171,694 results
-
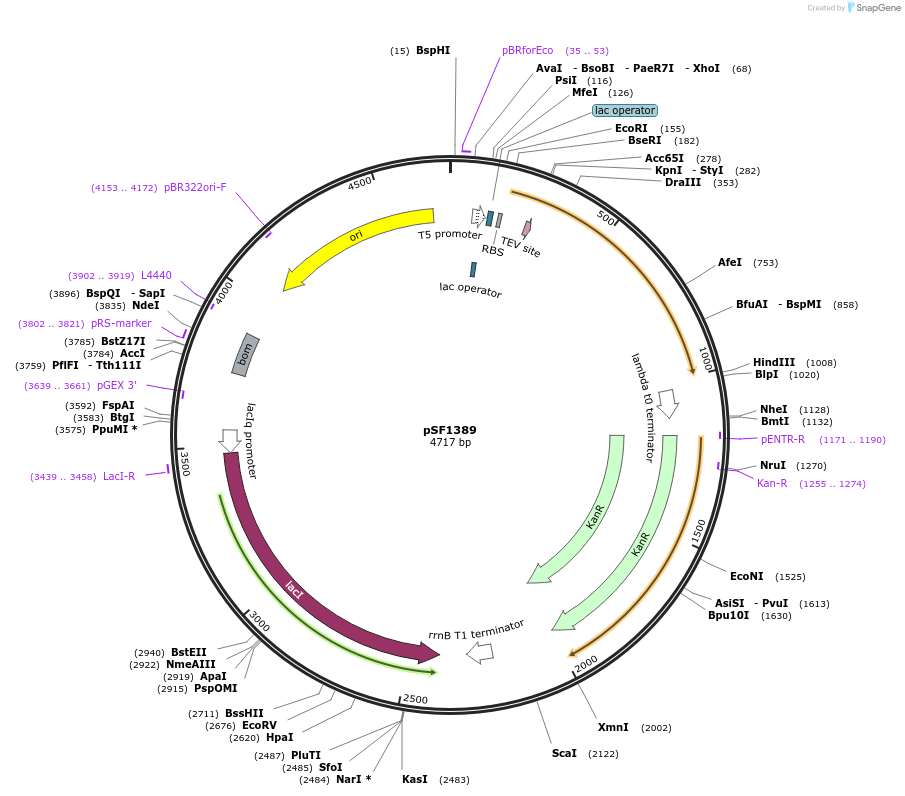
Plasmid#104962PurposeBacterial expression plasmid of bdSENP1 proteaseDepositorInsertSENP1
TagsHis14 tag and Tev protease cleavage siteExpressionBacterialPromoterlacUV5Available SinceJan. 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
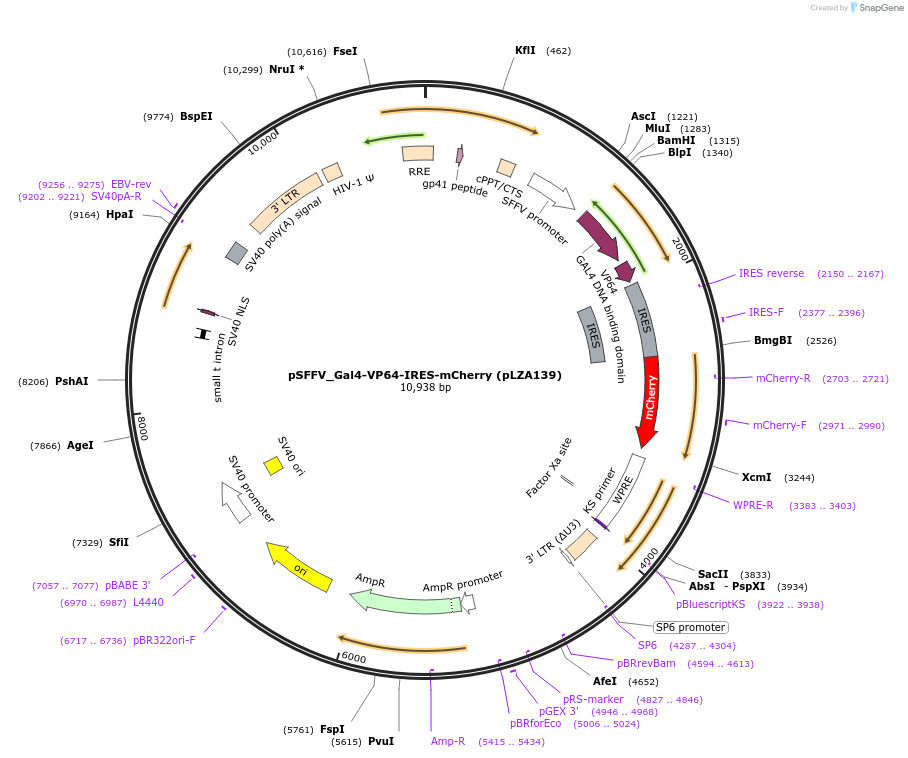
pSFFV_Gal4-VP64-IRES-mCherry (pLZA139)
Plasmid#203909PurposeEncodes wild type Gal4-VP64 transcription factor in mammalian cellsDepositorInsertGal4-VP64-IRES-mCherry
ExpressionMammalianPromoterSFFVAvailable SinceJuly 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
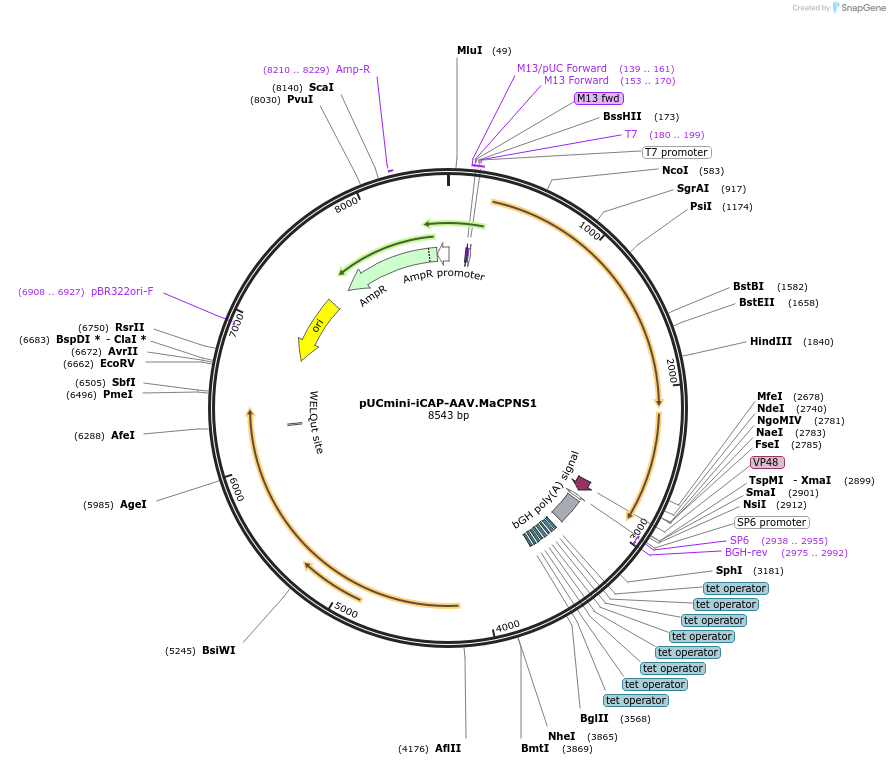
pUCmini-iCAP-AAV.MaCPNS1
Plasmid#185136Purposenon-standard AAV2 rep-AAV-MaCPNS1 cap plasmid with AAV cap expression controlled by a tTA-TRE amplification systemDepositorInsertSynthetic construct isolate AAV-MaCPNS1 VP1 gene
UseAAVExpressionMammalianPromoterp41Available SinceJune 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
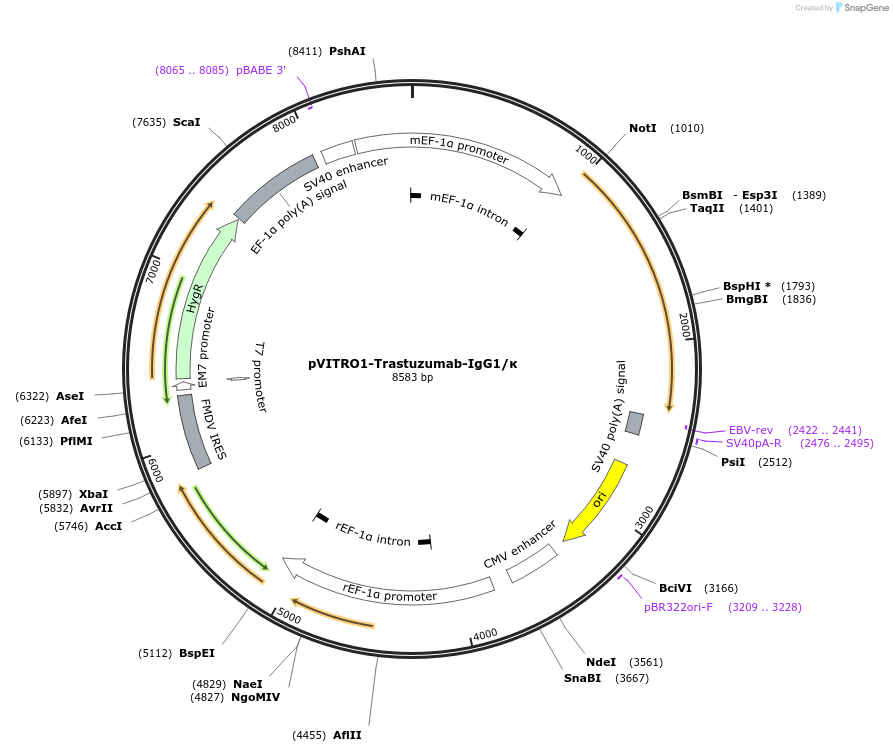
pVITRO1-Trastuzumab-IgG1/κ
Plasmid#61883PurposeExpresses HER2/neu receptor specific humanized IgG1/κ antibody isotype (Trastuzumab-IgG1/κ)DepositorInsertsHER2/neu receptor specific humanized Gamma 1 heavy chain expression cassette
HER2/neu receptor specific humanized kappa light chain expression cassette
ExpressionMammalianPromotermEF1 Prom and rEF1 PromAvailable SinceMarch 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
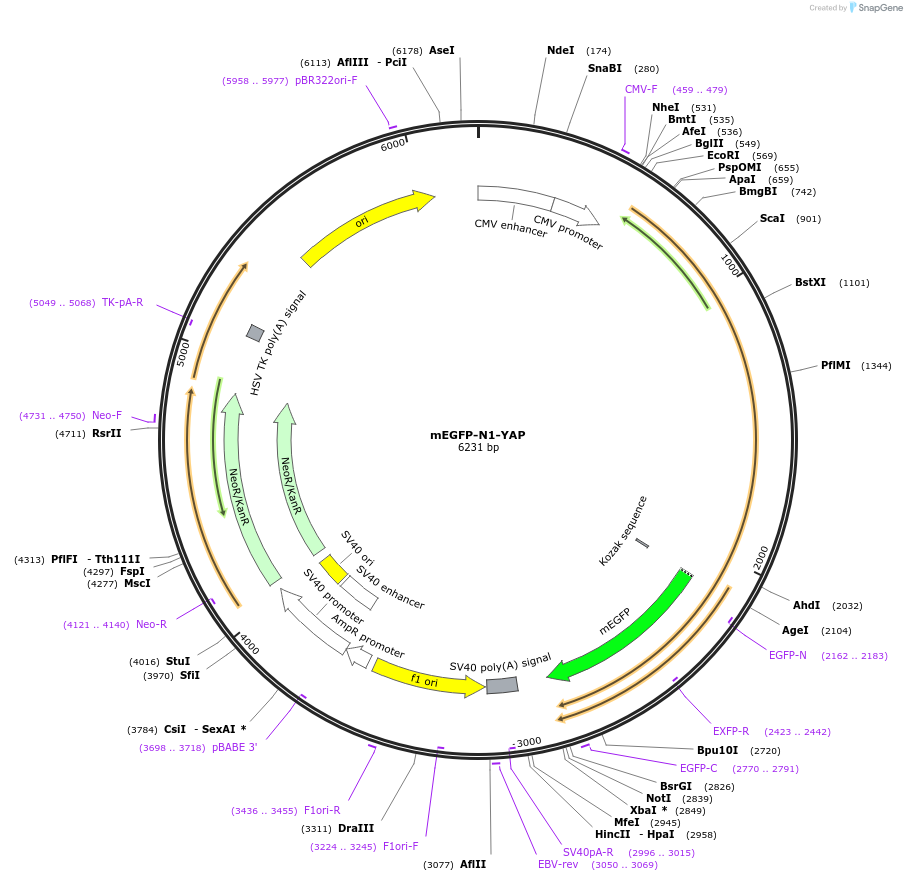
mEGFP-N1-YAP
Plasmid#166457PurposeExpresses fusion of mEGFP and YAPDepositorInsertmEGFP, YAP (YAP1 Human)
ExpressionMammalianAvailable SinceApril 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
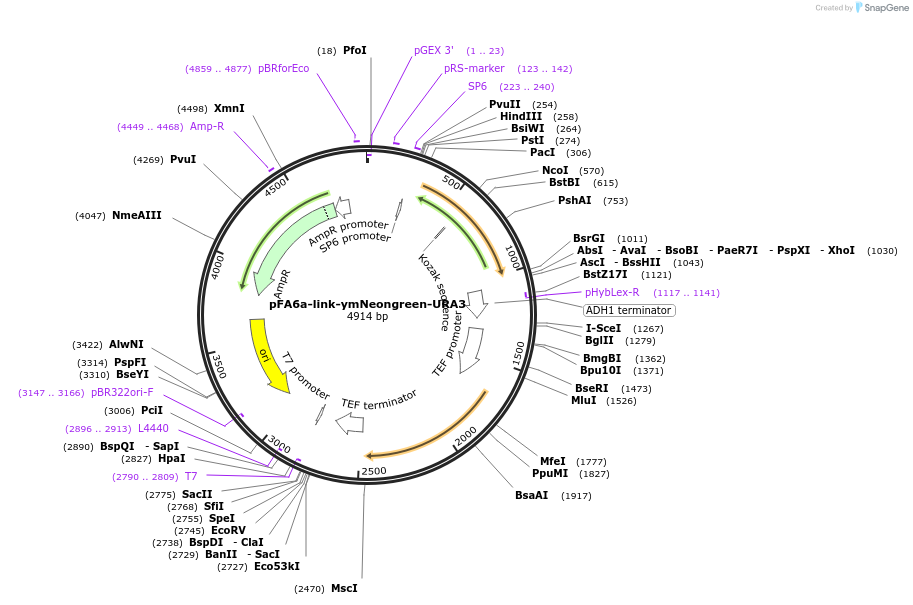
pFA6a-link-ymNeongreen-URA3
Plasmid#168059PurposeReplaces Addgene Plasmid 125703. Yeast tagging vector to create a gene fusion with yeast optimized mNeongreen with CaUra3 selection; Contains the linker region at the 5’ end of the FPDepositorInsertymNeonGreen
TagsymNeonGreenExpressionYeastAvailable SinceJune 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
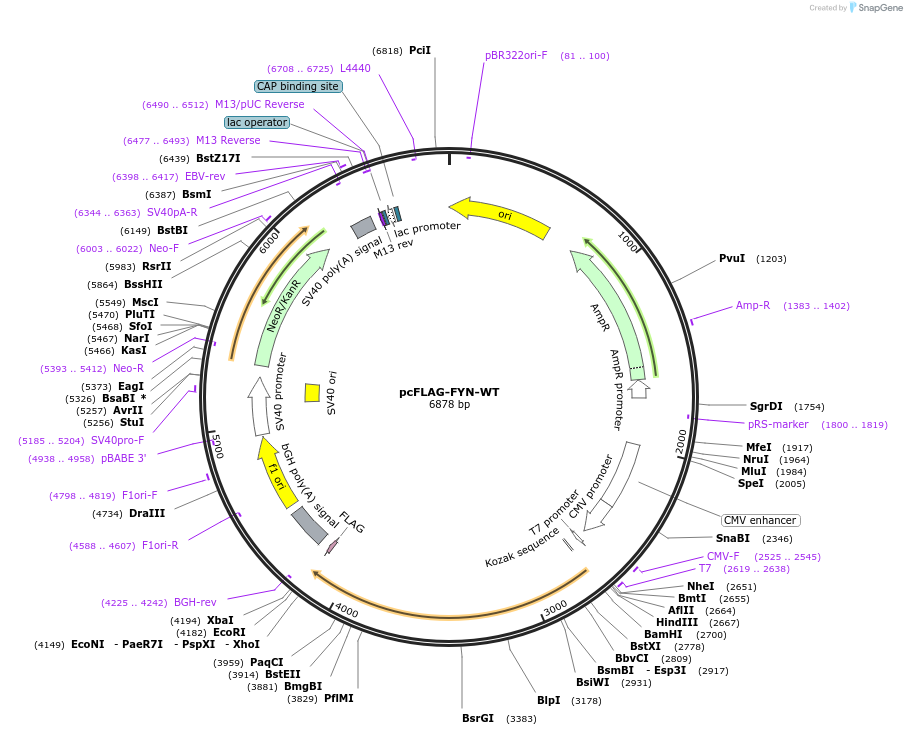
pcFLAG-FYN-WT
Plasmid#74509PurposeOverexpression of FYN in mammalian cellsDepositorAvailable SinceMay 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
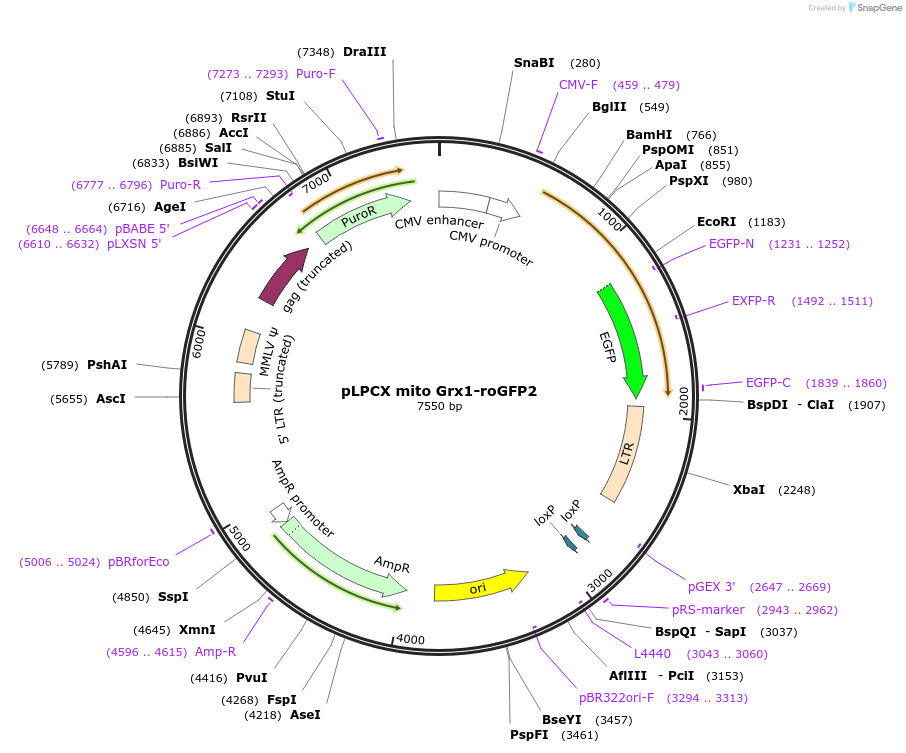
pLPCX mito Grx1-roGFP2
Plasmid#64977PurposeMammalian expression of mitochondrial Grx1-roGFP2 (retroviral vector)DepositorInsertGlutaredoxin-1 (GLRX Human)
UseRetroviralTagsroGFP2ExpressionMammalianMutationmitochondrial targeting sequenceAvailable SinceJune 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
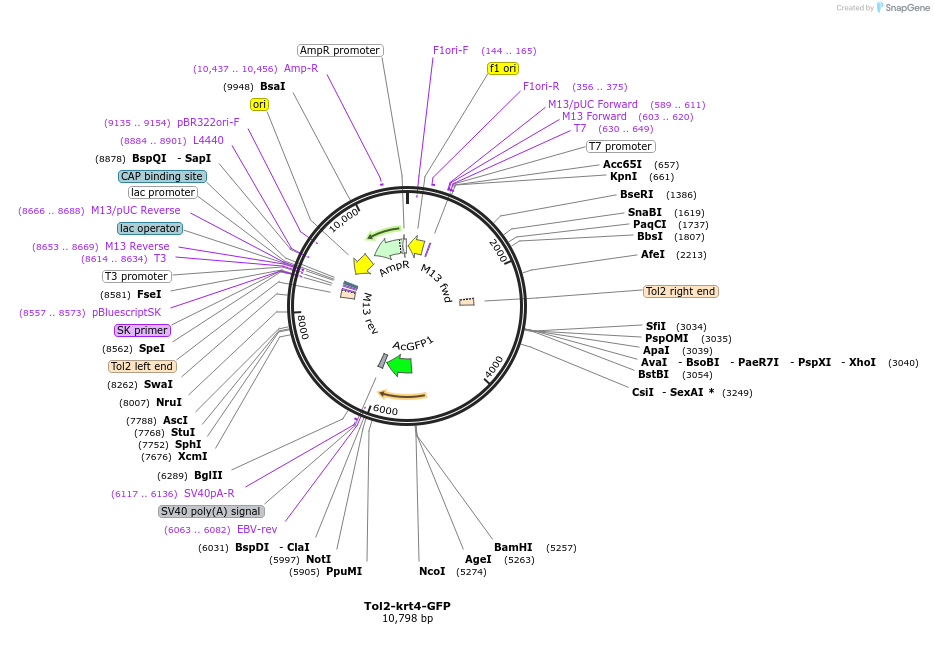
Tol2-krt4-GFP
Plasmid#97151Purposelabel apical epithelial cells in zebrafishDepositorInsertAcGFP
UseZebrafishAvailable SinceJuly 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
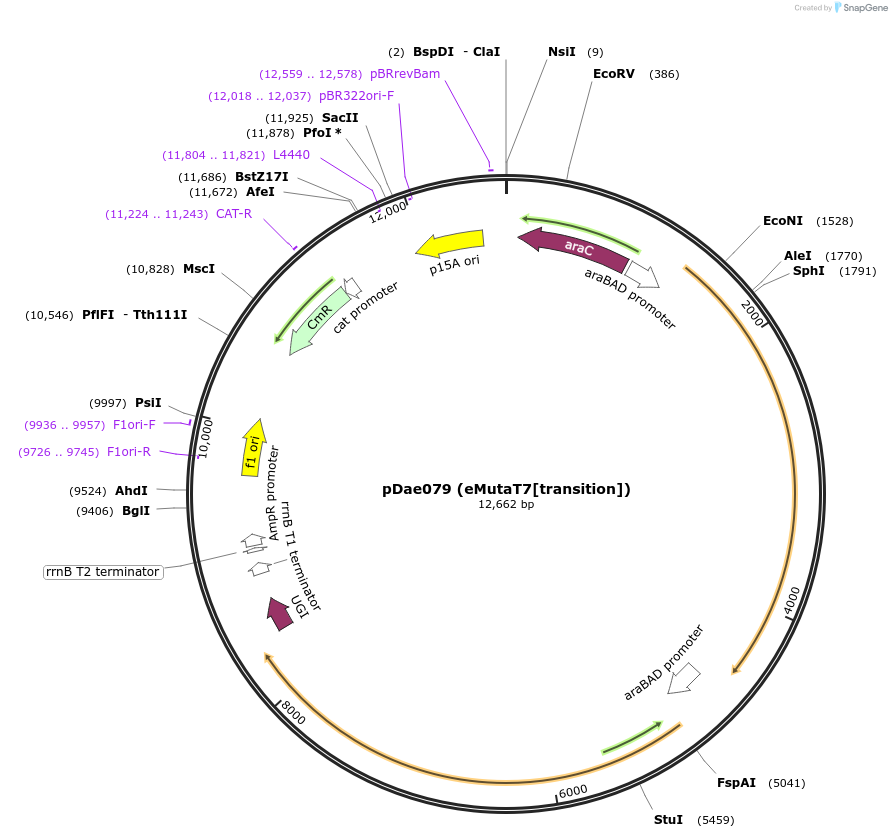
pDae079 (eMutaT7[transition])
Plasmid#187622PurposeExpresses eMutaT7[TadA-8e], eMutaT7[PmCDA1] and UGI for mutagenizing genes downstream from a T7 promoter in e.coliDepositorInsertsTadA-8e-T7RNAP
PmCDA1-T7RNAP
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
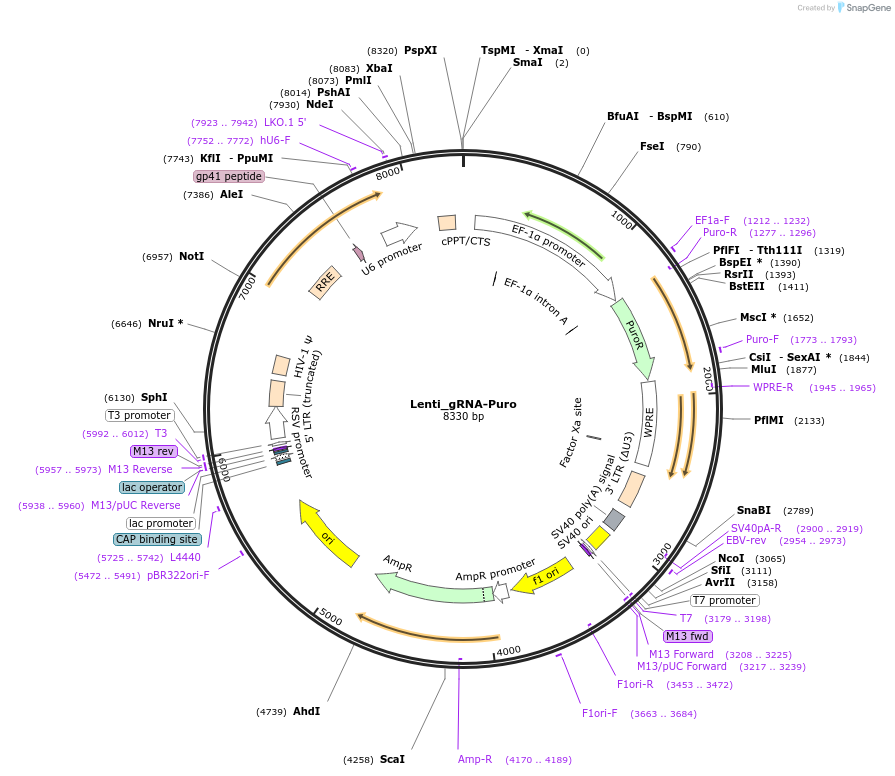
Lenti_gRNA-Puro
Plasmid#84752PurposeExpresses Cpf1 customizable sgRNA from U6 promoter and puromycin resistance from EF-1a promoterDepositorTypeEmpty backboneUseCRISPR and LentiviralExpressionMammalianPromoterhU6Available SinceDec. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
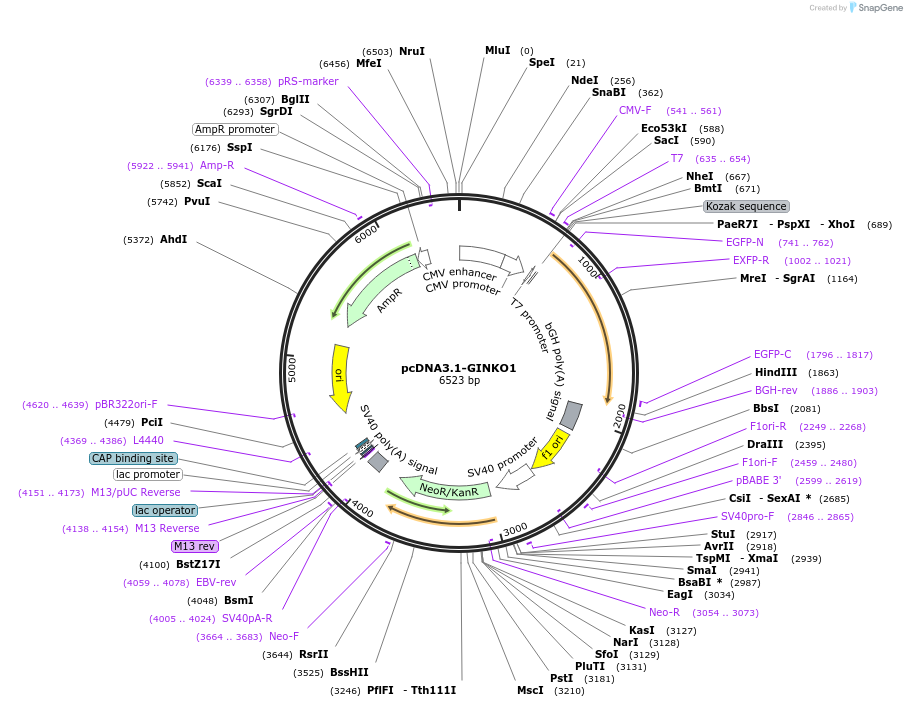
pcDNA3.1-GINKO1
Plasmid#113112PurposeMammalian expression of genetically encoded green fluorescent potassium ion indicator GINKO1DepositorInsertGINKO1
ExpressionMammalianPromoterCMVAvailable SinceOct. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
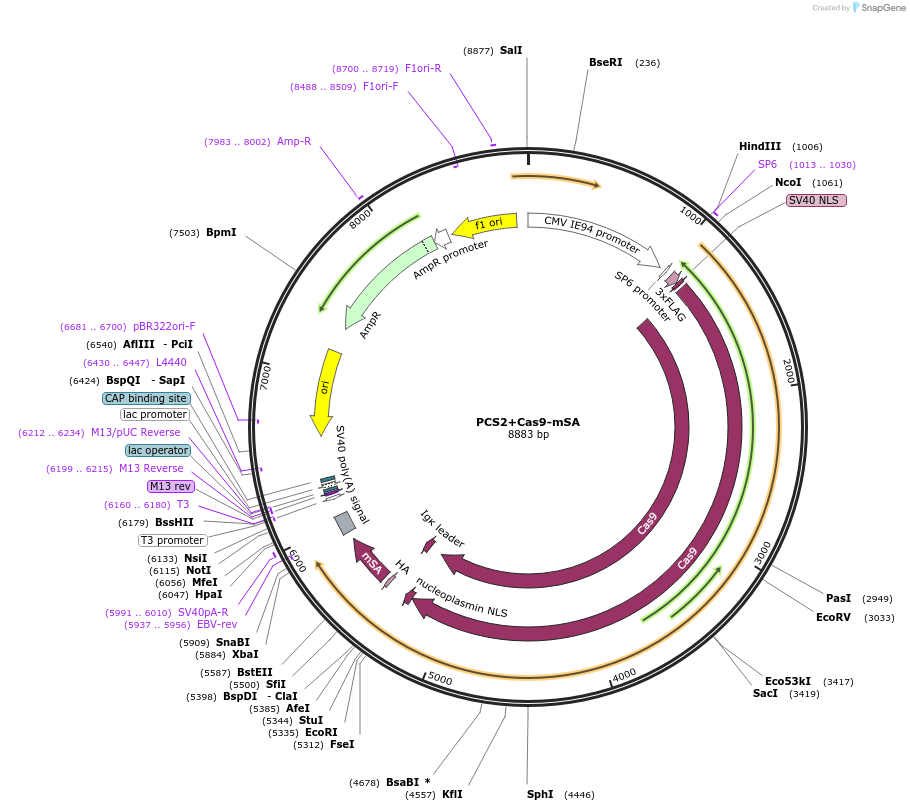
PCS2+Cas9-mSA
Plasmid#103882PurposeCas9 mSA for in vitro transcription for gene editing in Mouse EmbryosDepositorInsertCAS9-MSA
UseCRISPRTagsMSA (monomeric streptavidinMutationnonePromoterSP6Available SinceNov. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
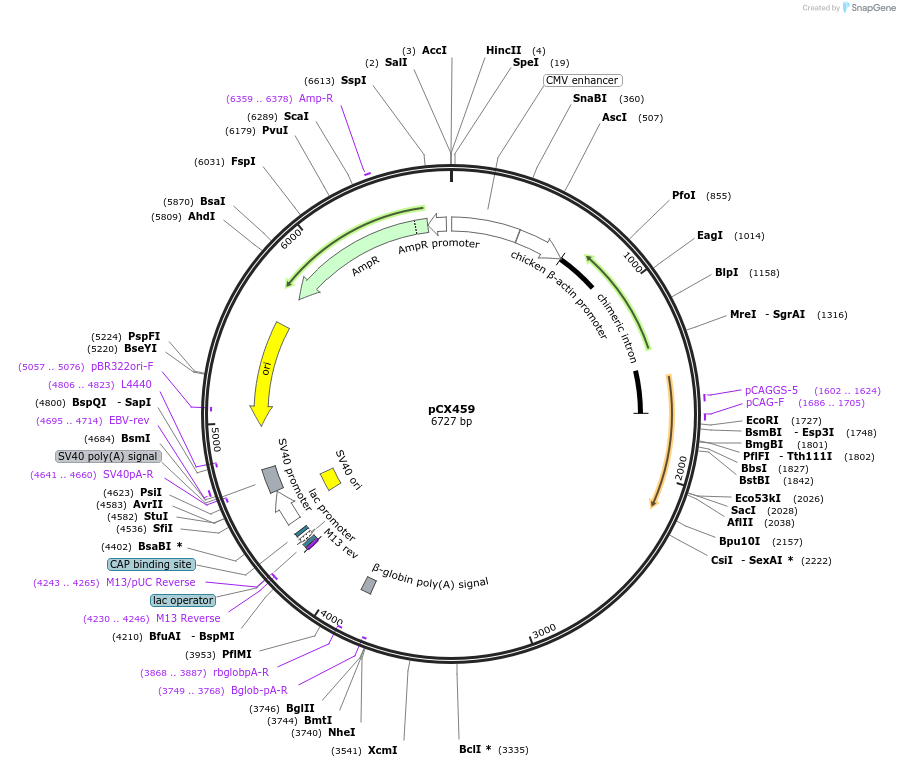
pCX459
Plasmid#229214PurposeCRISPR array plasmid for MAP1B (Spacers 1-24)DepositorInsertCRISPR array plasmid for MAP1B (Spacers 1-24)
ExpressionMammalianAvailable SinceFeb. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
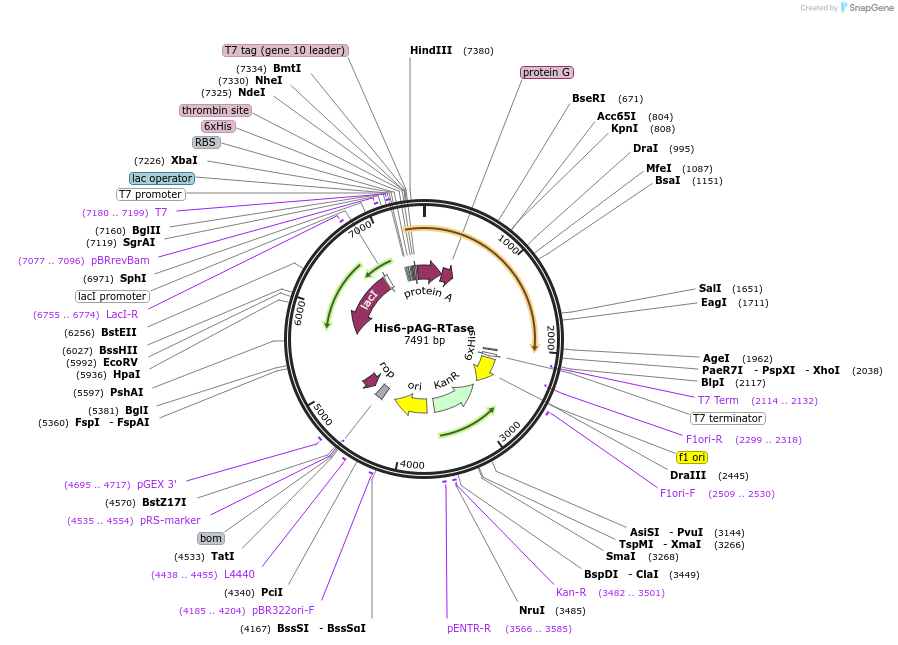
His6-pAG-RTase
Plasmid#214932PurposeExpress fusion proteins of Protein A/G and MMLV reverse transcriptase (25-497) in bacterial systemsDepositorInsertfusion protein of protein A/G and MMLV reverse transcriptase (25-497)
TagsHis6ExpressionBacterialMutationMMLV RTase (H8Y, D200N, T306K, W313F, T330P, D524…PromoterT7 promoterAvailable SinceApril 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
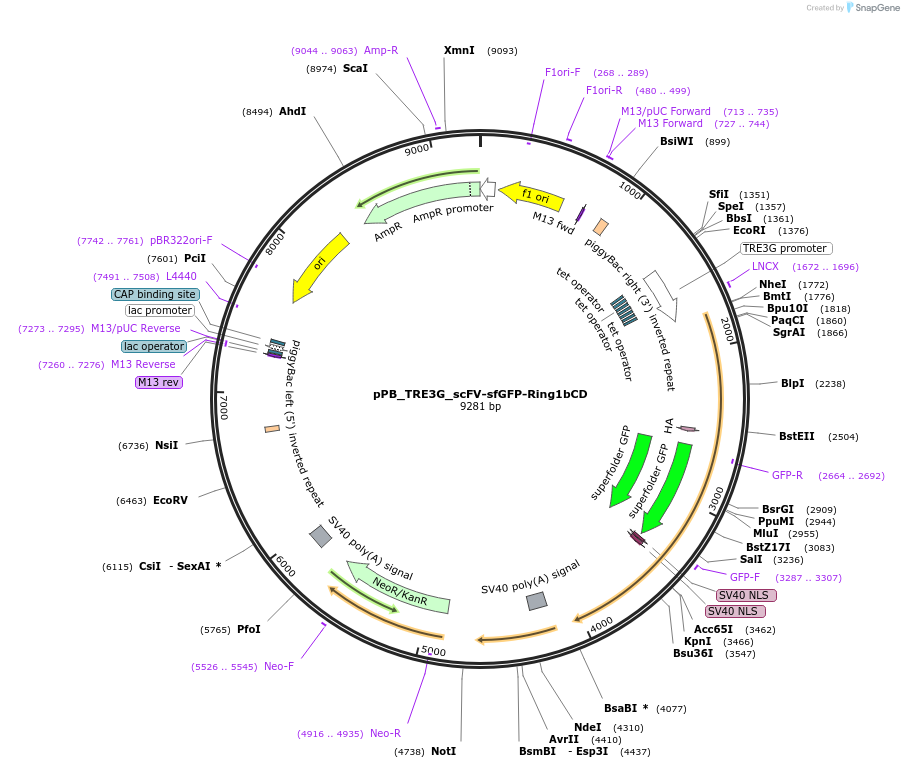
pPB_TRE3G_scFV-sfGFP-Ring1bCD
Plasmid#235574PurposeDox-inducible expression of the catalytic-domain (CD) of epigenetic effector Ring1b fused with scFV to programme H2AK119ub.DepositorInsertRing1b (Rnf2 Mouse)
UseCRISPRAvailable SinceApril 29, 2025AvailabilityAcademic Institutions and Nonprofits only -
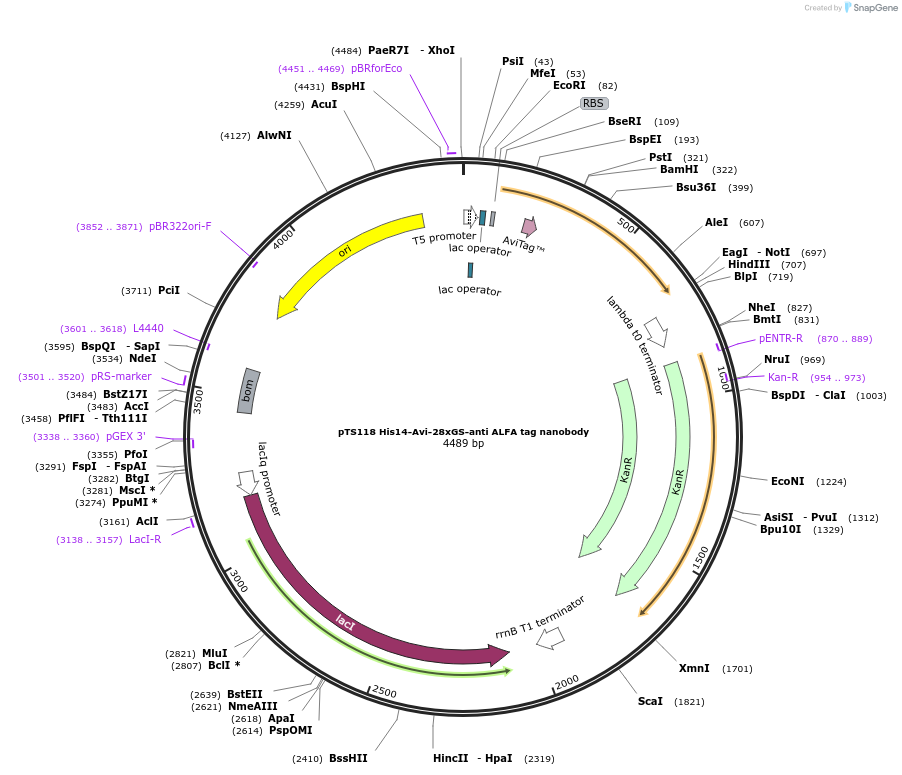
pTS118 His14-Avi-28xGS-anti ALFA tag nanobody
Plasmid#199371PurposeBacterial expression plasmid for anti-ALFA nanobody with an N-terminal His-tag and biotin acceptor peptide (Avi)DepositorInsertanti-ALFA nanobody
Tags14xHis-AviExpressionBacterialMutationWTPromoterT5-LacOAvailable SinceApril 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
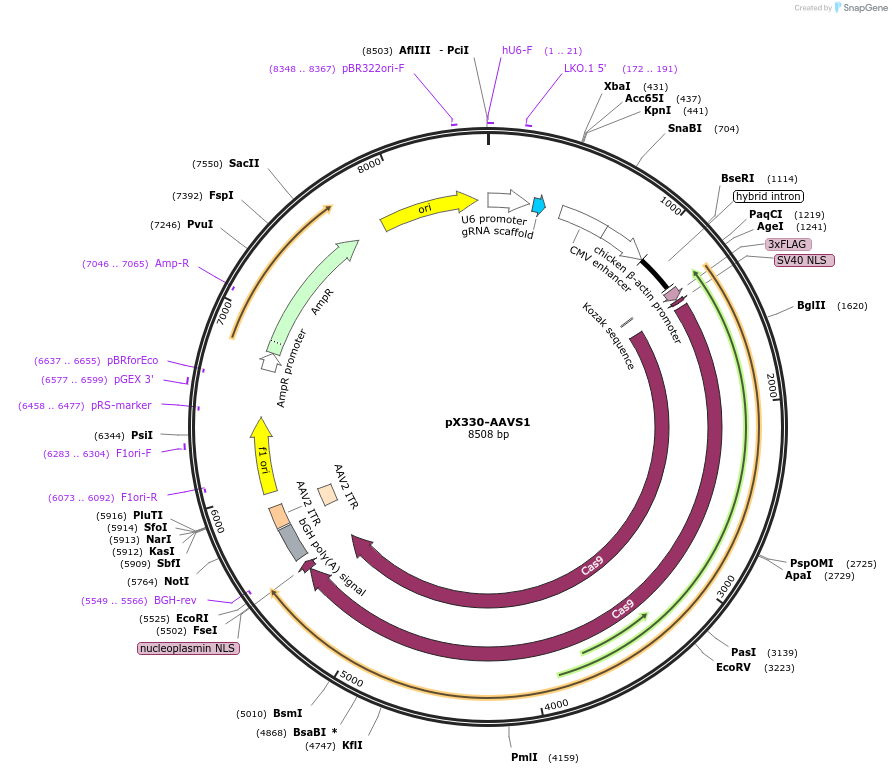
pX330-AAVS1
Plasmid#227272PurposeExpresses SpCas9 and a sgRNA targeting the AAVS1 loci for knock-in.DepositorAvailable SinceNov. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
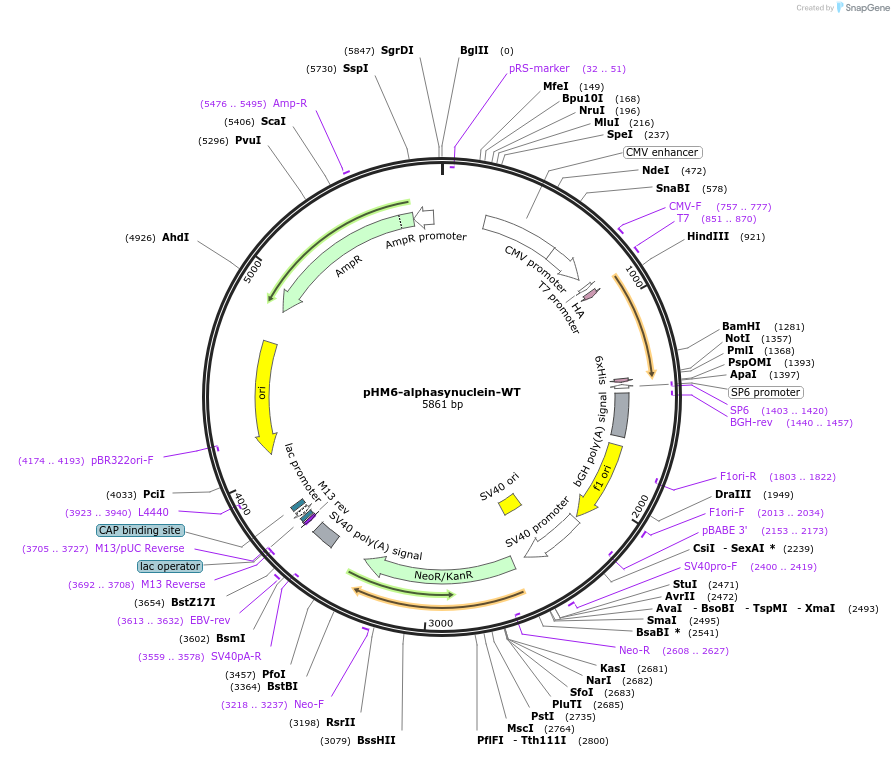
pHM6-alphasynuclein-WT
Plasmid#40824DepositorAvailable SinceMay 8, 2013AvailabilityAcademic Institutions and Nonprofits only -
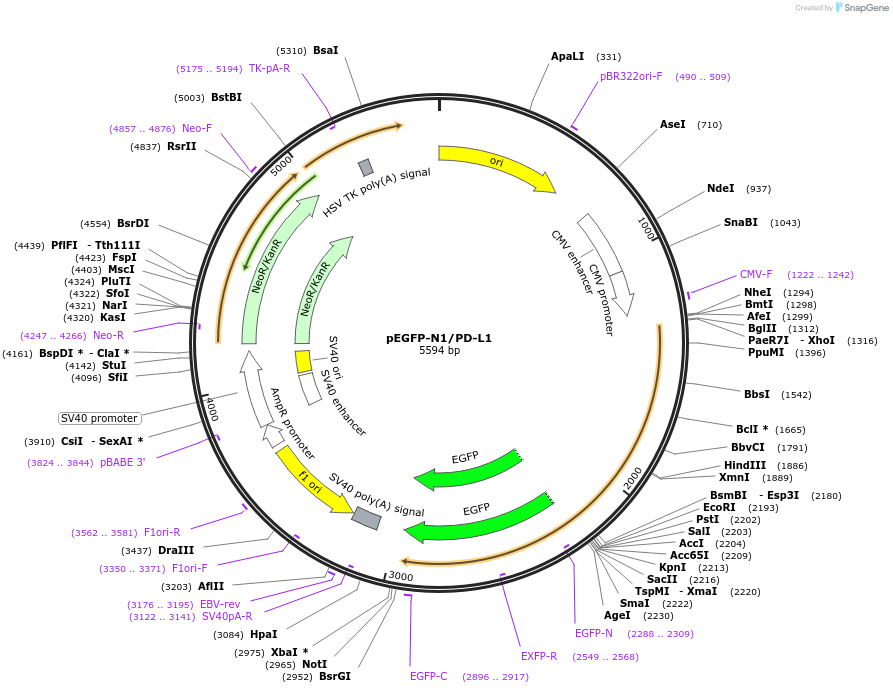
pEGFP-N1/PD-L1
Plasmid#121478Purposeexpression of human PD-L1 in mammalian cellsDepositorAvailable SinceFeb. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
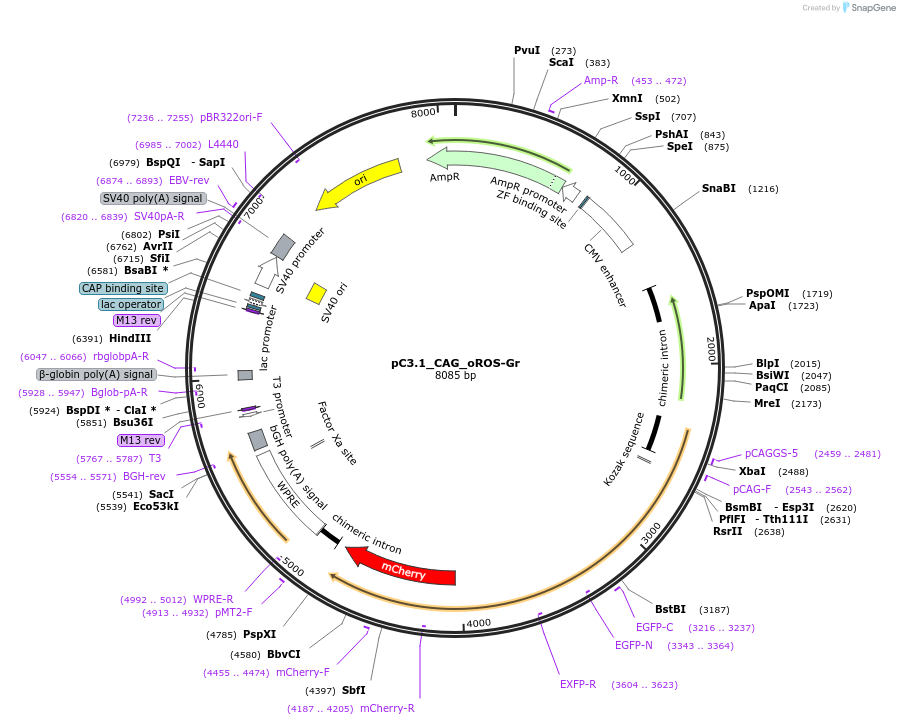
pC3.1_CAG_oROS-Gr
Plasmid#216113PurposeExpresses the genetically encoded, ratiometric hydrogen peroxide sensor oROS-Gr in mamalian cells.DepositorInsertCAG_oROS-Gr
ExpressionMammalianPromoterCAGAvailable SinceApril 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
pAAV-Syn-ChrimsonR-tdT (AAV Retrograde)
Viral Prep#59171-AAVrgPurposeReady-to-use AAV Retrograde particles produced from pAAV-Syn-ChrimsonR-tdT (#59171). In addition to the viral particles, you will also receive purified pAAV-Syn-ChrimsonR-tdT plasmid DNA. Syn-driven ChrimsonR-tdTomato expression for optogenetic neural activation. These AAV preparations are suitable purity for injection into animals.DepositorPromoterSynTagstdTomatoAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
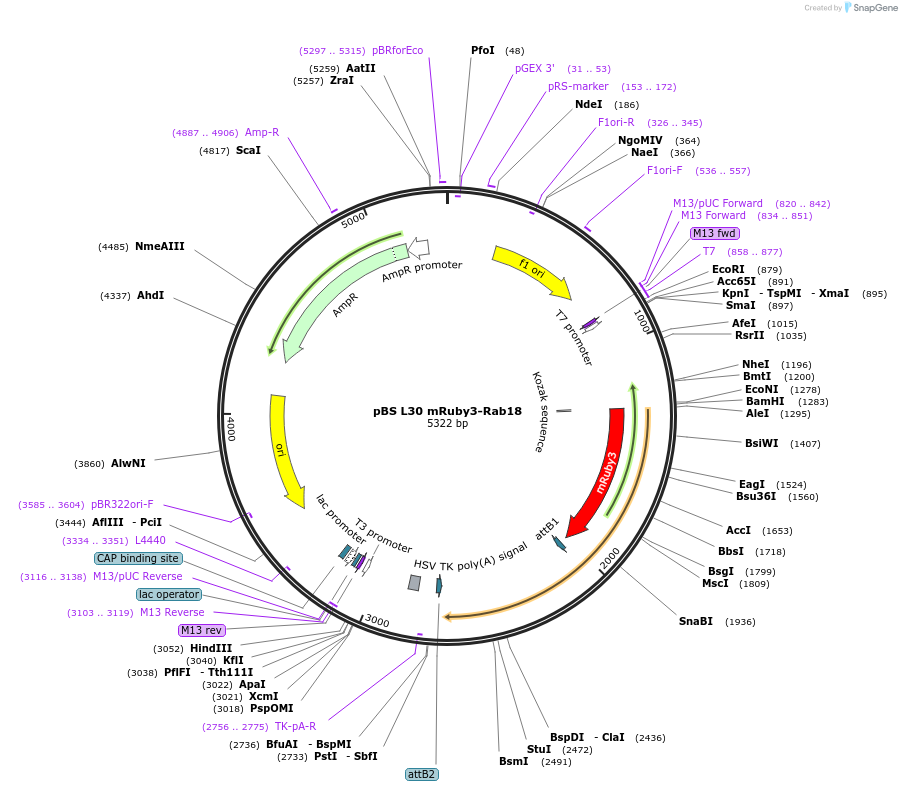
pBS L30 mRuby3-Rab18
Plasmid#166431PurposeRab18 fused to mRuby3 fluorescent protein (ex558/em592) tag in N-ter under control of a weak L30 promoter.DepositorInsertRab18
TagsmRuby3ExpressionMammalianPromoterL30 weak promoterAvailable SinceOct. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
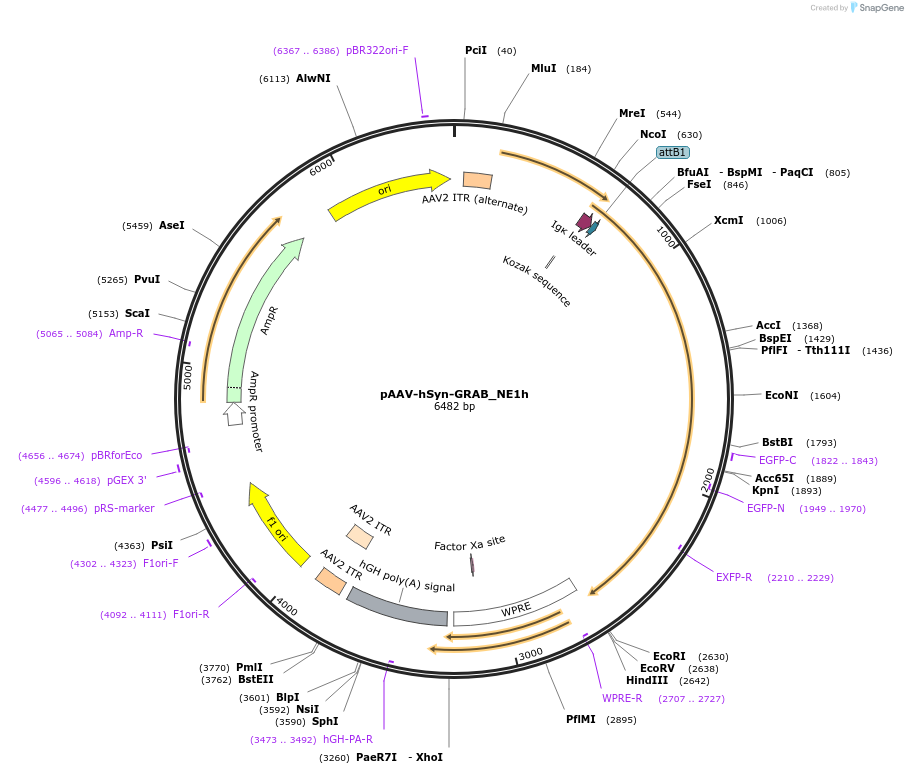
pAAV-hSyn-GRAB_NE1h
Plasmid#123309PurposeExpresses the genetically-encoded fluorescent norepinephrine(NE) sensor GRAB_NE1h in neuronsDepositorHas ServiceAAV Retrograde and AAV9InsertGPCR activation based NE sensor GRAB_NE1h
UseAAVPromoterhSynAvailable SinceApril 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
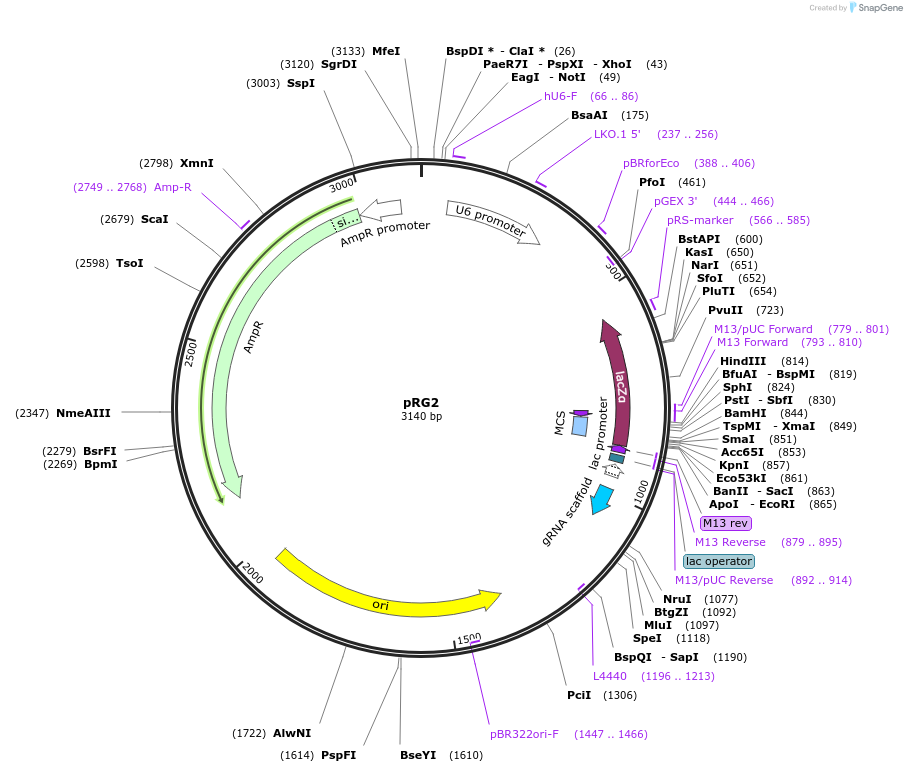
pRG2
Plasmid#104174PurposesgRNA expressionDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceDec. 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
AAV-hSyn-Soma-jGCaMP8s (AAV9)
Viral Prep#169256-AAV9PurposeReady-to-use AAV9 particles produced from AAV-hSyn-Soma-jGCaMP8s (#169256). In addition to the viral particles, you will also receive purified AAV-hSyn-Soma-jGCaMP8s plasmid DNA. hSyn-driven expression of soma-targeted calcium sensor GCaMP8s (more sensitive). These AAV preparations are suitable purity for injection into animals.DepositorAvailable SinceFeb. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
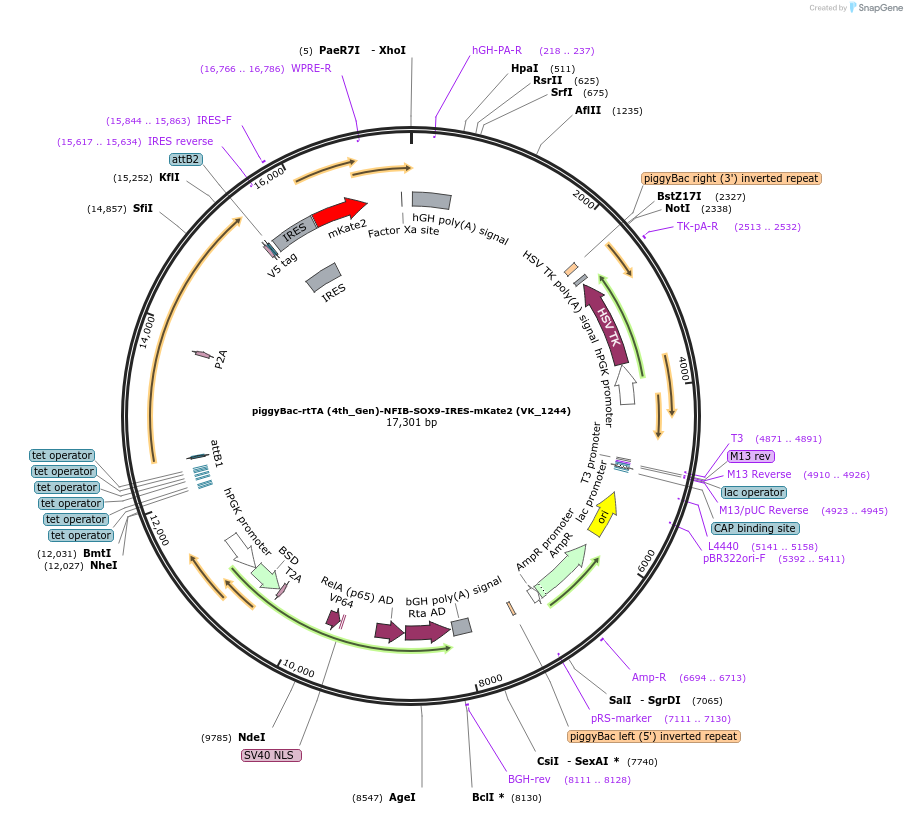
piggyBac-rtTA (4th_Gen)-NFIB-SOX9-IRES-mKate2 (VK_1244)
Plasmid#209082PurposeNFIB and SOX9 mediated astrocye inductionDepositorAvailable SinceDec. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
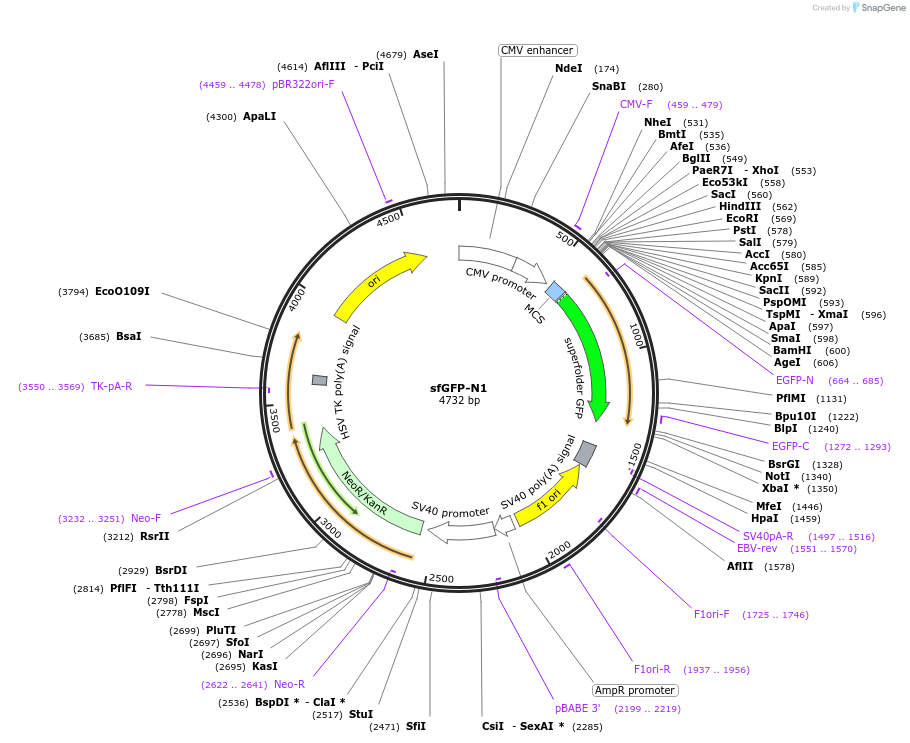
sfGFP-N1
Plasmid#54737PurposeLocalization: N1 Cloning Vector, Excitation: 485, Emission: 507DepositorTypeEmpty backboneTagssfGFPExpressionMammalianAvailable SinceJune 20, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
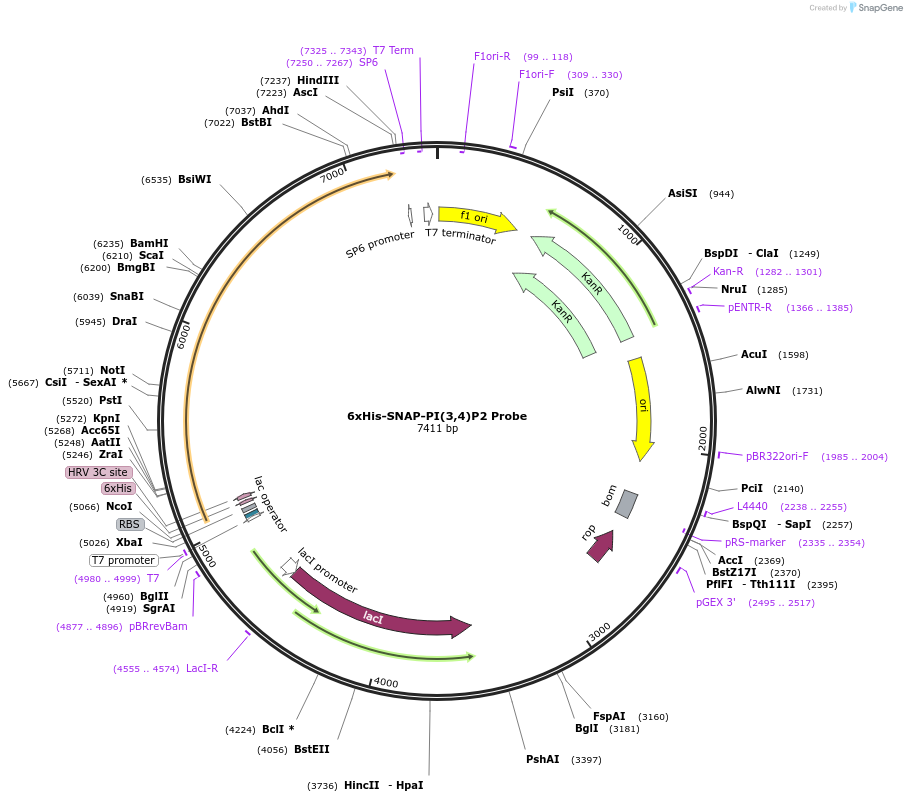
6xHis-SNAP-PI(3,4)P2 Probe
Plasmid#211511PurposeFor expression of recombinant biosensor for detection of Phosphatidylinositol-3,4-Bisphosphate (PI(3,4)P2) from bacteriaDepositorInsertTAPP1 cPHx3
Tags6xhis-SNAPExpressionBacterialAvailable SinceJan. 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
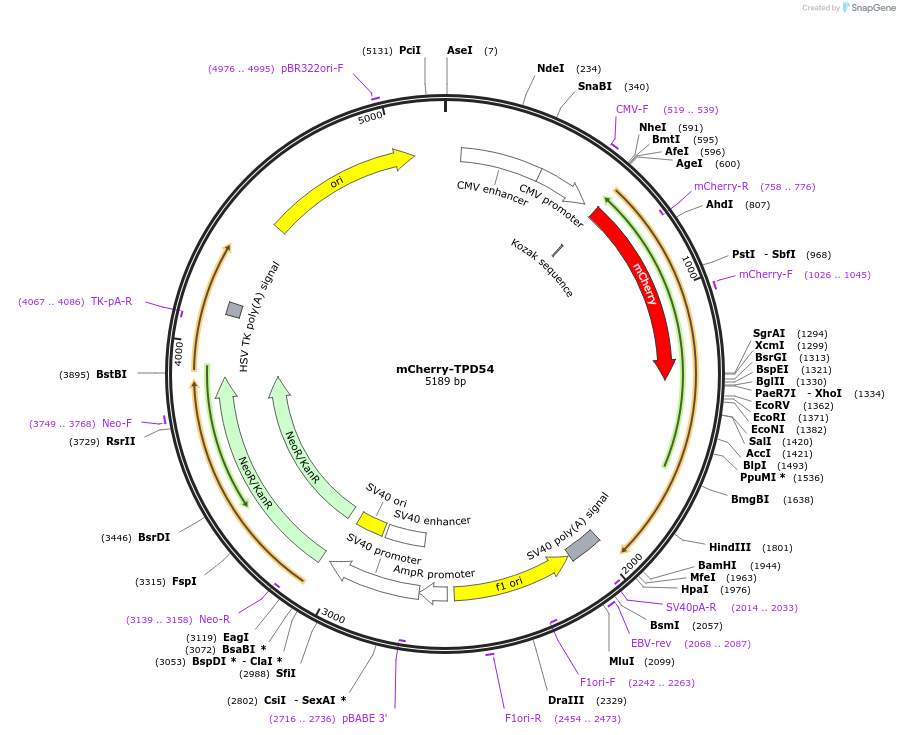
mCherry-TPD54
Plasmid#172449PurposeExpression of Tumor Protein D52 like 2 (TPD54/TPD52L2) with N-terminal mCherry tagDepositorAvailable SinceAug. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
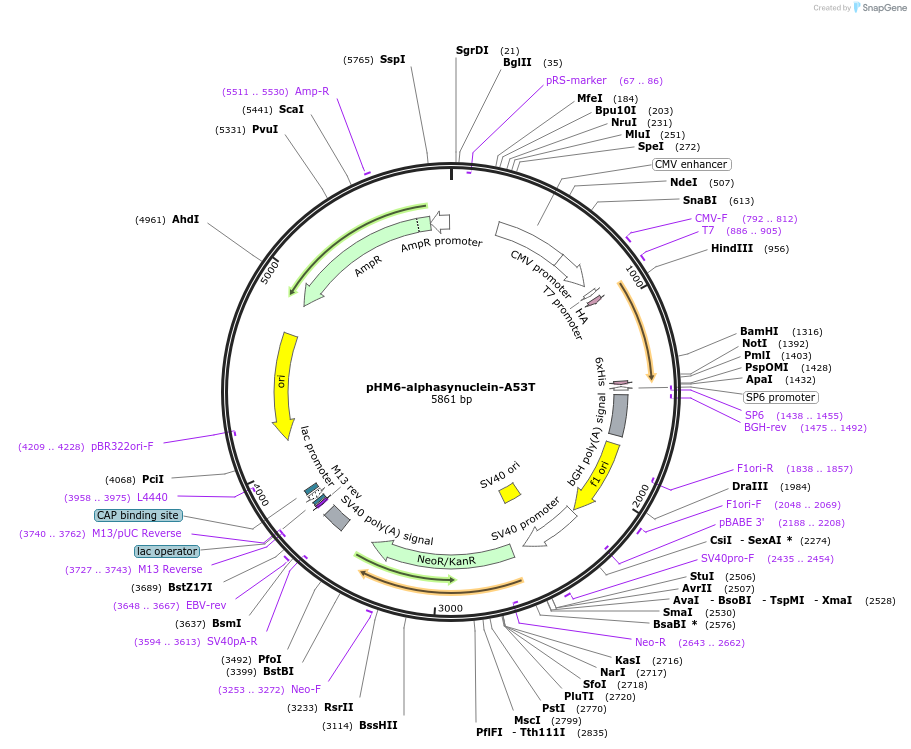
pHM6-alphasynuclein-A53T
Plasmid#40825DepositorAvailable SinceMay 8, 2013AvailabilityAcademic Institutions and Nonprofits only -
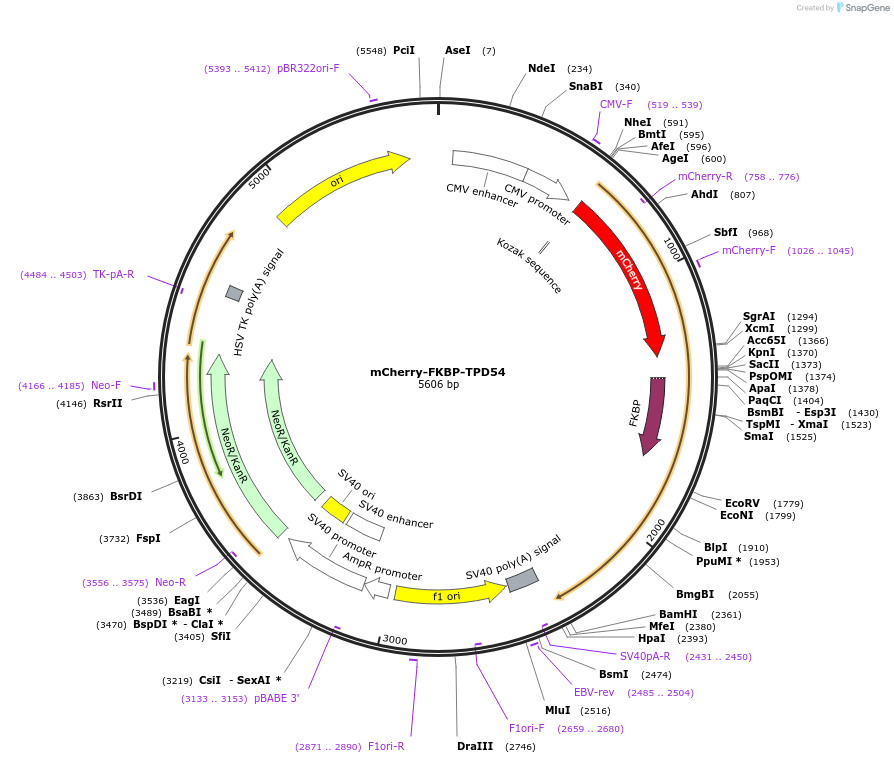
mCherry-FKBP-TPD54
Plasmid#172451PurposeExpression of Tumor Protein D52 like 2 (TPD54/TPD52L2) with N-terminal mCherry-FKBP tagDepositorAvailable SinceAug. 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
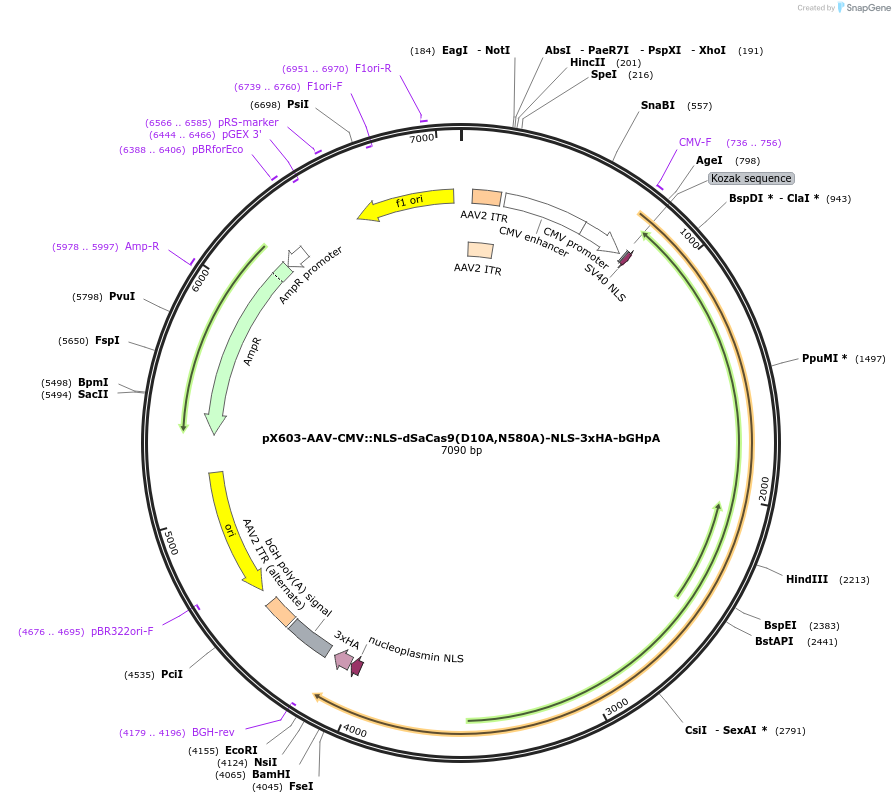
pX603-AAV-CMV::NLS-dSaCas9(D10A,N580A)-NLS-3xHA-bGHpA
Plasmid#61594PurposeA catalytically inactive SaCas9 (dSaCas9) with the D10A and N580A mutations.DepositorInserthSaCas9
UseAAV and CRISPRTags3xHA and NLSExpressionMammalianMutationD10A, N580AAvailable SinceJuly 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
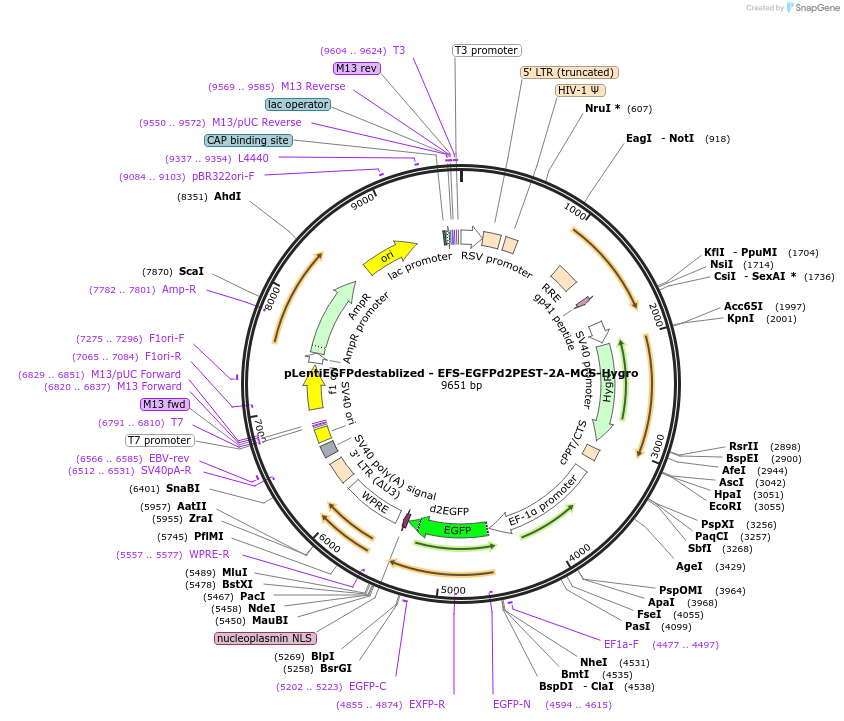
pLentiEGFPdestablized - EFS-EGFPd2PEST-2A-MCS-Hygro
Plasmid#138152PurposeExpresses eGFP in mammalian cells, contains a 3' MCS. Localized to the nucleusDepositorInserteGFP-MCS
UseLentiviralTags2PEST AND NLSAvailable SinceFeb. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
-
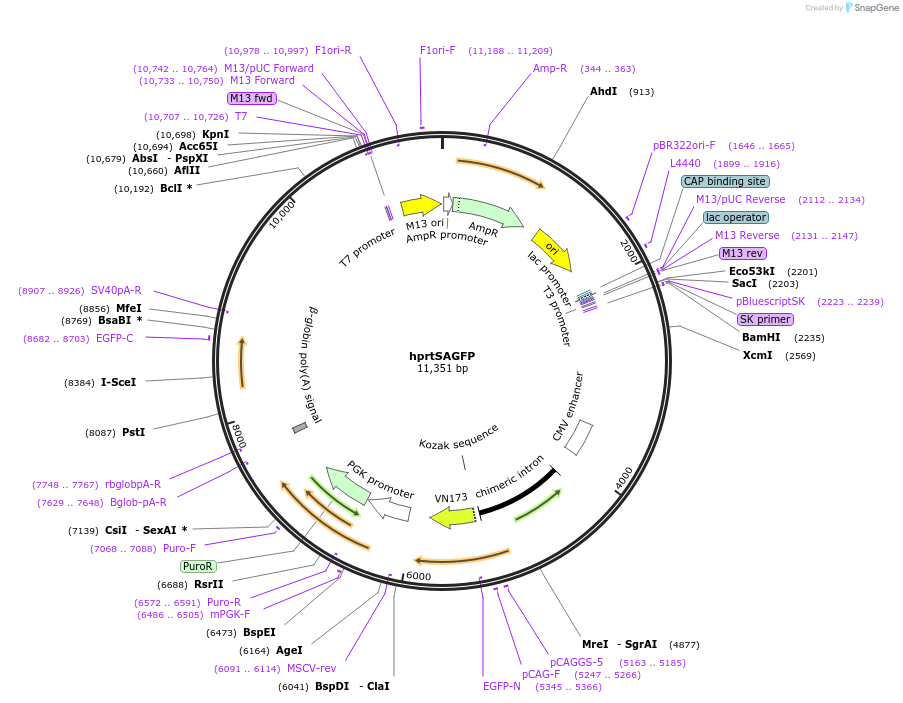
hprtSAGFP
Plasmid#41594PurposeIn vivo single-strand annealing substrate composed of two truncated GFP genes, separated by a drug selection marker, and flanked by mouse hprt targeting sequencesDepositorInsertSA-GFP reporter
UseMouse Targeting; Dsb repair reporter substrateExpressionMammalianAvailable SinceDec. 3, 2012AvailabilityAcademic Institutions and Nonprofits only -
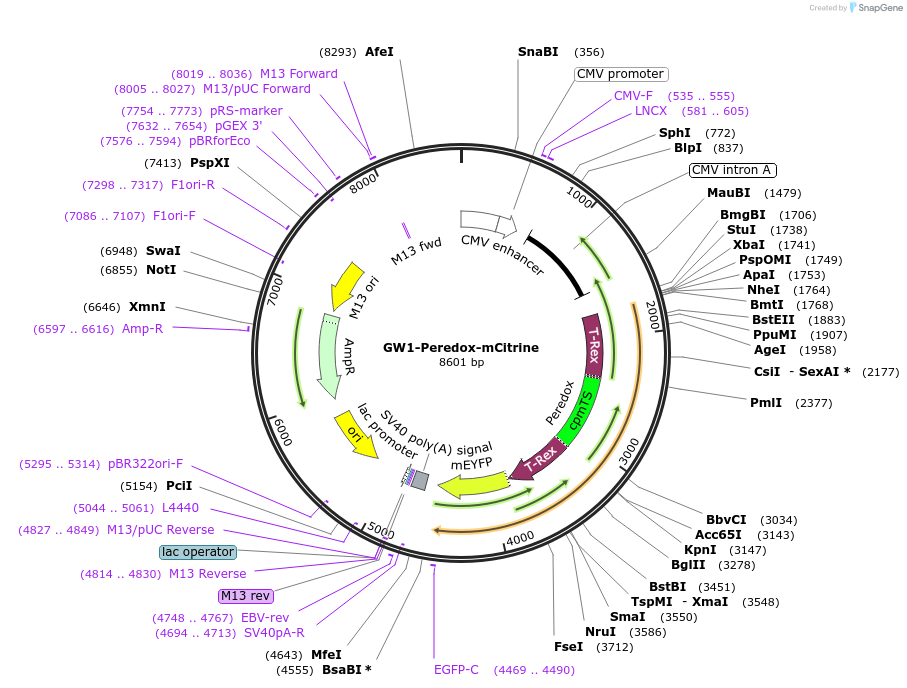
GW1-Peredox-mCitrine
Plasmid#32386DepositorInsertPeredox-mCitrine
ExpressionMammalianPromoterCMVAvailable SinceOct. 18, 2011AvailabilityAcademic Institutions and Nonprofits only -
pAAV-nEF-Con/Foff 2.0-NpHR3.3-EYFP (AAV8)
Viral Prep#137153-AAV8PurposeReady-to-use AAV8 particles produced from pAAV-nEF-Con/Foff 2.0-NpHR3.3-EYFP (#137153). In addition to the viral particles, you will also receive purified pAAV-nEF-Con/Foff 2.0-NpHR3.3-EYFP plasmid DNA. nEF-driven, Cre-dependent expression of NpHR3.3-EYFP (inhibited in presence of Flp) for optogenetic inhibition. These AAV preparations are suitable purity for injection into animals.DepositorPromoternEFTagsEYFP (Cre-dependent)Available SinceApril 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
pAAV_BiPVe3_ChR2_mCherry (AAV1)
Viral Prep#213941-AAV1PurposeReady-to-use AAV1 particles produced from pAAV_BiPVe3_ChR2_mCherry (#213941). In addition to the viral particles, you will also receive purified pAAV_BiPVe3_ChR2_mCherry plasmid DNA. Expression of mCherry-tagged channelrhodopsin-2 under the control of the PV+ basket cell-targeting enhancer E3. These AAV preparations are suitable purity for injection into animals.DepositorTagsmCherryAvailable SinceSept. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
psgRNA_EcoWG1
Pooled Library#131625PurposeBacterial inhibition library targeting every small RNA and ORF of the E. coli MG1655 genome. This library has higher inhibition efficiency than the psgRNA library.DepositorExpressionBacterialSpeciesEscherichia coliUseCRISPRAvailable SinceMay 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
-
-
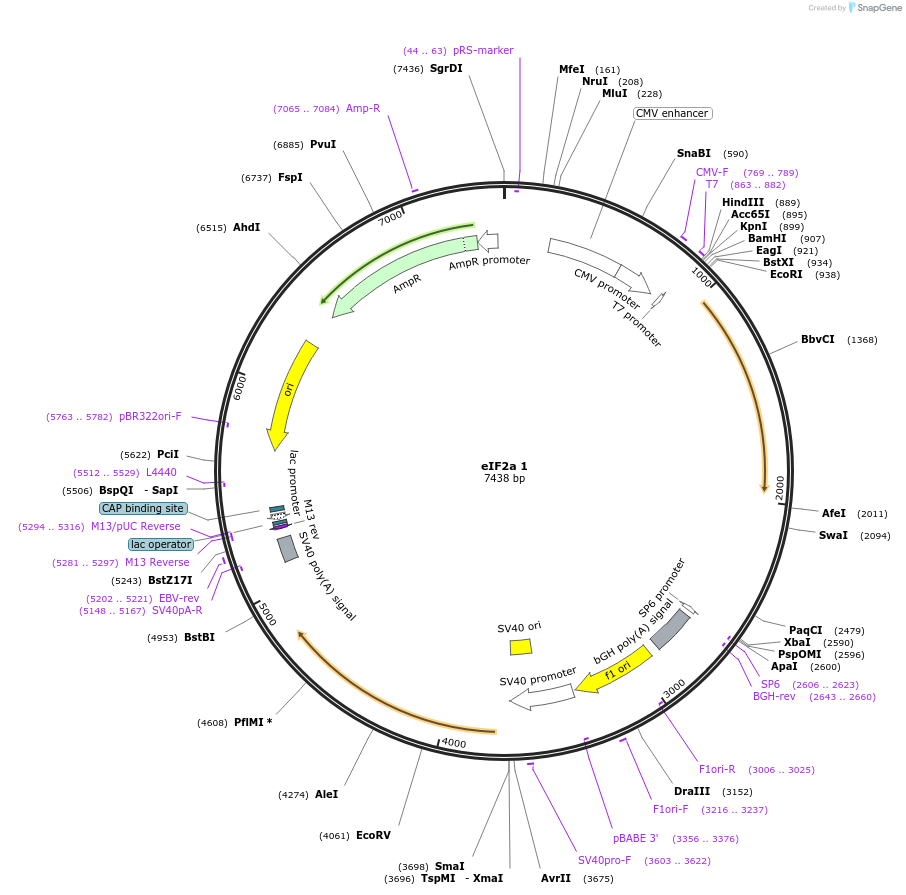
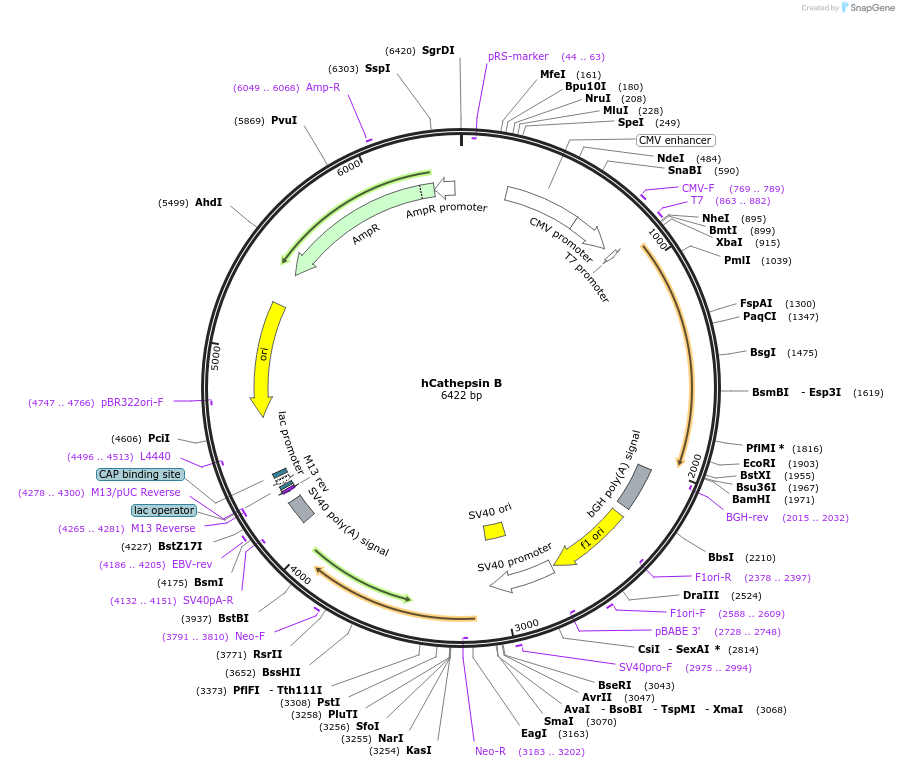
hCathepsin B
Plasmid#11249PurposeMammalian expression of human cathepsin BDepositorAvailable SinceMay 25, 2006AvailabilityIndustry, Academic Institutions, and Nonprofits -
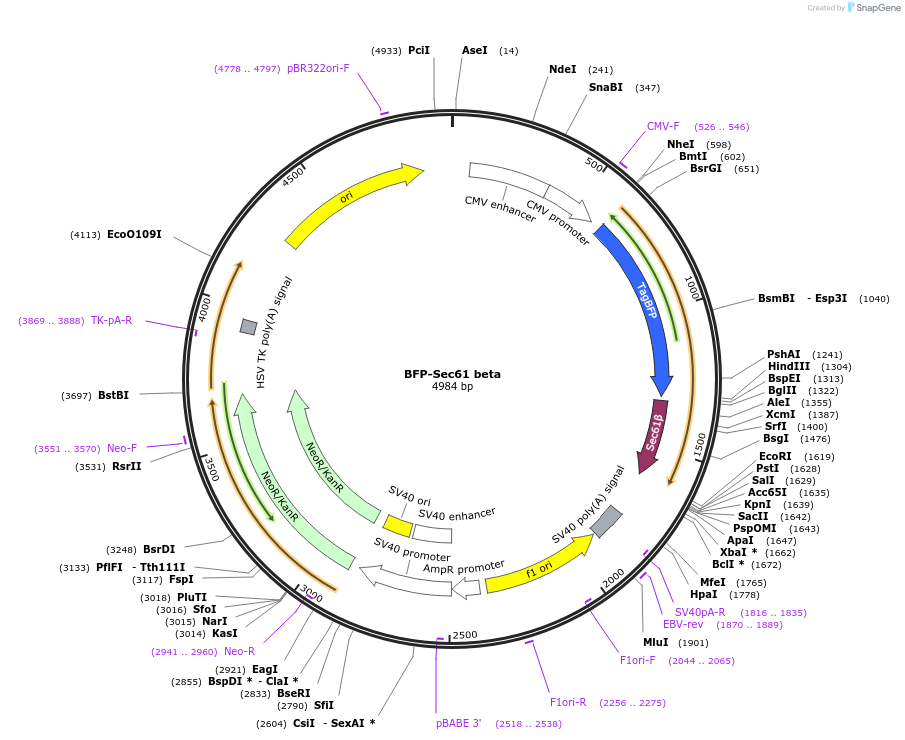
BFP-Sec61 beta
Plasmid#49154Purposeexpression of Sec61 beta fused to TagBFP, used as a general ER markerDepositorAvailable SinceMay 29, 2014AvailabilityAcademic Institutions and Nonprofits only -
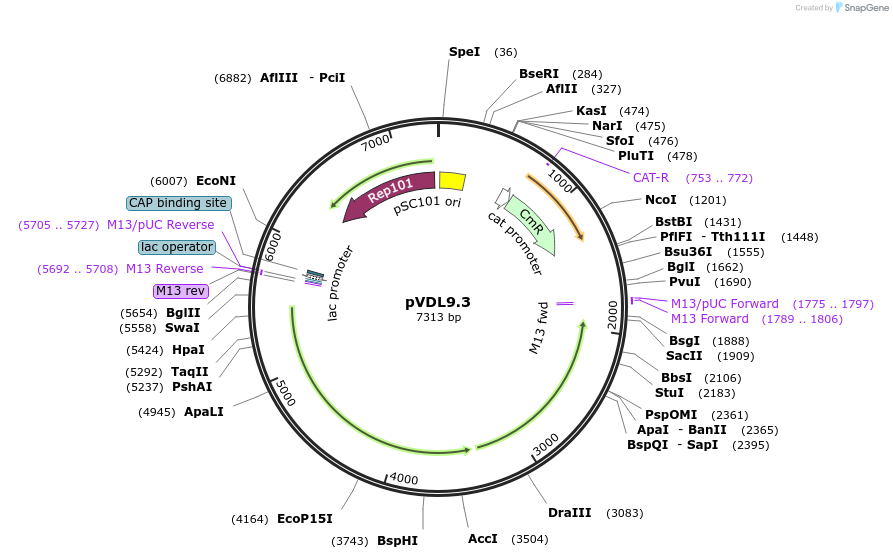
pVDL9.3
Plasmid#168299PurposeSecretion of polypeptides (nanobodies) in E. coli fused to C-terminal secretion signal of HlyADepositorInsertSegment of the hly operon, spanning the C-terminal part of hlyA, hlyB, and hlyD
ExpressionBacterialPromoterLacI-PlacAvailable SinceJune 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
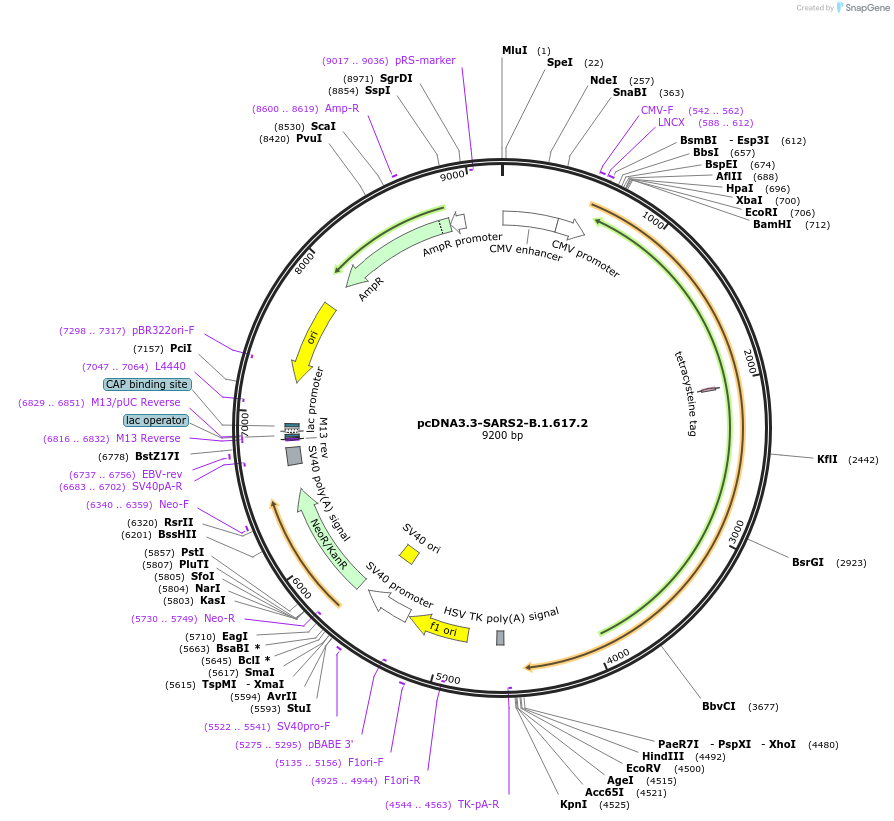
pcDNA3.3-SARS2-B.1.617.2
Plasmid#172320Purposeexpressing SARS-CoV-2 B.1.617.2 spike protein (delta strain) for pseudovirus production.DepositorInsertSpike of B.1.617.2 strain (S SARS-CoV-2)
ExpressionMammalianMutationT19R, 156G, 157-158del, L452R, T478K, D614G, P681…PromoterCMV-FAvailable SinceJuly 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
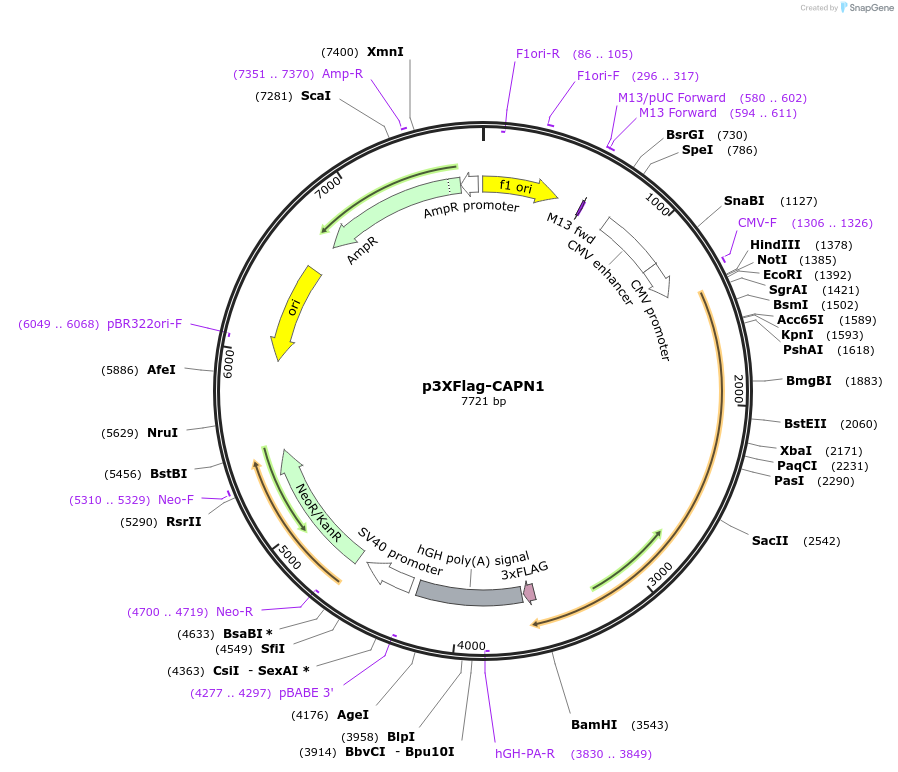
p3XFlag-CAPN1
Plasmid#60941PurposeExpresses calpain1 in mammalian cellsDepositorAvailable SinceDec. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
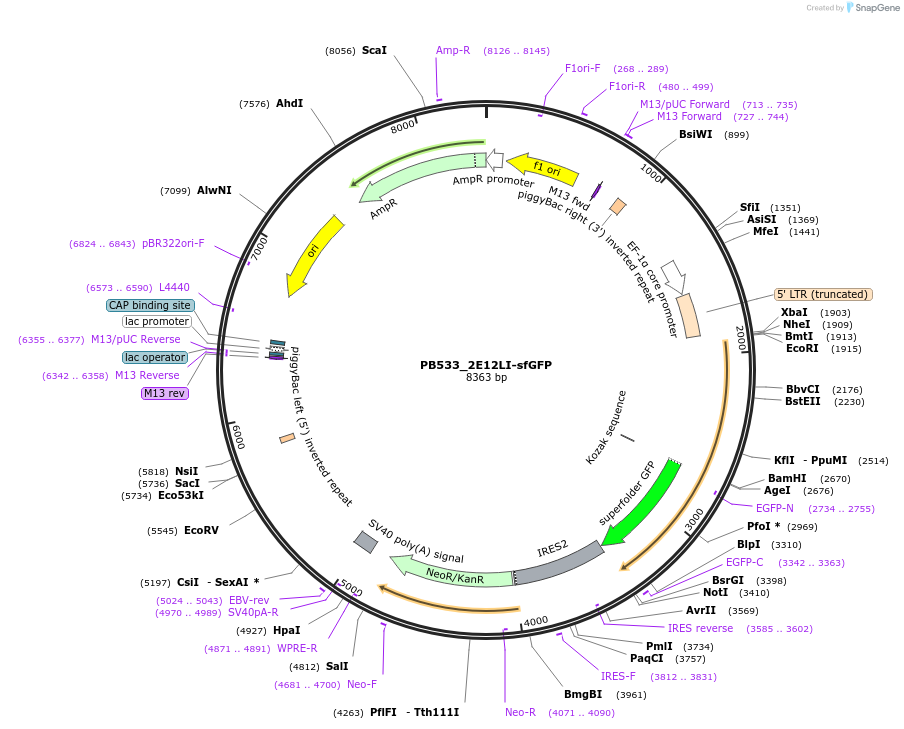
PB533_2E12LI-sfGFP
Plasmid#167527PurposeH3K27me3-mintbody expression vector for mammalian cellsDepositorInsert2E12LI-scFv
TagssfGFPExpressionMammalianAvailable SinceApril 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
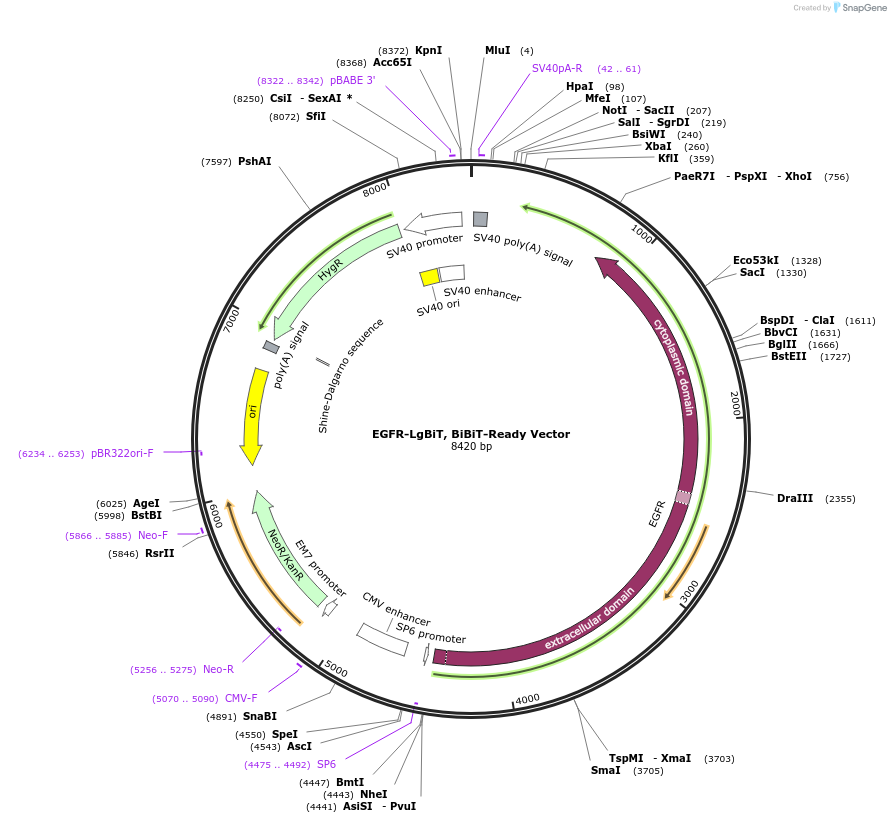
EGFR-LgBiT, BiBiT-Ready Vector
Plasmid#238755PurposeExpress EGFR-LgBiT Fusion Protein in Mammalian Cells under a CMV promoterDepositorAvailable SinceSept. 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
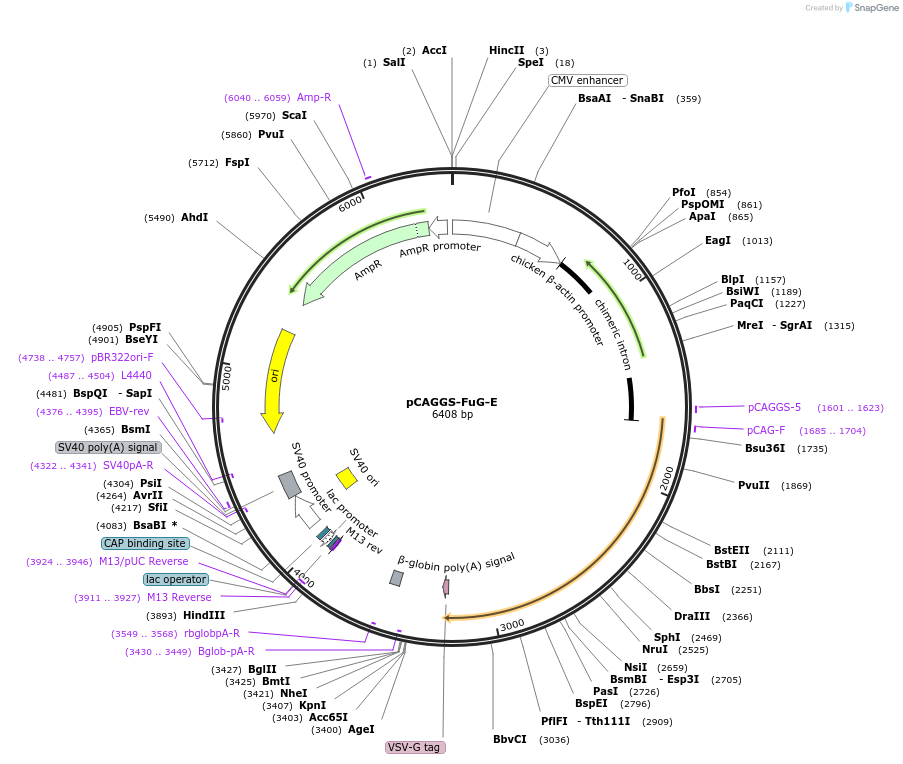
pCAGGS-FuG-E
Plasmid#67509PurposeThis is an envelope gene encoding plasmid to make retrogradely lentiviral vector. If you use this plasmid, you can make type E envelope coating virus particle with NeuRet.DepositorInsertFusion Glycoprotein type E
UseLentiviralTagsFusion protein of Vesicular stomatitis Indiana vi…PromoterCAGGSAvailable SinceJuly 31, 2015AvailabilityAcademic Institutions and Nonprofits only