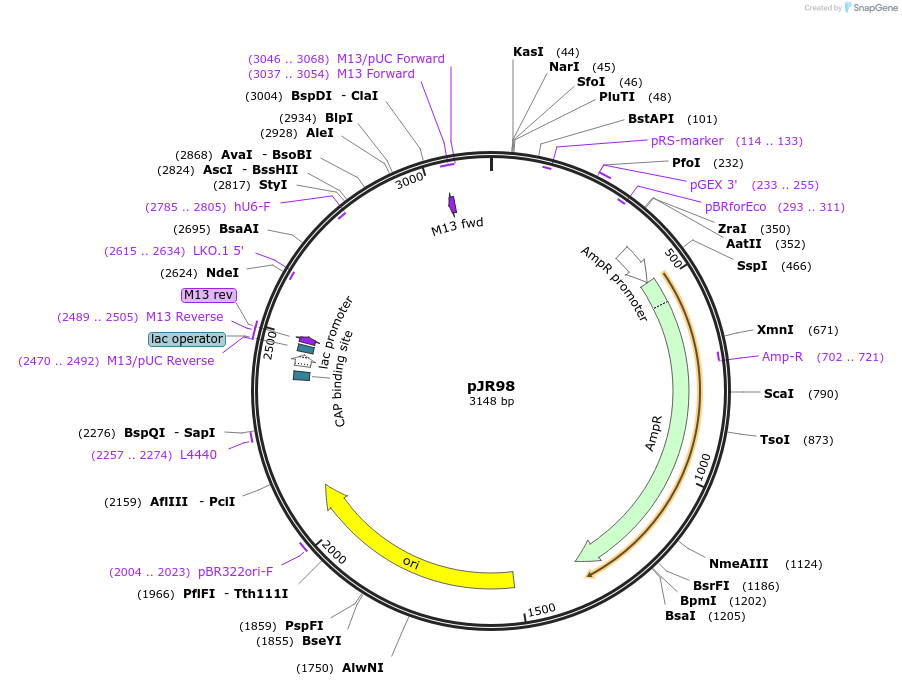
pJR98
Plasmid#187239PurposeCR3 constant region - hU6 sgRNA promoter flanked by BsmBI sitesDepositorInsertCR3 constant region - hU6 sgRNA promoter flanked by BsmBI sites
ExpressionBacterialAvailable SinceOct. 18, 2022AvailabilityAcademic Institutions and Nonprofits only