We narrowed to 7,396 results for: tet on
-
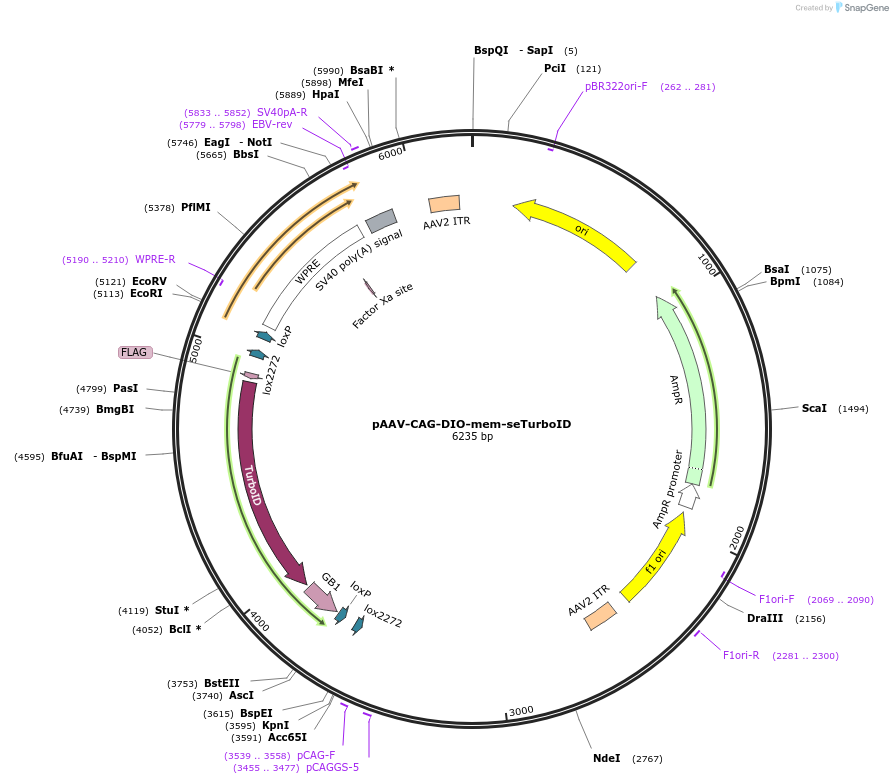
Plasmid#237887PurposeAAV vector for Cre-dependent expression of membrane-tethered seTurboIDDepositorInsertmem-seTurboID
UseAAV and Cre/LoxTagsFLAG tag and GAP43 palmitoylation signalExpressionMammalianPromoterCAGAvailable SinceAug. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
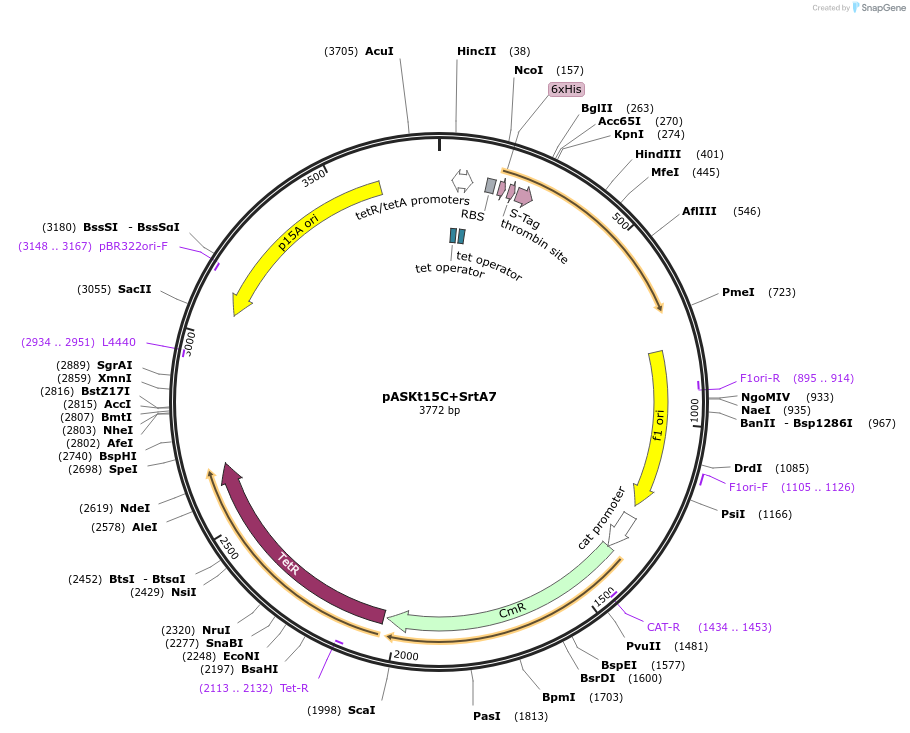
pASKt15C+SrtA7
Plasmid#64986PurposeExpresses the P94R/E105K/E108A/D160N/D165A/K190E/K196T mutant of Staphylococcus aureus sortase A in E. coliDepositorInsertsortase A
TagsHis6 tag and S tagExpressionBacterialMutationdeleted amino acids 1-59, P94R, E105K, E108A, D16…Promotertetracycline promoter/operatorAvailable SinceJune 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
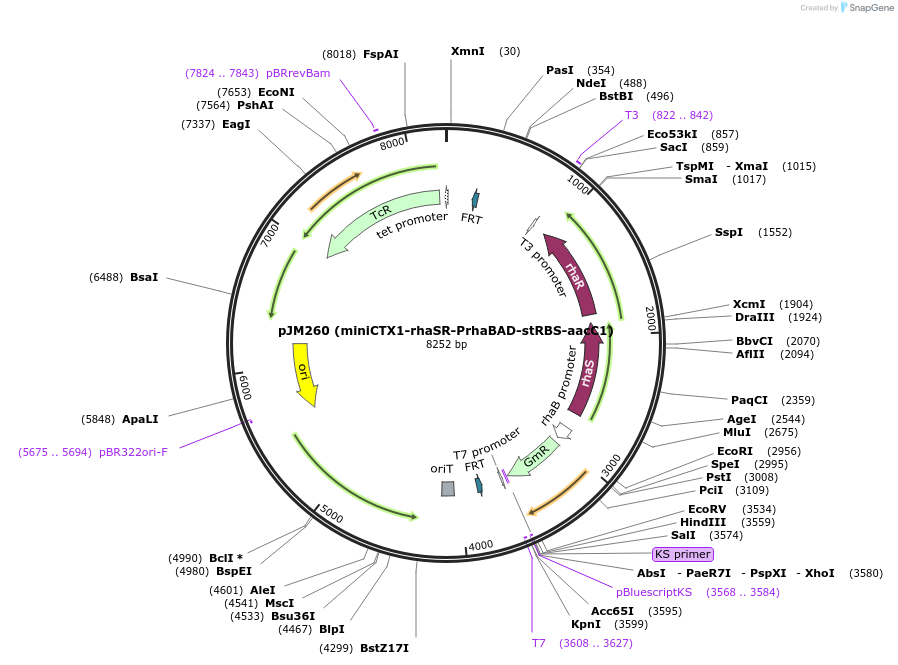
pJM260 (miniCTX1-rhaSR-PrhaBAD-stRBS-aacC1)
Plasmid#110572PurposeIntegration vector for Pseudomonas aeruginosa with rhaSR-PrhaBAD inducible promoter and StRBS-aacC1 insertDepositorInsertrhaSR-PrhaBAD-stRBS-aacC1
ExpressionBacterialPromoterrhaSR-PrhaBADAvailable SinceMay 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
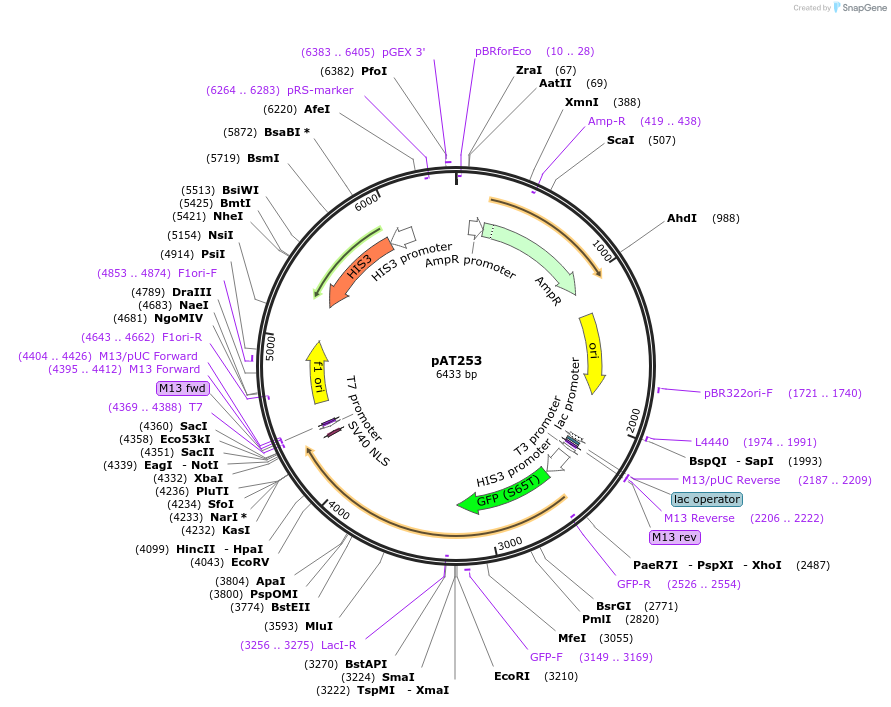
pAT253
Plasmid#40943DepositorInsertGFP-LacI**
TagsGFP(S65T, V163A) and SV40 NLSExpressionYeastMutationLacI**/LacI2G(4 mutations) (see comment below); l…PromoterHis3 promoter fragmentAvailable SinceFeb. 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
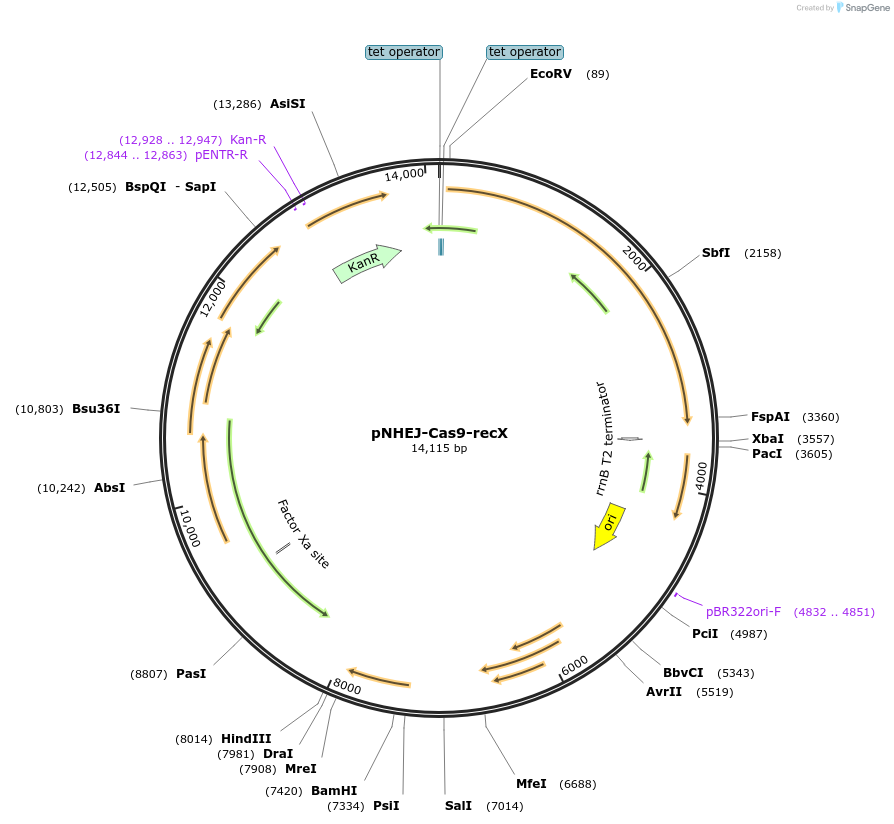
pNHEJ-Cas9-recX
Plasmid#158715PurposeCRISPR-assisted-NHEJ system used for genome editing in Mycobacterium smegmatis. Helper plasmid expresses Cas9, NHEJ machinery of M. marinum and recXDepositorInsertcas9
UseCRISPRExpressionBacterialPromoterTetOAvailable SinceJan. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
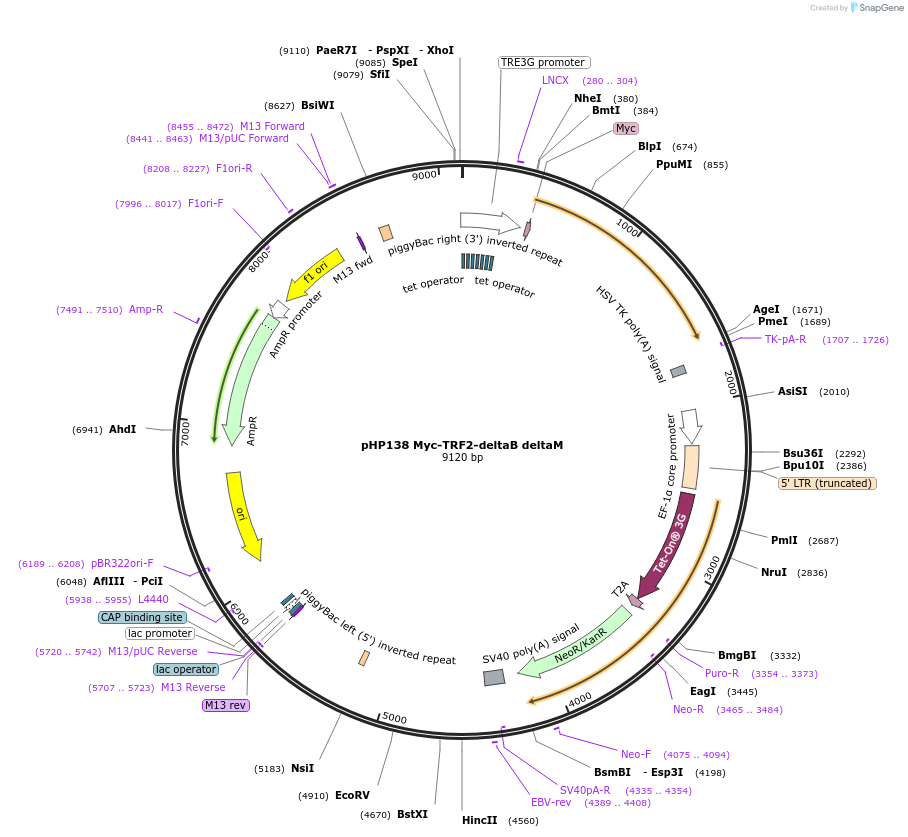
pHP138 Myc-TRF2-deltaB deltaM
Plasmid#164255Purposefor generation of mammalian cells with inducible expression of dominant negative TRF2 for induction of telomere crisis used together with any PiggyBac transposase plasmidDepositorInsertTRF2
TagsMycExpressionMammalianMutationdeletion of amino acids-86 and 496-542PromoterTetracycline inducible promotorAvailable SinceApril 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
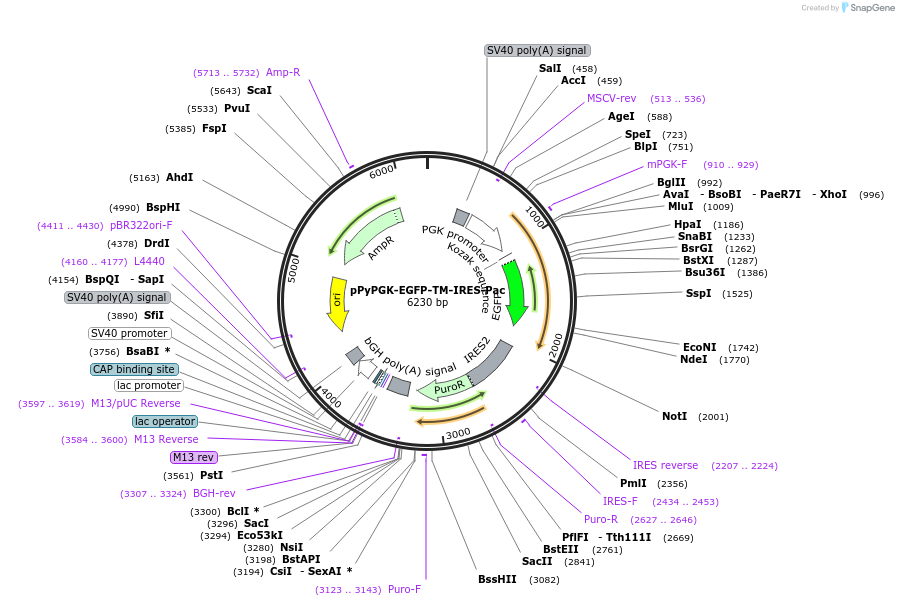
pPyPGK-EGFP-TM-IRES-Pac
Plasmid#183604PurposeMammalian expression vector for membrane-tethered EGFP from the mouse Pgk1 promoter. Confers puromycin resistance.DepositorInsertEGFP-TM
TagsHuman PDGFRB transmembrane domain and Mouse IgGK …ExpressionMammalianPromoterMouse Pgk1Available SinceJune 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
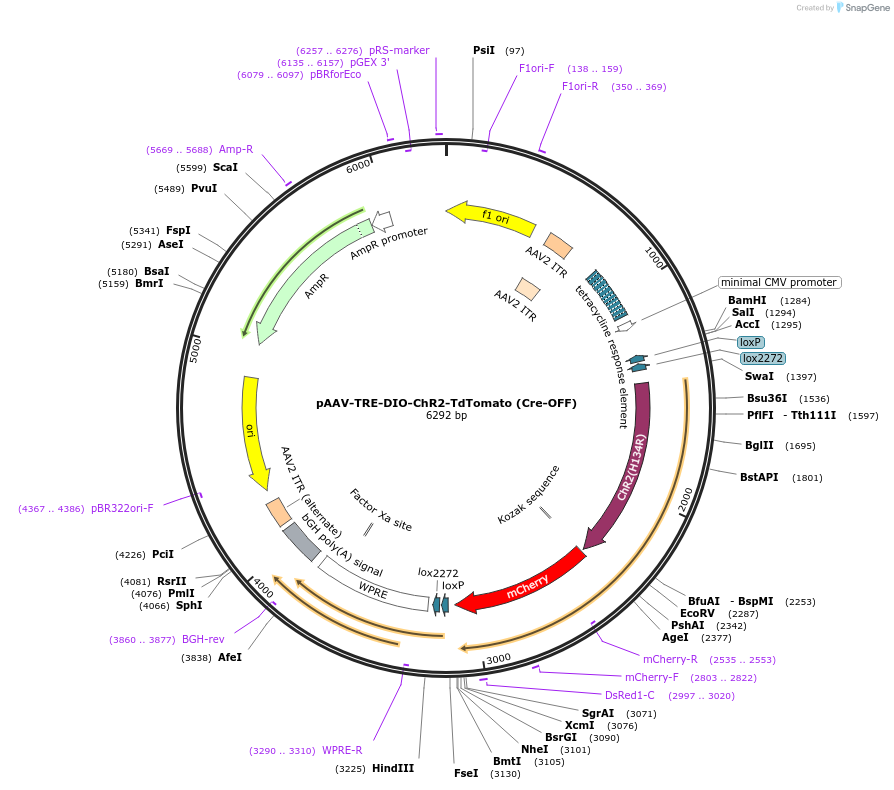
pAAV-TRE-DIO-ChR2-TdTomato (Cre-OFF)
Plasmid#166610PurposeEncodes Cre-inactivated ChR2-TdTomato under control of the TREDepositorInsertChR2-TdTomato
UseAAVMutationH134RPromotertetracycline response elementAvailable SinceApril 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
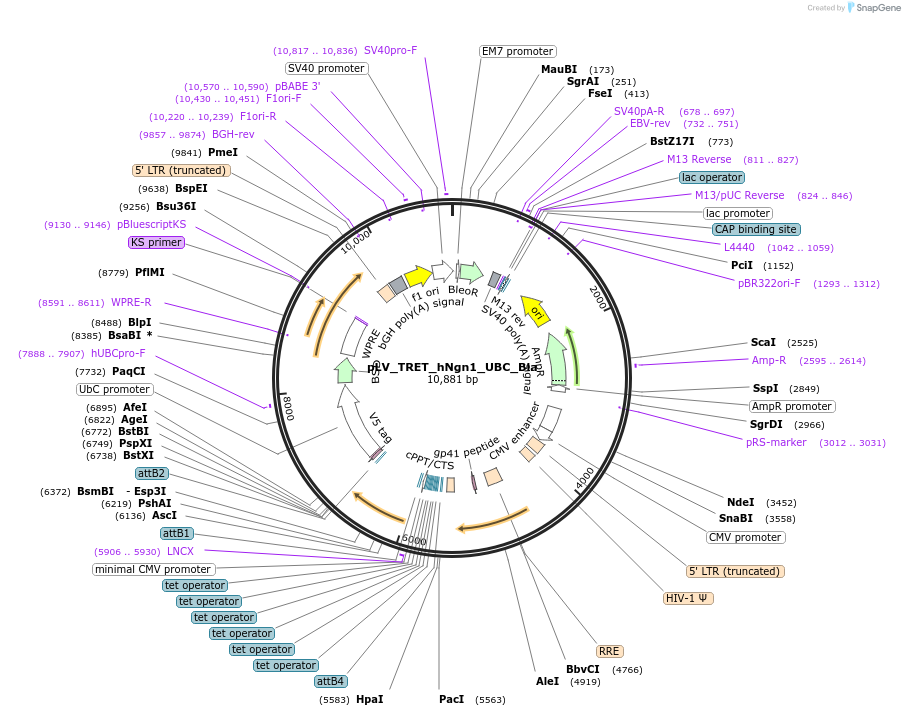
pLV_TRET_hNgn1_UBC_Bla
Plasmid#61473Purpose3rd generation lentiviral vector; TetON promoter driving human Ngn1; constitutive expression of Blasticidine selection markerDepositorInsertsUseLentiviralAvailable SinceFeb. 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
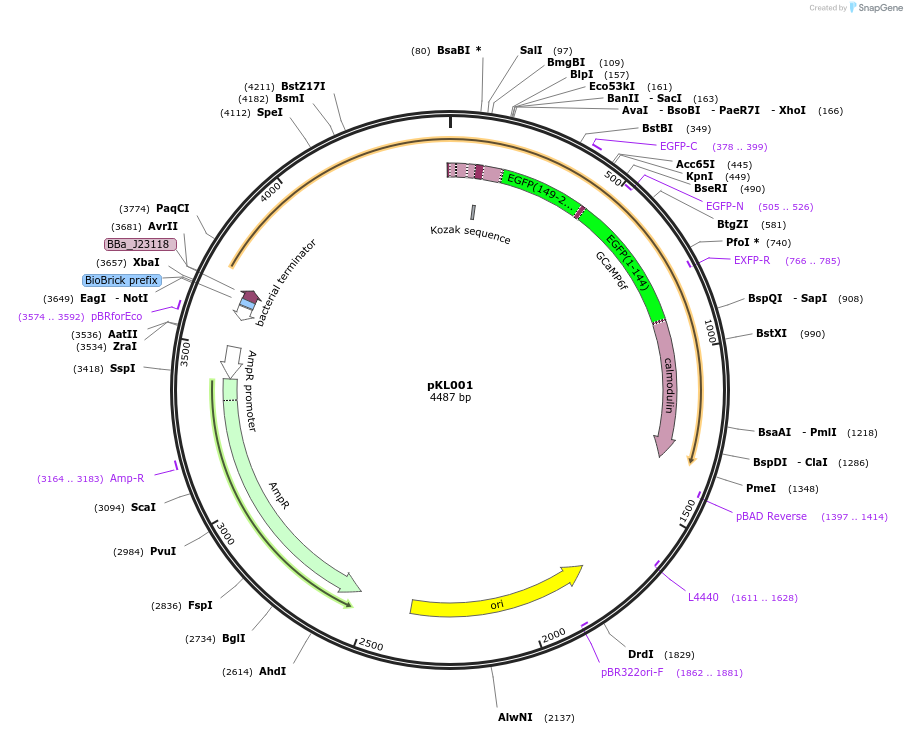
pKL001
Plasmid#98917PurposeConstitutively expresses the CaPR construct, the voltage indicator PROPS tethered to the calcium sensor GCaMP6f, under the Sigma 70 promoter in E. coli.DepositorInsertCaPR
ExpressionBacterialPromoterBBa_J23118 - Strong ConstitutiveAvailable SinceAug. 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
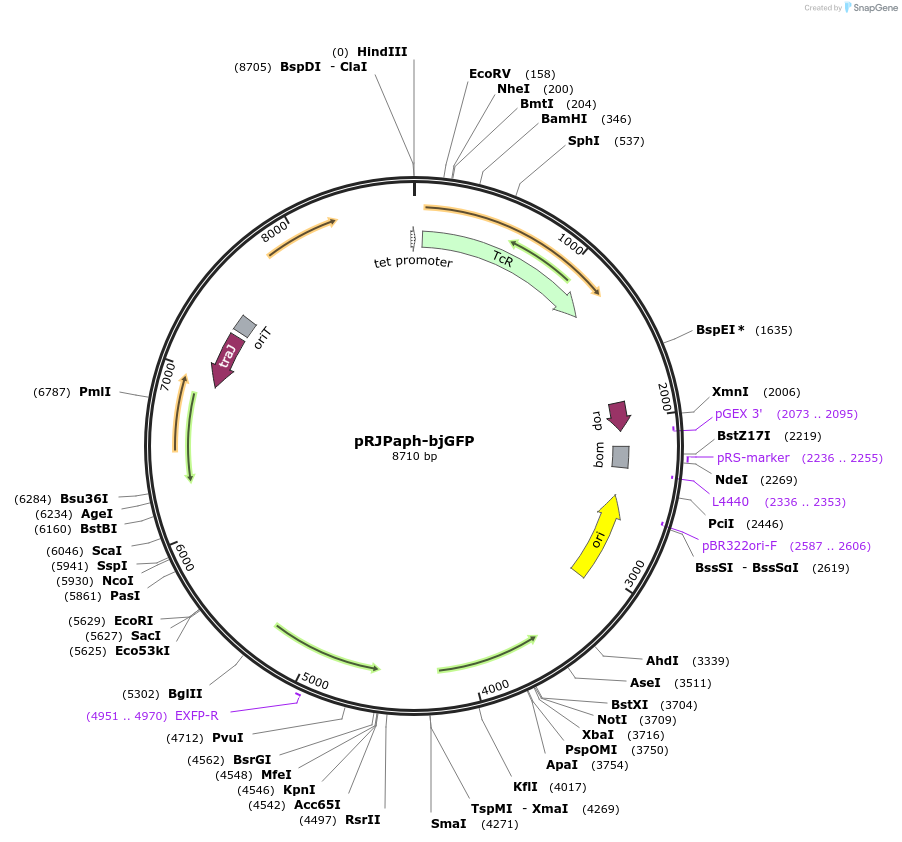
pRJPaph-bjGFP
Plasmid#67032PurposePlasmid for Paph-bjGFP integration downstream of B. diazoefficiens (B. japonicum) scoIDepositorInsertsB. diazoefficiens DNA from scoI downstream region (comprising blr1132')
bjGFP
UseConjugative bacterial vectorAvailable SinceSept. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
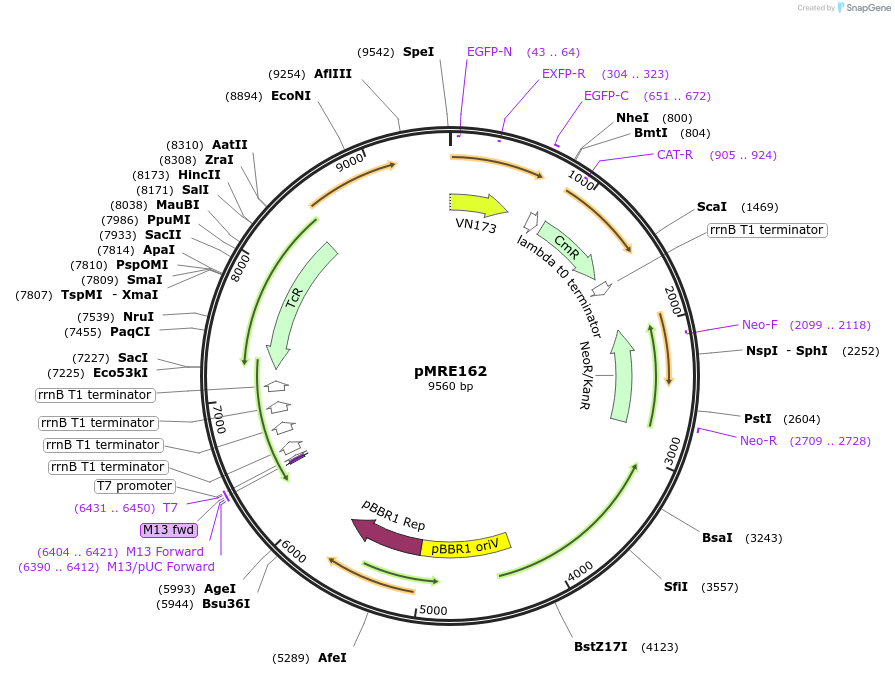
pMRE162
Plasmid#118510Purposebroad-host range plasmid to constitutively express fluorescent protein genes in bacteriaDepositorInsertsGFP2
ExpressionBacterialAvailable SinceOct. 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
mTagBFP2-CD81-10
Plasmid#55281PurposeLocalization: Tetraspanin/Membrane, Excitation: 399, Emission: 456DepositorAvailable SinceOct. 10, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
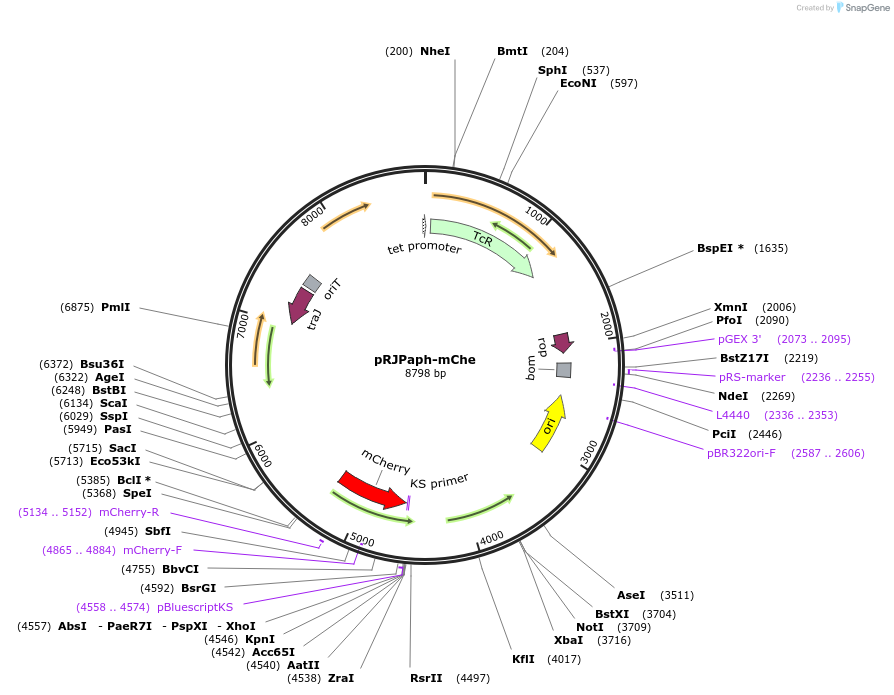
pRJPaph-mChe
Plasmid#67034PurposePlasmid for Paph-mCherry integration downstream of B. diazoefficiens (B. japonicum) scoIDepositorInsertsB. diazoefficiens DNA from scoI downstream region (comprising blr1132')
mCherry
UseConjugative bacterial vectorAvailable SinceOct. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
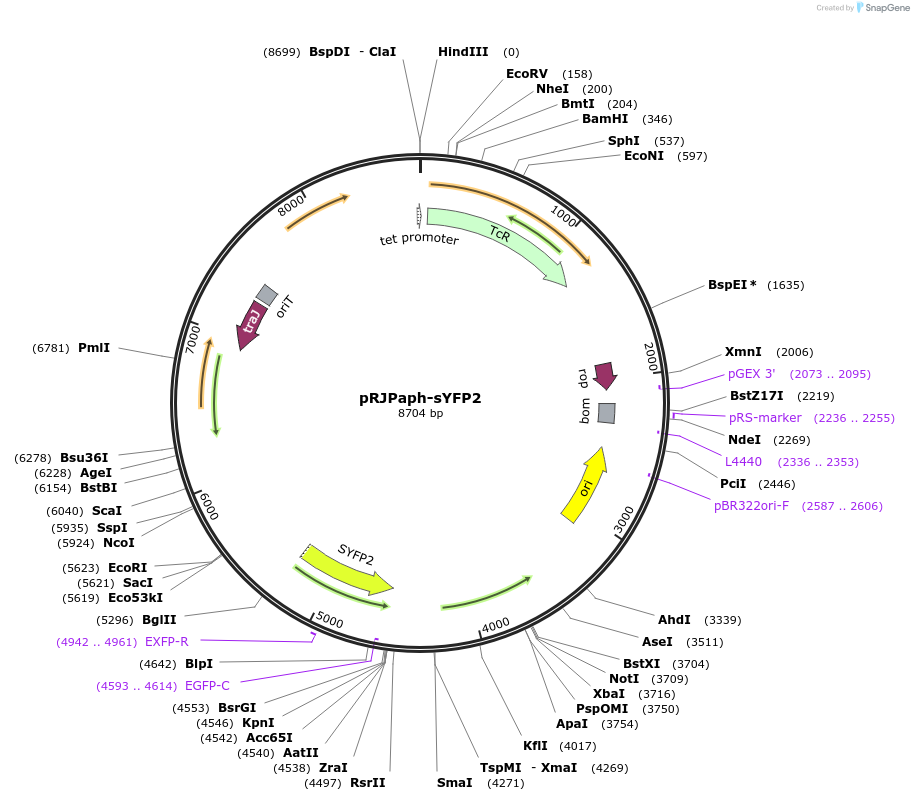
pRJPaph-sYFP2
Plasmid#67033PurposePlasmid for Paph-sYFP2 integration downstream of B. diazoefficiens (B. japonicum) scoIDepositorInsertsB. diazoefficiens DNA from scoI downstream region (comprising blr1132')
sYFP2
UseConjugative bacterial vectorAvailable SinceOct. 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
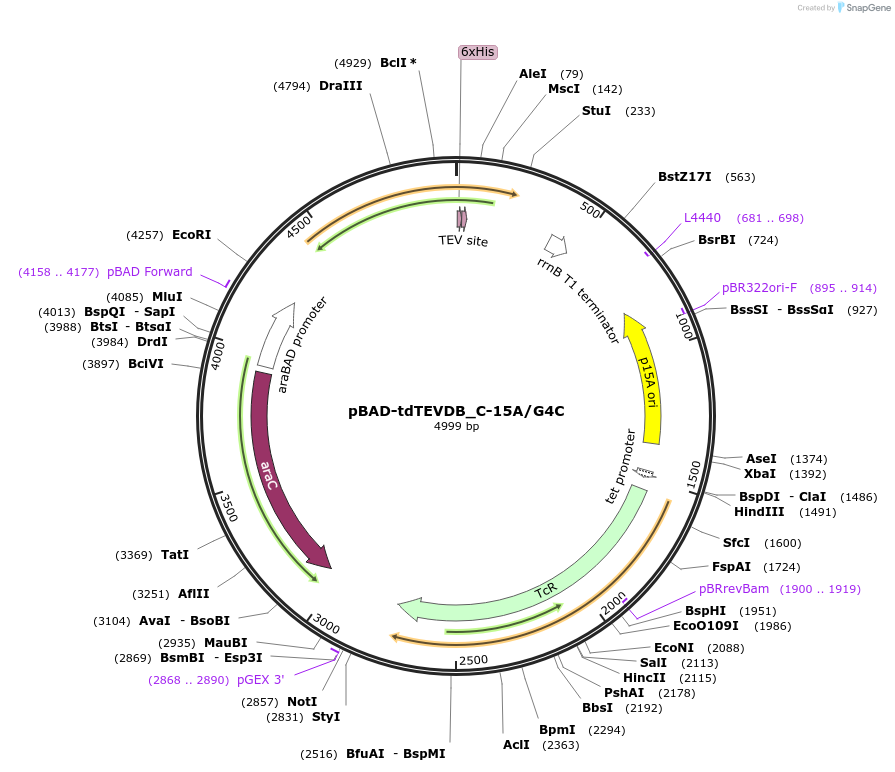
pBAD-tdTEVDB_C-15A/G4C
Plasmid#177205PurposePermuted td intron with evolved initiation sequence for the expression of circular mRNADepositorInsertPermuted GFP with evolved initiation sequence for circular mRNA
Tags6x HisExpressionBacterialMutationPermuted sfGFP after residue 52 & Changed Cyt…PromoteraraBADAvailable SinceSept. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
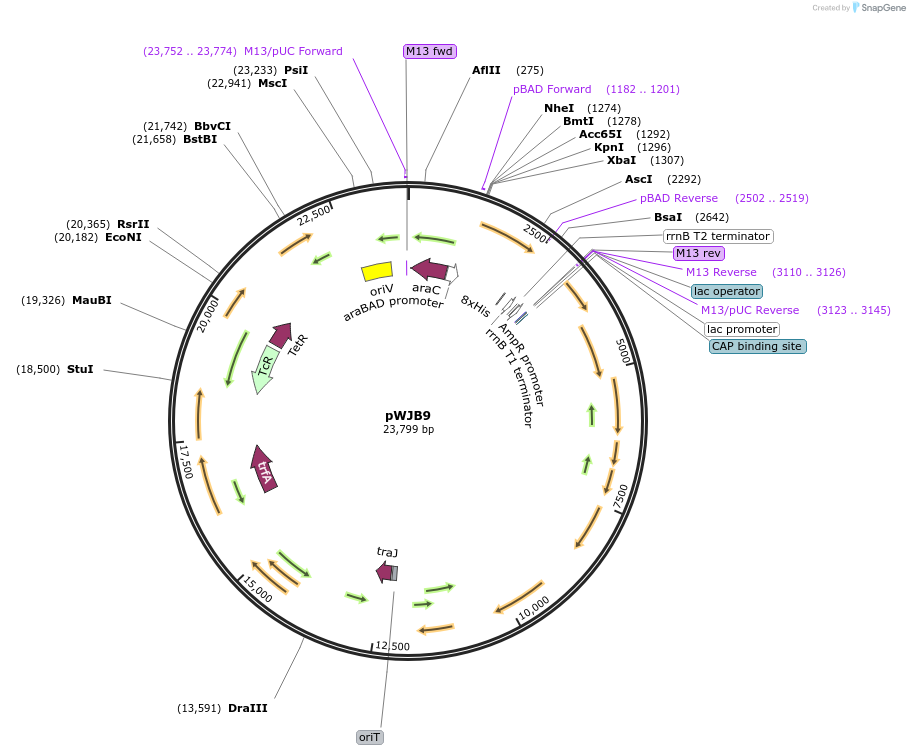
pWJB9
Plasmid#126650PurposeExpresses AdrA from a modified pLAFR3DepositorInsertadrA-8xHis/AdrA-8xHis including flanking promoter regions from pBAD30
Tags8xHisExpressionBacterialPromoterPBADAvailable SinceAug. 15, 2019AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1 3xFlag IFIT5
Plasmid#53556Purposemammalian expression of IFIT5DepositorAvailable SinceMay 29, 2014AvailabilityAcademic Institutions and Nonprofits only -
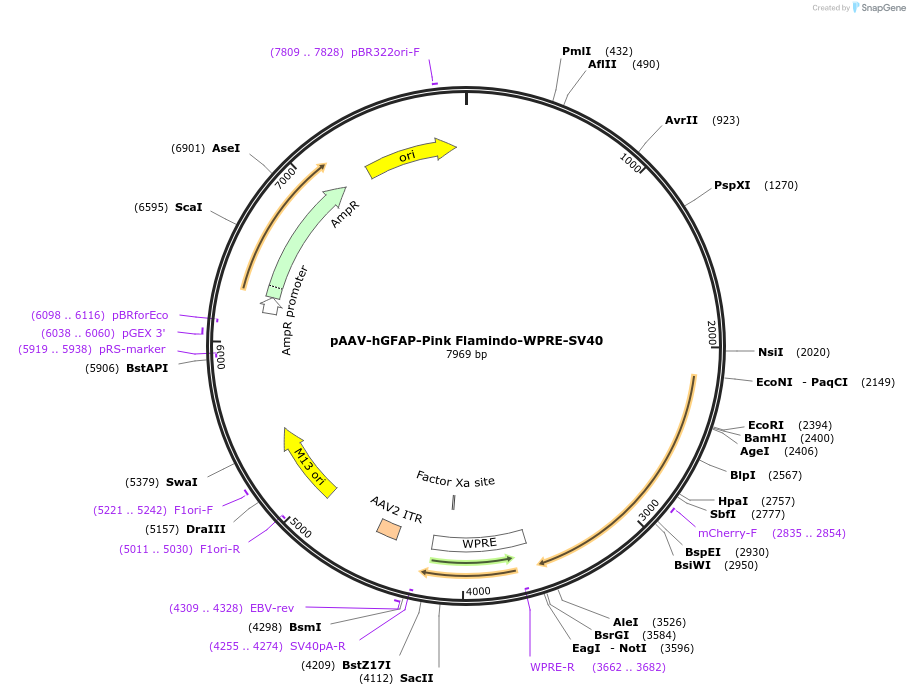
pAAV-hGFAP-Pink Flamindo-WPRE-SV40
Plasmid#178790PurposeAAV vector to express the red cAMP indicator in astrocytesDepositorInsertPink Flamindo
UseAAVPromoterhGFAPAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
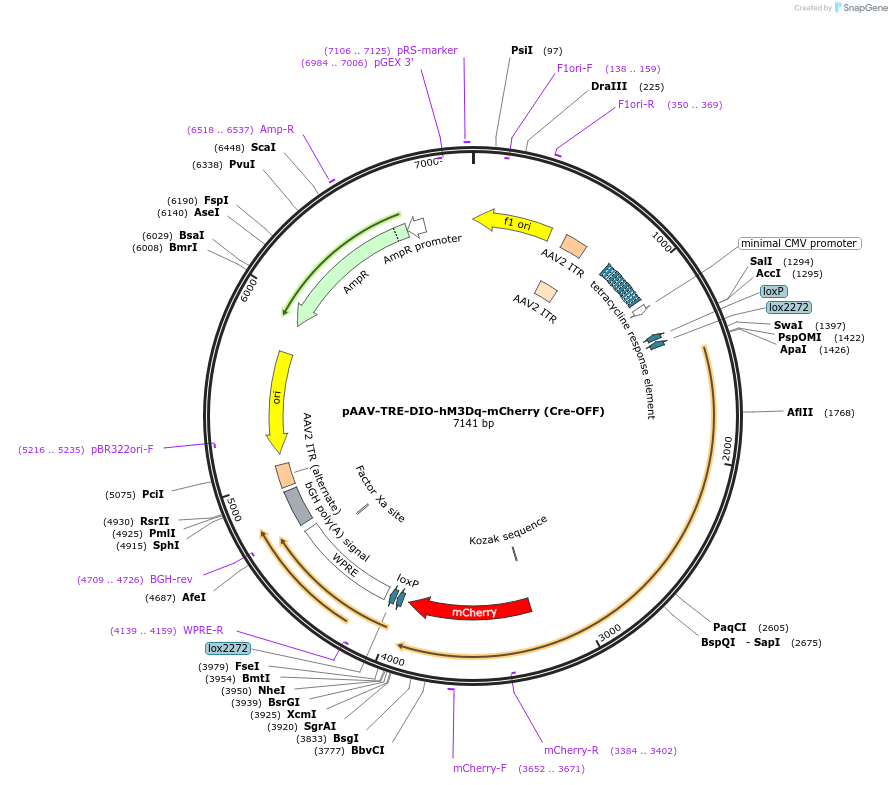
pAAV-TRE-DIO-hM3Dq-mCherry (Cre-OFF)
Plasmid#166609PurposeEncodes Cre-inactivated hM3Dq-mCherry under control of the TREDepositorInserthM3Dq-mCherry
UseAAVPromotertetracycline response elementAvailable SinceApril 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
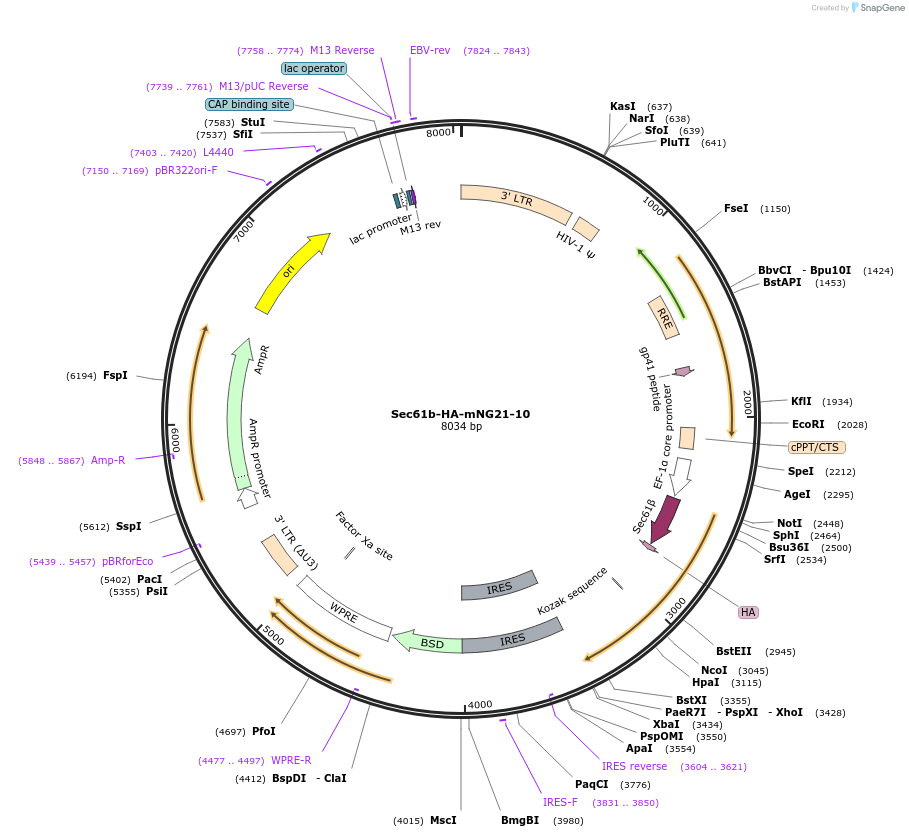
Sec61b-HA-mNG21-10
Plasmid#141213PurposeMembrane Tethered Split Fluorescent Reporter for ER RetragradeDepositorInsertSec61b-HA-mNG21-10
UseLentiviralTagsHAExpressionMammalianAvailable SinceJuly 31, 2020AvailabilityAcademic Institutions and Nonprofits only -
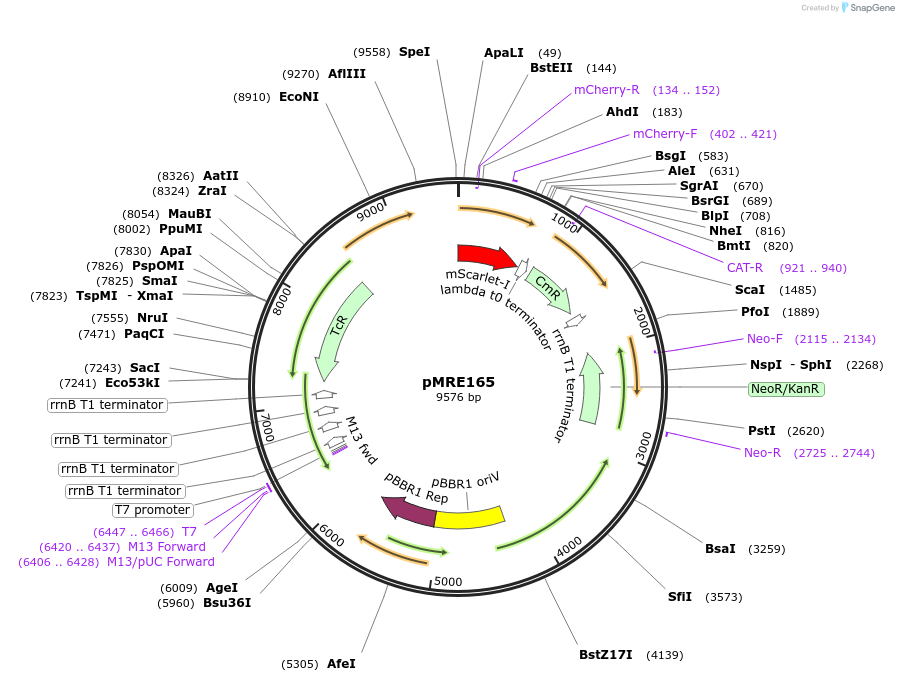
pMRE165
Plasmid#118513Purposebroad-host range plasmid to constitutively express fluorescent protein genes in bacteriaDepositorInsertmScarlet-I
ExpressionBacterialAvailable SinceOct. 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
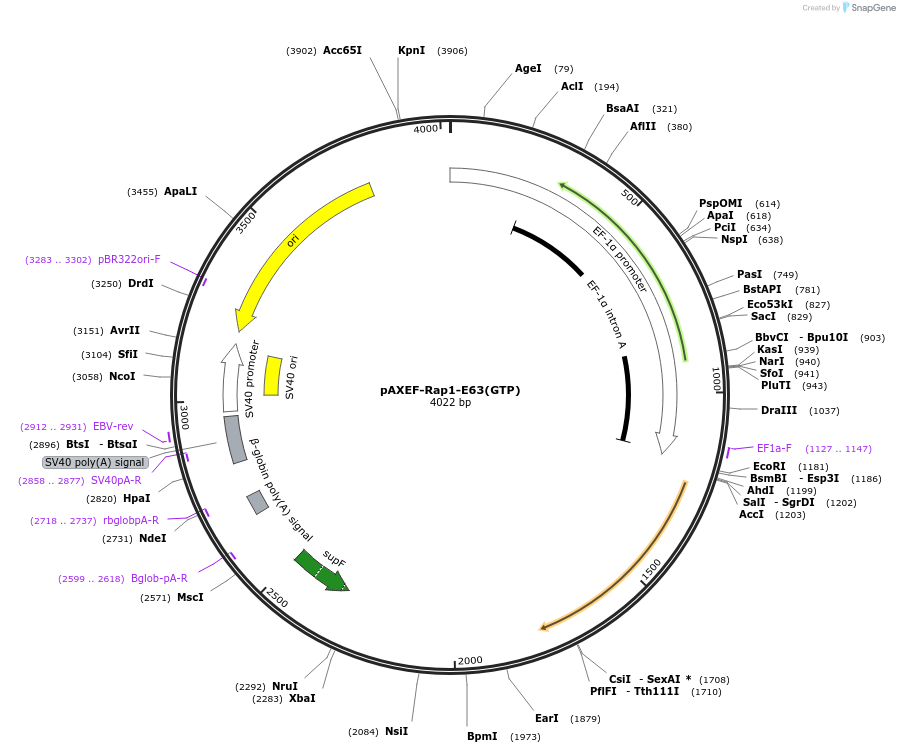
pAXEF-Rap1-E63(GTP)
Plasmid#32698DepositorInsertRAP1 (RAP1A Human)
ExpressionMammalianMutationE63: Consitutively GTP-boundPromoterEF promoterAvailable SinceDec. 5, 2011AvailabilityAcademic Institutions and Nonprofits only -
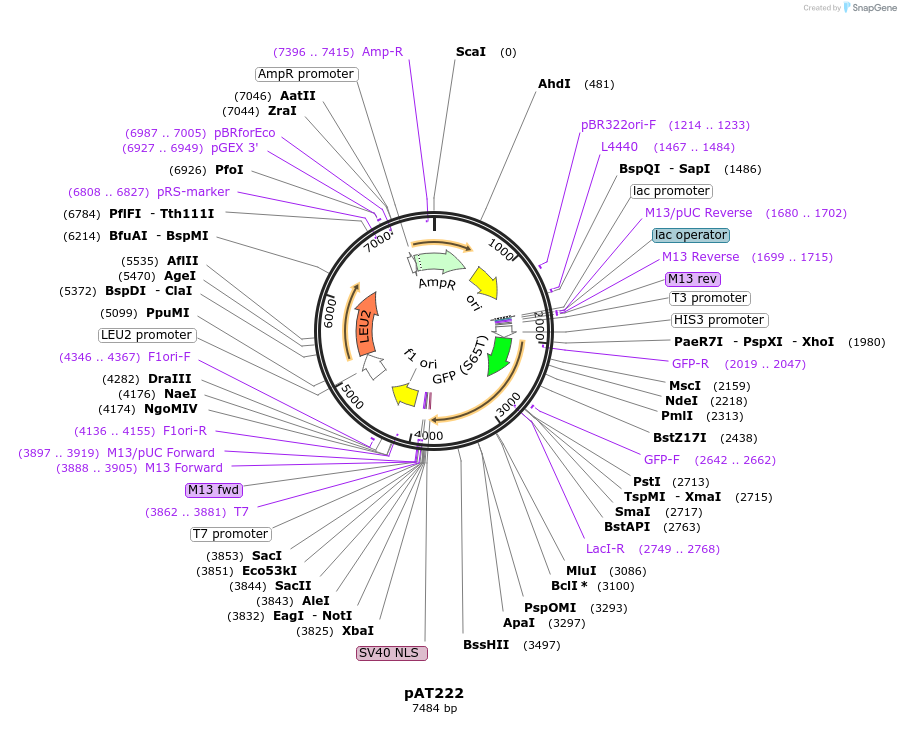
pAT222
Plasmid#40942DepositorInsertGFP-LacI**
TagsGFP(S65T, V163A) and SV40 NLSExpressionYeastMutationLacI**/LacI2G(4 mutations) (see comment below); l…PromoterHis3 promoter fragmentAvailable SinceFeb. 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
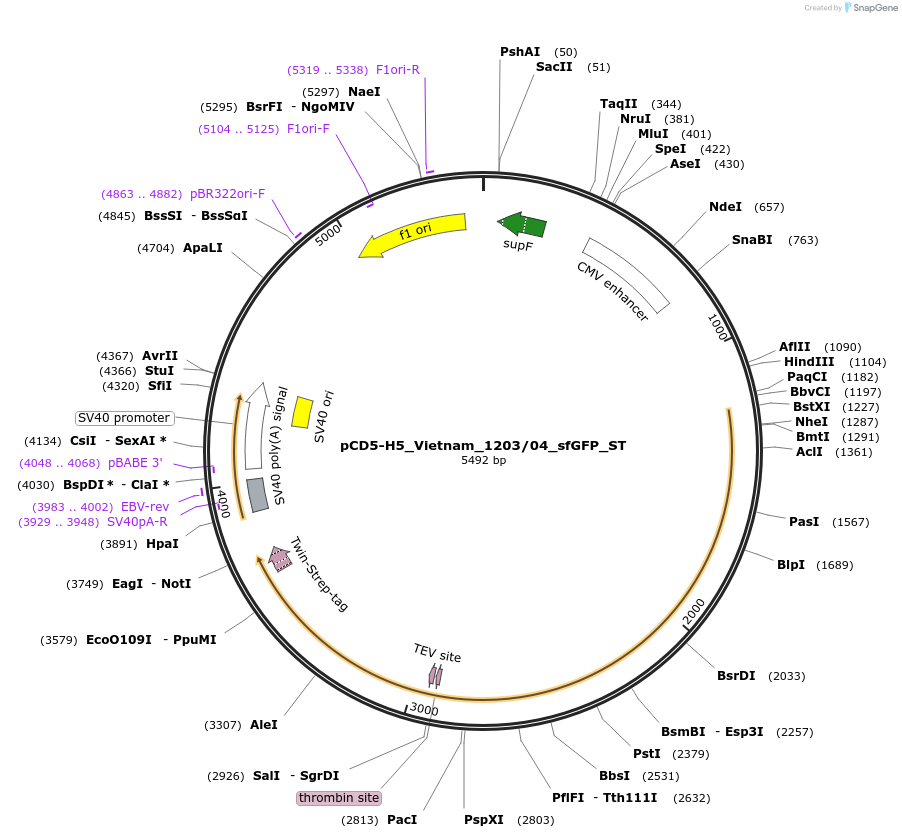
pCD5-H5_Vietnam_1203/04_sfGFP_ST
Plasmid#182546PurposeExpresses a sfGFP fused H5 protein of the prototype A/Vietnam/1203/04 virusDepositorInsertH5_Vietnam_1203_04
TagsGCN4-TEV-sfGFP-TwinStrepExpressionMammalianPromoterCMVAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
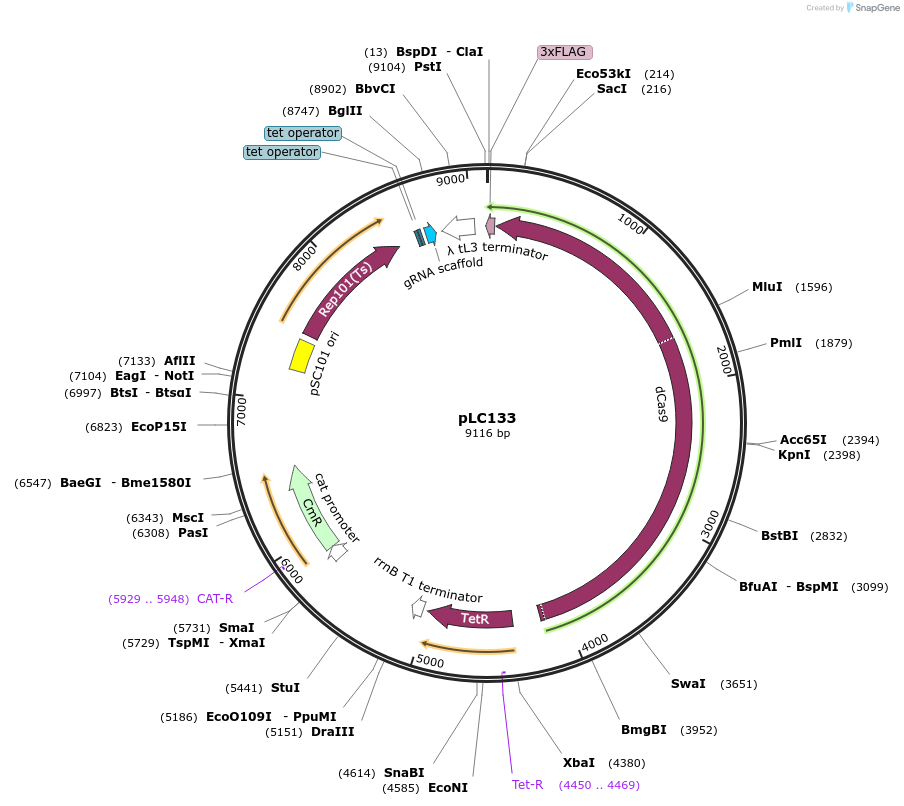
pLC133
Plasmid#198639PurposeExpression plasmid for dCas9 in E. coli used in Chip-seq experiments. dCas9 is tagged with a C-terminal 3x FLAG tag. A guide RNA can be cloned using BsaI restriction sites.DepositorInsertdCas9
Tags3x-FLAGExpressionBacterialPromoterpTetAvailable SinceApril 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
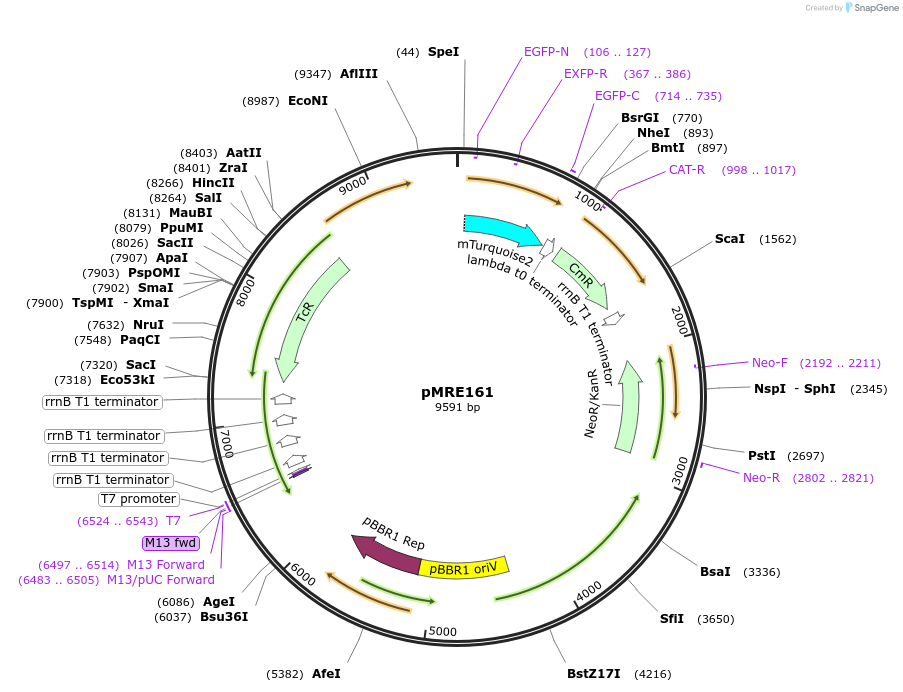
pMRE161
Plasmid#118509Purposebroad-host range plasmid to constitutively express fluorescent protein genes in bacteriaDepositorInsertmTurquoise2
ExpressionBacterialAvailable SinceNov. 19, 2018AvailabilityAcademic Institutions and Nonprofits only -
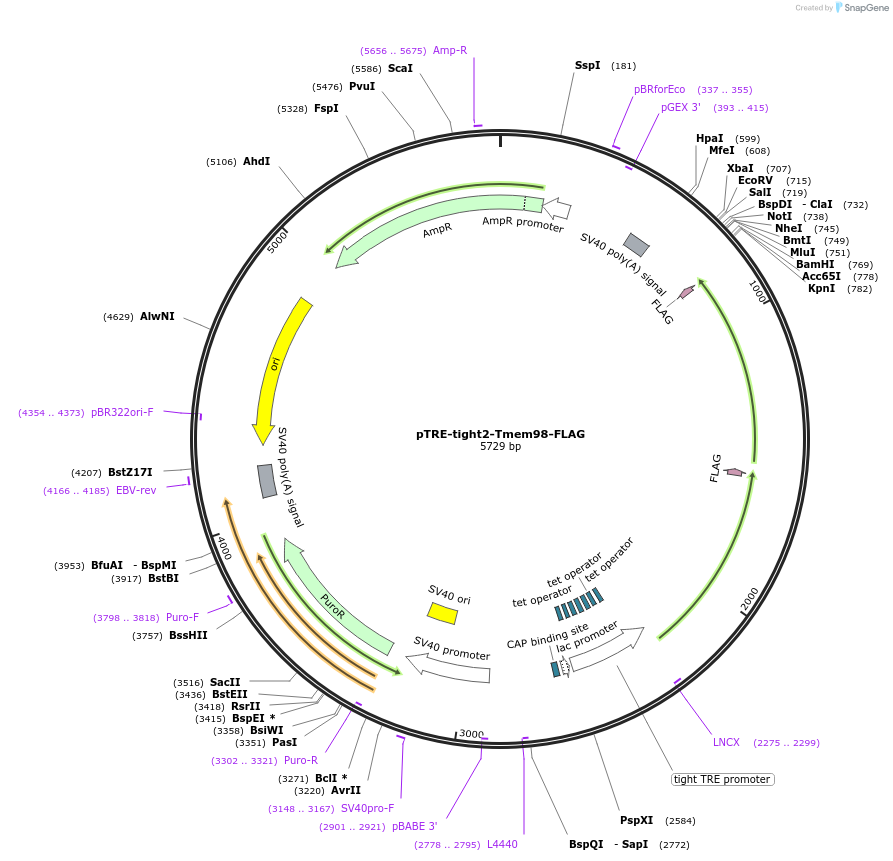
pTRE-tight2-Tmem98-FLAG
Plasmid#124506PurposeExpresses FLAG-tagged mouse TMEM98 in mammalian cells in a doxycycline-dependent manner (C-terminal tag)DepositorAvailable SinceMay 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
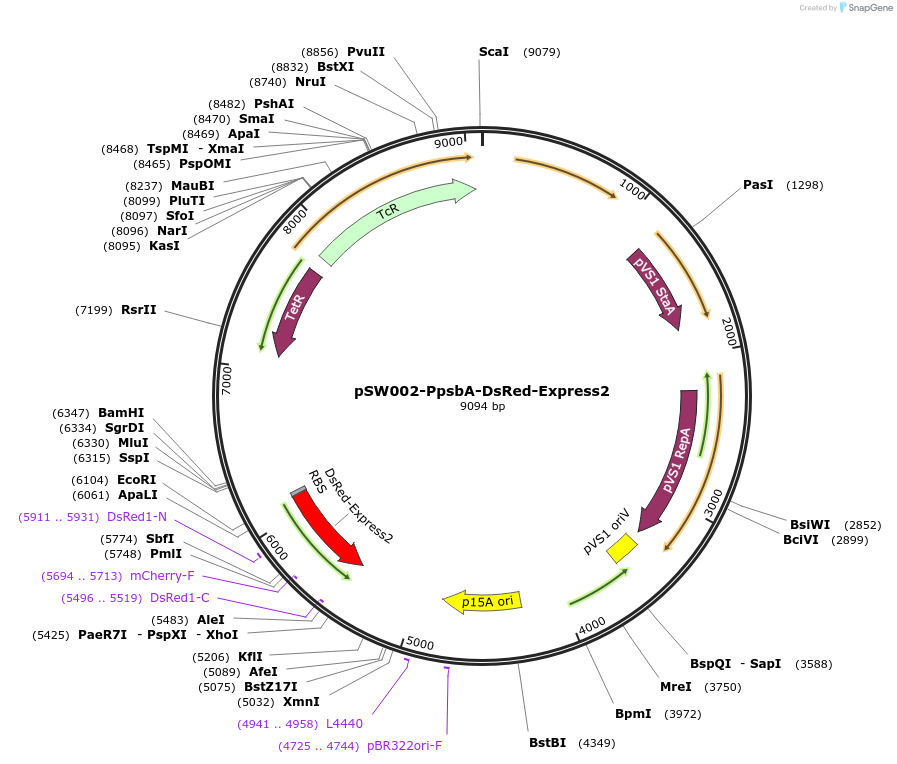
pSW002-PpsbA-DsRed-Express2
Plasmid#111257PurposeBroad host-range bacterial expression vector with constitutive PsbA promoter; for tagging bacteria with DsRed-Express2DepositorInsertDsRed-Express2
ExpressionBacterialPromoterPpsbAAvailable SinceAug. 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
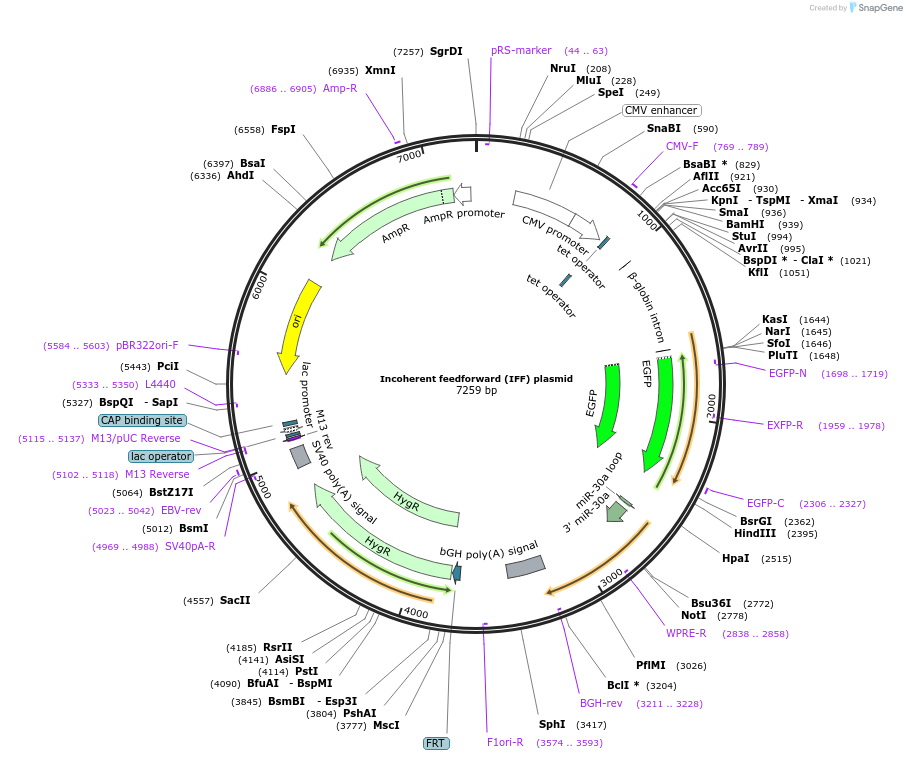
Incoherent feedforward (IFF) plasmid
Plasmid#169748PurposeStandalone IFF circuit plasmid that has the same implementation of the IFF subcircuit in Equalizer-L.DepositorInsertmiR(FF4) target-eGFP-miR(FF4) target-miR(FF4)
UseSynthetic BiologyExpressionMammalianPromoterCMV-tetO2Available SinceJune 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
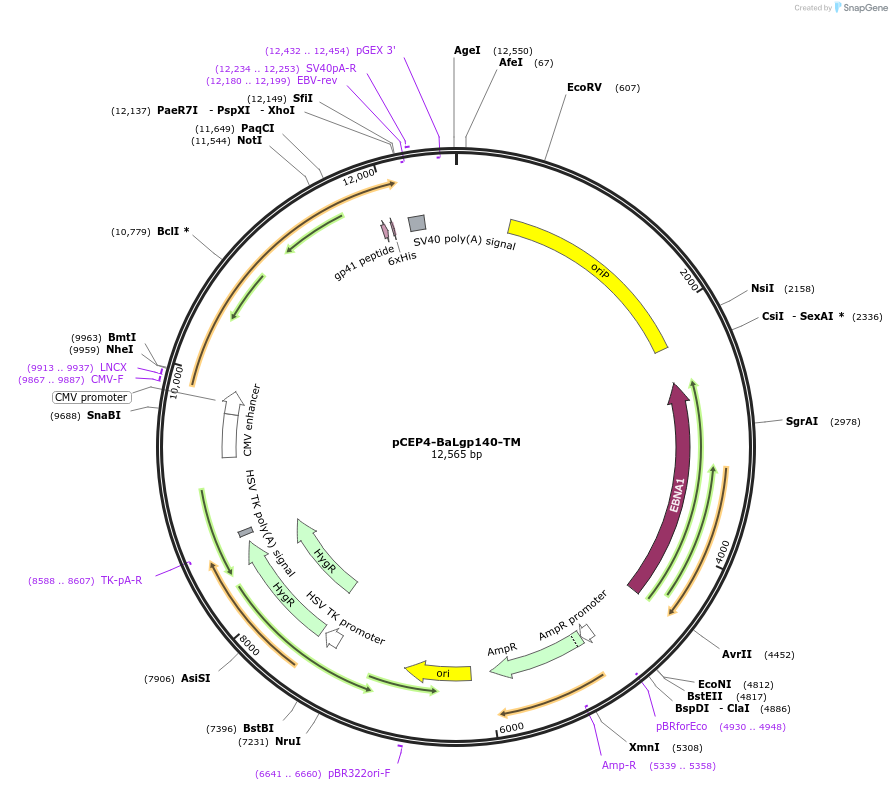
pCEP4-BaLgp140-TM
Plasmid#111843PurposeMammalian expression plasmid for extracellular gp140 from the BaL HIV-1 strain fused to a transmembrane tetherDepositorInsertHIV-1 (BaL) Env
TagsCD5 leader sequence and TM helix from MHC class IExpressionMammalianMutationCodon-optimized synthetic genePromoterCMVAvailable SinceMarch 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
pET28a(+) 6xHis-3xFlag-IFIT5
Plasmid#53561Purposebacterial expression of IFIT5DepositorAvailable SinceMay 29, 2014AvailabilityAcademic Institutions and Nonprofits only -
Dead Streptavidin-SpyTag
Plasmid#59548PurposepET21-DTag encodes Dead streptavidin (negligible biotin binding) bearing a C-terminal SpyTag, for generation of chimeric SpyAvidin tetramersDepositorInsertDead Streptavidin-SpyTag
TagsSpyTagExpressionBacterialMutationThe construct lacks final K of SpyTagPromoterT7Available SinceOct. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
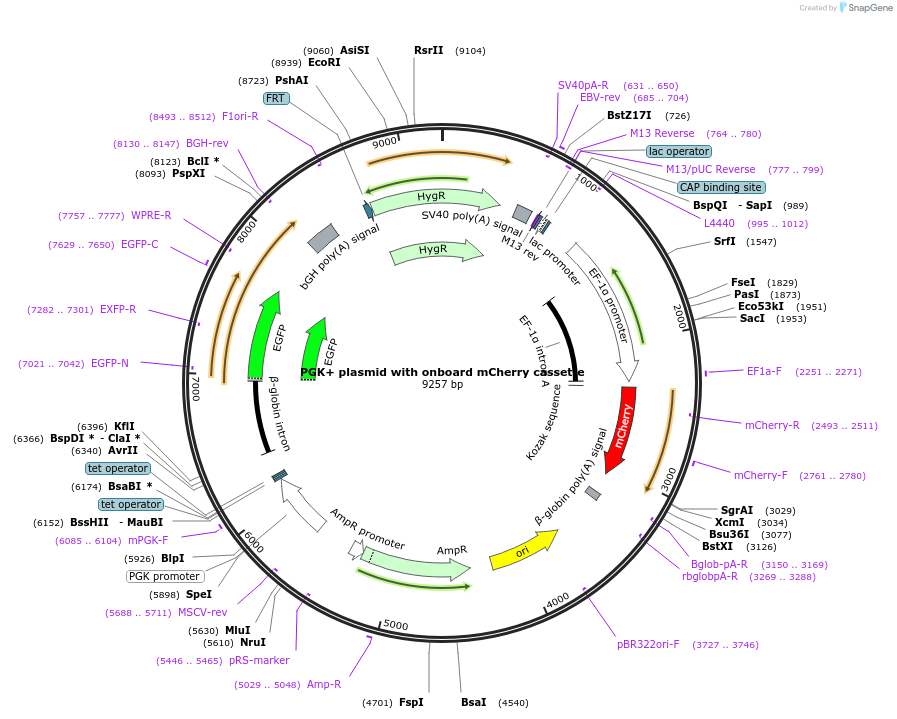
PGK+ plasmid with onboard mCherry cassette
Plasmid#169745PurposePGK+ plasmid that also encodes separate mCherry cassette. mCherry is used to monitor plasmid dosage.DepositorInserteGFP
UseSynthetic BiologyExpressionMammalianPromoterPGK-tetO2Available SinceJune 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
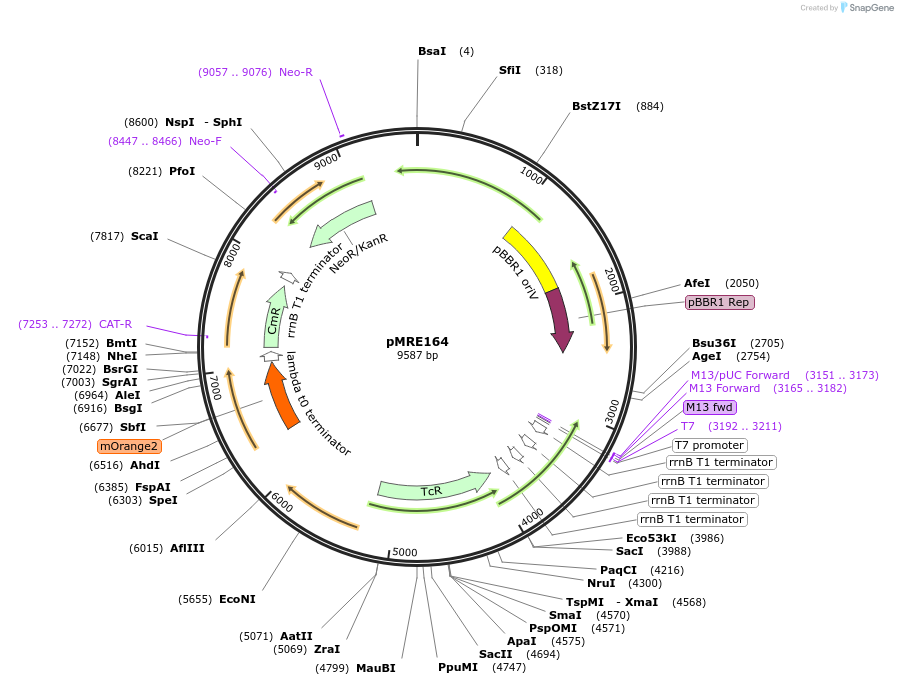
pMRE164
Plasmid#118512Purposebroad-host range plasmid to constitutively express fluorescent protein genes in bacteriaDepositorInsertmOrange2
ExpressionBacterialAvailable SinceOct. 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
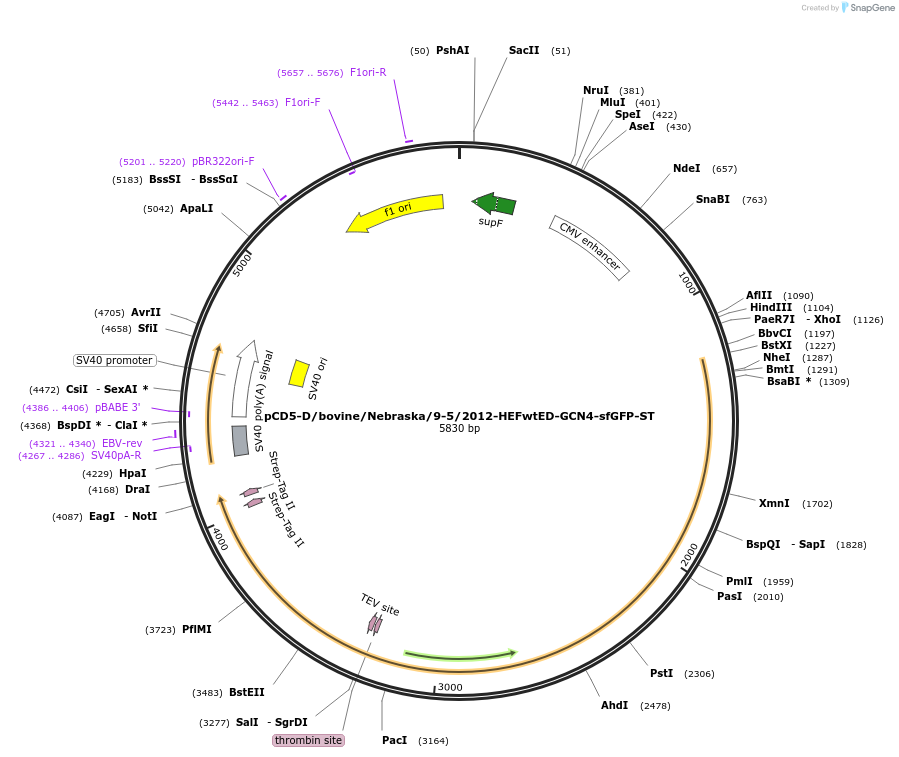
pCD5-D/bovine/Nebraska/9-5/2012-HEFwtED-GCN4-sfGFP-ST
Plasmid#175019PurposeExpresses influenza D virus trimeric hemagglutinin esterase fusion protein fused to a sfGFPDepositorInsertD/bovine/Nebraska/9-5/2012
TagsGCN4-TEV-sfGFP-TwinStrepExpressionMammalianPromoterCMVAvailable SinceJan. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
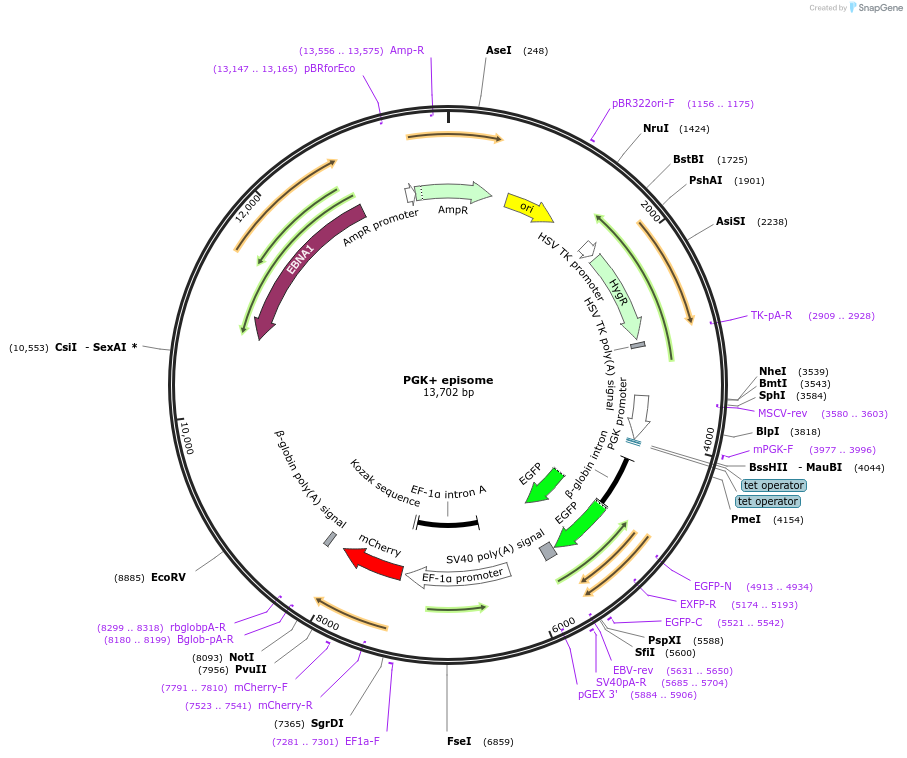
PGK+ episome
Plasmid#170041PurposePGK+ circuit on an episomal vector. This plasmid encodes eGFP to report circuit output levels. This plasmid also encodes a separate mCherry expression cassette to monitor plasmid dosage.DepositorInserteGFP
UseSynthetic BiologyExpressionMammalianPromoterPGK-tetO2Available SinceOct. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
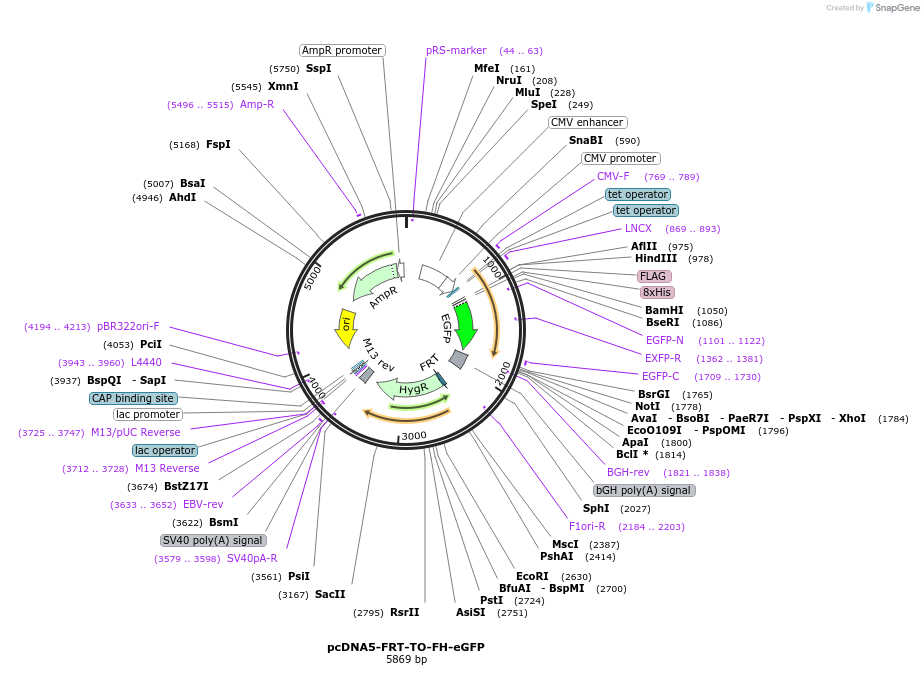
pcDNA5-FRT-TO-FH-eGFP
Plasmid#157734Purposemammalian expression of N-terminally Flag-His8 tagged eGFP under control of a tetracycline-inducible promoterDepositorInsertFH-eGFP
TagsFlag-His8ExpressionMammalianAvailable SinceAug. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
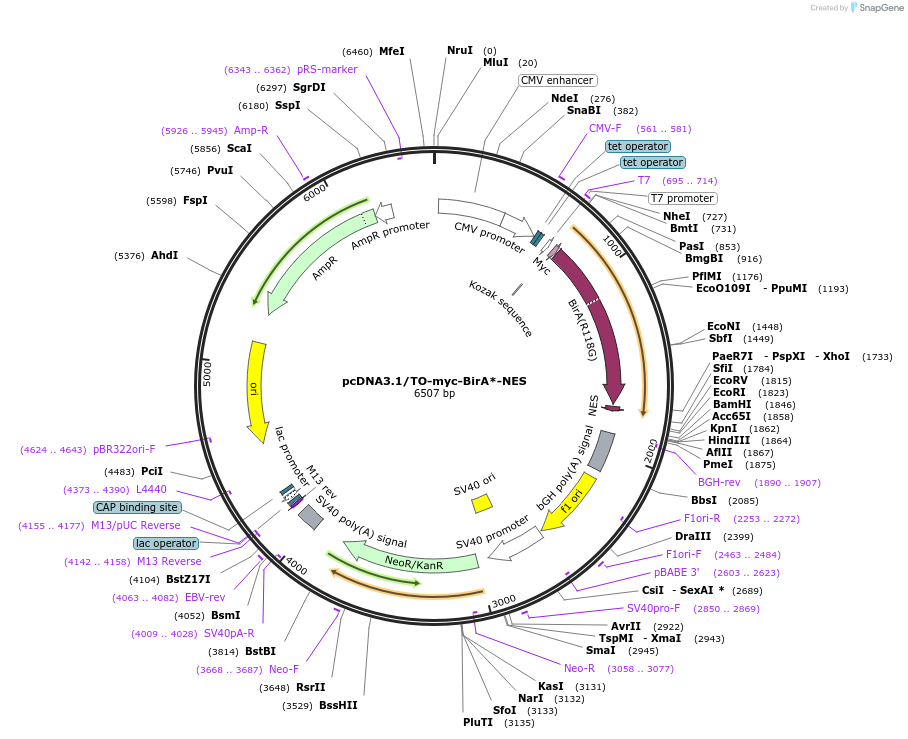
pcDNA3.1/TO-myc-BirA*-NES
Plasmid#112714PurposeFor tetracycline-inducible expression of myc-BirA*-NES in mammalian cells. For BioID studies.DepositorInsertHIV Rev nuclear export signal (NES)
ExpressionMammalianAvailable SinceJune 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
CMV+ episome
Plasmid#169742PurposeCMV+ circuit on an episomal vector. This plasmid encodes eGFP to report circuit output levels. This plasmid also encodes a separate mCherry expression cassette to monitor plasmid dosage.DepositorInserteGFP
UseSynthetic BiologyExpressionMammalianPromoterpCMV-tetO2Available SinceOct. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
pAluYNF1
Plasmid#50933PurposeExpresses dimeric AluY RNA in mammalian cells.DepositorInsertAluYNF1
ExpressionMammalianPromoter7SL RNA promoter (pol III)Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
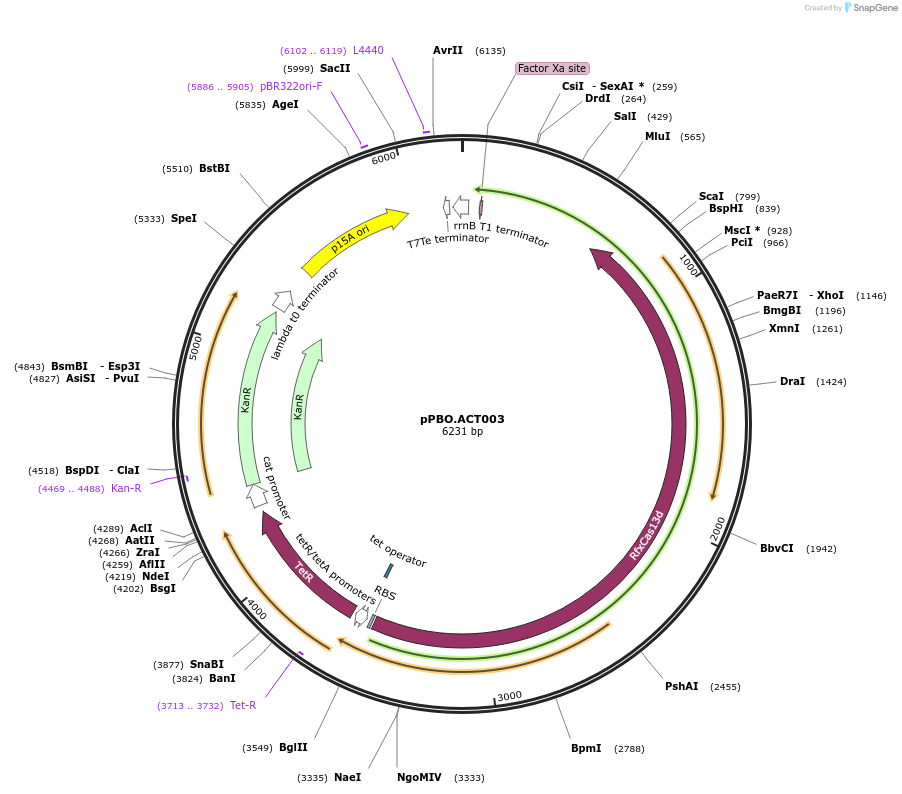
pPBO.ACT003
Plasmid#182713PurposeaTc inducible dCasRx-IF3DepositorInsertdCasRx
UseCRISPRTags3x(GGGS)-Escherichia Coli Initiation Factor 3ExpressionBacterialMutationCatalytically deactivated R295A, H300A, R849A, H8…PromoterpTetAvailable SinceAug. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
mEos2-CD9-10
Plasmid#57356PurposeLocalization: Tetraspanin/Membrane, Excitation: 505 / 569, Emission: 516 / 581DepositorAvailable SinceFeb. 5, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
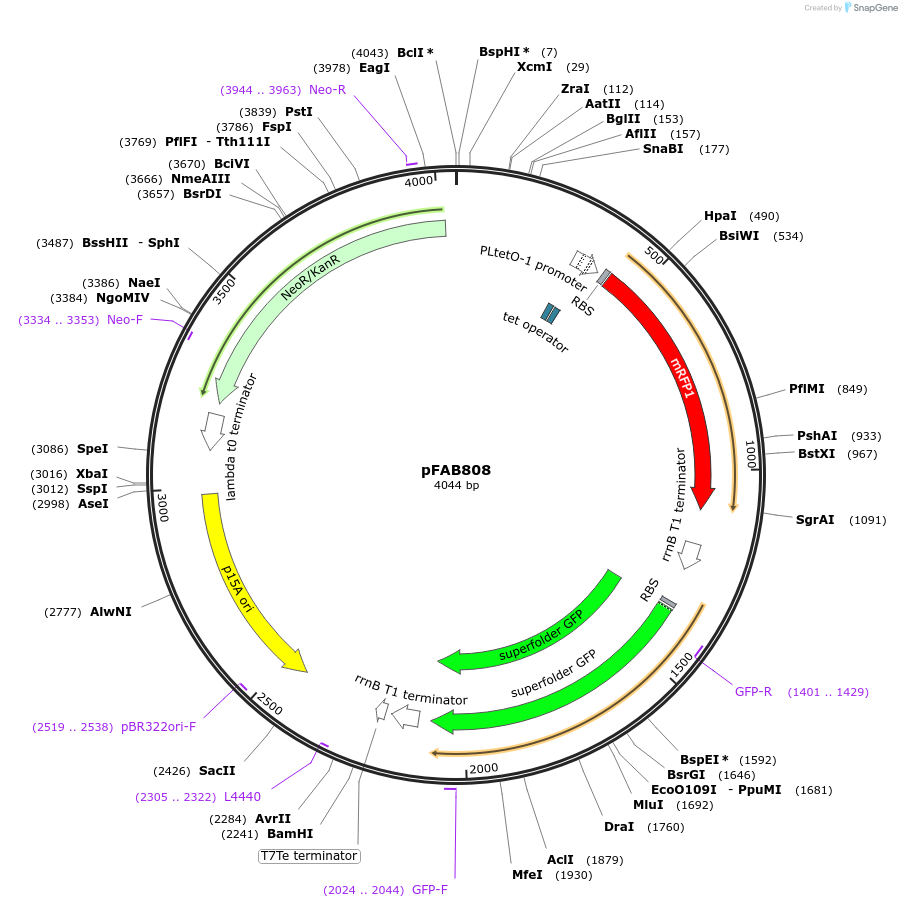
pFAB808
Plasmid#47853PurposeBIOFAB reporter plasmid for measuring rrnB T1 termination efficiencyDepositorInsertrrnB T1 terminator
UseSynthetic BiologyExpressionBacterialPromoterpLTetO8Available SinceDec. 3, 2013AvailabilityAcademic Institutions and Nonprofits only -
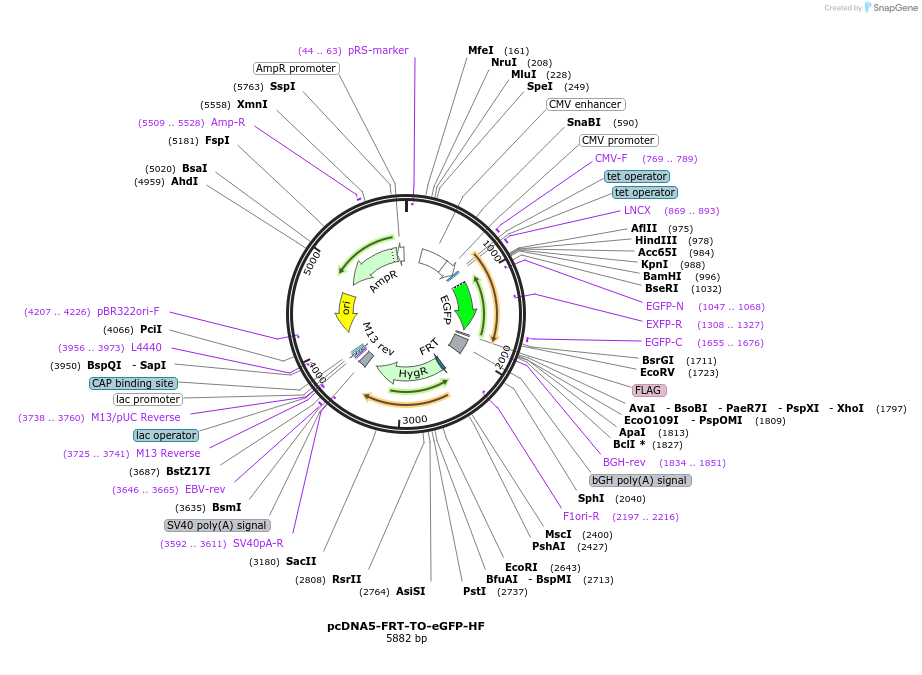
pcDNA5-FRT-TO-eGFP-HF
Plasmid#157735Purposemammalian expression of C-terminally His8-Flag tagged eGFP under control of a tetracycline-inducible promoterDepositorInserteGFP-HF
TagsHis8-FlagExpressionMammalianAvailable SinceAug. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
CMV+ plasmid
Plasmid#169740PurposeUnregulated "open-loop" CMV promoter plasmid. This plasmid encodes eGFP to report circuit output levels. This plasmid has additional sequences in the UTRs to increase gene expression levels.DepositorInserteGFP
UseSynthetic BiologyExpressionMammalianPromoterpCMV-tetO2Available SinceJune 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
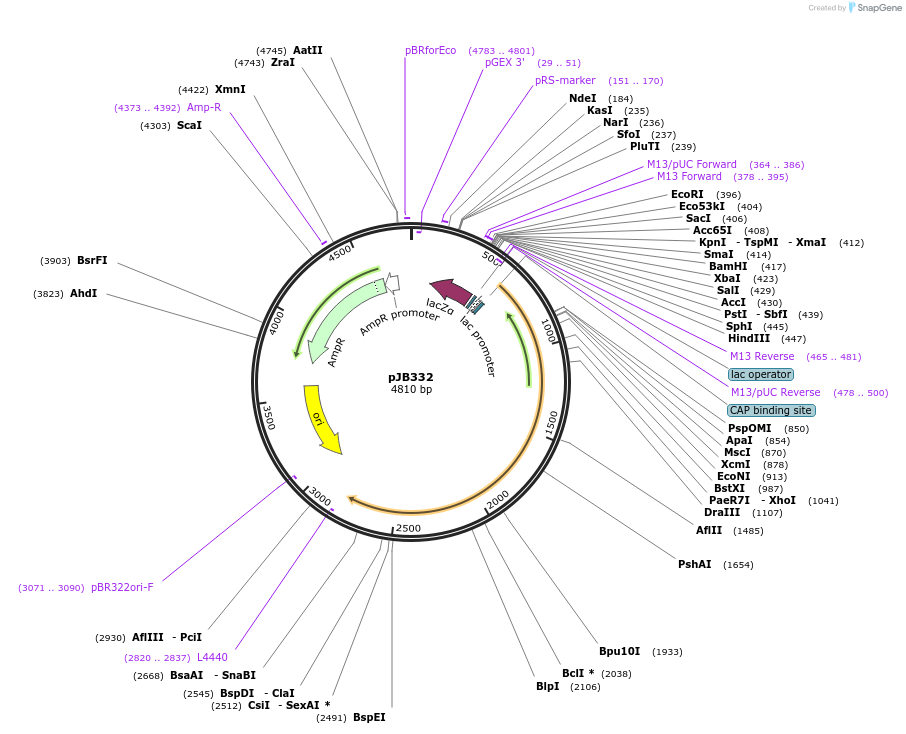
pJB332
Plasmid#115949PurposeORF insert for replicon MoCloDepositorInsertLvl0 TetR-Gly9-DDX6 (contains SapI)
UseSynthetic BiologyAvailable SinceNov. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
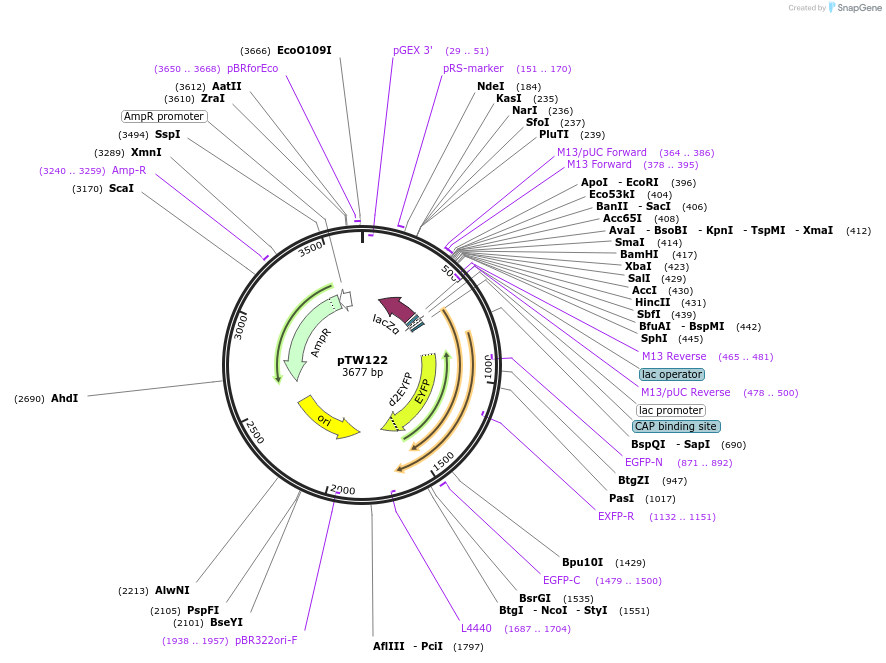
pTW122
Plasmid#115962PurposeORF insert for replicon MoCloDepositorInsertLvl0 5'-2xTetApt-mVenus-PEST
UseSynthetic BiologyAvailable SinceMay 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
mCHOP10.pCDNA1
Plasmid#21899DepositorInsertCHOP10 (Ddit3 Mouse)
ExpressionMammalianAvailable SinceSept. 10, 2009AvailabilityAcademic Institutions and Nonprofits only