We narrowed to 19,819 results for: Cre
-
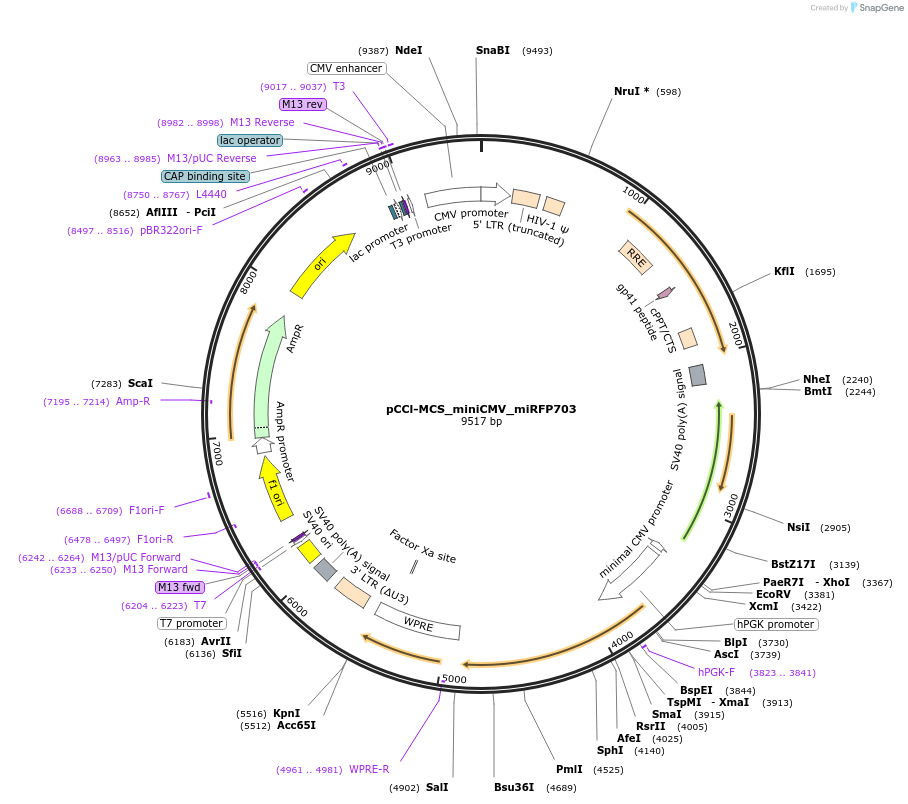
Plasmid#134985PurposeThe plasmid includes a miRFP703 gene driven by a minimal CMV promoter. Cis-regulatory elements can be cloned into the single EcoRV site. This plasmid can be used for generating a lentiviral vectorDepositorInsertmiRFP703
UseLentiviralPromoterminCMVAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
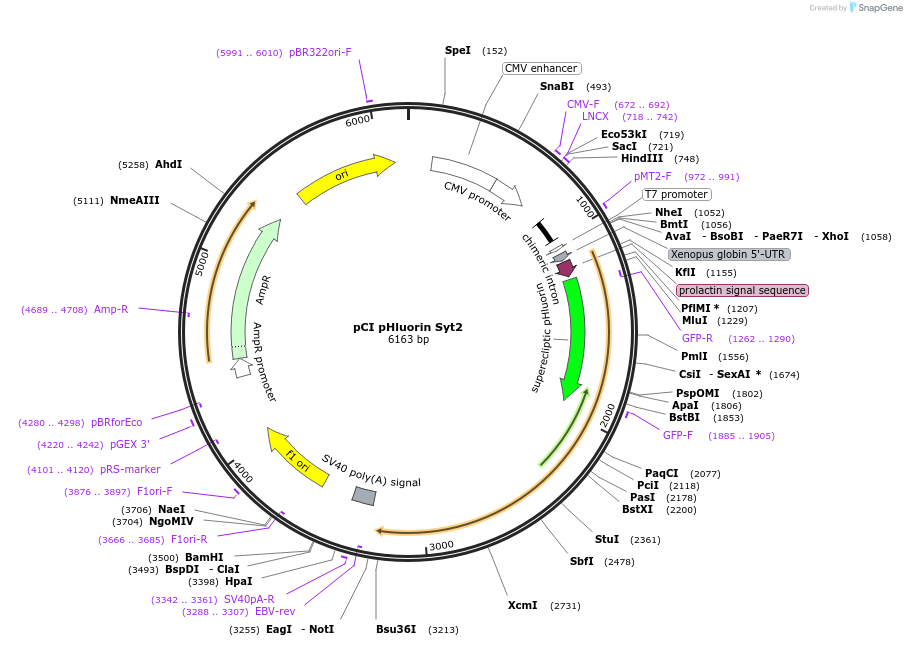
pCI pHluorin Syt2
Plasmid#184503PurposeMammalian expression of Synaptotagmin 2 with pH-sensitive fluorescent proteinDepositorAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
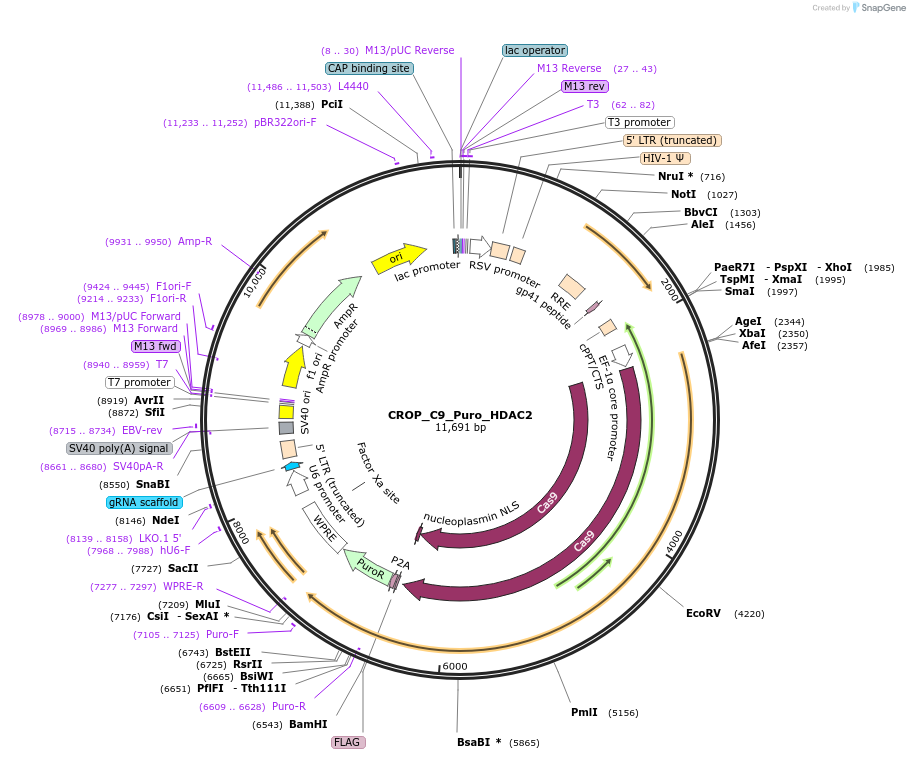
CROP_C9_Puro_HDAC2
Plasmid#183298PurposeAll-in-One CRISPRko system with a guide RNA that targets HDAC2 geneDepositorInsertHDAC2
UseLentiviralAvailable SinceMay 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
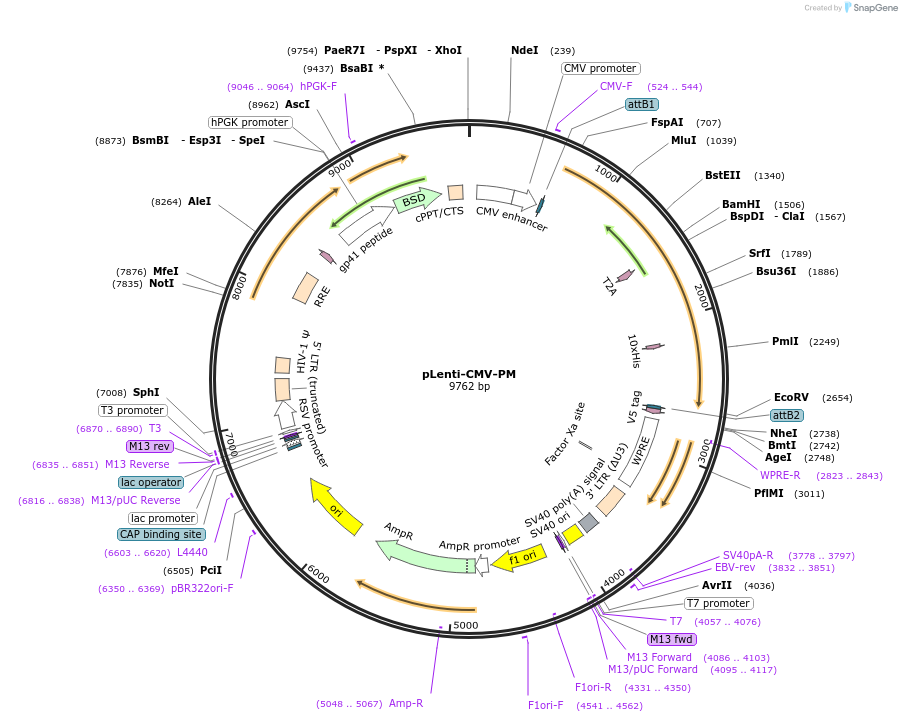
pLenti-CMV-PM
Plasmid#197386PurposeTo constitutively express Pdx1-Mafa and in cells (Blasticidin resistant).DepositorInsertPDX1-T2A-MAFA
UseLentiviralExpressionMammalianPromoterCMVAvailable SinceFeb. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
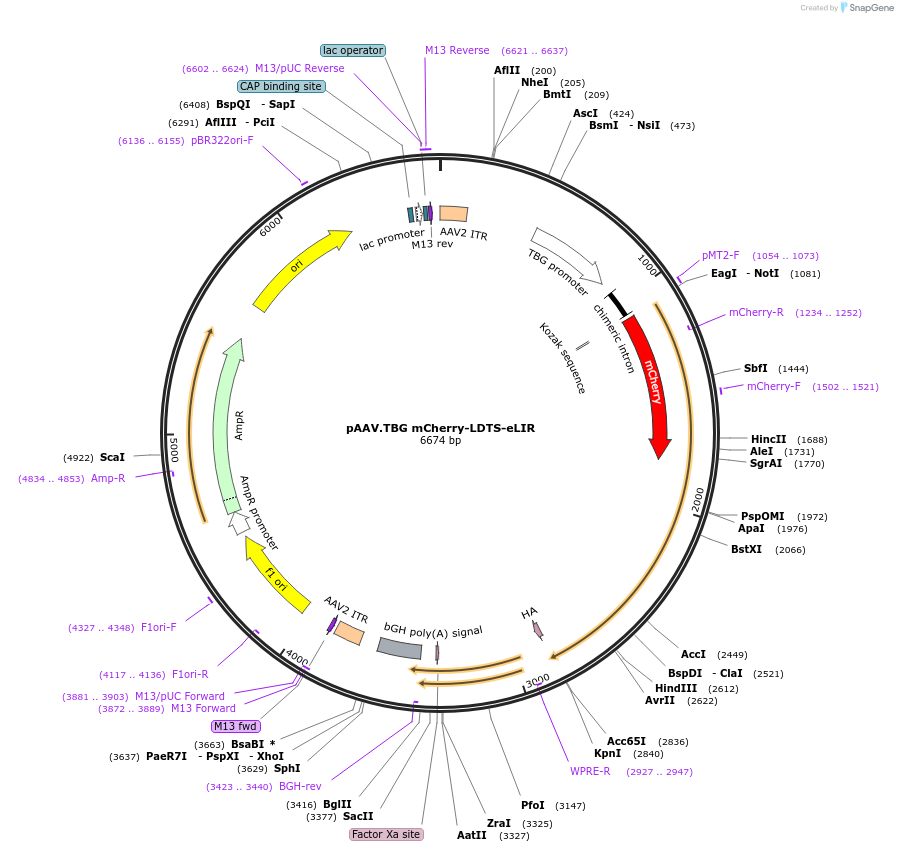
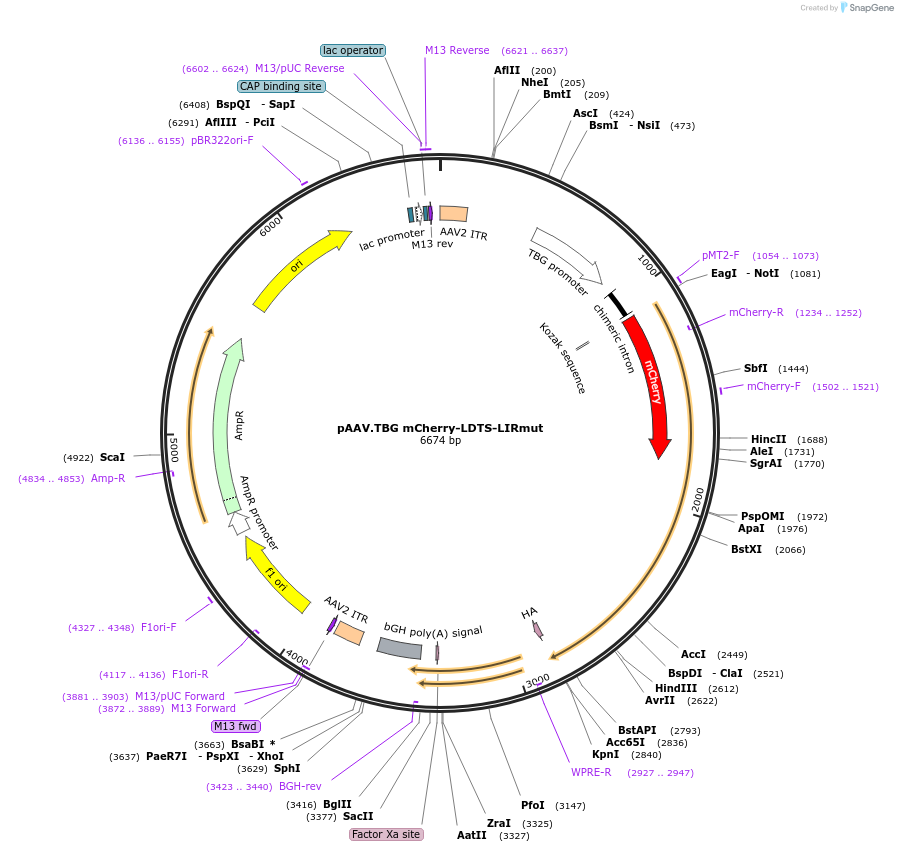
pAAV.TBG mCherry-LDTS-eLIR
Plasmid#189002Purposelipophagy-inducing adaptor protein, AAVDepositorInsertLDTS-eLIR, mCherry
UseAAVTagsHAAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
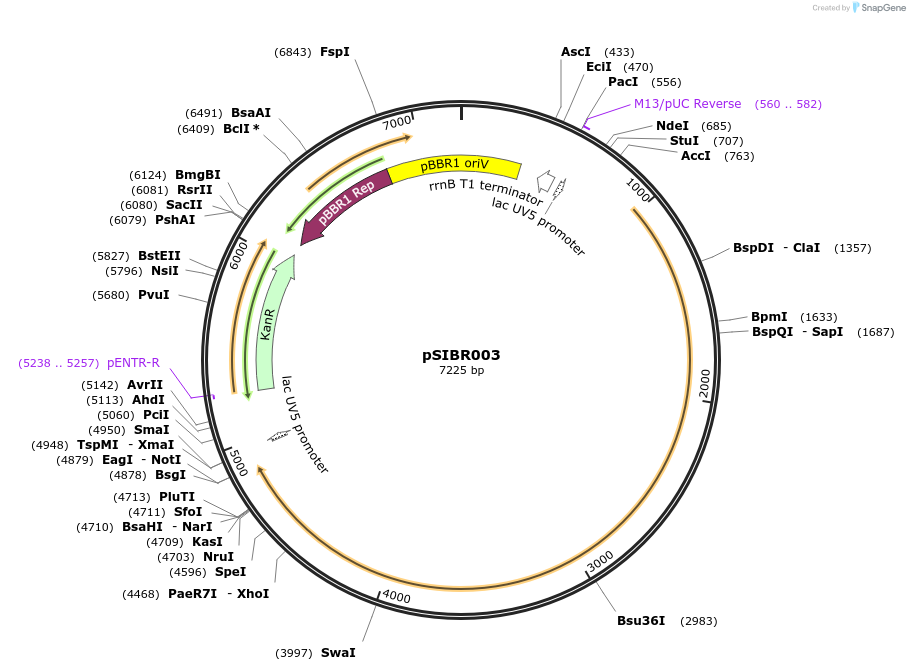
pSIBR003
Plasmid#177666PurposeSIBR Intron 3 (Int3) interrupts the CDS of FnCas12aDepositorTypeEmpty backboneUseCRISPRExpressionBacterialPromoterlacUV5Available SinceNov. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
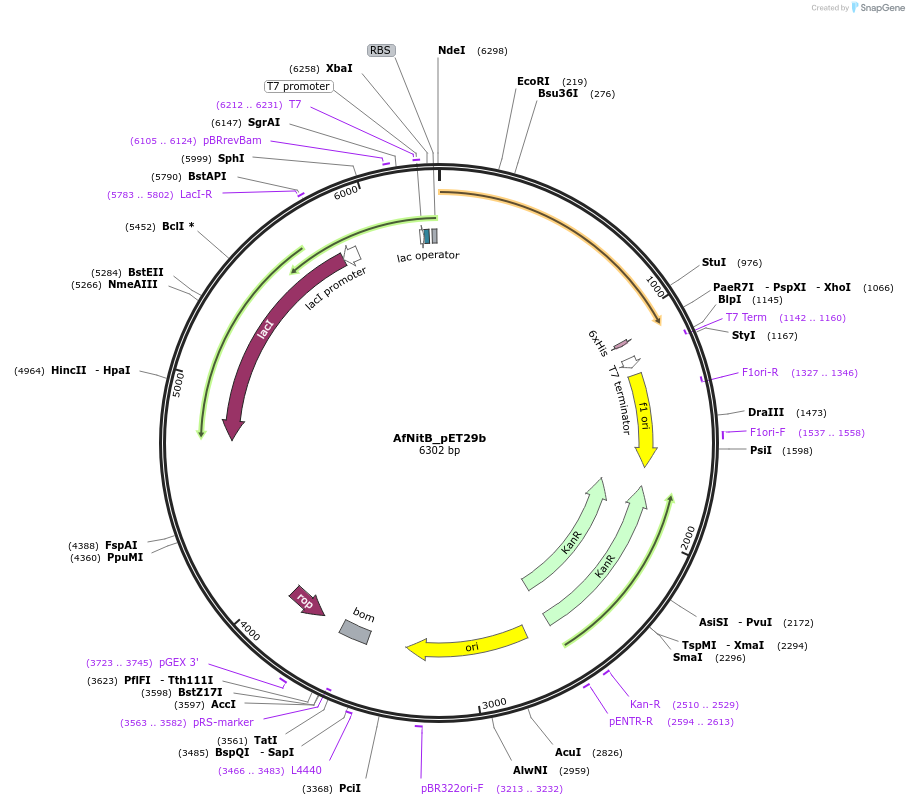
AfNitB_pET29b
Plasmid#215401PurposeExpression construct for AfNitB gene, codon optimized for expression in E. coliDepositorInsertAfNitB
ExpressionBacterialAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
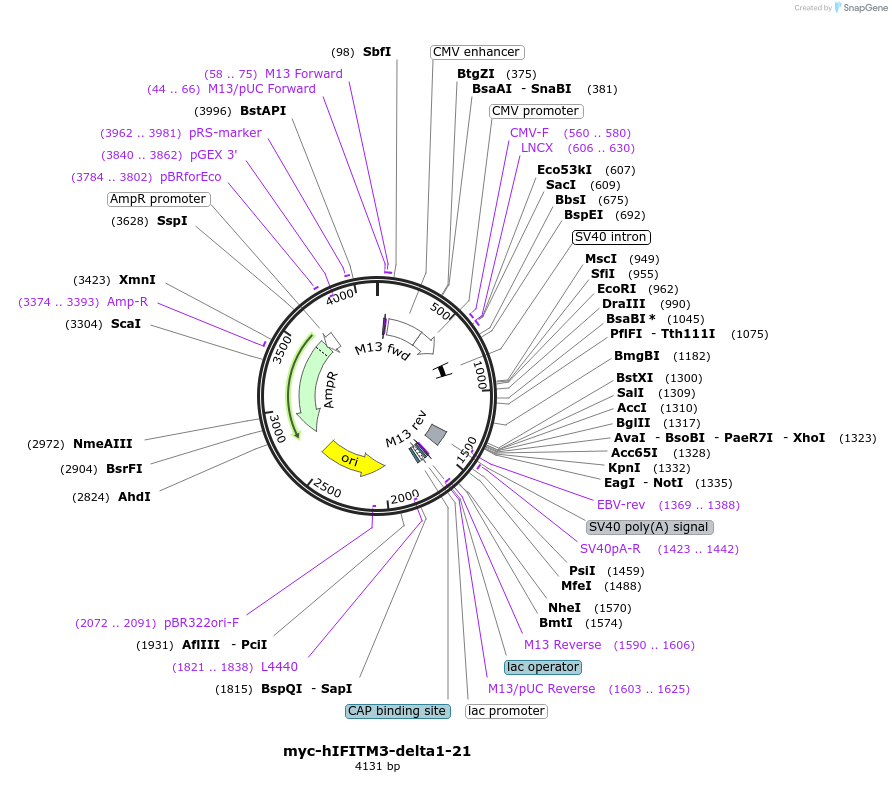
myc-hIFITM3-delta1-21
Plasmid#104365PurposeExpresses human IFITM3-delta1-21 with an N-terminal myc tag in mammalian cells.DepositorInsertIFITM3 (IFITM3 Human)
TagsmycExpressionMammalianMutationdeleted amino acids 1-21PromoterCMVAvailable SinceDec. 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
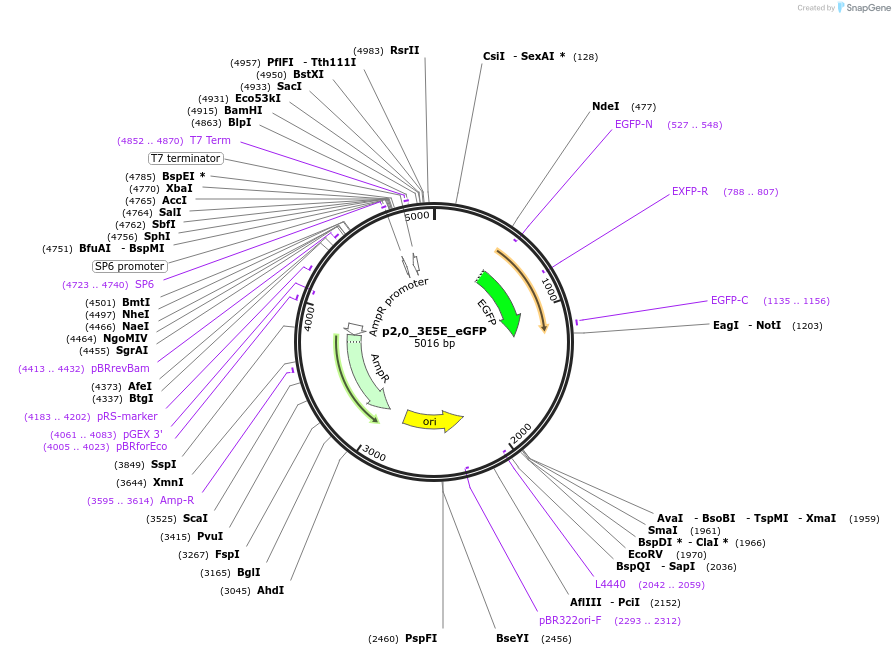
p2,0_3E5E_eGFP
Plasmid#69359PurposeEncodes for Ebola Minigenome with eGFP reporter gene under control of a T7 RNA polymerase promoterDepositorInsert3E5E eGFP minigenome
UseT7PromoterT7Available SinceNov. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
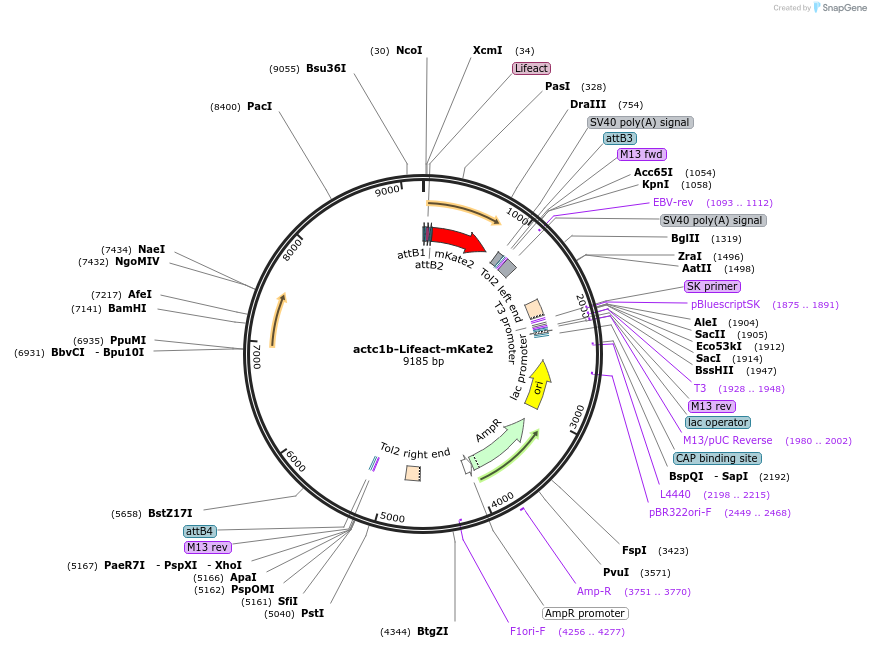
actc1b-Lifeact-mKate2
Plasmid#109493PurposeMuscle specific zebrafish expression of mKate2 fused to the actin binding lifeact sequenceDepositorInsertactc1b-Lifeact-mKate2
UseZebrafish plasmidsAvailable SinceFeb. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
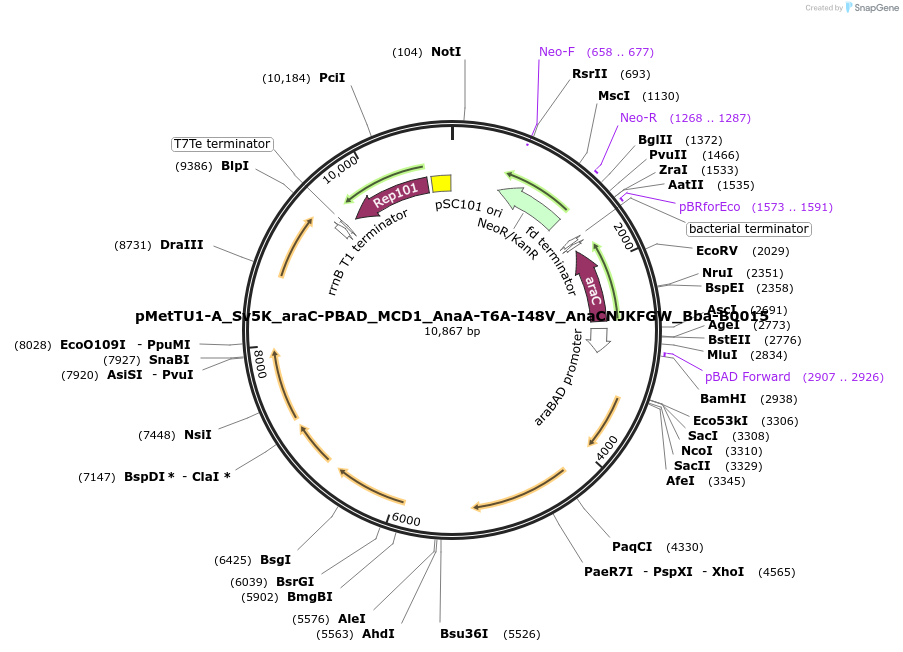
pMetTU1-A_Sv5K_araC-PBAD_MCD1_AnaA-T6A-I48V_AnaCNJKFGW_Bba-B0015
Plasmid#202023PurposeExpresses Anabaena flos-aquae ARG cluster (GvpA T6A-I48V mutant) in E. coliDepositorInsertAnabaena flos-aquae ARG cluster (GvpA T6A-I48V mutant)
ExpressionBacterialMutationGvpA T6A-I48VAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
-
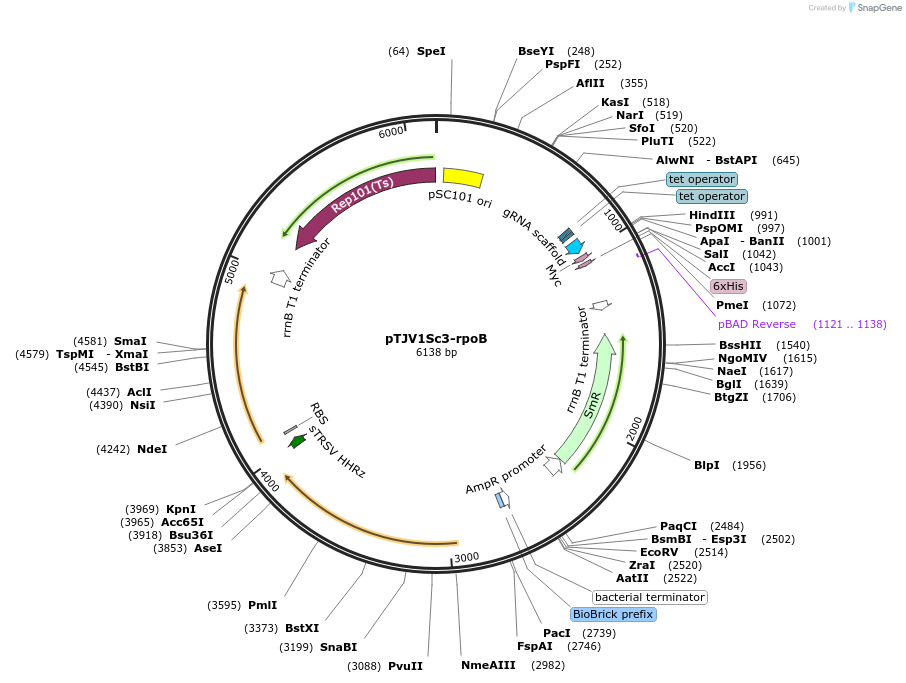
pTJV1Sc3-rpoB
Plasmid#166983PurposeGenerates ssDNA in vivo targeting E. coli rpoB for recombineering w/ EcRecT recombinase; temp. sensitive pSC101 ori and sgRNA for Cas9 counterselection; aada for spectinomycin resistanceDepositorAvailable SinceMay 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
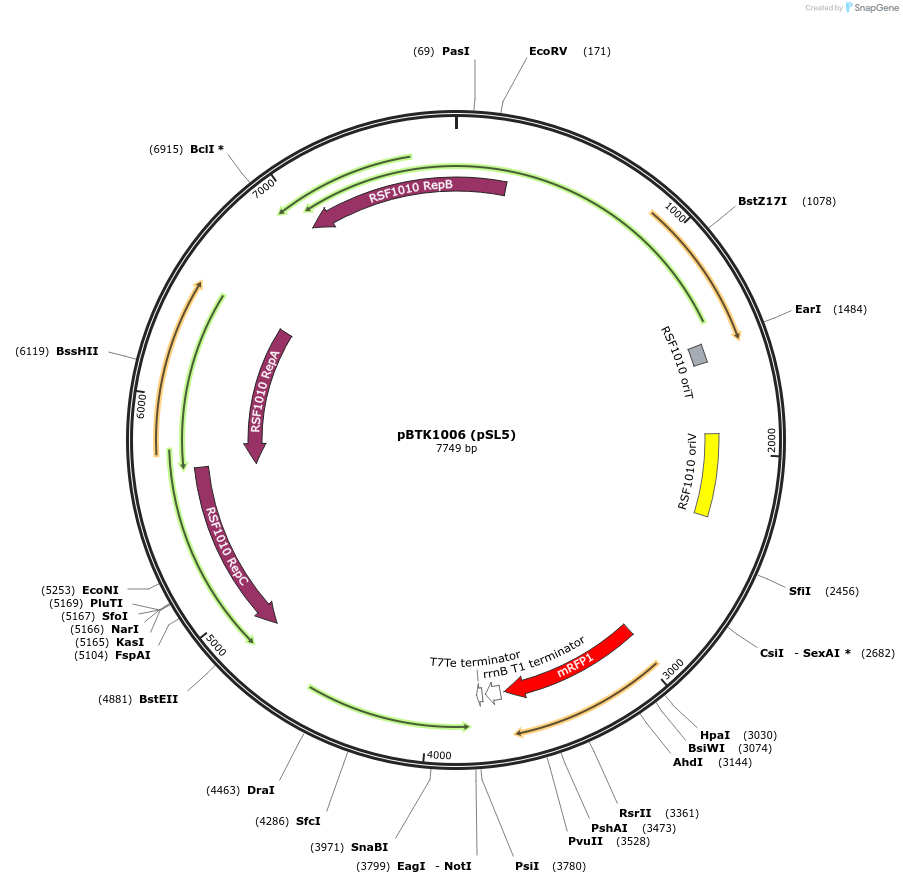
pBTK1006 (pSL5)
Plasmid#191001PurposeExpresses red chromoprotein on a broad host range RSF1010 origin plasmidDepositorInsertred chromoprotein
UseSynthetic BiologyExpressionBacterialPromoterCP25Available SinceOct. 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
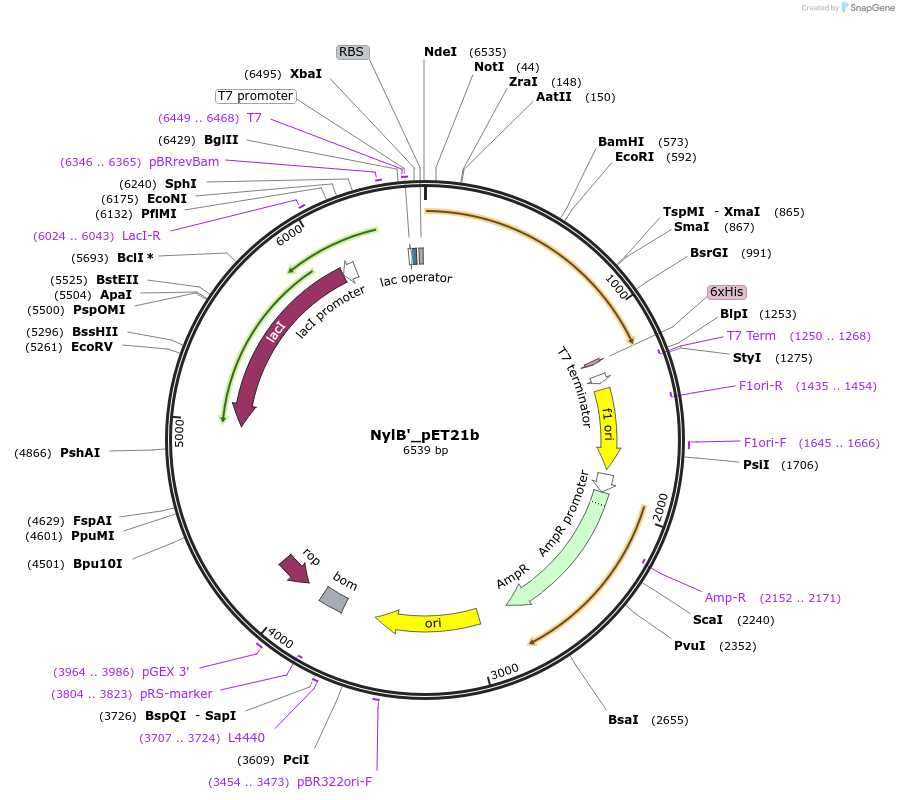
NylB'_pET21b
Plasmid#215416PurposeExpression construct for NylB' gene, codon optimized for expression in E. coliDepositorInsertNylB'
ExpressionBacterialAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
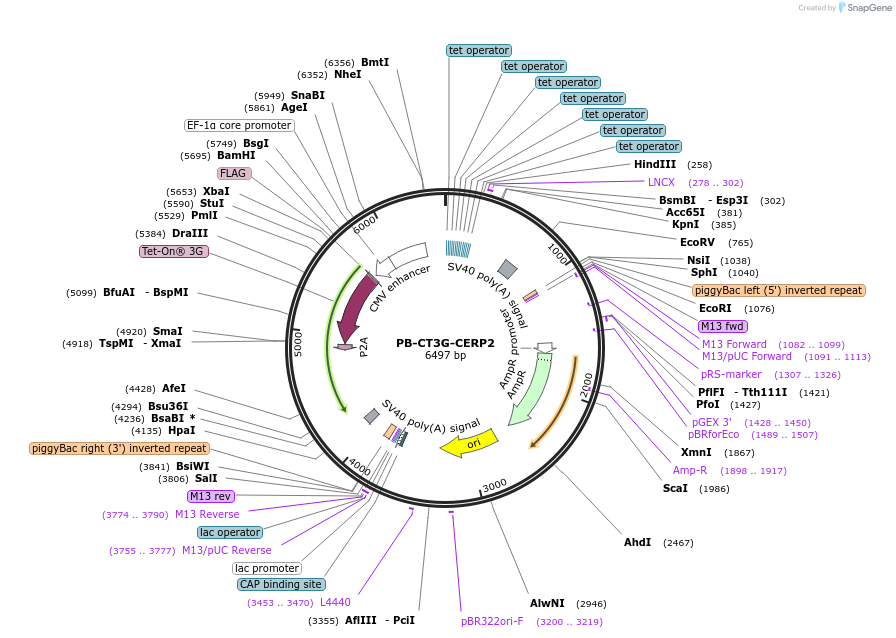
PB-CT3G-CERP2
Plasmid#175499PurposeAll-in-one piggyBac transposon vector for mCMV+dox-inducible expression of Golden Gate cloned elements (CMVe-hEF1a-driven rtTA and puromycin resistance)DepositorTypeEmpty backboneUseSynthetic BiologyExpressionMammalianPromoterTRE3G-mCMVAvailable SinceOct. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
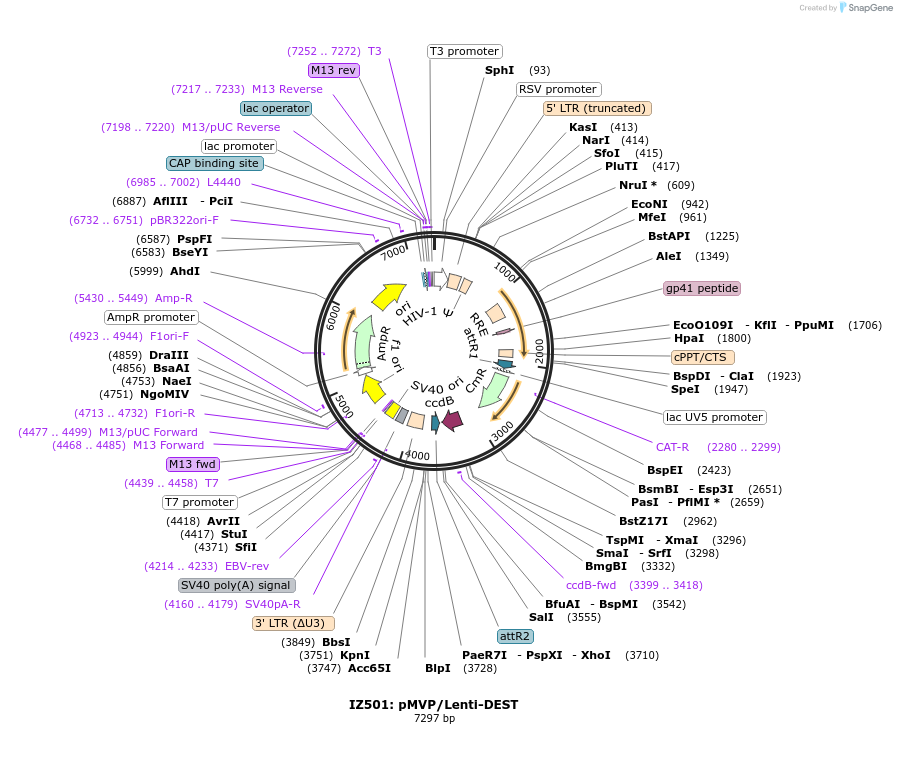
IZ501: pMVP/Lenti-DEST
Plasmid#121848PurposepMVP destination vector, empty Lentivirus vector backboneDepositorTypeEmpty backboneUseLentiviral; Pmvp gateway destination vectorAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
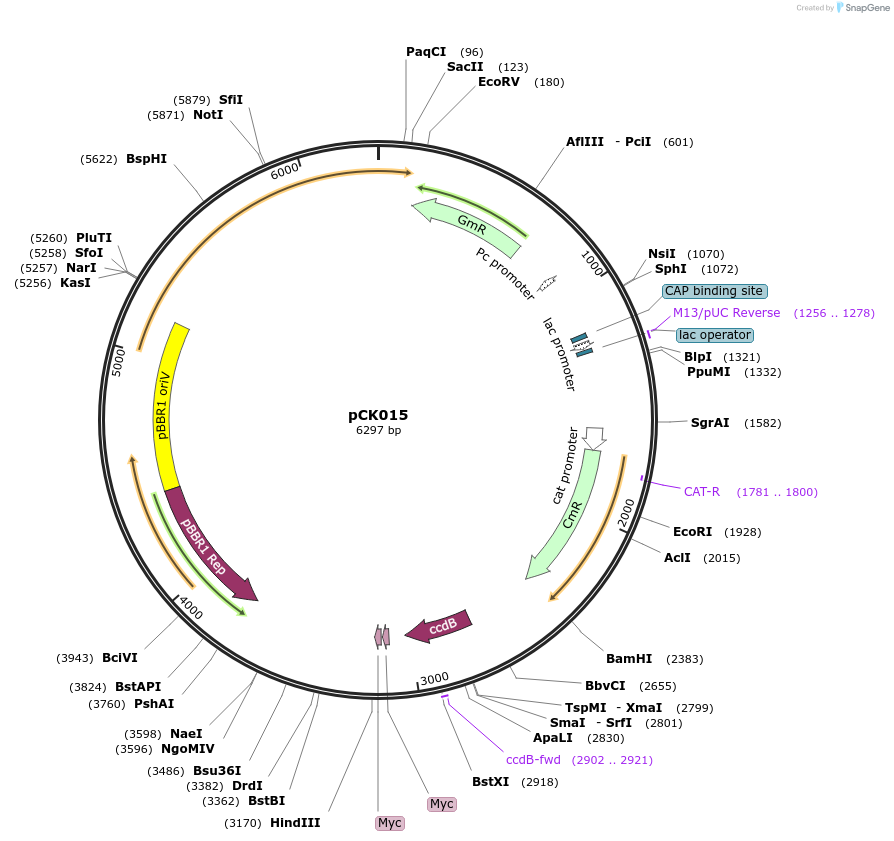
pCK015
Plasmid#105347PurposeBacterial translocation fo proteins into plant cellsDepositorInsertsecretion signal, ccdB cassette
UseBacterial secretionMutationBsaI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
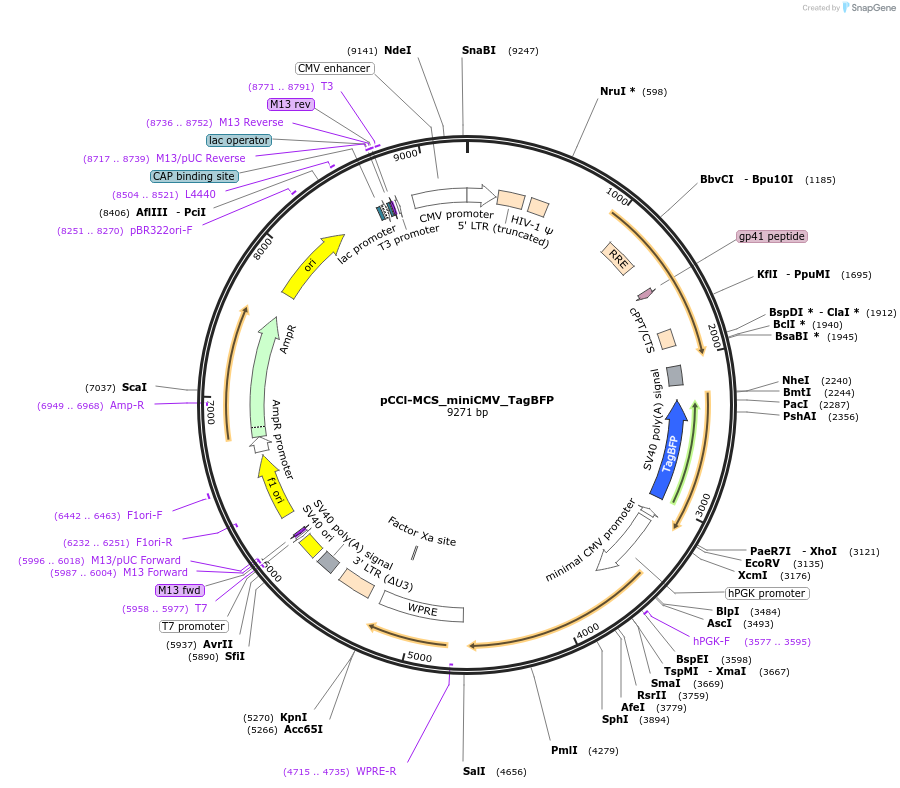
pCCl-MCS_miniCMV_TagBFP
Plasmid#134986PurposeThe plasmid includes a TagBFP gene driven by a minimal CMV promoter. Cis-regulatory elements can be cloned into the single EcoRV site. This plasmid can be used for generating a lentiviral vectorDepositorInsertTagBFP
UseLentiviralPromoterminCMVAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
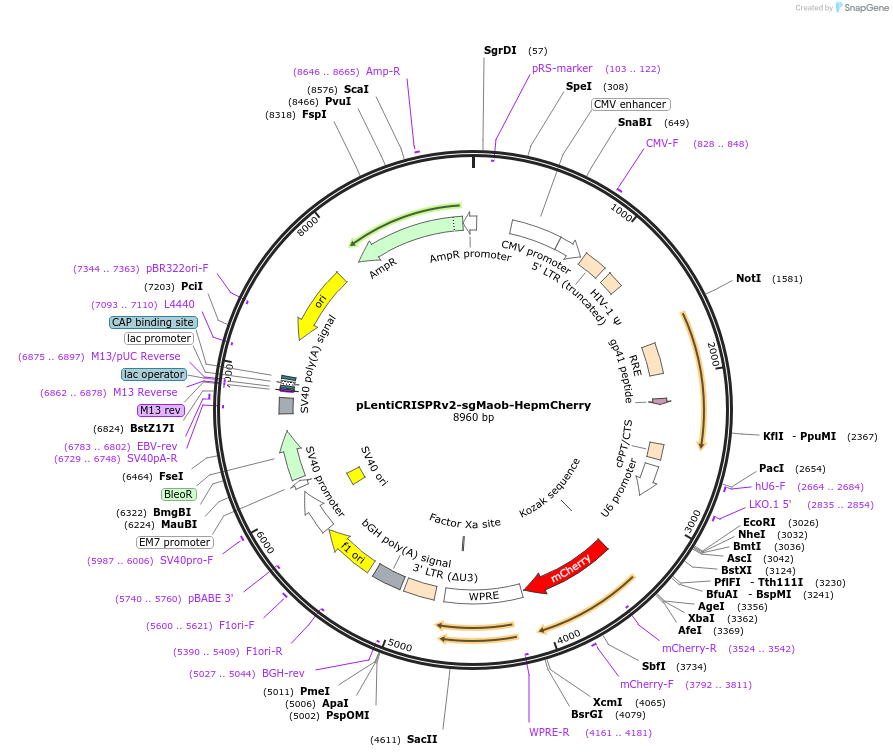
pLentiCRISPRv2-sgMaob-HepmCherry
Plasmid#192829PurposeExpresses sgRNA targeting Maob and mCherry from hepatocyte-specific promoterDepositorInsertmCherry
UseCRISPR and LentiviralExpressionMammalianPromoterU6 for sgRNA and hepatocyte-specific promoter (HS…Available SinceNov. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
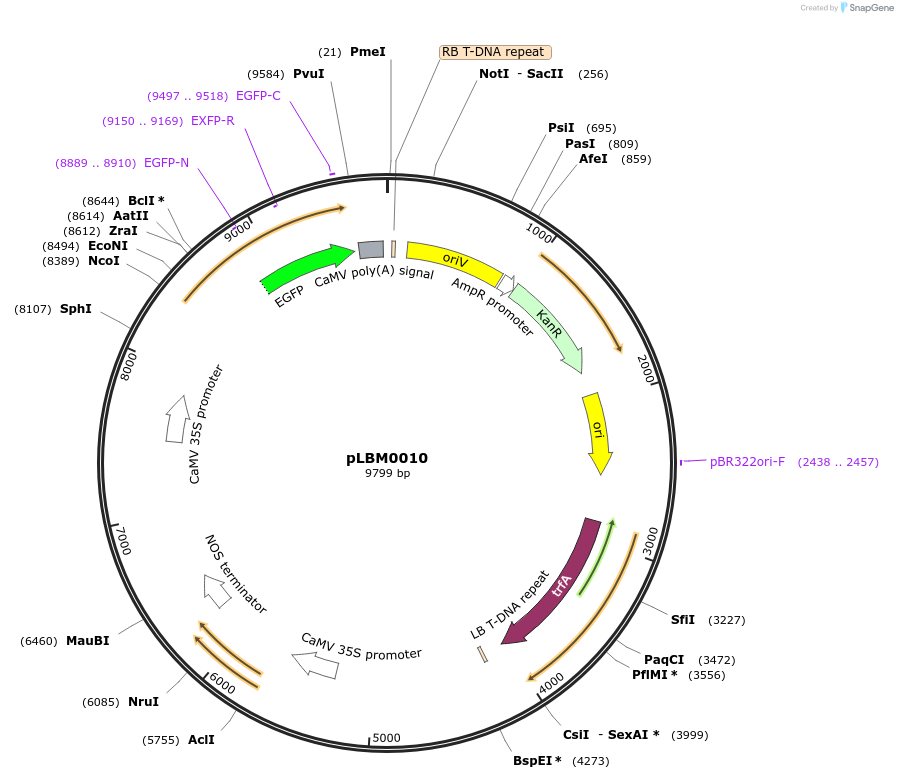
pLBM0010
Plasmid#172816PurposeLevel 2 module containing HSP21-GFPDepositorInsert[35S-P19][35S-CTPSSU-mHSP21-GFP]
UseSynthetic BiologyAvailable SinceApril 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
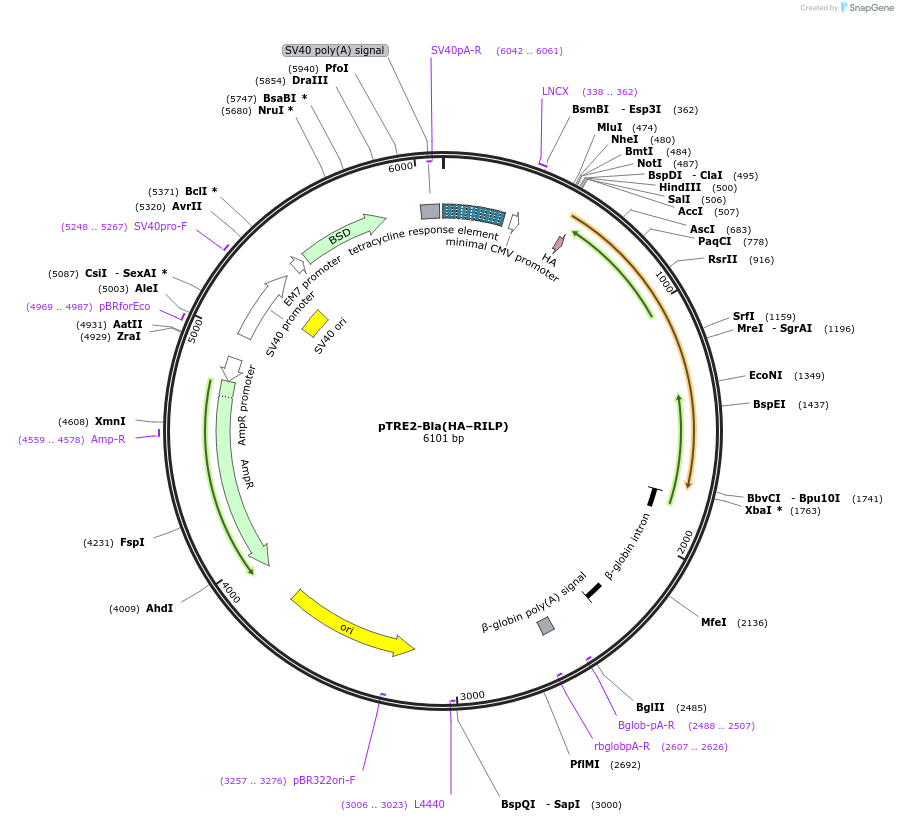
pTRE2-Bla(HA–RILP)
Plasmid#102425PurposeHA tag fused to the N-terminus of RILP for the expression in mammalian cells.DepositorInsertRab interacting lysosomal protein (RILP Human)
TagsHAExpressionMammalianPromoterTet-responsiveAvailable SinceOct. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
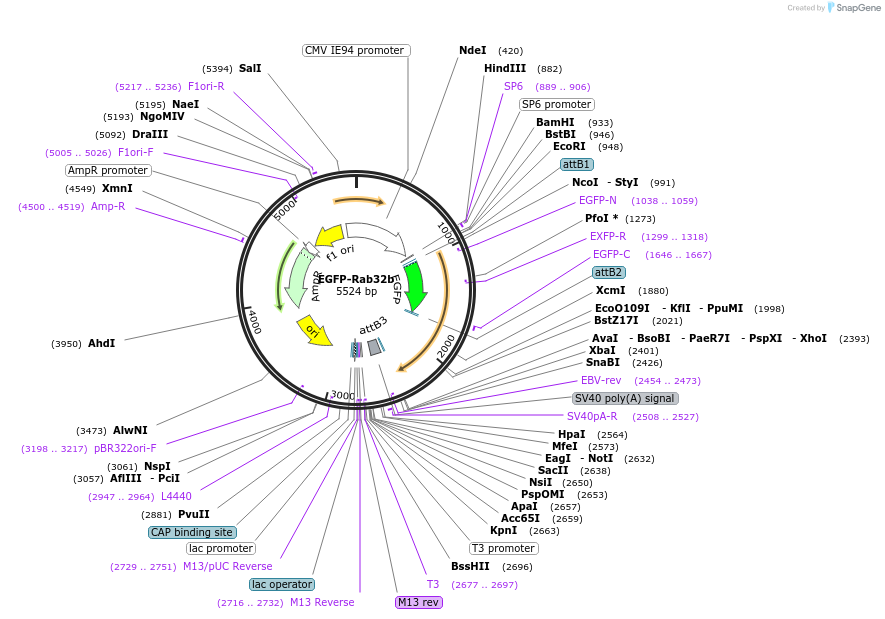
EGFP-Rab32b
Plasmid#140885PurposeExpression of zebrafish GFP-tagged Rab32b in mammalian cells.DepositorAvailable SinceFeb. 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
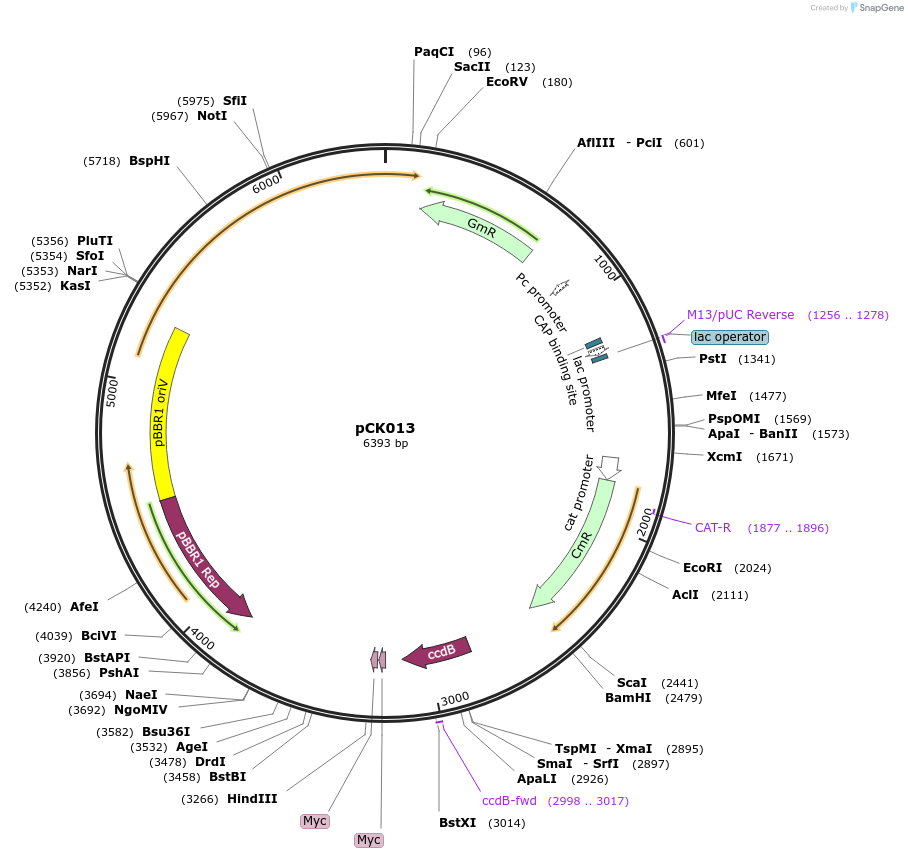
pCK013
Plasmid#105345PurposeBacterial translocation fo proteins into plant cellsDepositorInsertsecretion signal, ccdB cassette
UseBacterial secretionMutationBsaI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
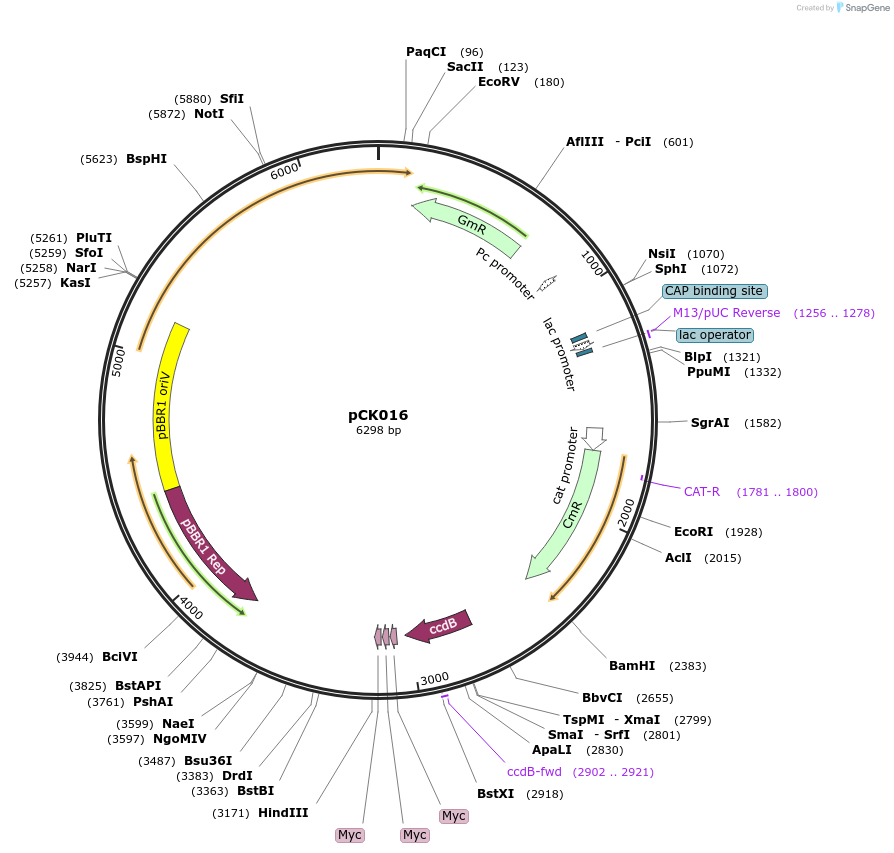
pCK016
Plasmid#105348PurposeBacterial translocation for proteins into plant cellsDepositorInsertsecretion signal, ccdB cassette
UseBacterial secretionMutationBsaI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
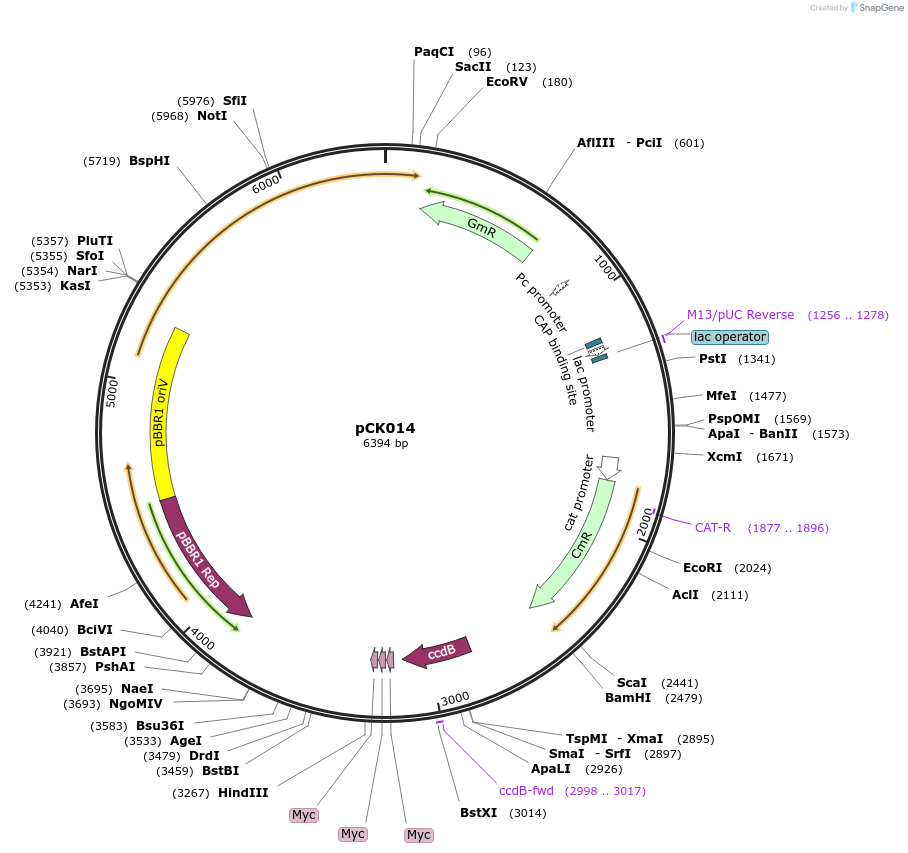
pCK014
Plasmid#105346PurposeBacterial translocation fo proteins into plant cellsDepositorInsertsecretion signal, ccdB cassette
UseBacterial secretionMutationBsaI restriction sites removedAvailable SinceMay 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
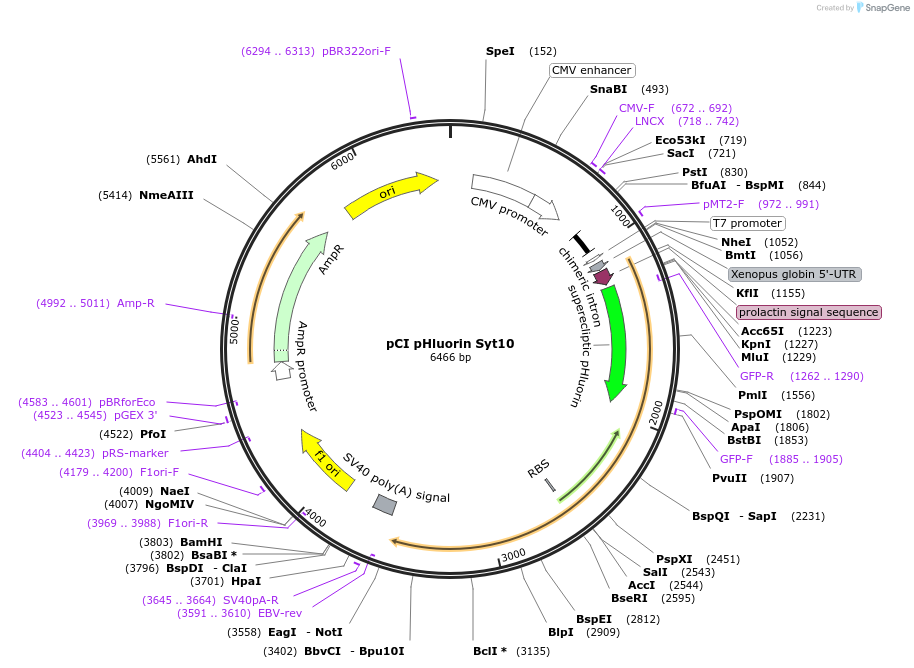
pCI pHluorin Syt10
Plasmid#184511PurposeMammalian expression of Synaptotagmin 10 with pH-sensitive fluorescent proteinDepositorAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAd NeuroD-I-nGFP
Plasmid#19414DepositorAvailable SinceNov. 10, 2008AvailabilityAcademic Institutions and Nonprofits only -
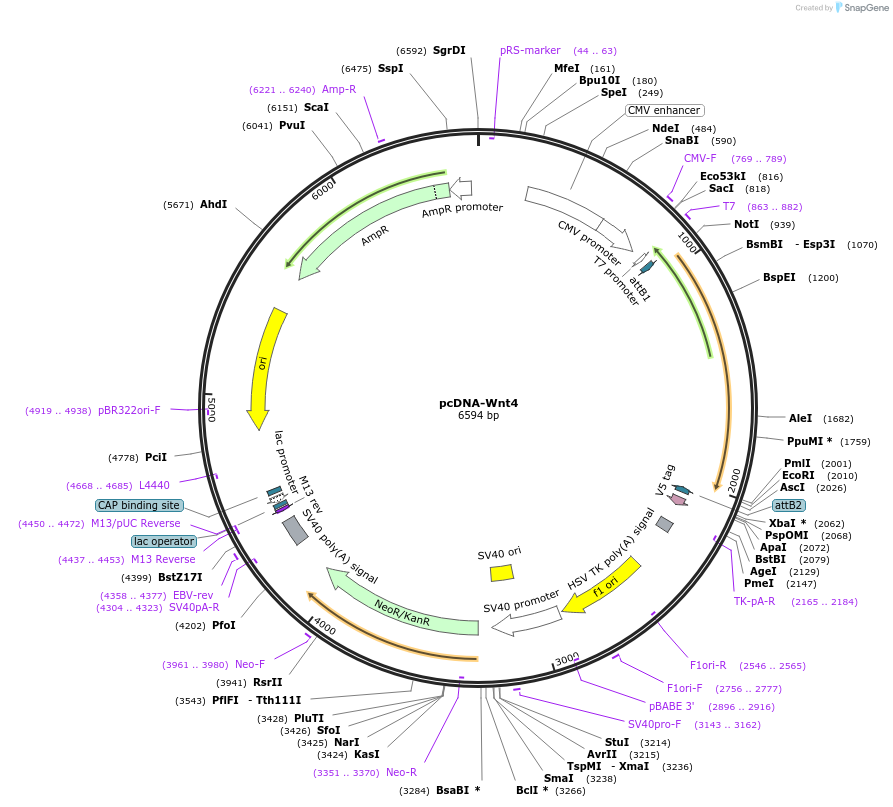
pcDNA-Wnt4
Plasmid#35910DepositorAvailable SinceMay 7, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAd 2B Ngn3-I-nGFP
Plasmid#19410DepositorAvailable SinceNov. 10, 2008AvailabilityAcademic Institutions and Nonprofits only -
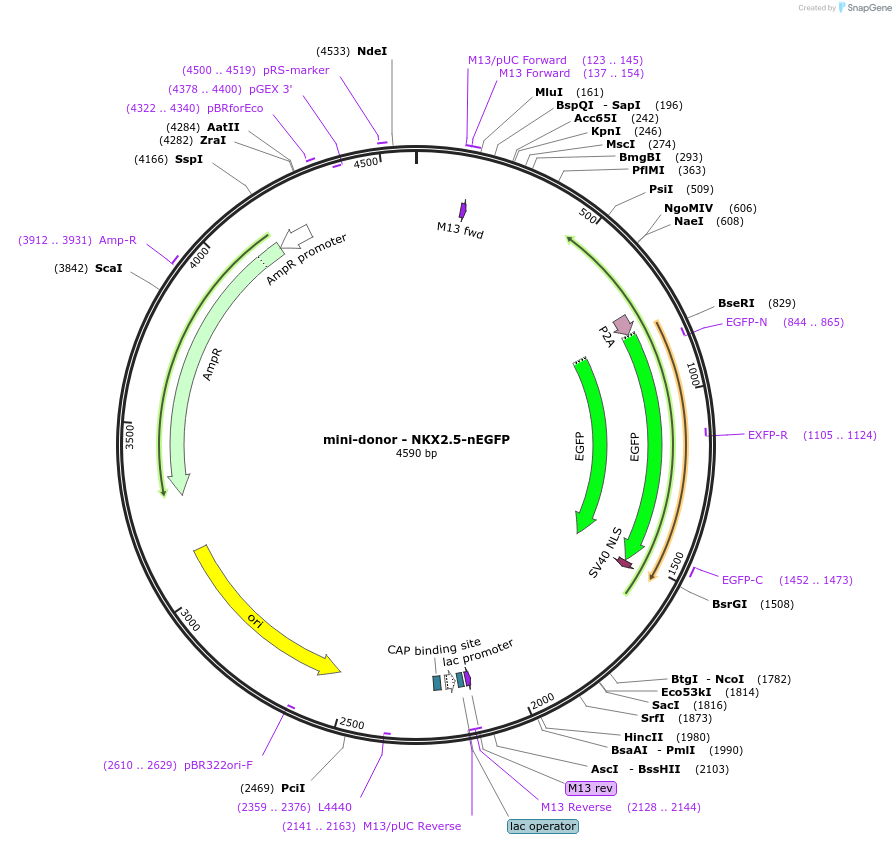
mini-donor - NKX2.5-nEGFP
Plasmid#209287Purposethis mini-donor knockin vector can be used to knock nuclear EGFP into NKX2.5 locusDepositorInsertEGFP-nls
UseKnockin donor vectorAvailable SinceApril 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
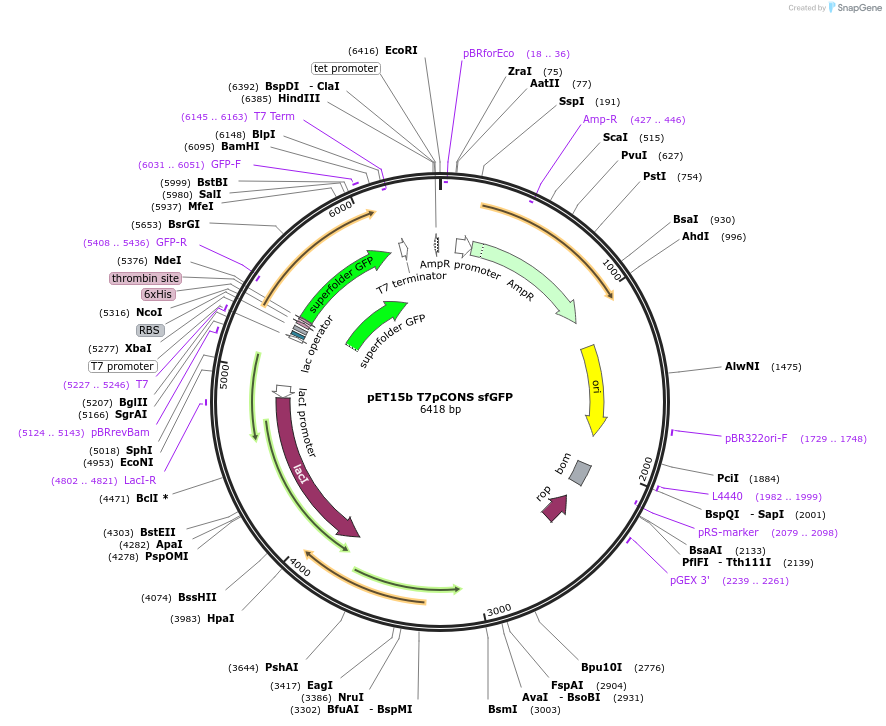
pET15b T7pCONS sfGFP
Plasmid#154467PurposepET15b with a design flaw in the T7 promoter fixed. Contains the coding sequence for sfGFPDepositorInsertsuper folder GFP
TagsHis-thrombinExpressionBacterialPromoterT7Available SinceJuly 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
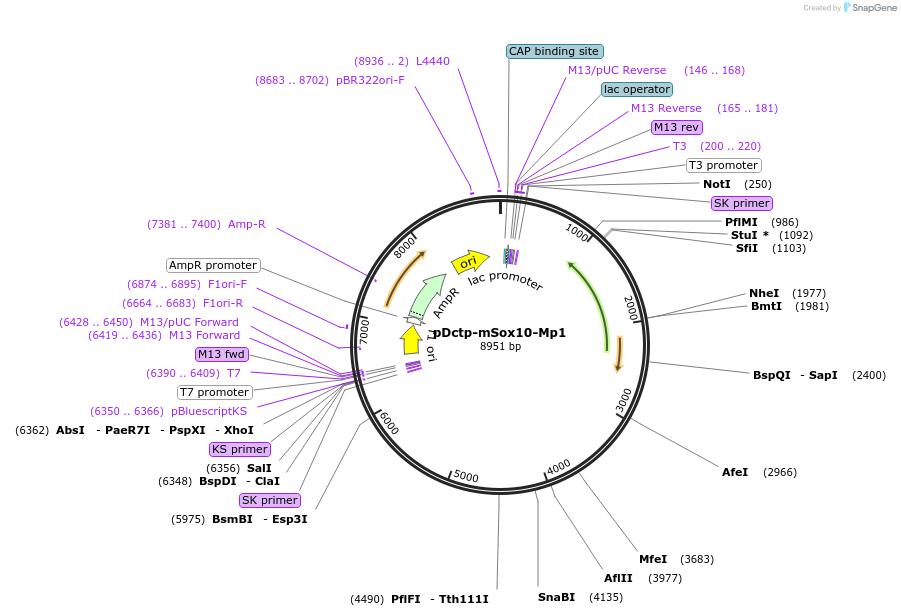
pDctp-mSox10-Mp1
Plasmid#13513DepositorAvailable SinceDec. 8, 2006AvailabilityAcademic Institutions and Nonprofits only -
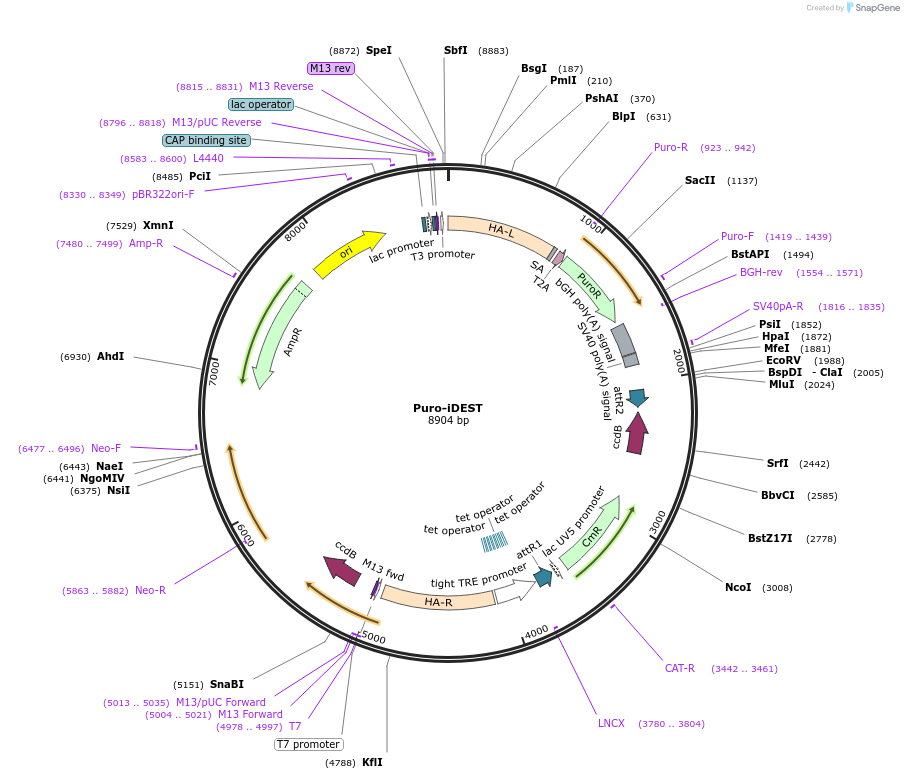
Puro-iDEST
Plasmid#75336PurposeGATEWAY destination vector for site-specific integration into the human AAVS1/PPP1R12C locus for doxycycline-inducible gene over-expressionDepositorInsertTRE-DEST
ExpressionMammalianPromoterTRE-tightAvailable SinceJune 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
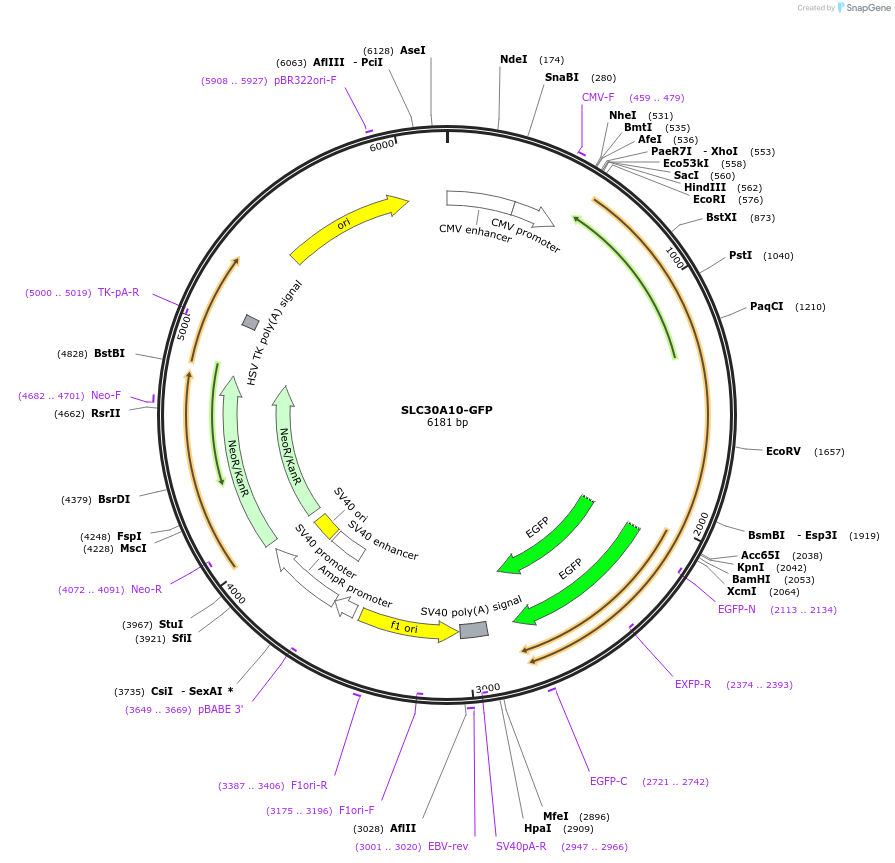
SLC30A10-GFP
Plasmid#104381PurposeExpresses SLC30A10-GFP wild-type in mammalian cellsDepositorAvailable SinceDec. 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
HUW-TetO-hNanog
Plasmid#25700DepositorAvailable SinceSept. 16, 2010AvailabilityAcademic Institutions and Nonprofits only -
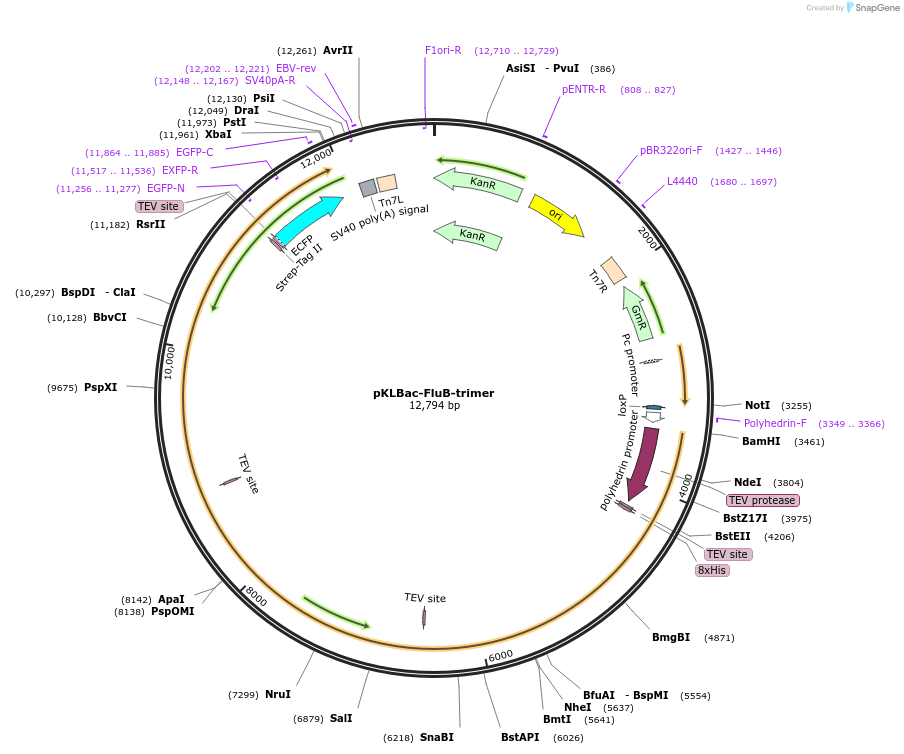
pKLBac-FluB-trimer
Plasmid#229859PurposeInfluenza B/Memphis/13/03 polymerase heterotrimer (self-cleaving polyprotein)DepositorInsertsPA, Influenza B (B/Memphis/13/03)
PB1, Influenza B (B/Memphis/13/03)
PB2, Influenza B (B/Los Angeles/1/02)
Tags2x-Streptag and 8xHis-tagExpressionInsectAvailable SinceFeb. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
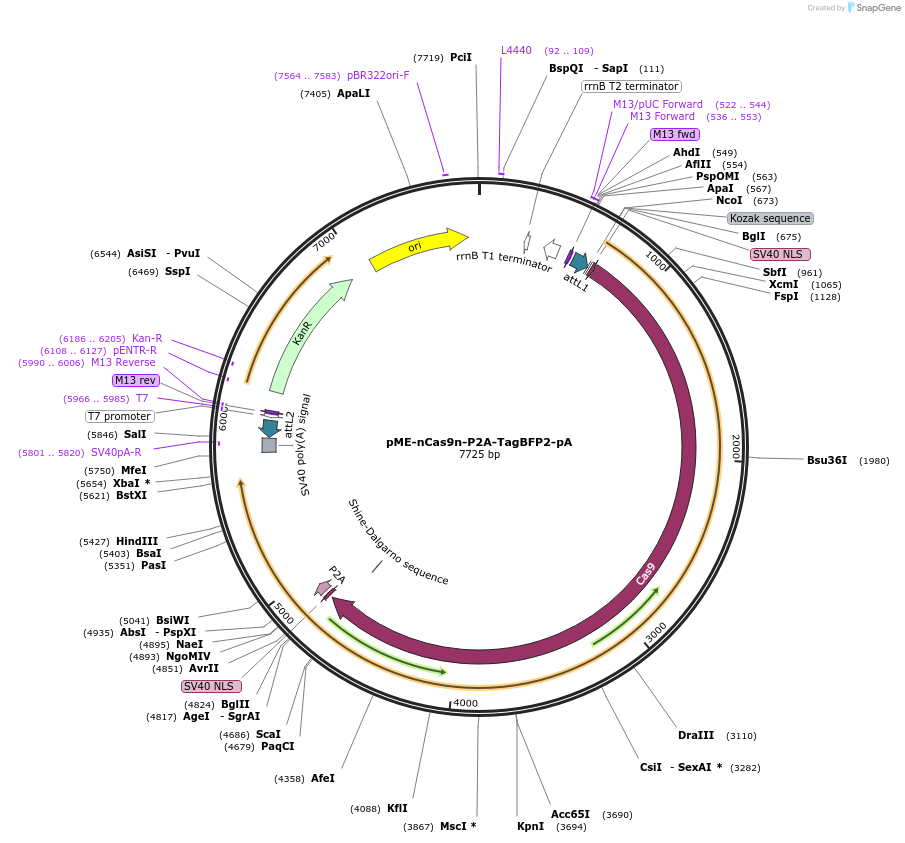
pME-nCas9n-P2A-TagBFP2-pA
Plasmid#176026PurposeMultisite gateway vector for expression of nCas9n with blue reporter. Parton lab clone ENTDepositorInsertnCas9n-P2A-TagBFP2
UseGateway entry cloneAvailable SinceOct. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3-HA-BRAT1
Plasmid#62802PurposeExpresses human BRAT1 with an N-terminal HA tag in mammalian cellsDepositorAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
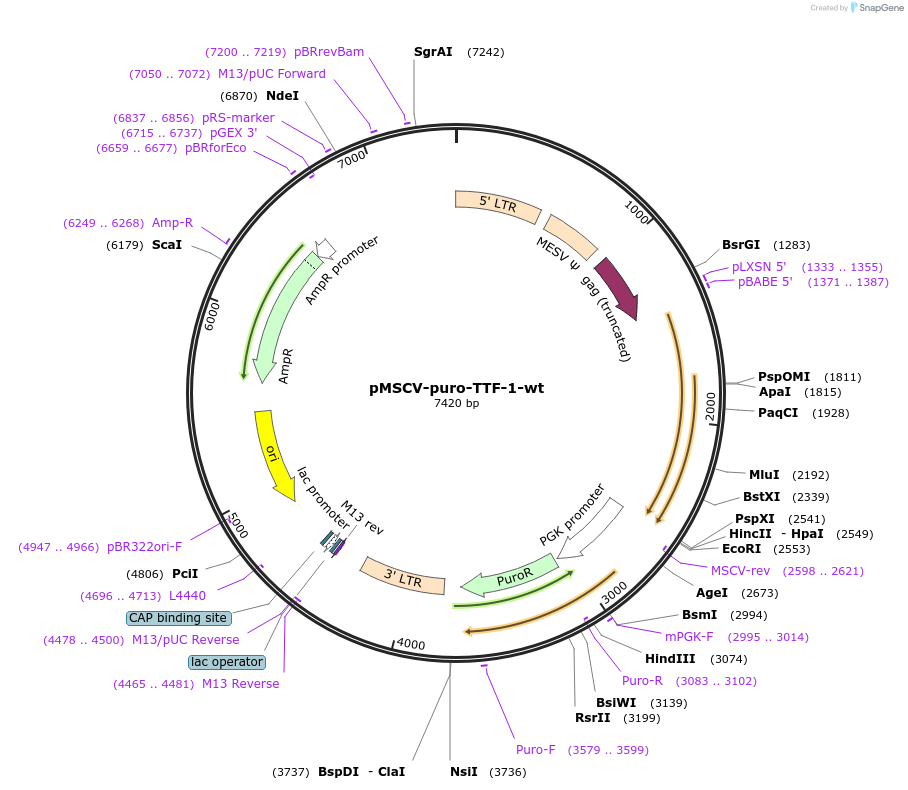
pMSCV-puro-TTF-1-wt
Plasmid#74990PurposeRetroviral vector holding the human wild-type full-length TTF-1 (NKX2-1) cDNADepositorInsertTTF-1 (TTF1 Human)
UseRetroviralExpressionMammalianMutationwild-type, full-length, without 3' UTRAvailable SinceMay 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
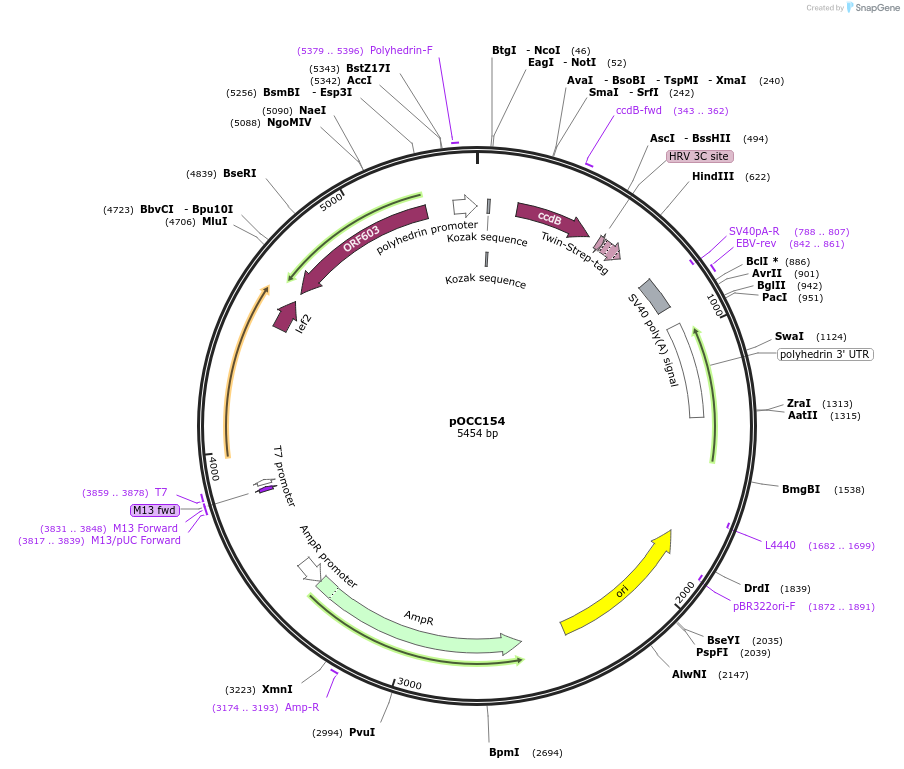
pOCC154
Plasmid#118902Purposeshuttle vector for baculovirus production, using FlashBac bacmid; for recombinant protein with C-terminal TwinStrep, cleavable with 3CDepositorInsertNcoI-NotI-ccdB-AscI-3C-TwinStrep-stop-HindIII cassette
TagsTwinStrep, cleavable with 3C proteaseExpressionInsectPromoterpolHAvailable SinceJan. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
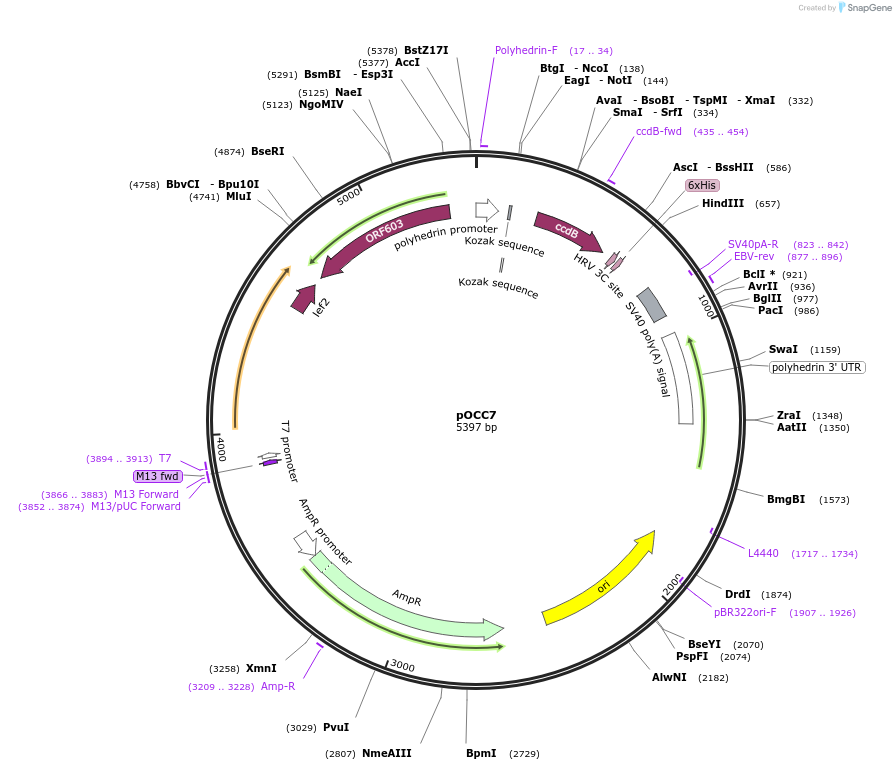
pOCC7
Plasmid#118876Purposeshuttle vector for baculovirus production, using FlashBac bacmid; for recombinant protein with C-terminal HIS6 tag, cleavable with 3CDepositorInsertNcoI-NotI-ccdB-AscI-3C-HIS6-stop-HindIII cassette
TagsHIS6, cleavable with 3C proteaseExpressionInsectPromoterpolHAvailable SinceJan. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
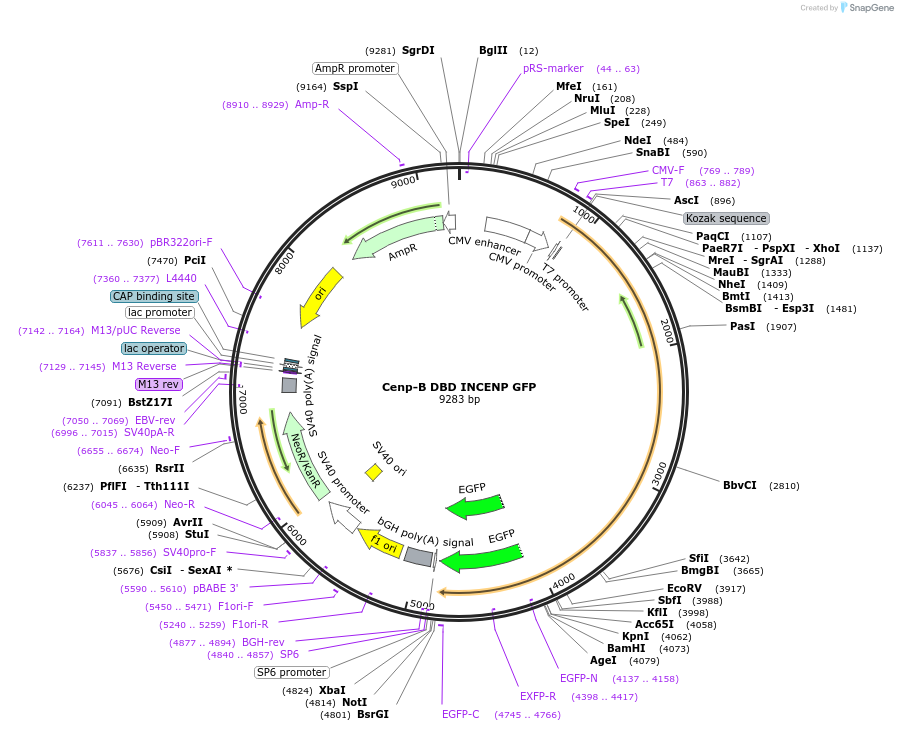
Cenp-B DBD INCENP GFP
Plasmid#45237PurposeTarget INCENP to centromeres via CENP-B DNA binding domain (DBD)DepositorInsertCenp-B-DBD, INCENP-Δcen, GFP
TagsDNA binding domain of CENP-B and GFPExpressionMammalianMutationCEN box deleted from INCENP (aa1-46)Available SinceSept. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
pAAV.TBG mCherry-LDTS-LIRmut
Plasmid#189003Purposelipophagy-inducing adaptor protein, AAV, negative controlDepositorInsertLDTS-LIRmut, mCherry
UseAAVTagsHAAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
HIR Y1158,1162,1163F
Plasmid#24054DepositorAvailable SinceDec. 16, 2010AvailabilityAcademic Institutions and Nonprofits only -
pRK-flag-USP19 C506S
Plasmid#78584PurposeExpression flag-USP19 C506S in mammalian cellsDepositorInsertflag-USP19 C506S (USP19 Human)
TagsflagExpressionMammalianMutationmutate 506 cysteine residue to serinePromoterCMVAvailable SinceJune 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
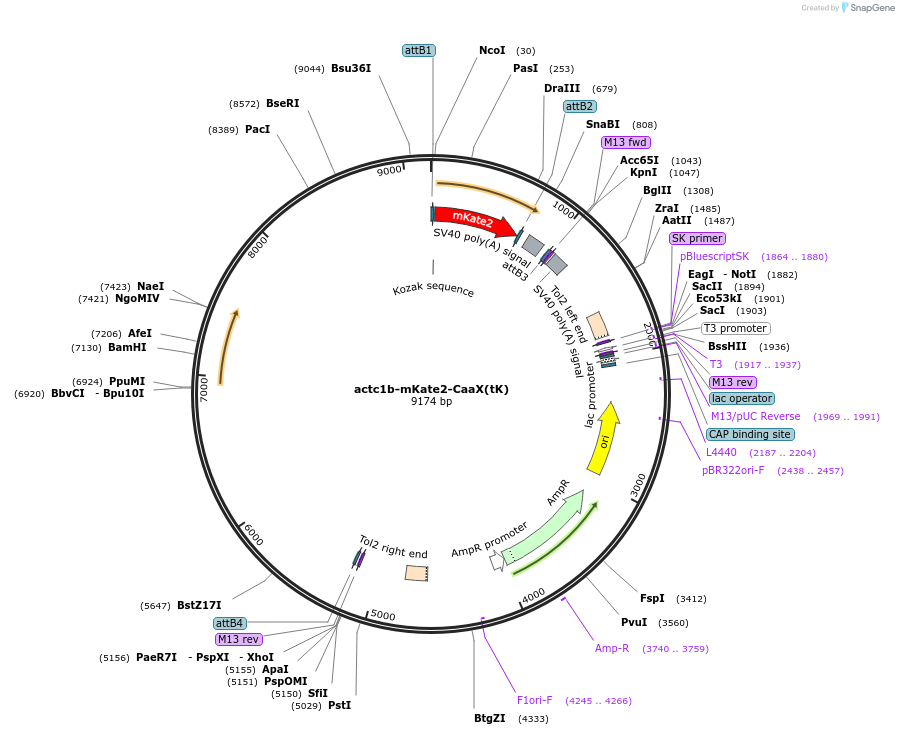
actc1b-mKate2-CaaX(tK)
Plasmid#109488PurposeMuscle specific zebrafish expression of mKate2 fused to the CaaX domain from KrasDepositorInsertactc1b-mKate2-CaaX(tK)
UseZebrafish plasmidsAvailable SinceFeb. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
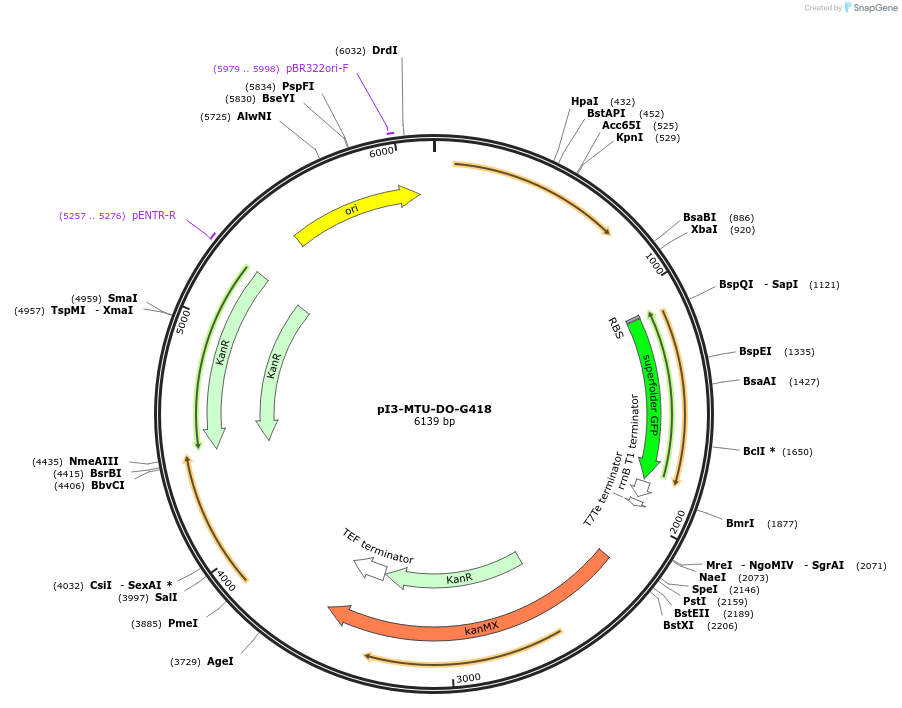
pI3-MTU-DO-G418
Plasmid#160023PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). G418 selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
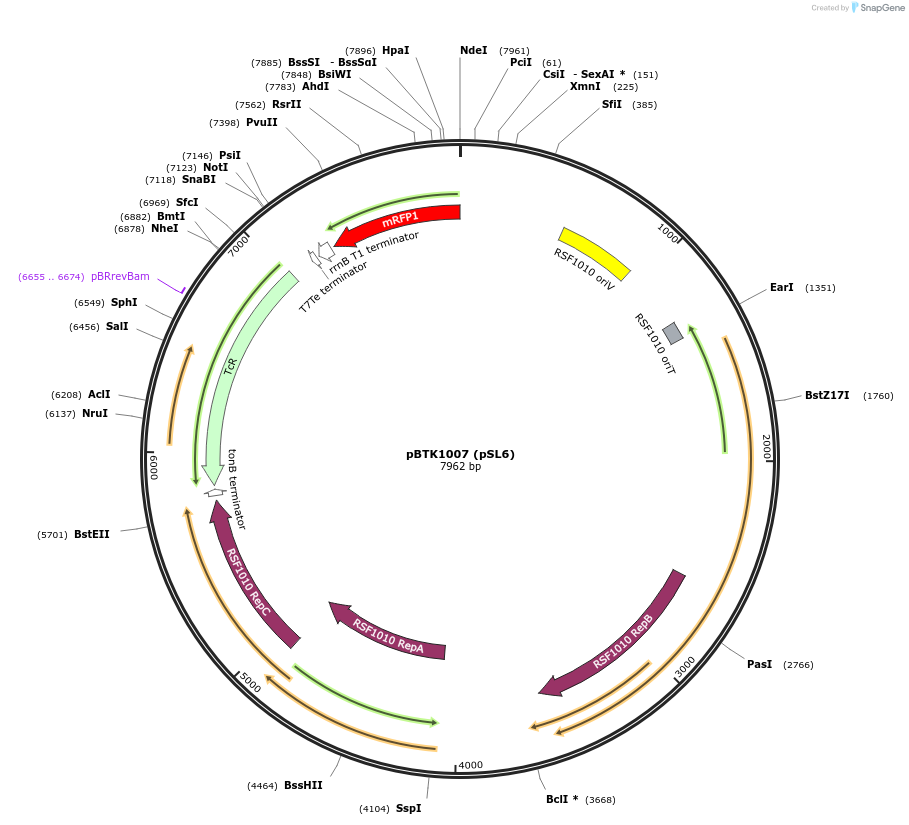
pBTK1007 (pSL6)
Plasmid#191002PurposeExpresses red chromoprotein on a broad host range RSF1010 origin plasmidDepositorInsertred chromoprotein
UseSynthetic BiologyExpressionBacterialPromoterCP25Available SinceOct. 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
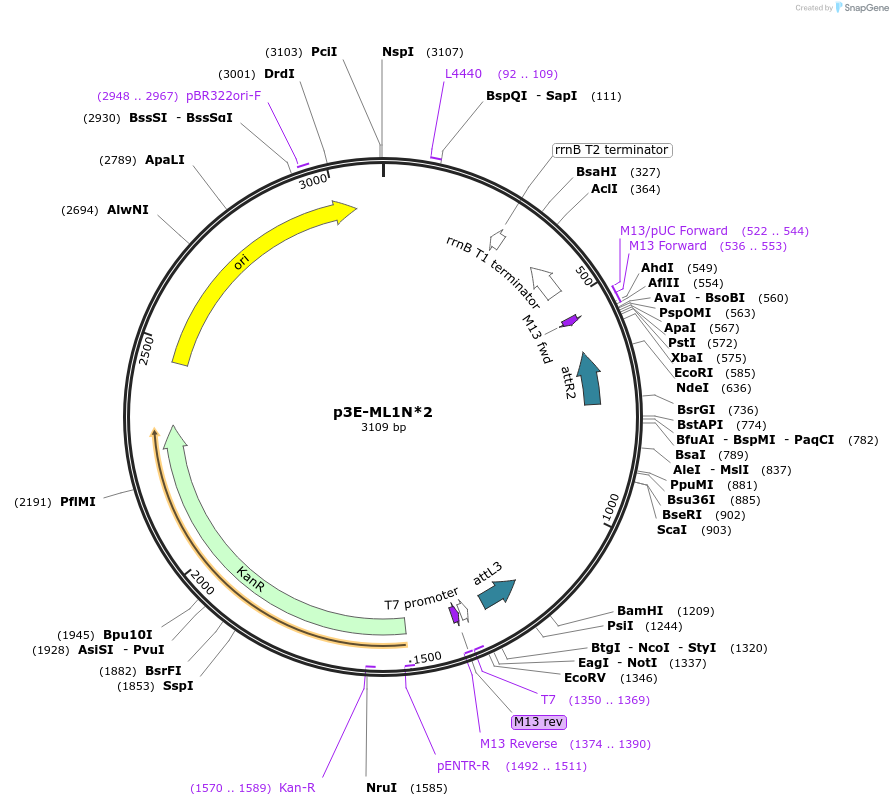
p3E-ML1N*2
Plasmid#67678PurposeMultisite gateway entry clone for adding C-terminal fusions of ML1N*2DepositorInsertML1N*2
UseGateway multisite 3' entry cloneAvailable SinceDec. 2, 2015AvailabilityAcademic Institutions and Nonprofits only