169,450 results
-
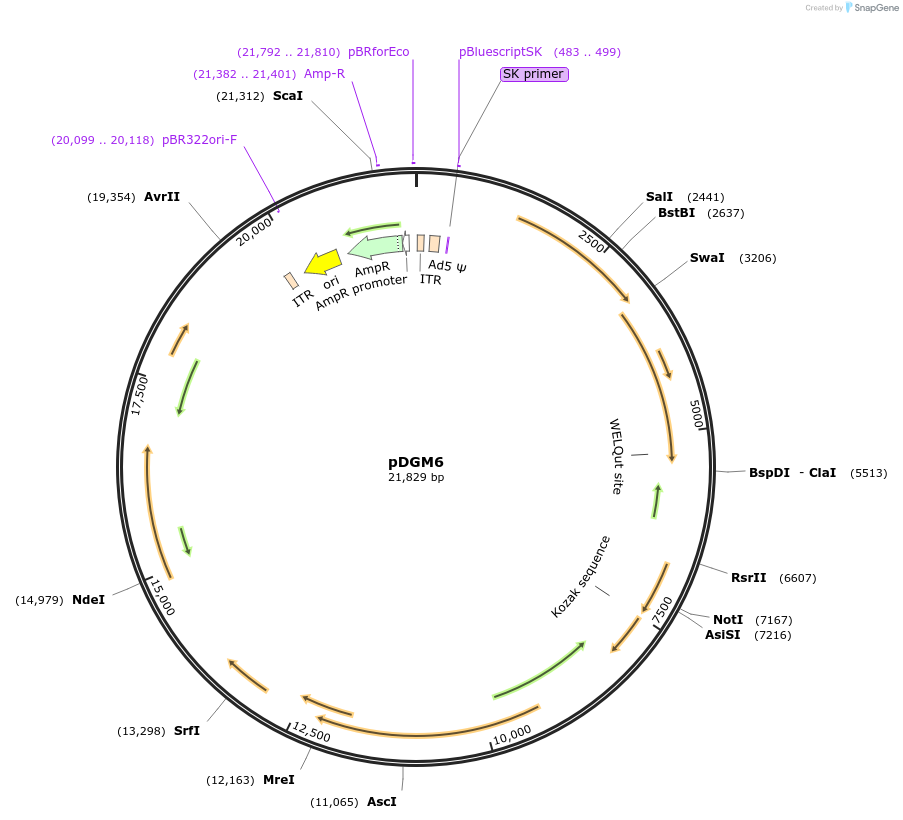
Plasmid#110660PurposeAdeno-Associated Virus (AAV) serotype 6; contains the AAV6 cap genes, AAV2 rep genes, and adenovirus helper genes. Good for in vivo transduction of muscle, lung, liver and other tissues.DepositorInsertAAV6 cap genes, AAV2 rep genes, adenovirus helper genes
UseAAVAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
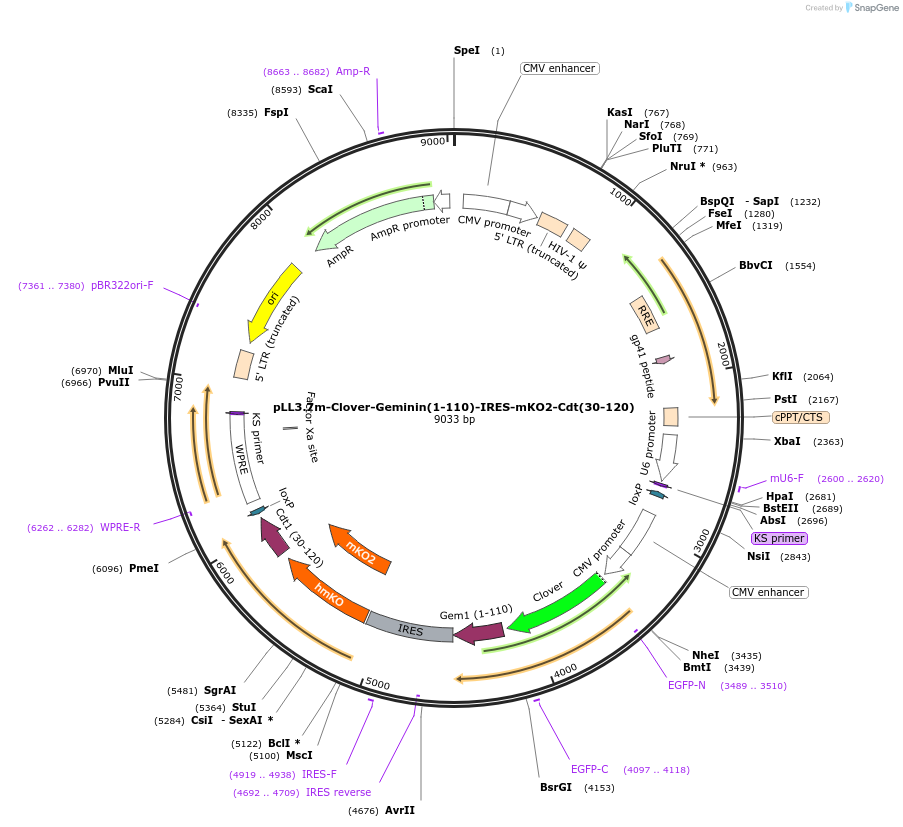
pLL3.7m-Clover-Geminin(1-110)-IRES-mKO2-Cdt(30-120)
Plasmid#83841PurposeFluorescent probe for G1/S transition and M/G1 transitionDepositorUseLentiviralExpressionMammalianPromoterCMVAvailable SinceJan. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
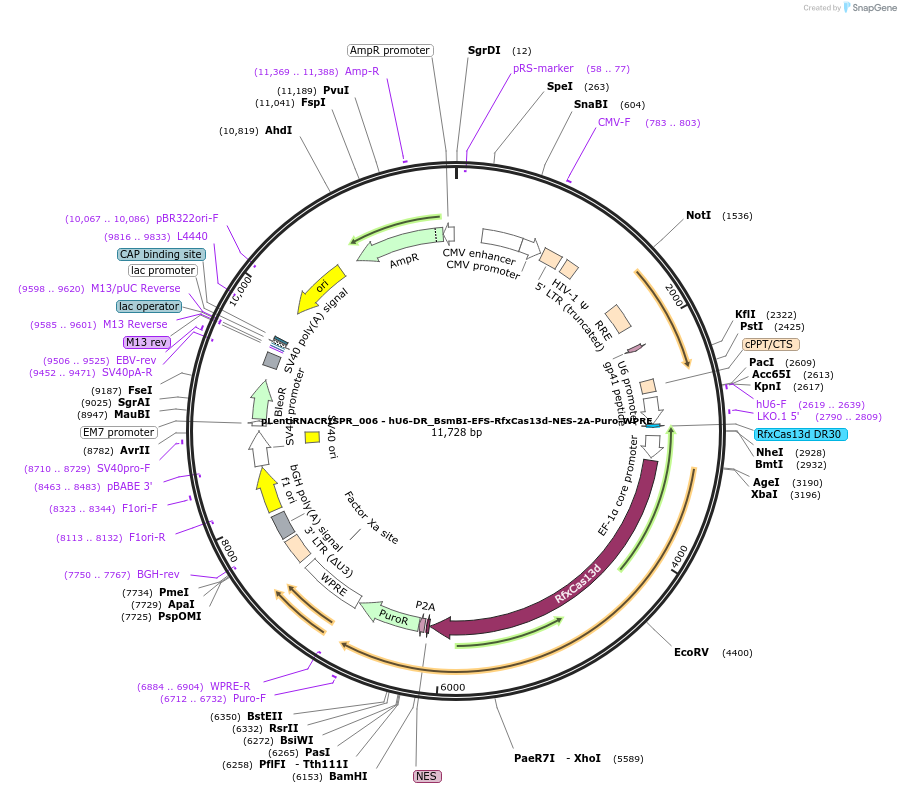
pLentiRNACRISPR_006 - hU6-DR_BsmBI-EFS-RfxCas13d-NES-2A-Puro-WPRE
Plasmid#138148PurposeExpresses RfxCas13d in mammalian cells, localized in the cytoplasm. For cloning of guide RNAs compatible with RfxCas13d. Contains a 5' direct repeat. Clone using BsmBI. F overhang aaac. R overhang aaaaDepositorInsertCasRx
UseLentiviralTagsNESAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
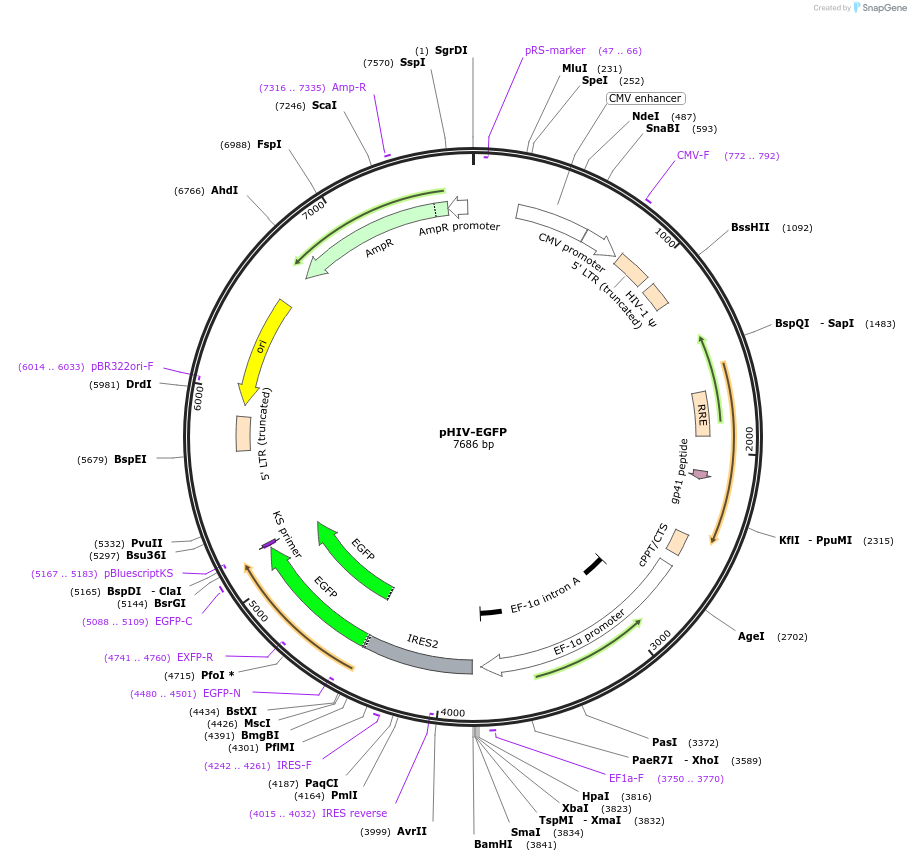
pHIV-EGFP
Plasmid#21373Purposeself-inactivating lentiviral plasmid for co-expression of your gene of interest and EGFPDepositorHas ServiceCloning Grade DNATypeEmpty backboneUseLentiviralExpressionMammalianAvailable SinceJune 19, 2009AvailabilityAcademic Institutions and Nonprofits only -
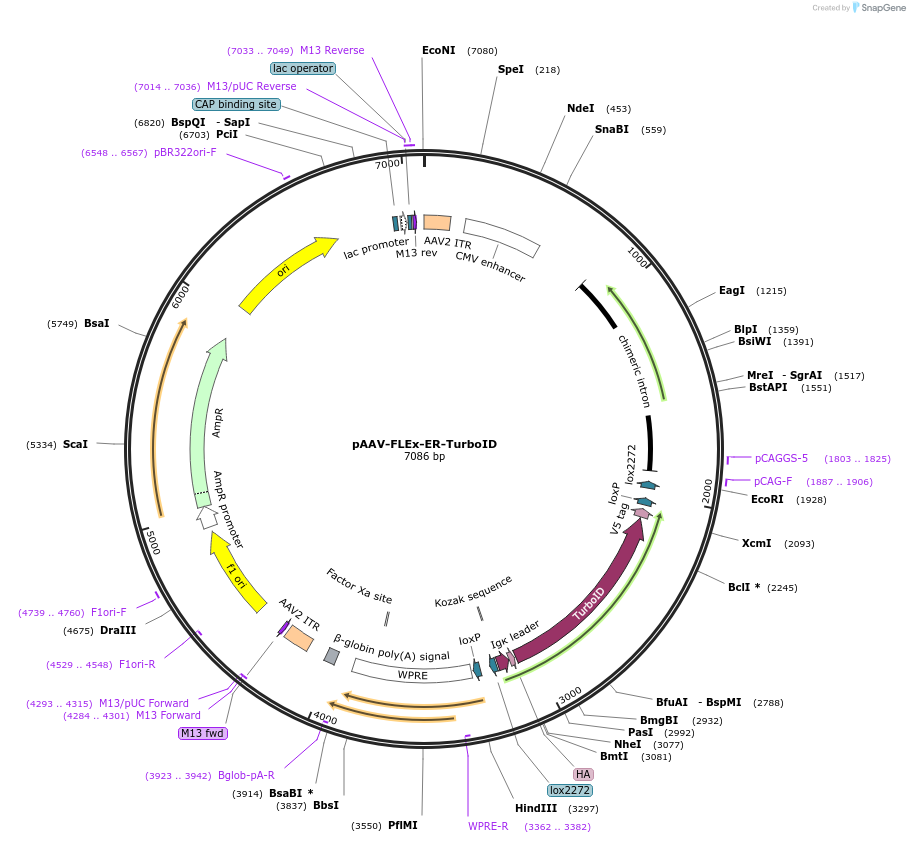
pAAV-FLEx-ER-TurboID
Plasmid#160857PurposeCre recombinase-dependent expression of ER TurboID, N-term HA tag, C-term V5 tag, in AAV expression plasmid. Amp selectionDepositorHas ServiceAAV8InsertER TurboID
UseAAVTagsHA and V5PromoterCAGAvailable SinceNov. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
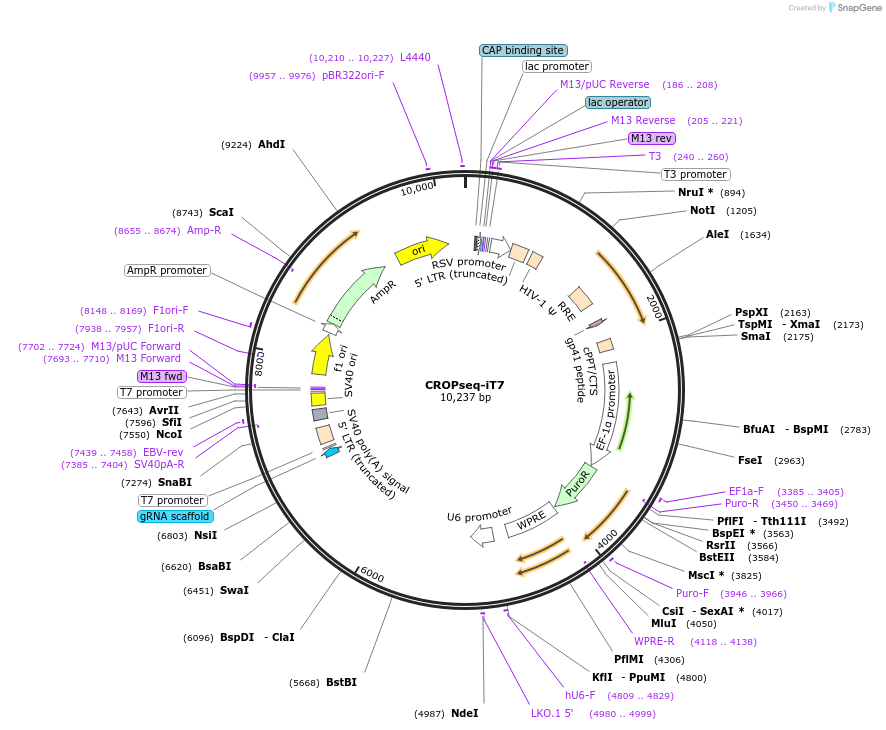
CROPseq-iT7
Plasmid#211699PurposeNIS-Seq-compatible lentiviral vector for expressing sgRNAsDepositorTypeEmpty backboneUseCRISPR and LentiviralExpressionMammalianAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
pAAV-Ef1a-fDIO-GCaMP6s (AAV8)
Viral Prep#105714-AAV8PurposeReady-to-use AAV8 particles produced from pAAV-Ef1a-fDIO-GCaMP6s (#105714). In addition to the viral particles, you will also receive purified pAAV-Ef1a-fDIO-GCaMP6s plasmid DNA. EF1a-driven, Flp recombinase-dependent expression of GCaMP6s calcium sensor. These AAV preparations are suitable purity for injection into animalsDepositorPromoterEf1a, Human elongation factor-1 alphaAvailable SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
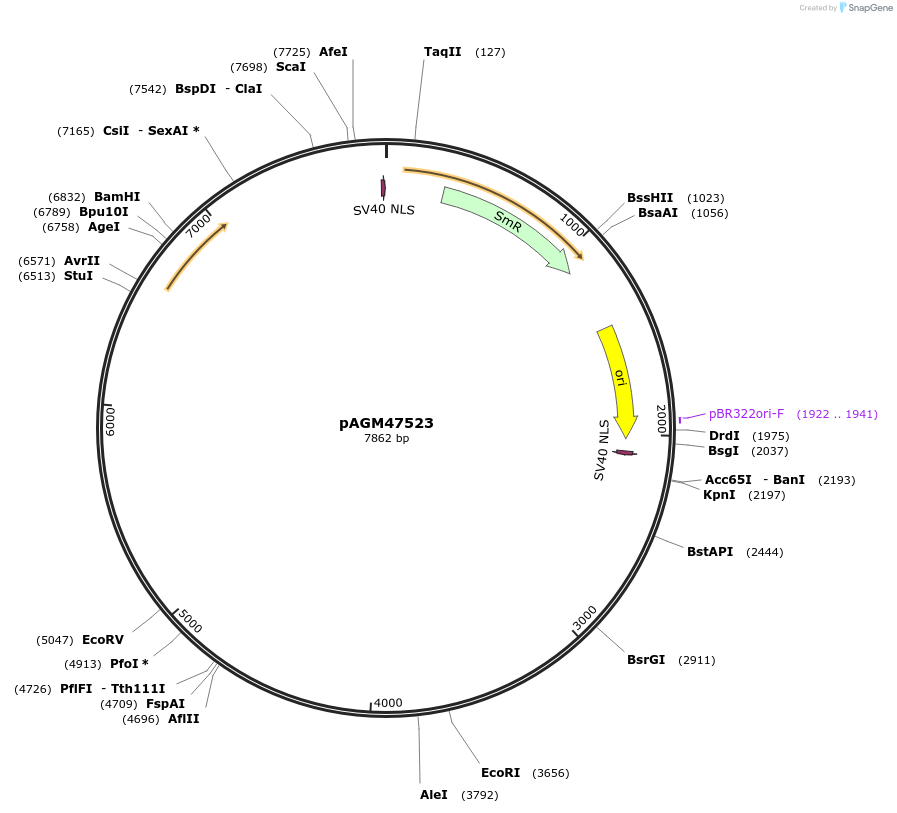
pAGM47523
Plasmid#153221PurposeLevel 0 zCas9i vectorDepositorInsertSpCas9 with introns and 2 NLS
UseSynthetic BiologyAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
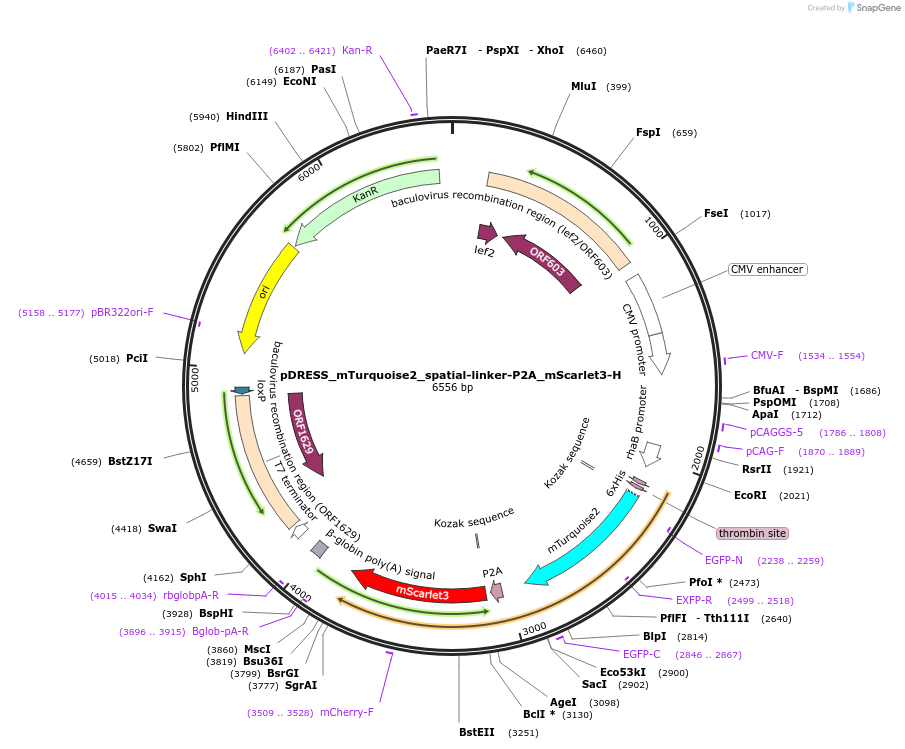
pDRESS_mTurquoise2_spatial-linker-P2A_mScarlet3-H
Plasmid#242971PurposeProduces an mTurquoise-antiFRET-mScarlet3 fusion protein in bacteria and in mamallian cells it produces 1:1 unfused mTurquoise2 and mScarlet3-H FPs for ratiometric analysisDepositorInsertmScarlet3-H
ExpressionBacterial and MammalianPromoterCMV & RhaAvailable SinceNov. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
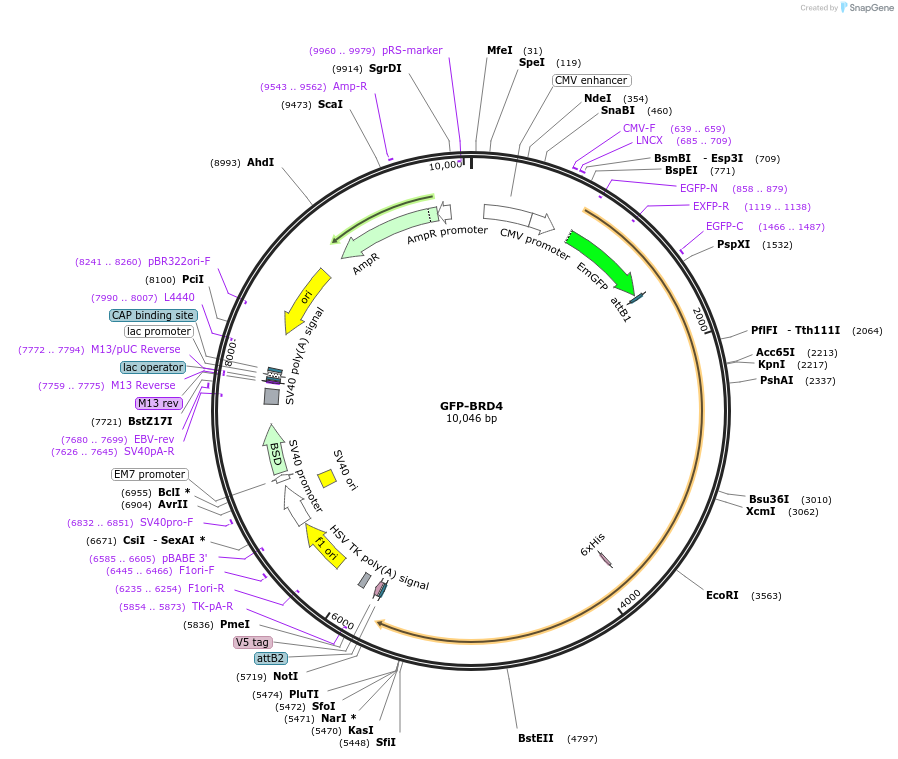
GFP-BRD4
Plasmid#65378PurposeGFP-tagged BRD protein, which can be used to be expressed in mammalian cells.DepositorAvailable SinceJune 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
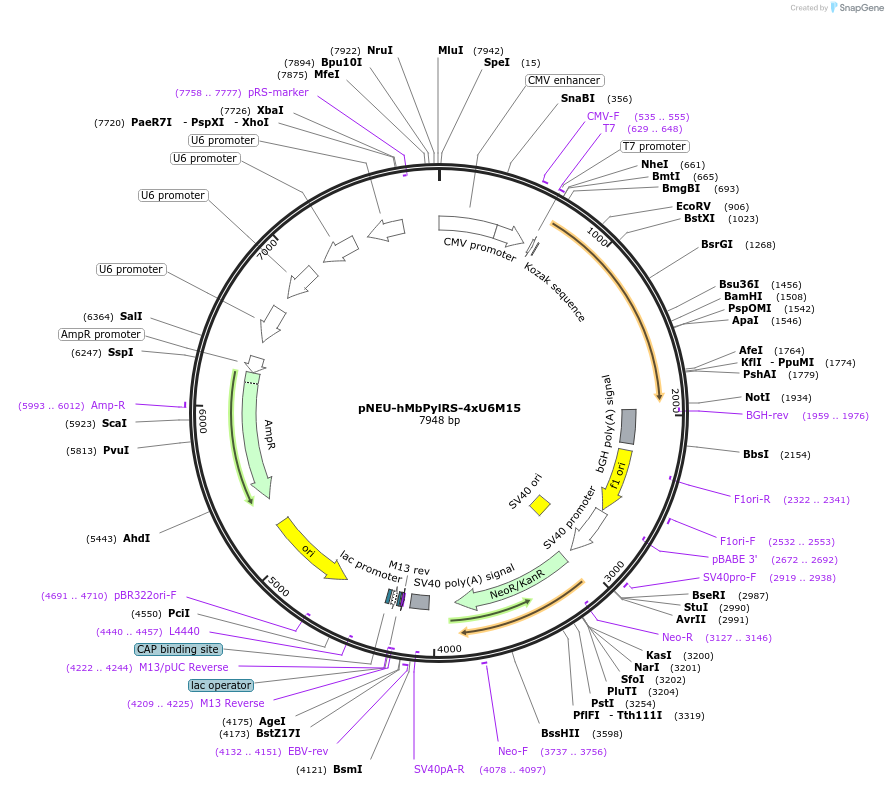
pNEU-hMbPylRS-4xU6M15
Plasmid#105830PurposePlasmid encoding for the Pyrrolysine translational pair that enables efficent incoporation of Pyl-like click amino acids in response to the amber codon into proteins in mammalian cellsDepositorInsertsHumanized MbPylRS
tRNAM15
ExpressionMammalianMutation(Y271A, Y349F)MbPylRSPromoterCMV and U6Available SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
pNP3425 [pNP2922 - ISCro4 Enhanced bRNA (TBL4, WT + DBL3, WT) + ISCro4 Recombinase (WT)]
Plasmid#247262PurposeExpress ISCro4 recombinase and bridge RNADepositorInsertpNP2922 - ISCro4 bRNA1-179 C60U, A61G, U73C, dU86, A87G, dU88, ins179(rc(101-111) (TBL4+DBL3)
ExpressionMammalianMutationC60U, A61G, U73C, dU86, A87G, dU88, ins179(rc(101…Available SinceOct. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
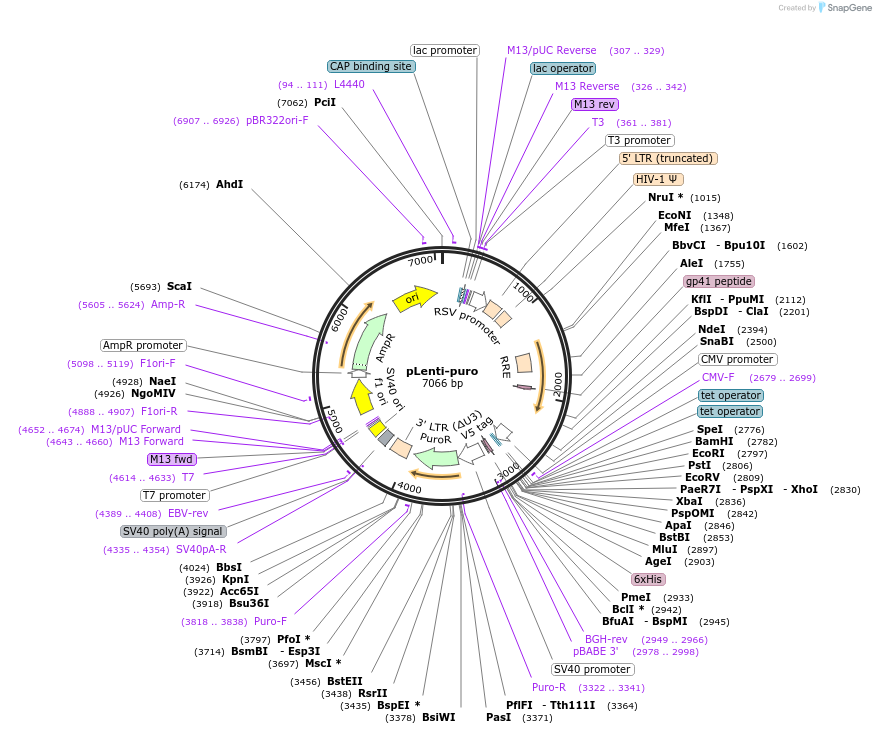
pLenti-puro
Plasmid#39481PurposeTetracycline inducible 3rd generation lentiviral vectorDepositorHas ServiceCloning Grade DNATypeEmpty backboneUseLentiviralTagsV5 and 6XHisExpressionMammalianPromoterCMVAvailable SinceSept. 13, 2012AvailabilityAcademic Institutions and Nonprofits only -
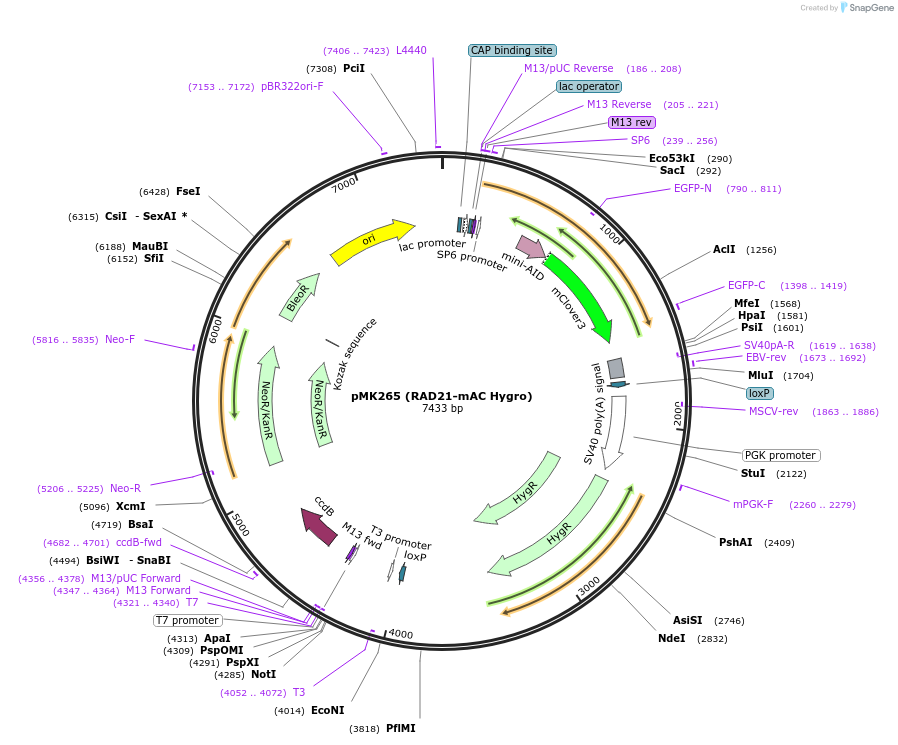
pMK265 (RAD21-mAC Hygro)
Plasmid#140539PurposeRAD21 tagging with mAID-CloverDepositorAvailable SinceNov. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
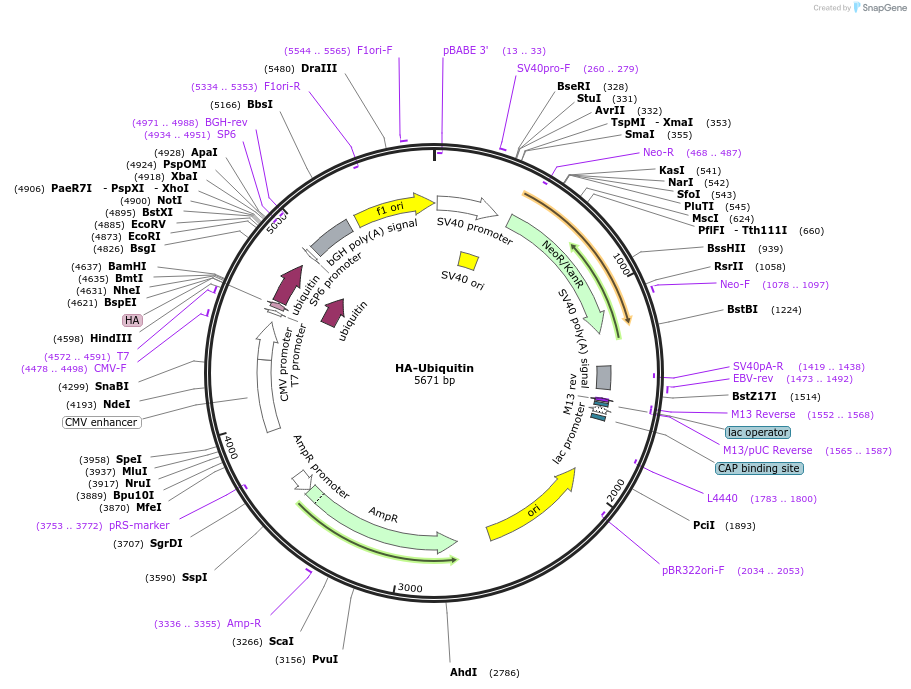
HA-Ubiquitin
Plasmid#18712PurposeMammalian expression of human ubiquitin with HA tagDepositorInsertubiquitin (UBB Human)
TagsHAExpressionMammalianMutationThe HA-Ubiquitin insert is from the UBB gene. The…Available SinceJune 20, 2008AvailabilityAcademic Institutions and Nonprofits only -
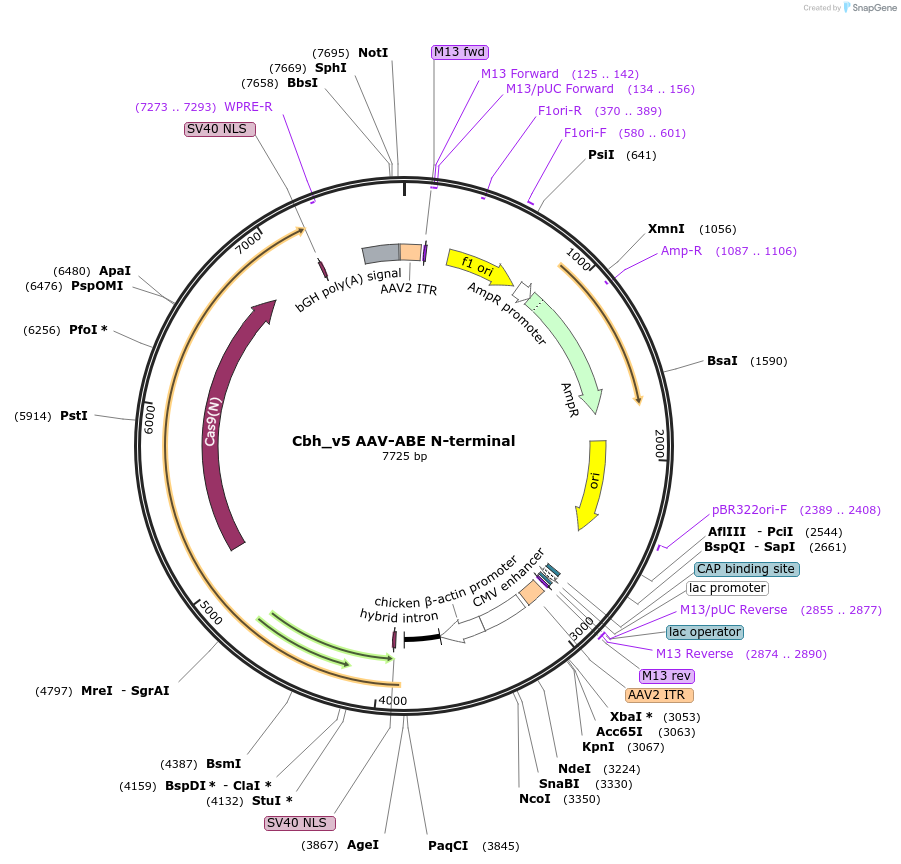
Cbh_v5 AAV-ABE N-terminal
Plasmid#137177PurposeAAV genome: expresses the N-terminal of v5 AAV-ABE from the Cbh promoterDepositorInsertv5 AAV-ABE N-terminal
UseAAVMutationCas9 D10APromoterCbhAvailable SinceJan. 30, 2020AvailabilityAcademic Institutions and Nonprofits only -
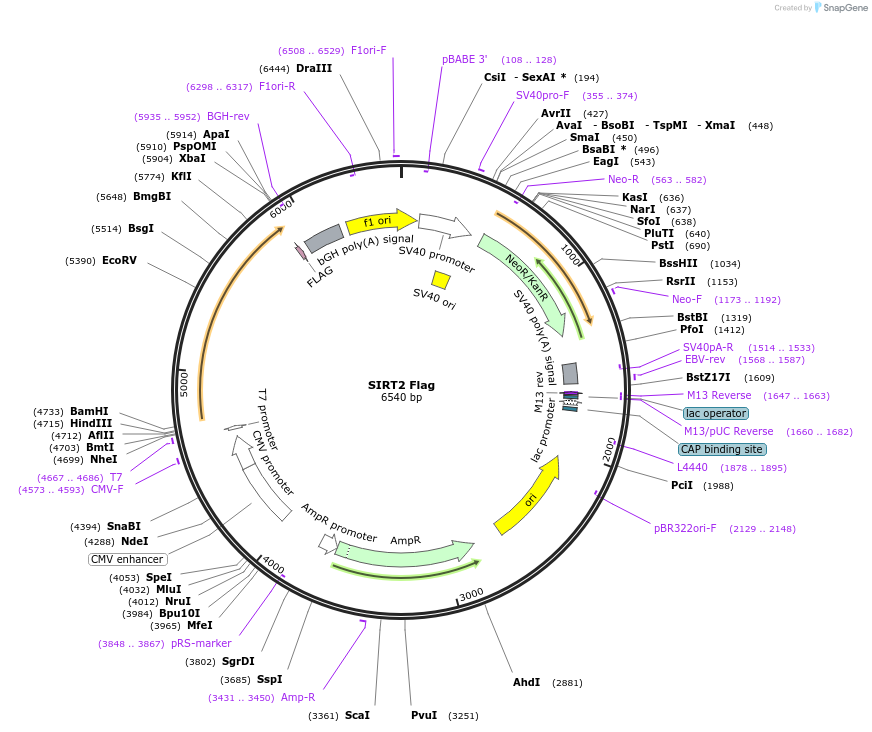
SIRT2 Flag
Plasmid#13813PurposeMammalian expression of human SIRT2 with flag tagDepositorAvailable SinceFeb. 19, 2007AvailabilityAcademic Institutions and Nonprofits only -
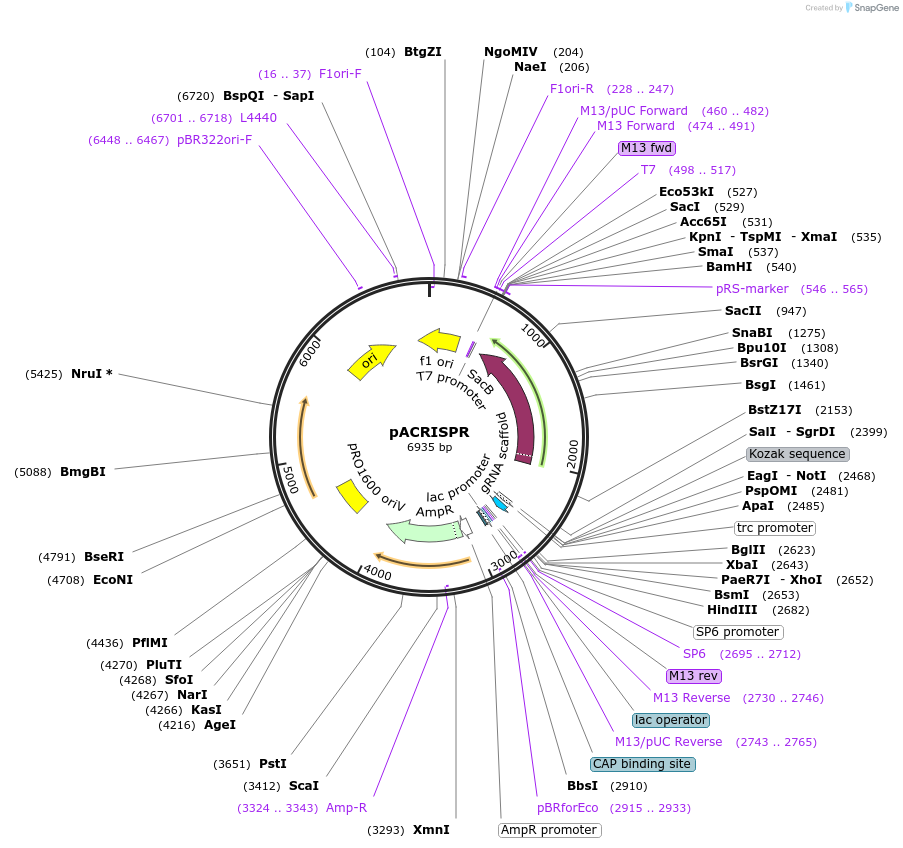
pACRISPR
Plasmid#113348PurposeA sgRNA expression plasmid for genome editing in Pseudomonas aeruginosaDepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionBacterialAvailable SinceAug. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
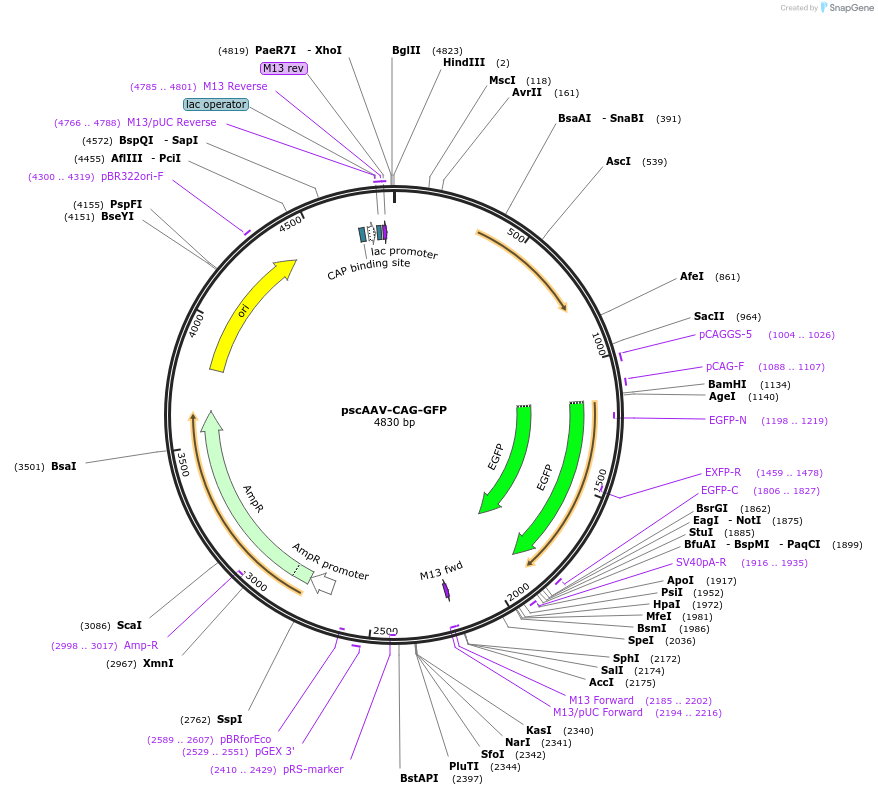
pscAAV-CAG-GFP
Plasmid#83279Purposerecombinant AAV vector packaging self-complementary GFP under the CAG promoterDepositorInsertenhanced green fluorescent protein
UseAAVExpressionMammalianPromoterCAGAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
pAAV/D377Y-mPCSK9 (AAV8)
Viral Prep#58376-AAV8PurposeReady-to-use AAV8 particles produced from pAAV/D377Y-mPCSK9 (#58376). In addition to the viral particles, you will also receive purified pAAV/D377Y-mPCSK9 plasmid DNA. hAAT-driven expression of mutant (D377Y) murine PCSK9. These AAV preparations are suitable purity for injection into animals.DepositorPromoterHCRApoE/hAATAvailable SinceJuly 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
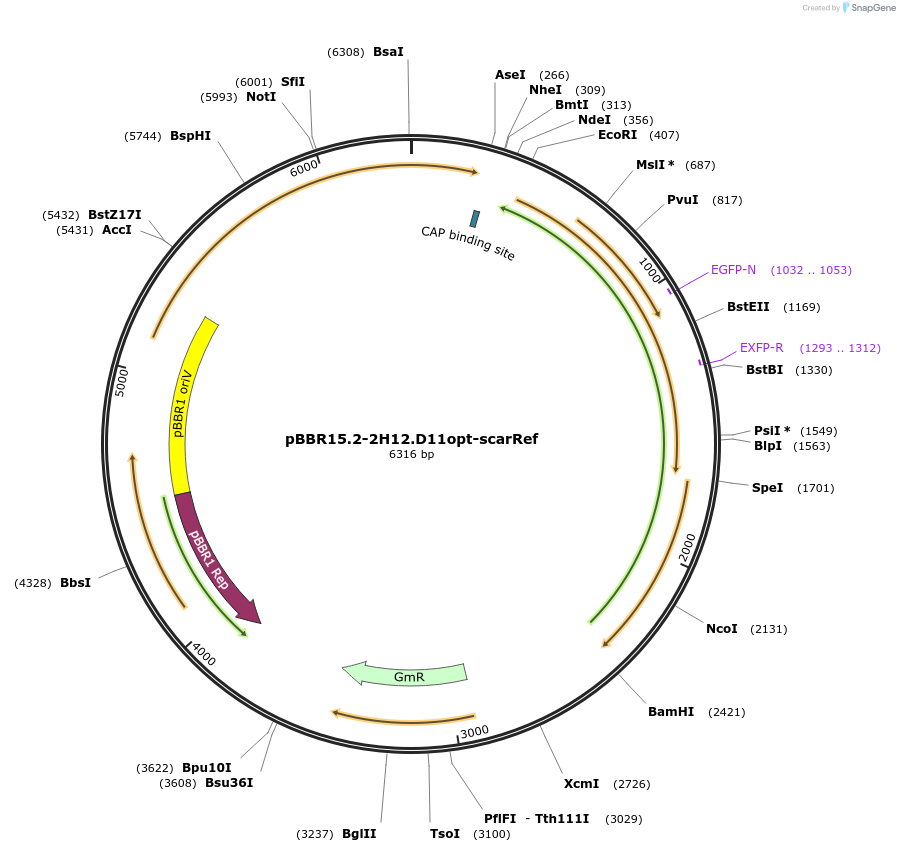
pBBR15.2-2H12.D11opt-scarRef
Plasmid#221150PurposecdGreen2.1 (optimized for Pseudomonas GC-content) expressed from a constitutive promoter in an operon with mScarlet-I (downstream) as a reference FP (Internal Lab ID: UJ11241)DepositorInsertscdGreen2.1
mScarlet-I
ExpressionBacterialMutationN-terminus changed: starts with MSKKYGEAVIKE ...PromoterConstitutive and None (same as cdGreen2.1)Available SinceJune 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
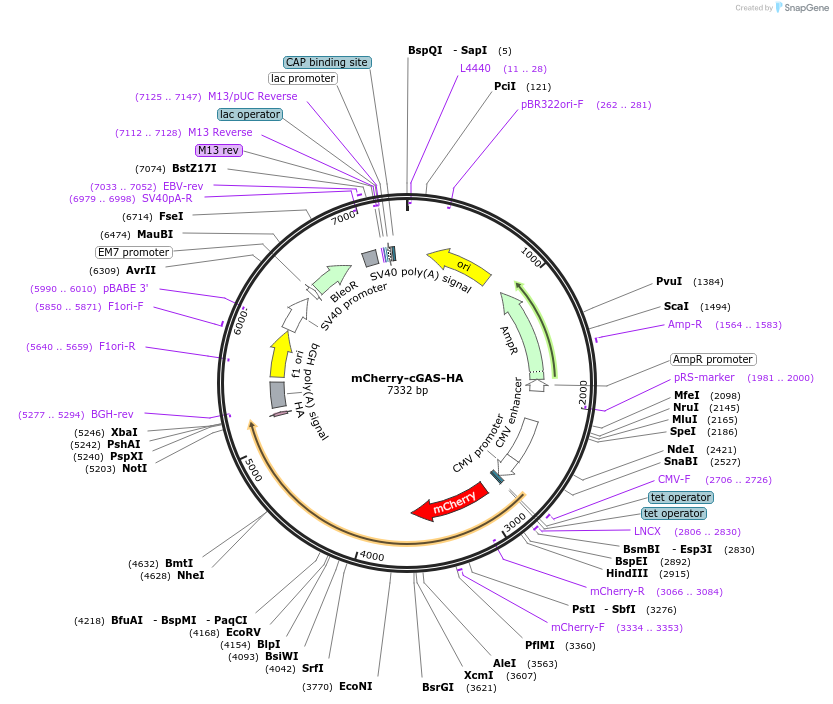
mCherry-cGAS-HA
Plasmid#231895PurposeExpression of cyclic GMP/AMP synthase (cGAS), as a nuclear rupture markerDepositorAvailable SinceFeb. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
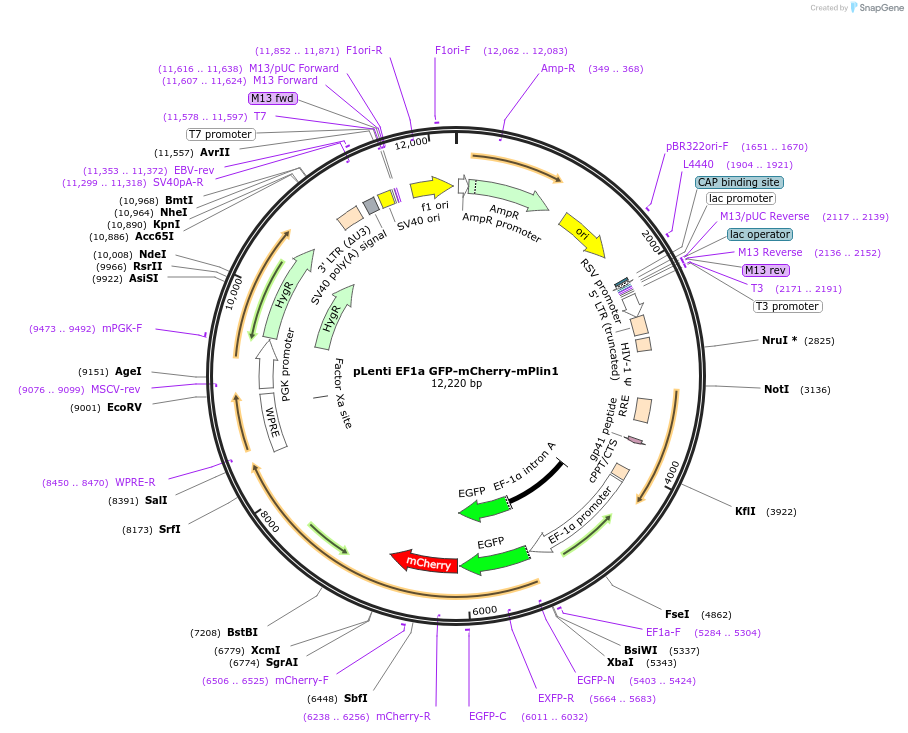
pLenti EF1a GFP-mCherry-mPlin1
Plasmid#189005PurposeFluorescent reportor for lipophagyDepositorInsertGFP, mChery, Plin1 (Plin1 Mouse)
UseLentiviralAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
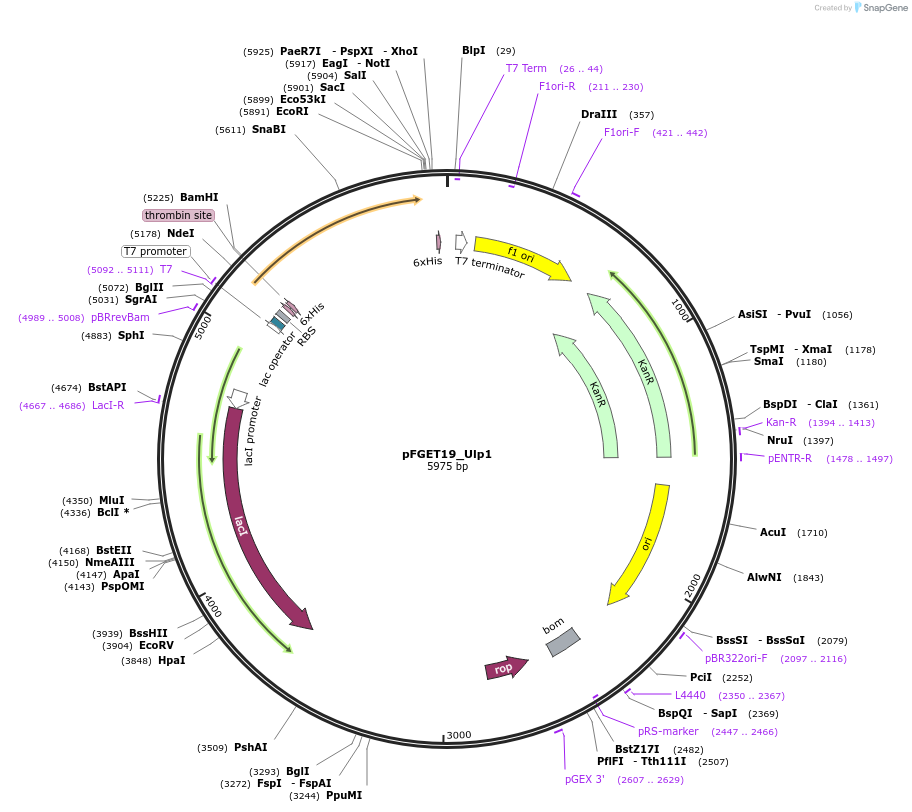
pFGET19_Ulp1
Plasmid#64697PurposeH6-Ulp1 from pHYRS52 into pET-28b(+)DepositorInsertULP1 (ULP1 Saccharomyces cerevisiae, Budding Yeast)
Tags6xHisExpressionBacterialMutationUlp1(403-621)PromoterT7Available SinceSept. 11, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAAV-Ef1a-DIO hChR2(E123T/T159C)-EYFP (AAV5)
Viral Prep#35509-AAV5PurposeReady-to-use AAV5 particles produced from pAAV-Ef1a-DIO hChR2(E123T/T159C)-EYFP (#35509). In addition to the viral particles, you will also receive purified pAAV-Ef1a-DIO hChR2(E123T/T159C)-EYFP plasmid DNA. EF1a-driven, Cre-dependent, humanized channelrhodopsin E123T/T159C mutant fused to EYFP for optogenetic activation. These AAV preparations are suitable purity for injection into animals.DepositorPromoterEF1aTagsEYFP (Cre-dependent)Available SinceFeb. 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
AAV-EF1a-BbChT (AAV9)
Viral Prep#45186-AAV9PurposeReady-to-use AAV9 particles produced from AAV-EF1a-BbChT (#45186). In addition to the viral particles, you will also receive purified AAV-EF1a-BbChT plasmid DNA. Brainbow construct encoding mCherry and mTFP, both in reverse orientation between mutant Lox sites. These AAV preparations are suitable purity for injection into animals.DepositorPromoterEF1aTagsmCherry, mTFP, or noneAvailable SinceApril 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
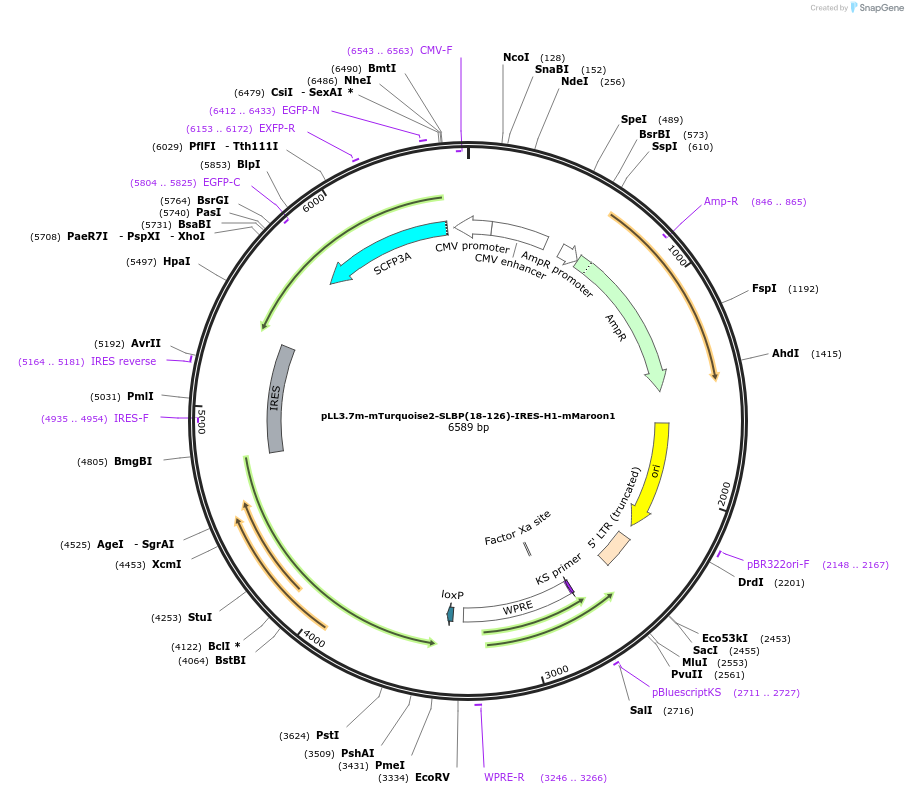
pLL3.7m-mTurquoise2-SLBP(18-126)-IRES-H1-mMaroon1
Plasmid#83842PurposeFluorescent probe for S/G2 cell cycle transition and G2/M transitionDepositorUseLentiviralExpressionMammalianPromoterCMVAvailable SinceNov. 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
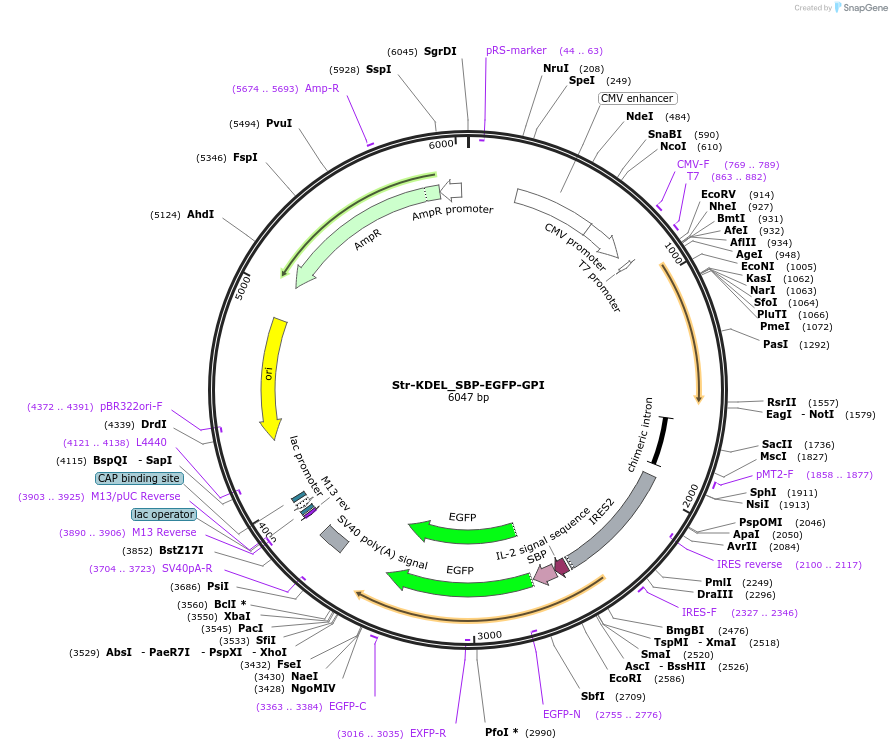
Str-KDEL_SBP-EGFP-GPI
Plasmid#65294Purposesynchronize trafficking of EGFP-GPI from the ER (RUSH system)DepositorInsertGPI anchor
TagsEGFP and IL-2 Signal Sequence and Streptavidin Bi…ExpressionMammalianPromoterCMVAvailable SinceJune 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
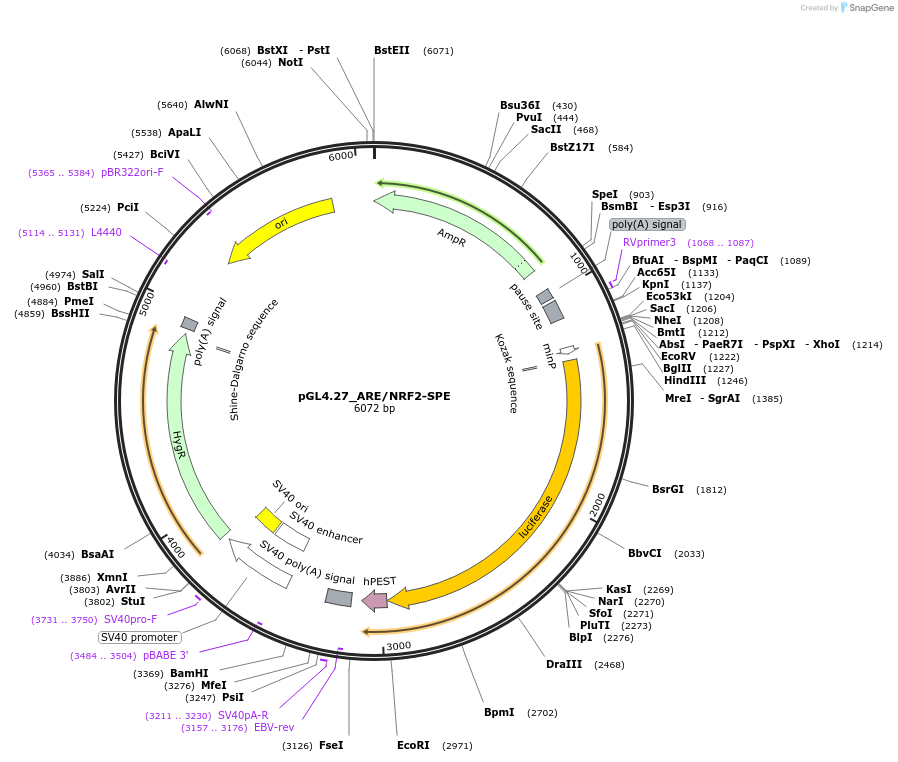
pGL4.27_ARE/NRF2-SPE
Plasmid#177775PurposeLuciferase reporter for ARE/NRF2DepositorInsertNRF2 (NFE2L2 Human)
UseLuciferaseAvailable SinceApril 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
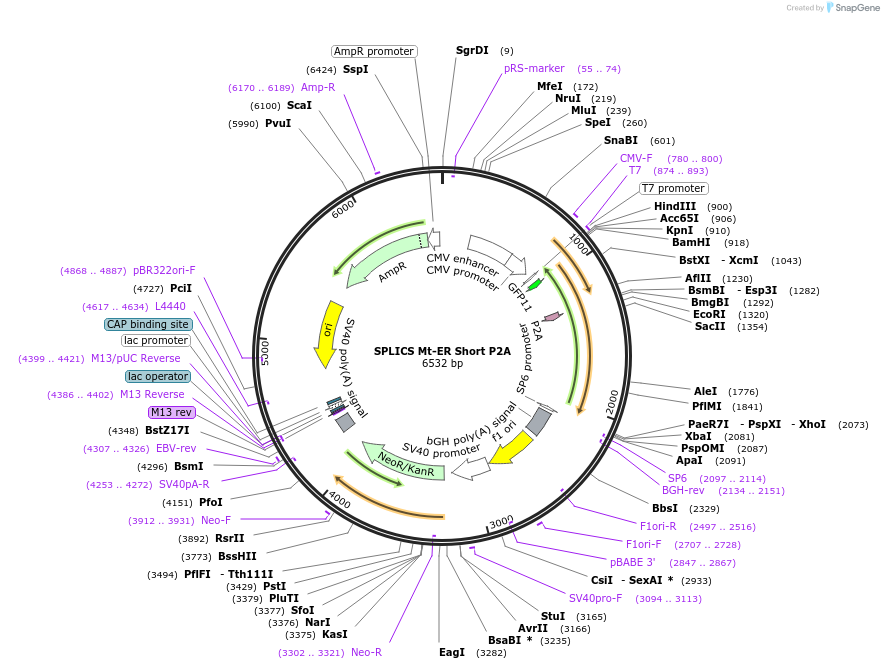
SPLICS Mt-ER Short P2A
Plasmid#164108PurposeDetect the short Mitochondria-Endoplasmic reticulum contactDepositorInsertSplit GFP Mt-ER Short P2A
ExpressionMammalianAvailable SinceJan. 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
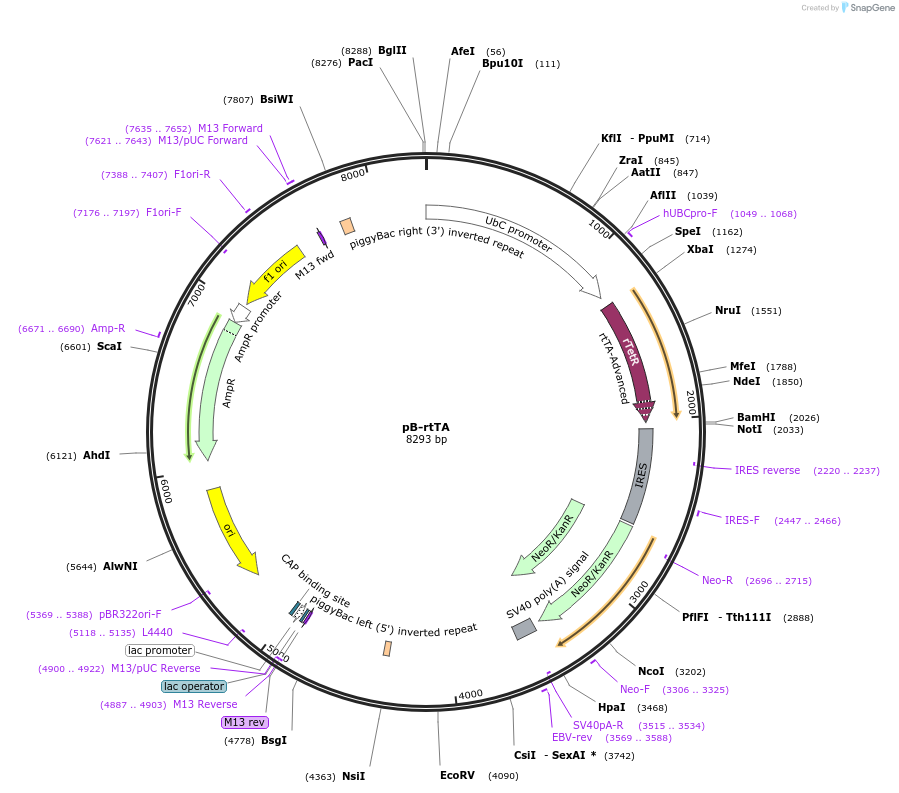
pB-rtTA
Plasmid#126034PurposePiggyBac cargo vector expressing rtTA. Also confers resistance to G418.DepositorInsertrtTA3-IRES-Neo cassette cloned from Addgene Plasmid #25735
UsePiggybacExpressionMammalianPromoterhUbiCAvailable SinceJune 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
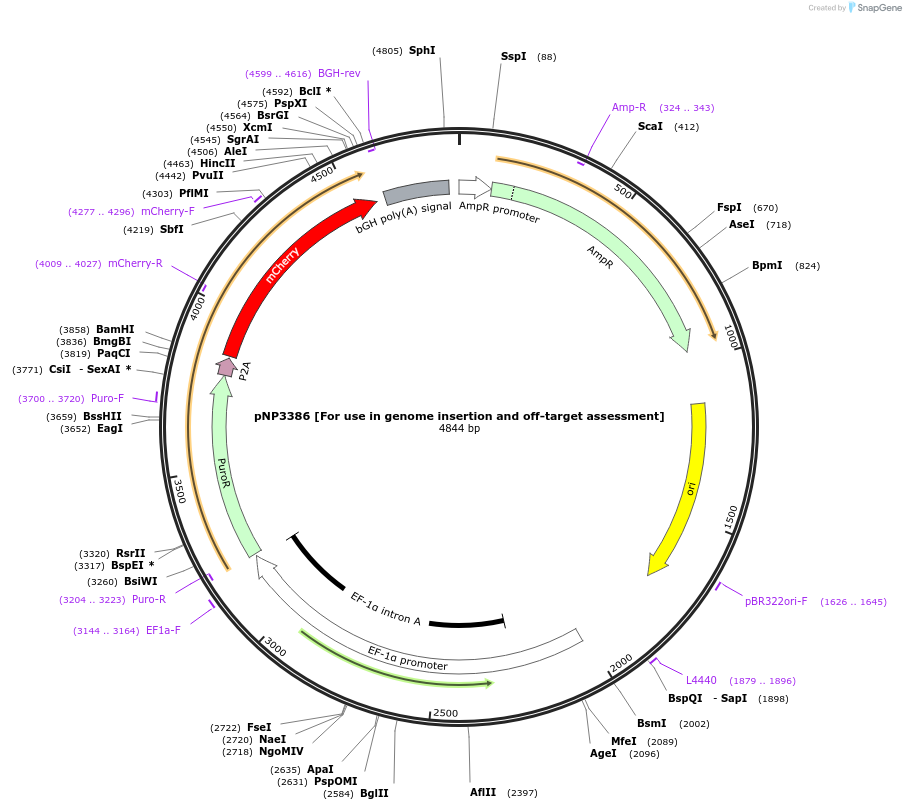
pNP3386 [For use in genome insertion and off-target assessment]
Plasmid#247258PurposeExample plasmid for bridge recombinase mediated insertion and off-target assessment with puromycin selection — must clone in UMIDepositorInsertISCro4 Recognition Site-UMI-Ef1a-PuroR-P2A-mCherry (Plasmid for Insertion Site Mapping [Example, Clone in Sequence + UMI])
ExpressionMammalianAvailable SinceOct. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
pENN.AAV.CamKII 0.4.Cre.SV40 (AAV5)
Viral Prep#105558-AAV5PurposeReady-to-use AAV5 particles produced from pENN.AAV.CamKII 0.4.Cre.SV40 (#105558). In addition to the viral particles, you will also receive purified pENN.AAV.CamKII 0.4.Cre.SV40 plasmid DNA. CamKII-driven Cre expression. These AAV preparations are suitable purity for injection into animals.DepositorPromoterCaMKIIAvailable SinceAug. 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
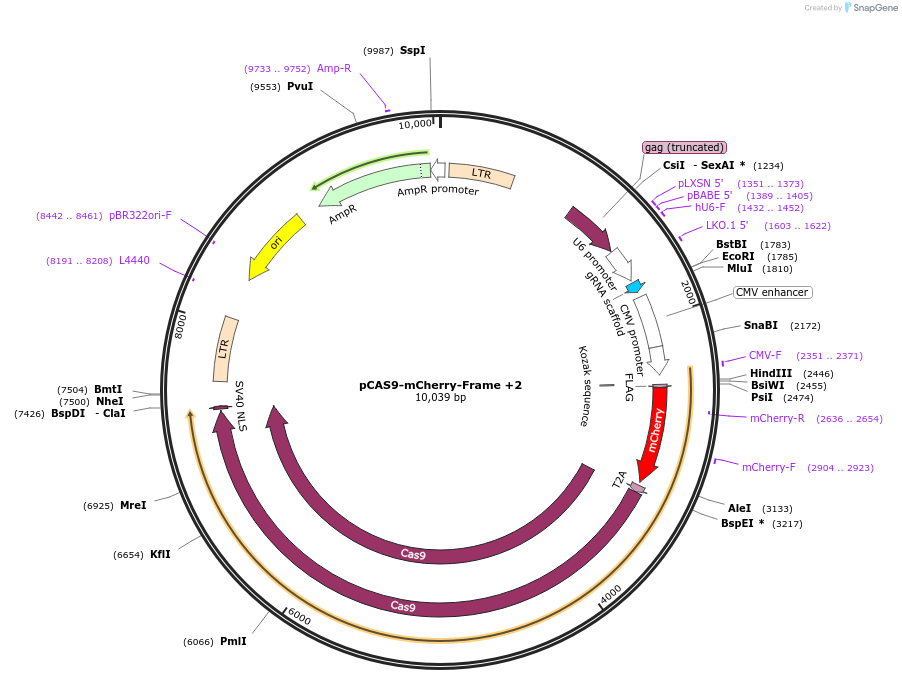
pCAS9-mCherry-Frame +2
Plasmid#66941PurposeCRISPaint frame selector +2DepositorInsertgRNA frame +2
Available SinceSept. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
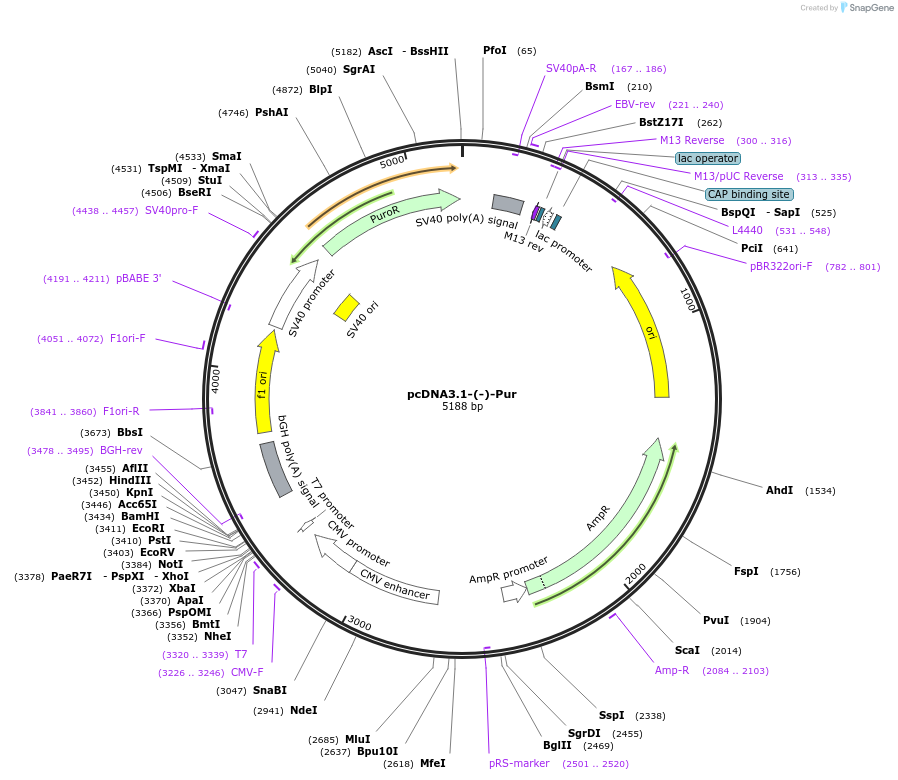
pcDNA3.1-(-)-Pur
Plasmid#200458PurposeEukaryotic expression vectorDepositorTypeEmpty backboneExpressionMammalianAvailable SinceMay 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
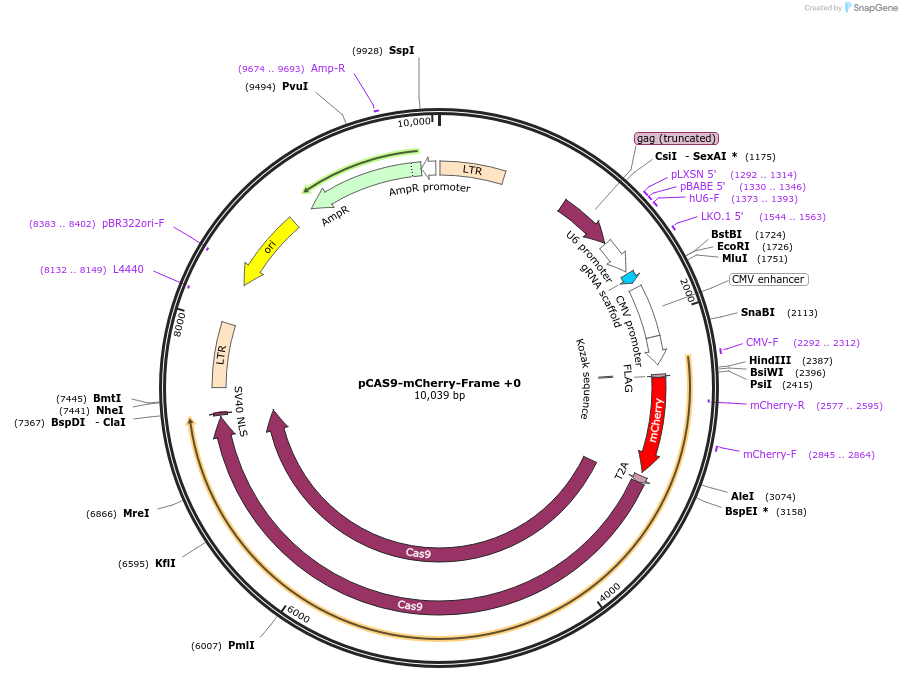
pCAS9-mCherry-Frame +0
Plasmid#66939PurposeCRISPaint frame selector +0DepositorInsertgRNA frame +0
Available SinceSept. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
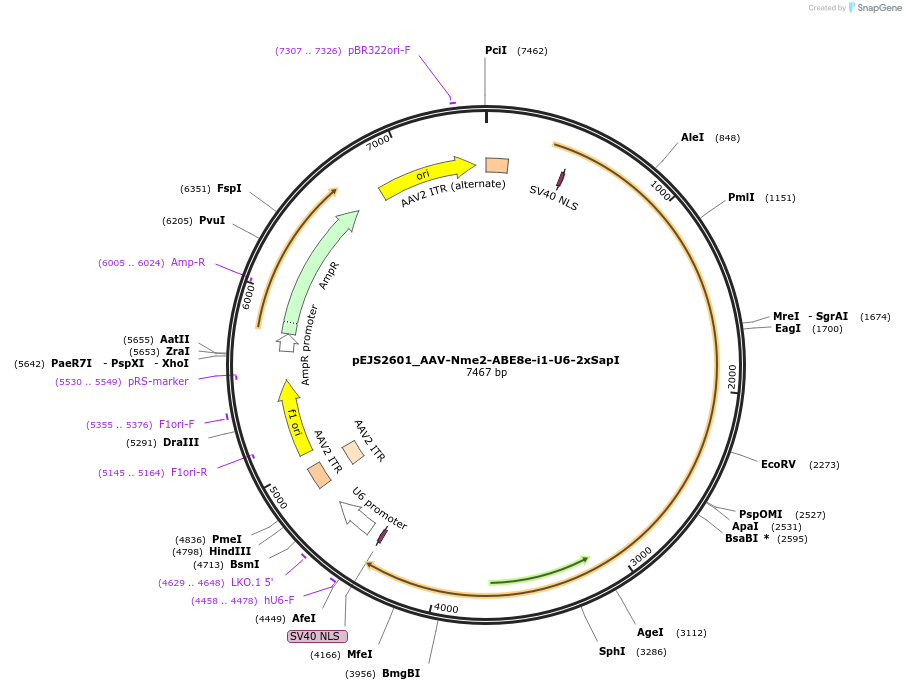
pEJS2601_AAV-Nme2-ABE8e-i1-U6-2xSapI
Plasmid#201683PurposeAll-in-one AAV plasmid expressing Nme2-ABE8e-i1 and U6 sgRNA cassette. SapI enables cloning new spacers.DepositorInsertNme2-ABE8e-i1
UseAAV and CRISPRExpressionMammalianPromoterU1aAvailable SinceJan. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
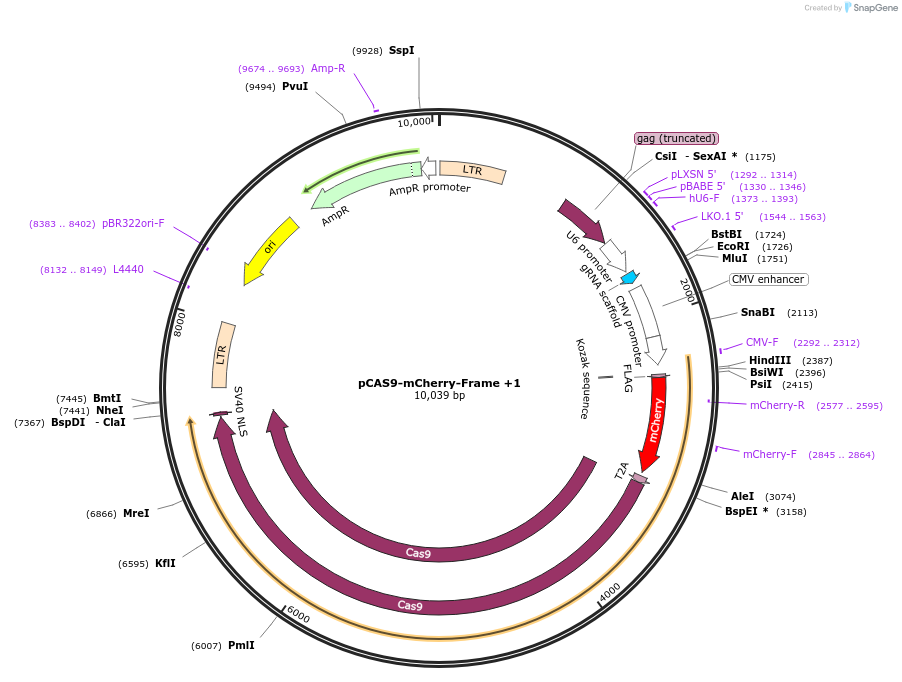
pCAS9-mCherry-Frame +1
Plasmid#66940PurposeCRISPaint frame selector +1DepositorInsertgRNA frame +1
Available SinceSept. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
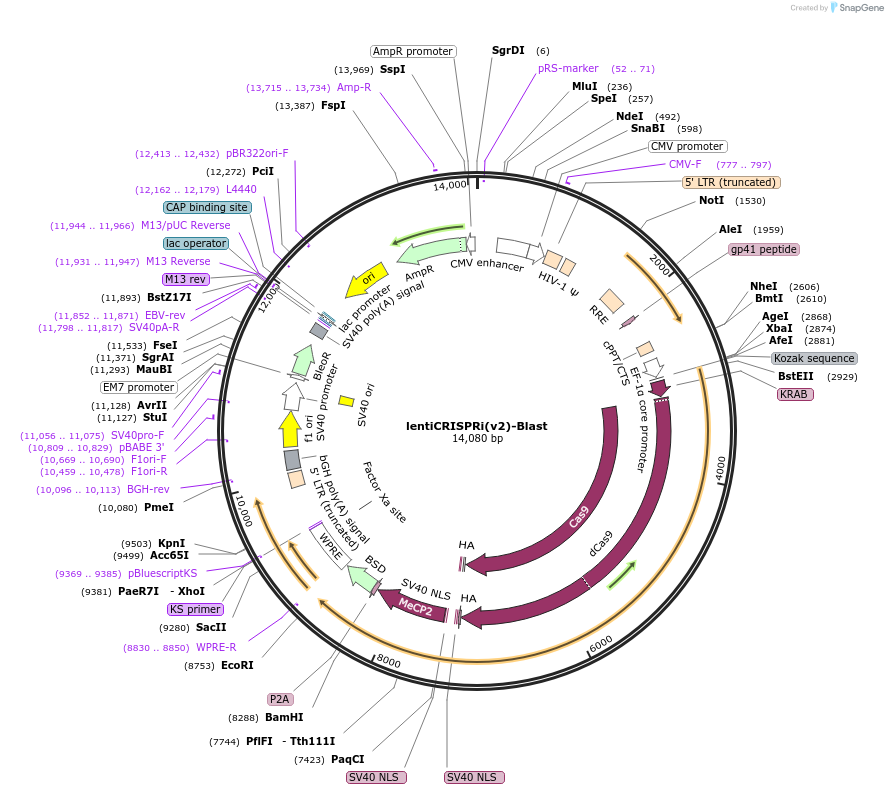
lentiCRISPRi(v2)-Blast
Plasmid#170068PurposeExpresses dCas9 repressor KRAB-dCas9-MeCP2 and blasticidin resistanceDepositorInsertKRAB-dCas9-MeCP2
UseCRISPR and LentiviralTags2A and 2xHA tagExpressionMammalianPromoterEFS-NSAvailable SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
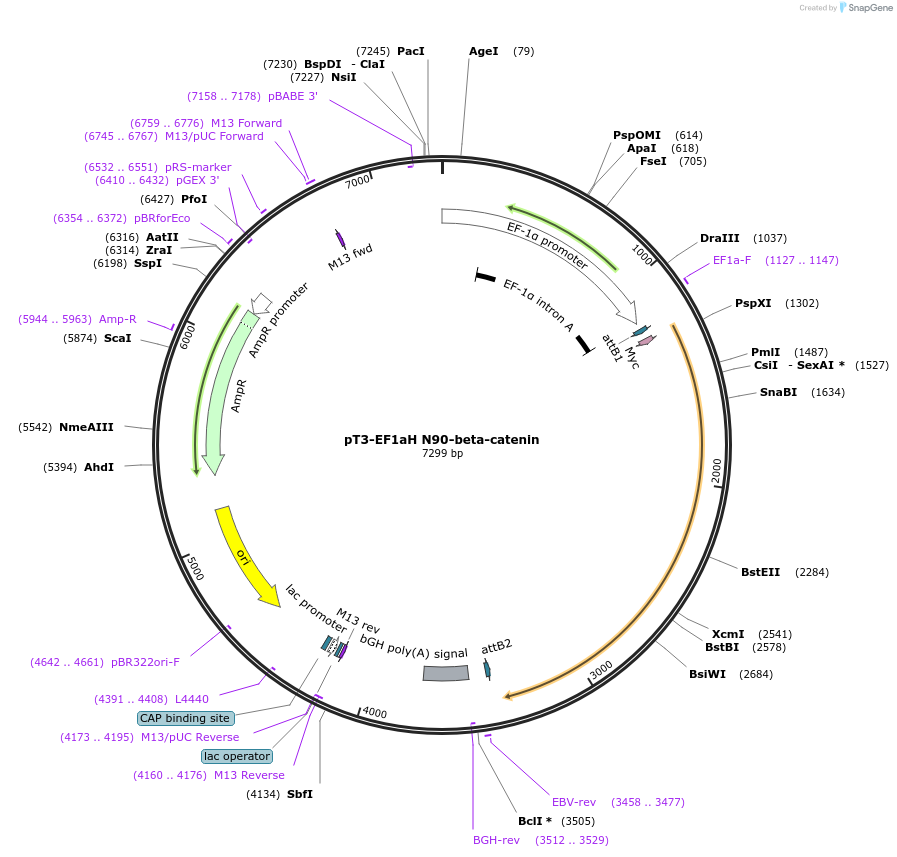
pT3-EF1aH N90-beta-catenin
Plasmid#86499PurposeMammalian expression of Myc tagged N90 beta-cateninDepositorInsertbeta-catenin (CTNNB1 Human)
TagsMycExpressionMammalianMutationDeletion of amino acids 1-90; constitutively acti…PromoterEF1aAvailable SinceFeb. 16, 2017AvailabilityAcademic Institutions and Nonprofits only