171,789 results
-
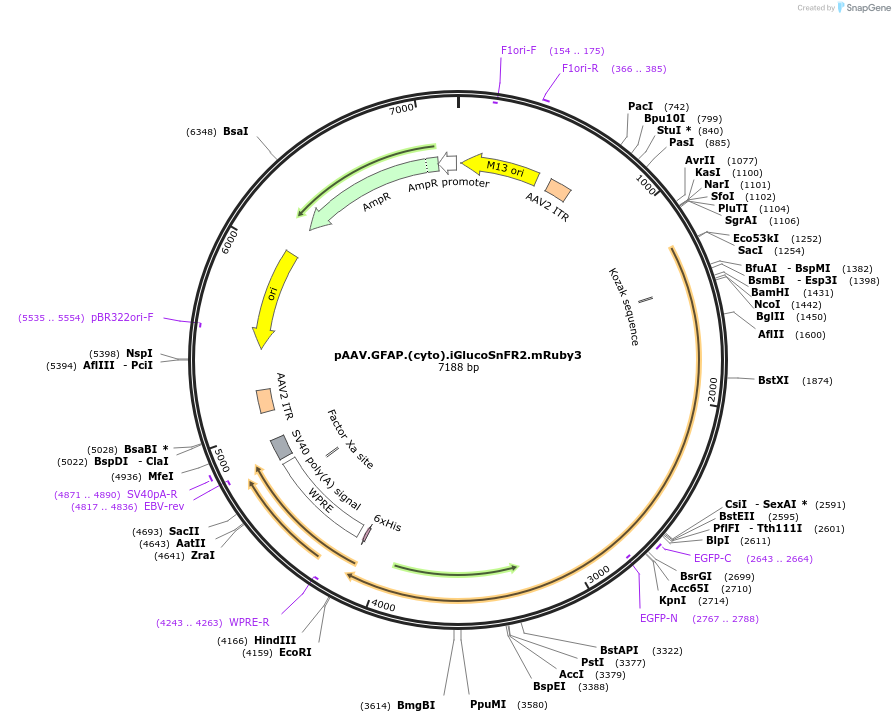
Plasmid#244084PurposeCytosolic expression of green glucose sensor with non-responsive mRuby3DepositorInsertiGlucoSnFR2.mRuby3
UseAAVTagsmRuby3Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
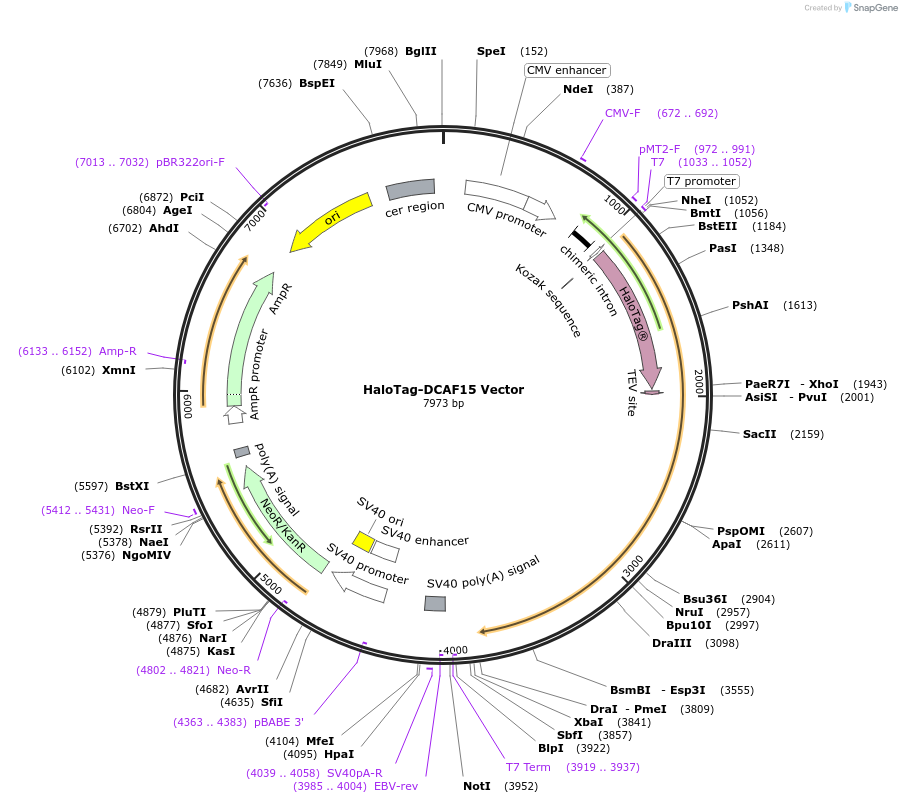
HaloTag-DCAF15 Vector
Plasmid#238731PurposeExpress HaloTag-DCAF15 Fusion Protein in Mammalian Cells under a CMV promoterDepositorHas ServiceDNAInsertDCAF15 (DCAF15 Human)
TagsHaloTag (R)ExpressionMammalianMutationNonePromoterCMVAvailable SinceSept. 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
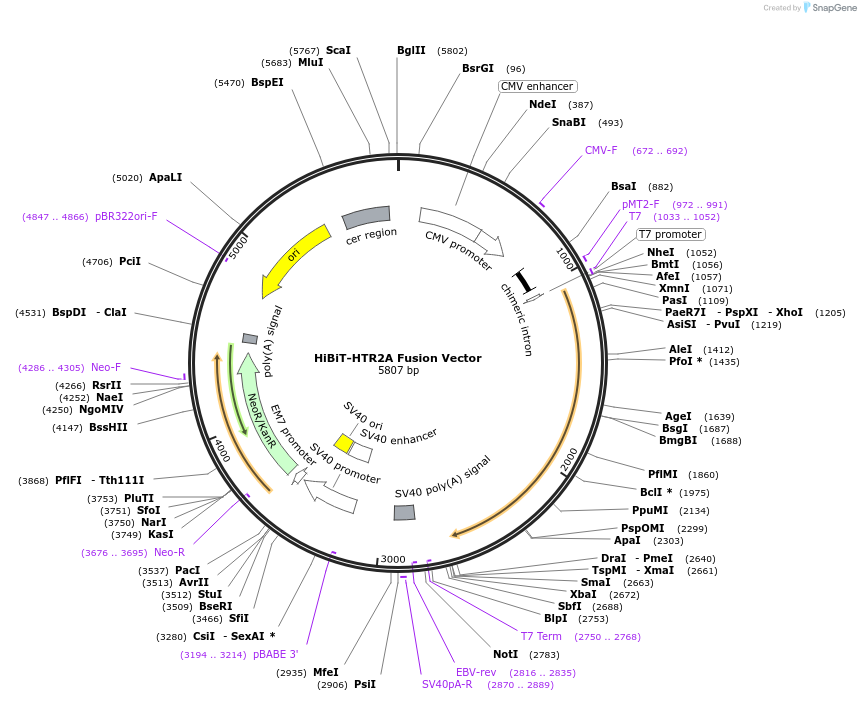
HiBiT-HTR2A Fusion Vector
Plasmid#238585PurposeExpress HiBiT-HTR2A Fusion Protein in Mammalian Cells under a CMV promoterDepositorHas ServiceDNAInsertHTR2A (HTR2A Human)
TagsHiBiTExpressionMammalianMutationNonePromoterCMVAvailable SinceSept. 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
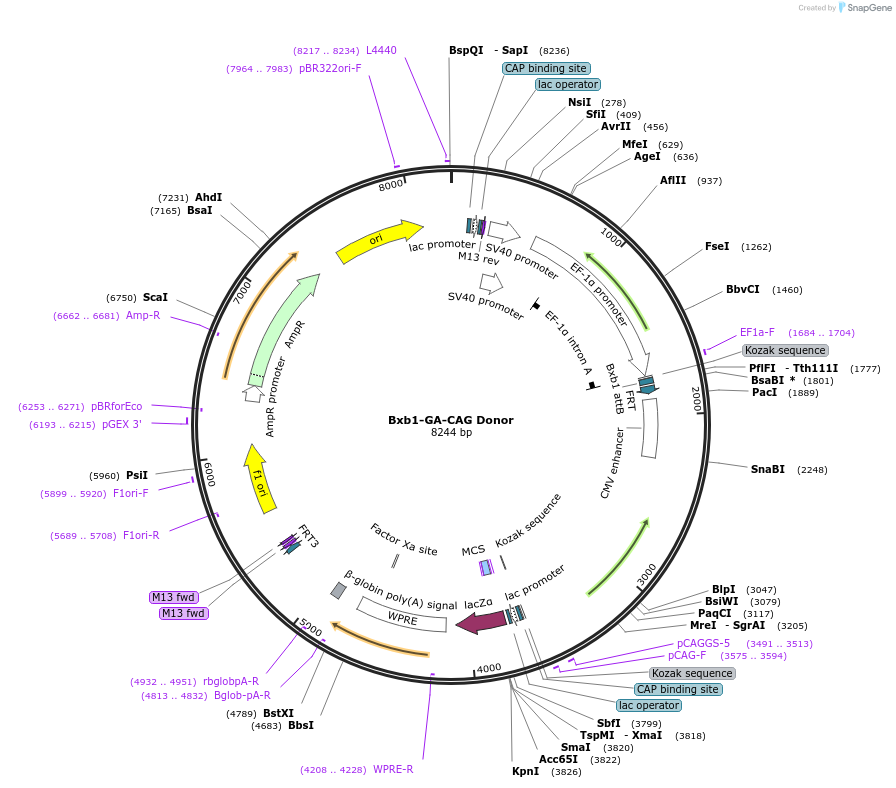
Bxb1-GA-CAG Donor
Plasmid#241374PurposeBxb1-GA donor plasmid with CAG promoter, WPRE and polyA signal for constitutive transgene expression (empty)DepositorTypeEmpty backboneExpressionMammalianAvailable SinceSept. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
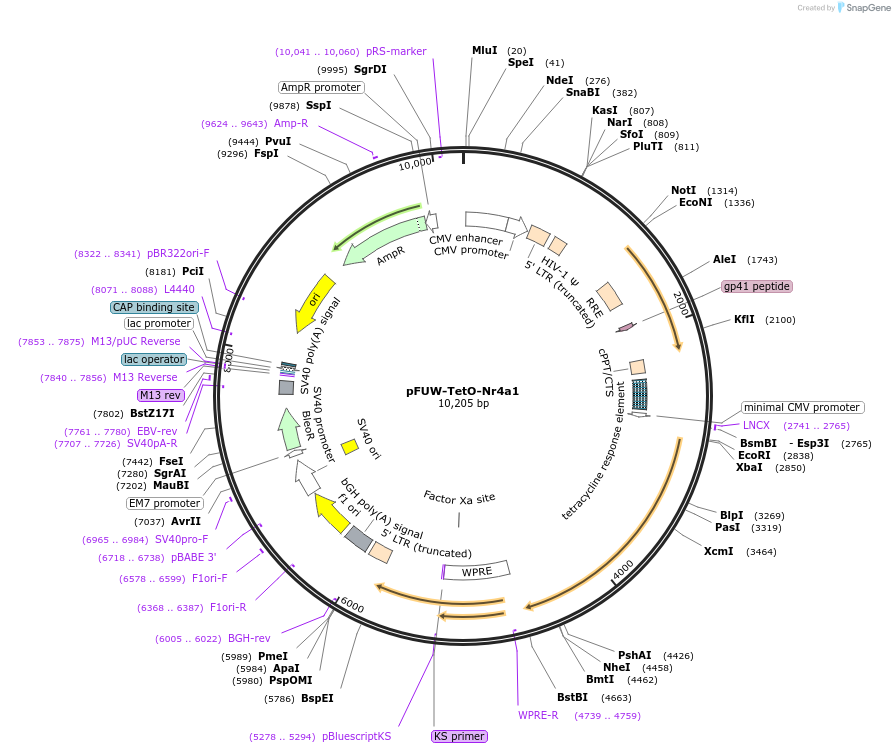
pFUW-TetO-Nr4a1
Plasmid#207194Purposedoxycycline-inducible expression of Mouse Nr4a1 in mammalian cellsDepositorInsertpFUW-TetO-NR4A1
UseLentiviralExpressionMammalianPromoterCMVAvailable SinceAug. 29, 2025AvailabilityAcademic Institutions and Nonprofits only -
-
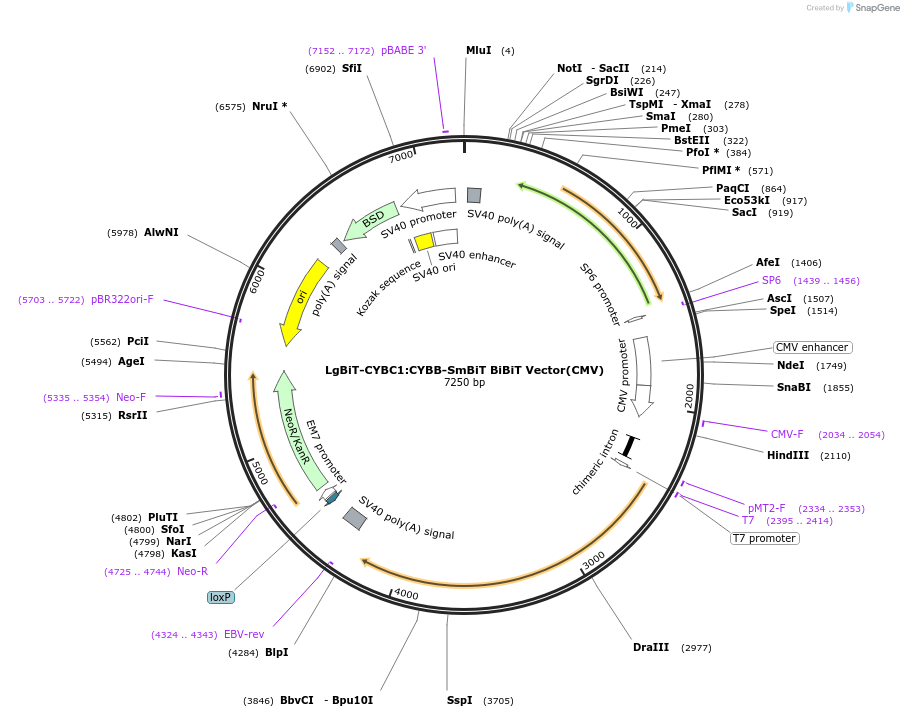
LgBiT-CYBC1:CYBB-SmBiT BiBiT Vector(CMV)
Plasmid#237084PurposeExpress LgBiT-CYBC1:CYBB-SmBiT Fusion Protein in Mammalian Cells under a CMV promoterDepositorHas ServiceDNATagsLgBiT and SmBiTExpressionMammalianMutationNonePromoterCMVAvailable SinceJuly 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
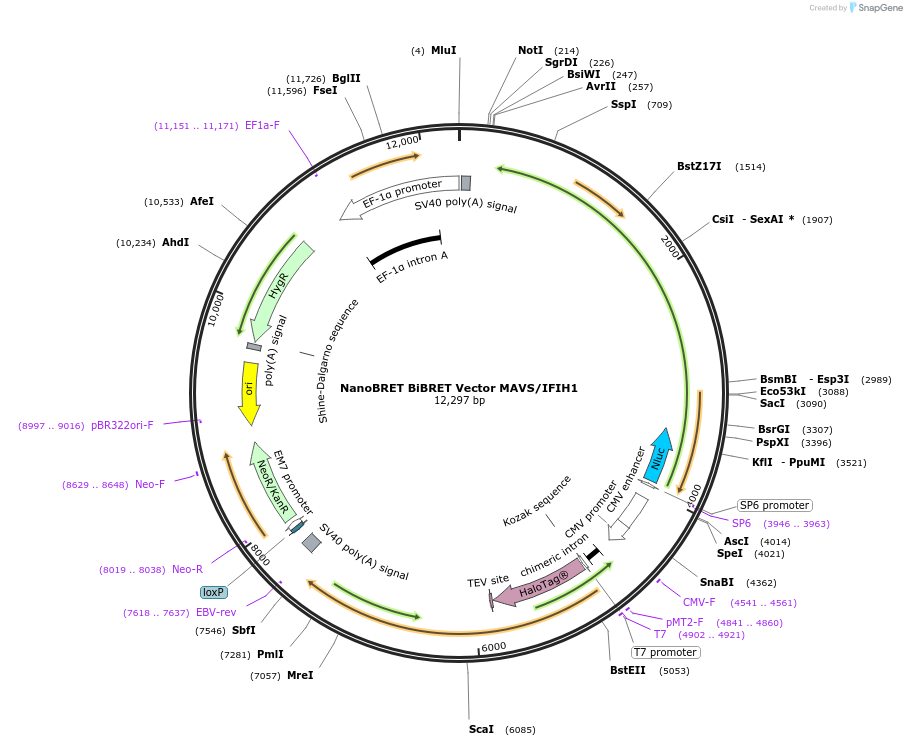
NanoBRET BiBRET Vector MAVS/IFIH1
Plasmid#237022PurposeExpress HaloTag-MAVS/NanoLuc(R)-IFIH1 Fusion Proteins in Mammalian Cells under a CMV promoterDepositorHas ServiceDNATagsHaloTag (R) and NanoLuc (R)ExpressionMammalianMutationNonePromoterCMVAvailable SinceJuly 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
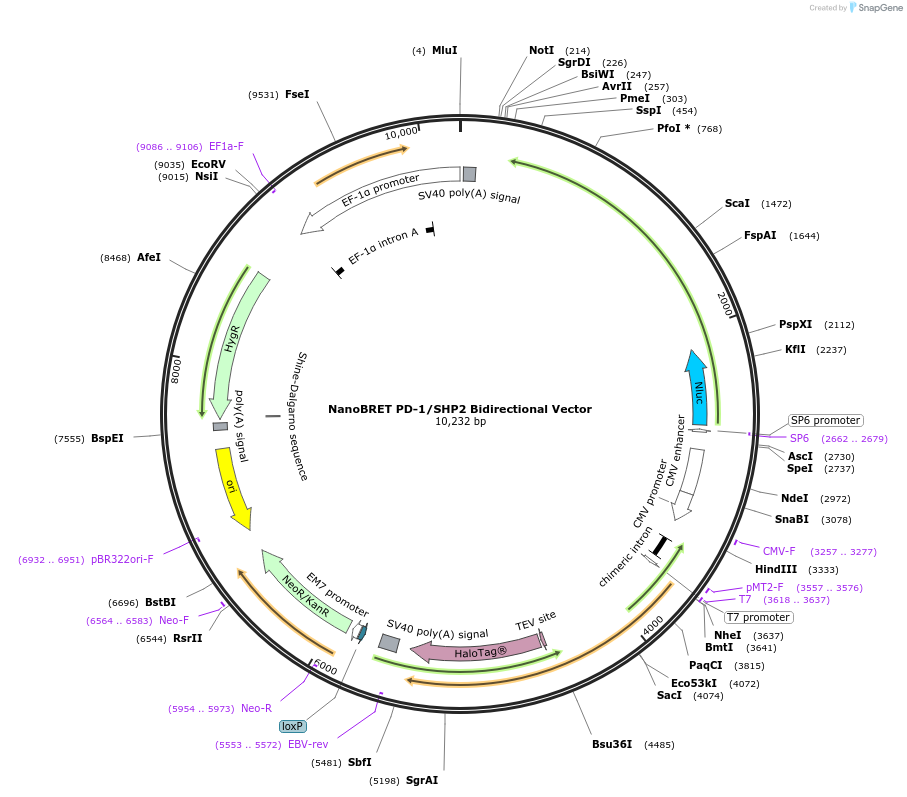
NanoBRET PD-1/SHP2 Bidirectional Vector
Plasmid#237016PurposeExpress NanoLuc(R)-PD1/SHP2-HaloTag Fusion Proteins in Mammalian Cells under a CMV promoterDepositorHas ServiceDNATagsHaloTag (R) and NanoLuc (R)ExpressionMammalianMutationNonePromoterCMVAvailable SinceJuly 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
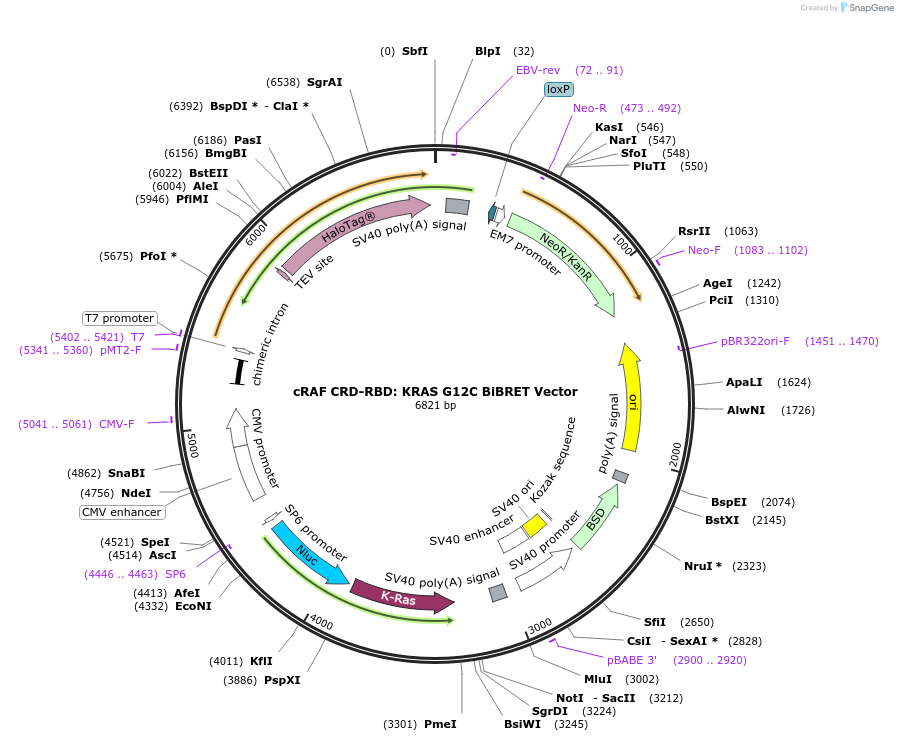
cRAF CRD-RBD: KRAS G12C BiBRET Vector
Plasmid#236872PurposeExpress cRAF CRD-RBD: KRAS G12C in Mammalian Cells under a CMV promoterDepositorHas ServiceDNATagsHaloTag (R) and NanoLuc (R)ExpressionMammalianMutationG12CPromoterCMVAvailable SinceJuly 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
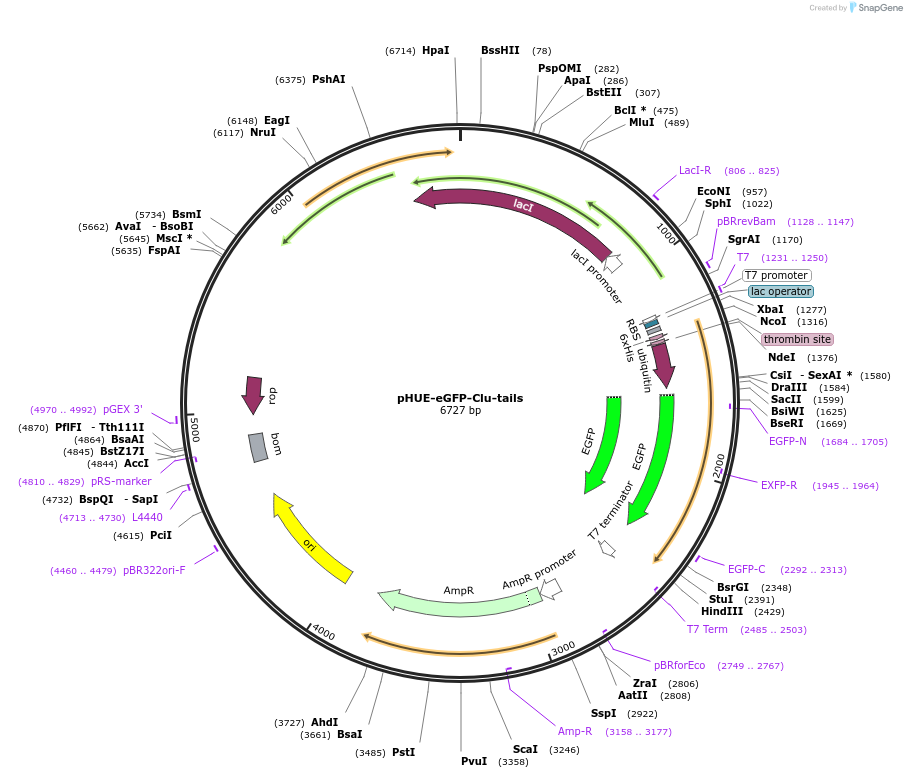
pHUE-eGFP-Clu-tails
Plasmid#215397PurposeExpression of a His6-Ubiquitin-Clu(228-238)-eGFP-Clu(204-227) fusion proteinDepositorInsertClu(228-238)-eGFP-Clu(204-227) (CLU Human)
TagsHis6-UbiquitinExpressionBacterialPromoterT7Available SinceMay 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
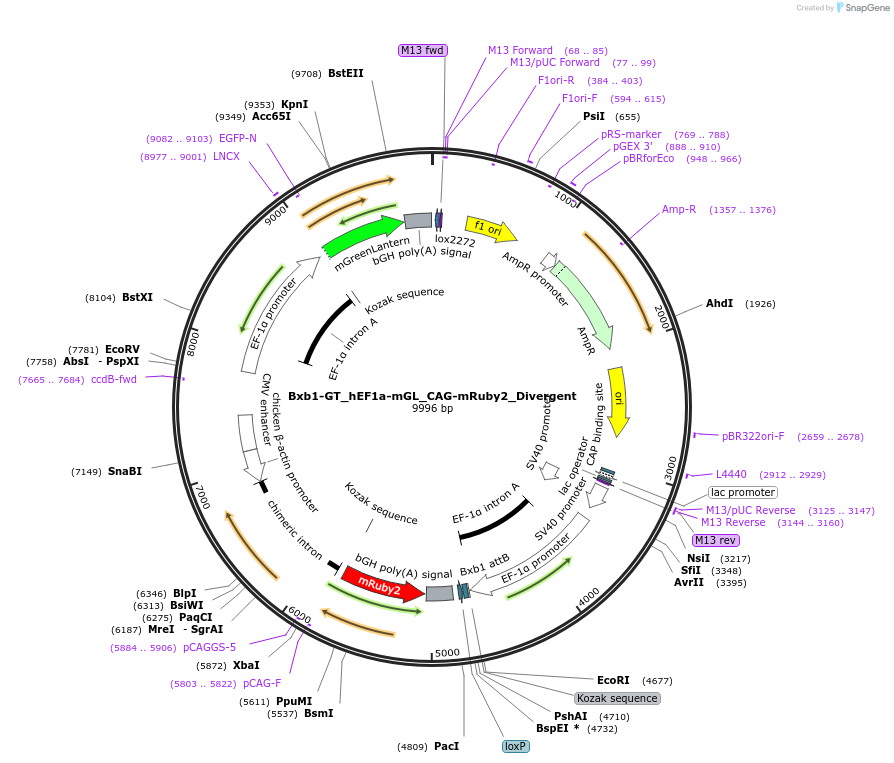
Bxb1-GT_hEF1a-mGL_CAG-mRuby2_Divergent
Plasmid#229800PurposeBxb1-GT donor plasmid with divergent syntax for constitutively expressing mRuby2 and mGreenLantern by the respective promoters CAG and hEF1aDepositorInsertmGreenLantern; mRuby2
UseSynthetic BiologyExpressionMammalianPromoterhEF1a and CAGAvailable SinceFeb. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
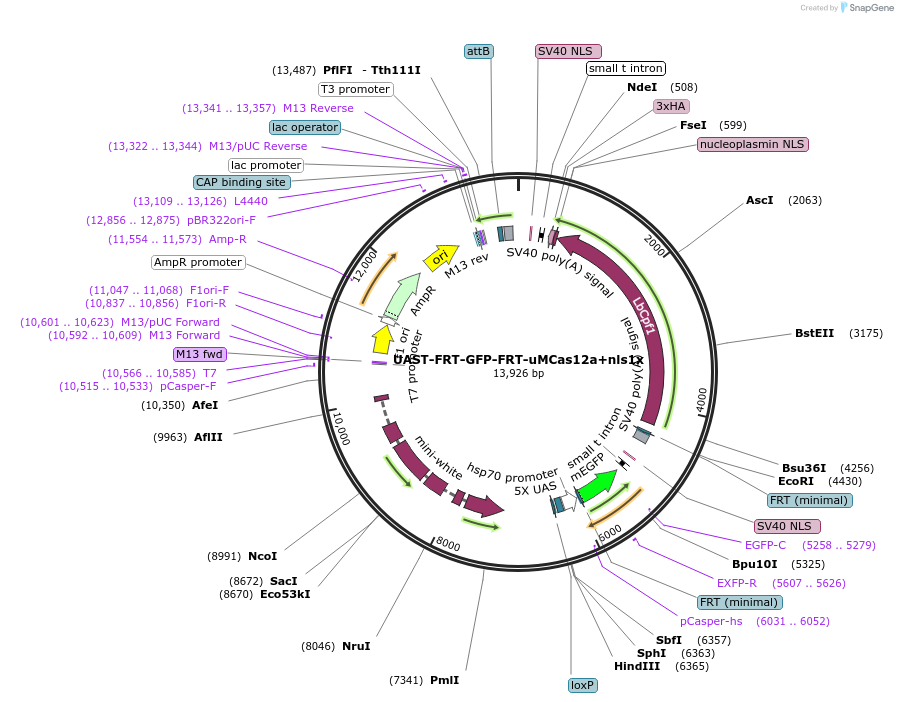
UAST-FRT-GFP-FRT-uMCas12a+nls1x
Plasmid#230908PurposeDouble inducible expression of Cas12a+ with Gal4 and Flp.DepositorInsertCas12a+
UseCRISPRExpressionInsectAvailable SinceFeb. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
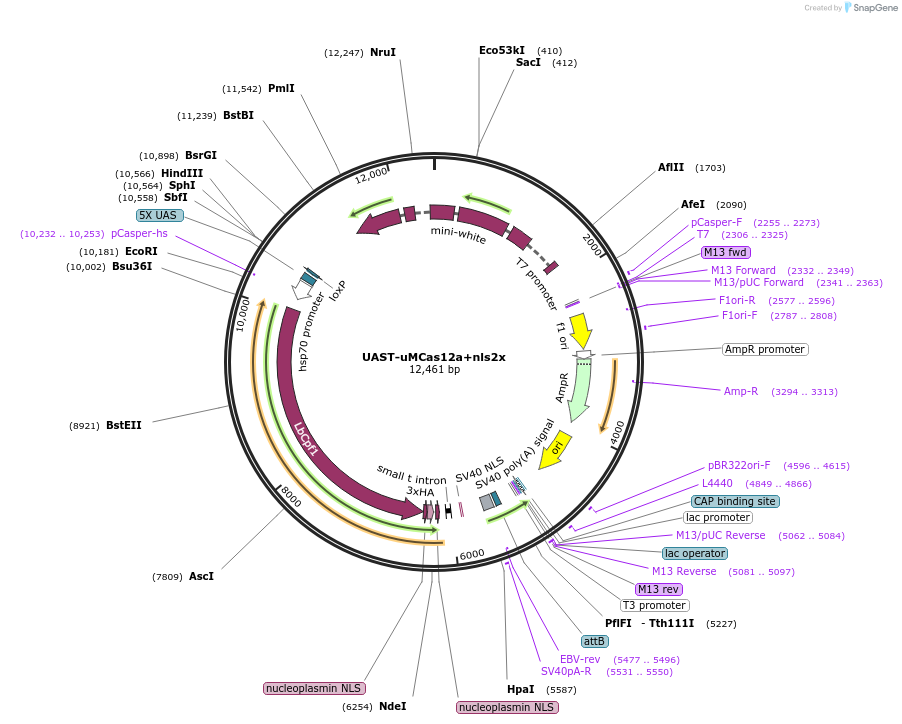
UAST-uMCas12a+nls2x
Plasmid#230909Purposeinducible expression of Cas12a+ with Gal4.DepositorInsertCas12a+
UseCRISPRAvailable SinceFeb. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
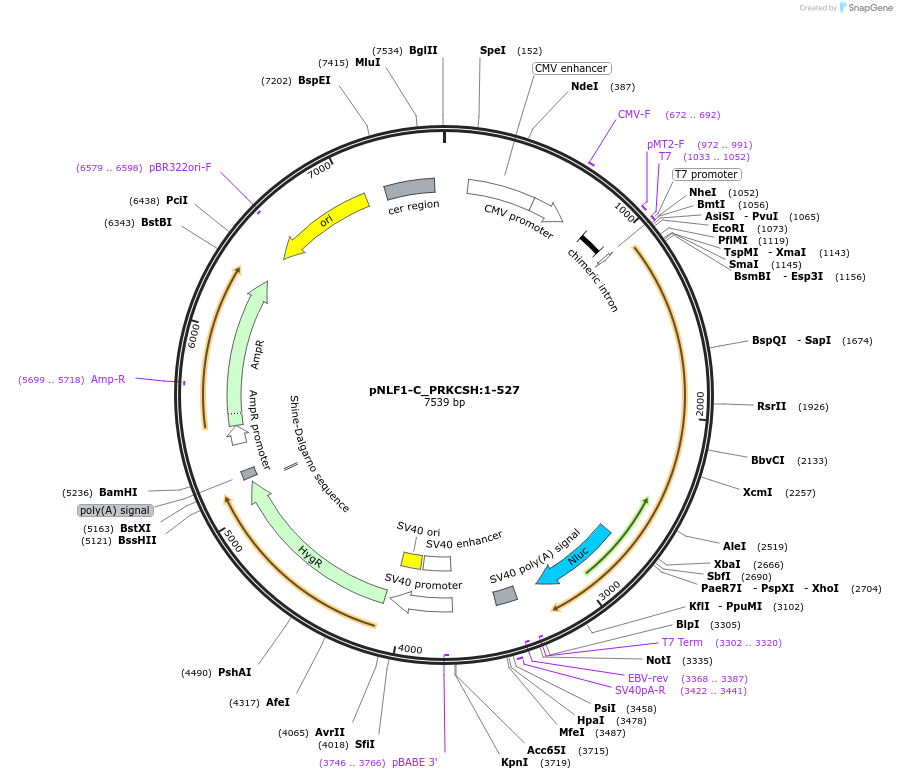
pNLF1-C_PRKCSH:1-527
Plasmid#211091PurposeMammalian expression of full-length human PRKCSH. C-terminal NanoLuc luciferase tag.DepositorInsertPRKCSH:M1-L527 (PRKCSH Human)
TagsNanoLuc luciferaseExpressionMammalianMutation333-333: missing. A291T natural variant (VAR_028…PromoterCMVAvailable SinceMarch 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
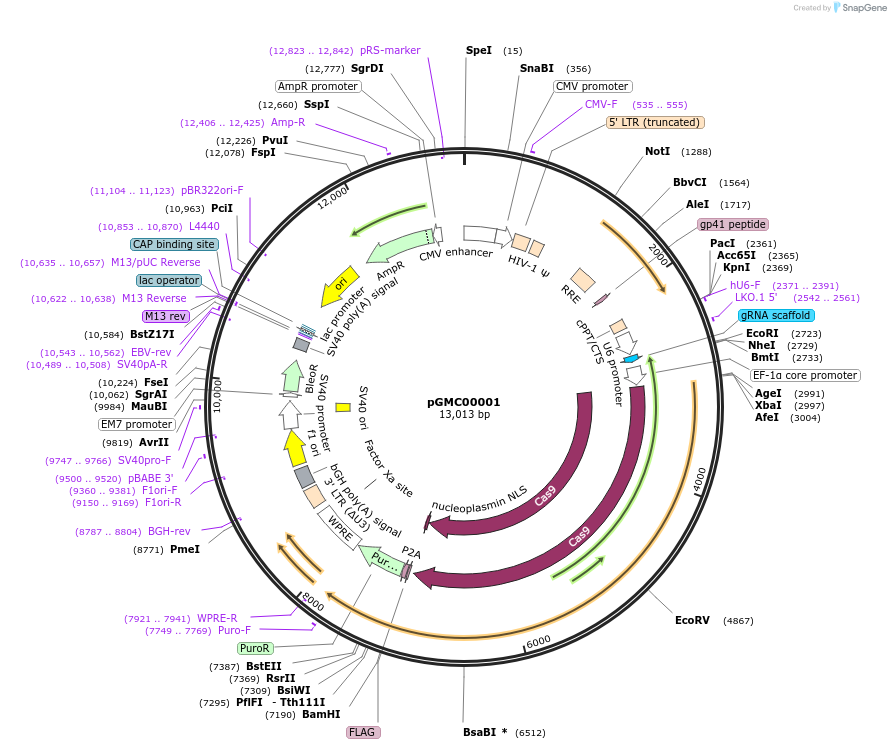
pGMC00001
Plasmid#166718PurposesgRNA against human BAP1DepositorAvailable SinceJune 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
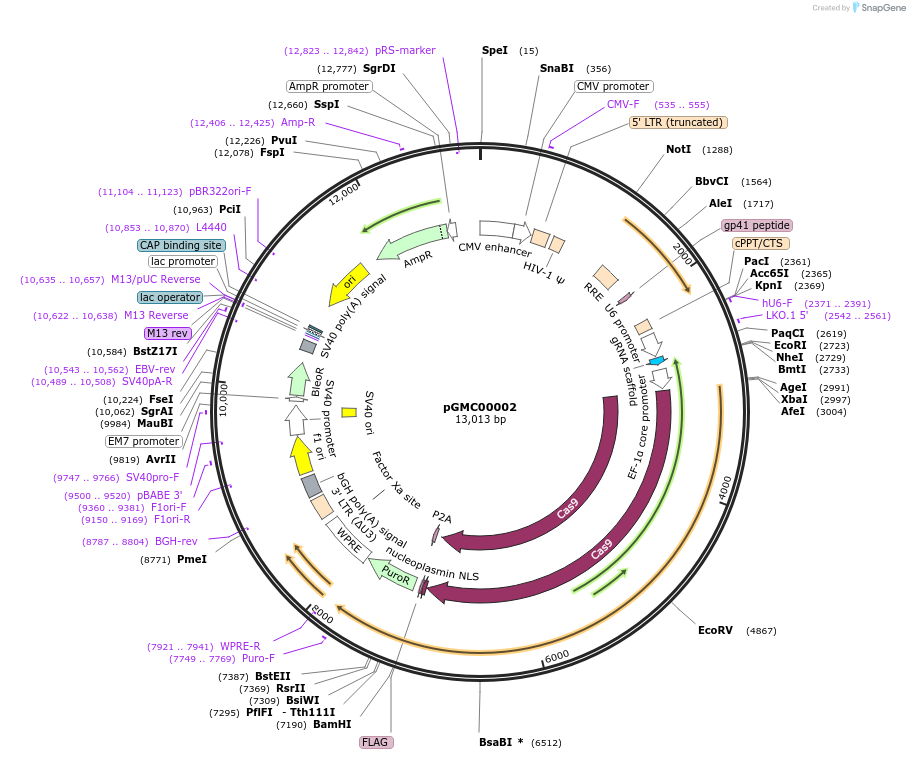
pGMC00002
Plasmid#166719PurposesgRNA against human BAP1DepositorAvailable SinceJune 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
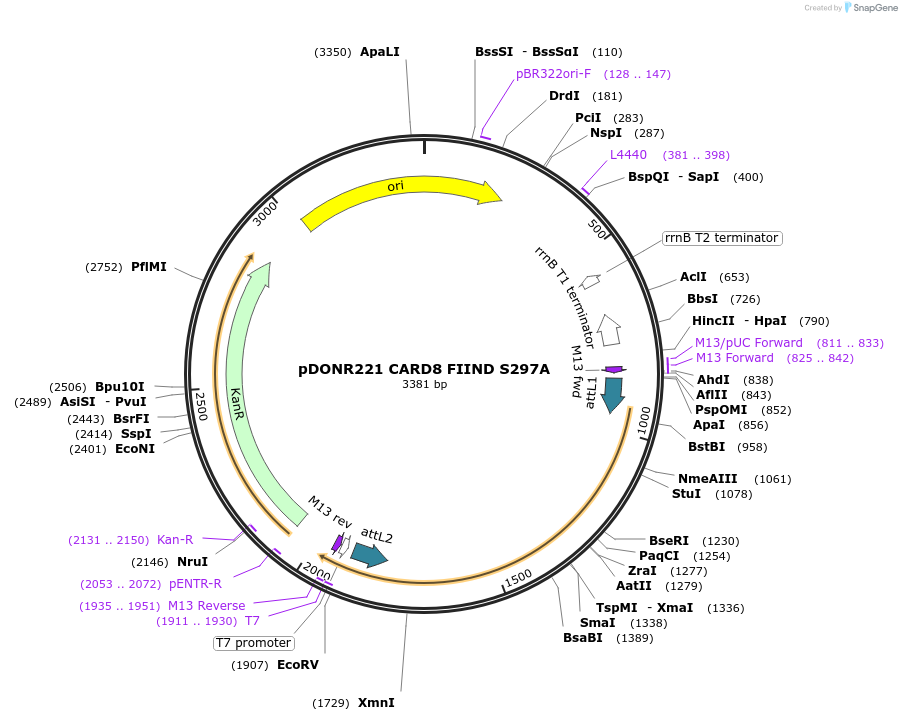
pDONR221 CARD8 FIIND S297A
Plasmid#169966PurposepDONR221 for Gateway shuttling. Contains the autoprocessing-deficient CARD8 FIIND (L162M-P434, no stop codon)DepositorAvailable SinceMay 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
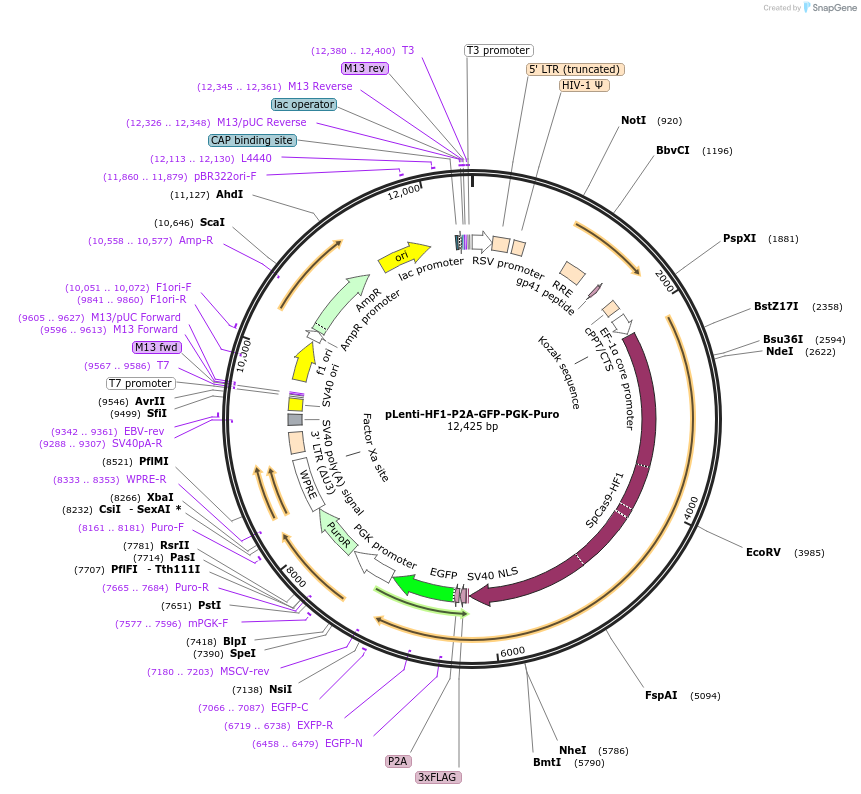
pLenti-HF1-P2A-GFP-PGK-Puro
Plasmid#110863PurposeLentiviral vector for constitutive expression of Cas9-HF1-P2A-GFP (not codon optimized)DepositorInsertCas9-HF1
UseLentiviralTags3X FLAGMutationN497A, R661A, Q695A, Q926APromoterEF1sAvailable SinceJune 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
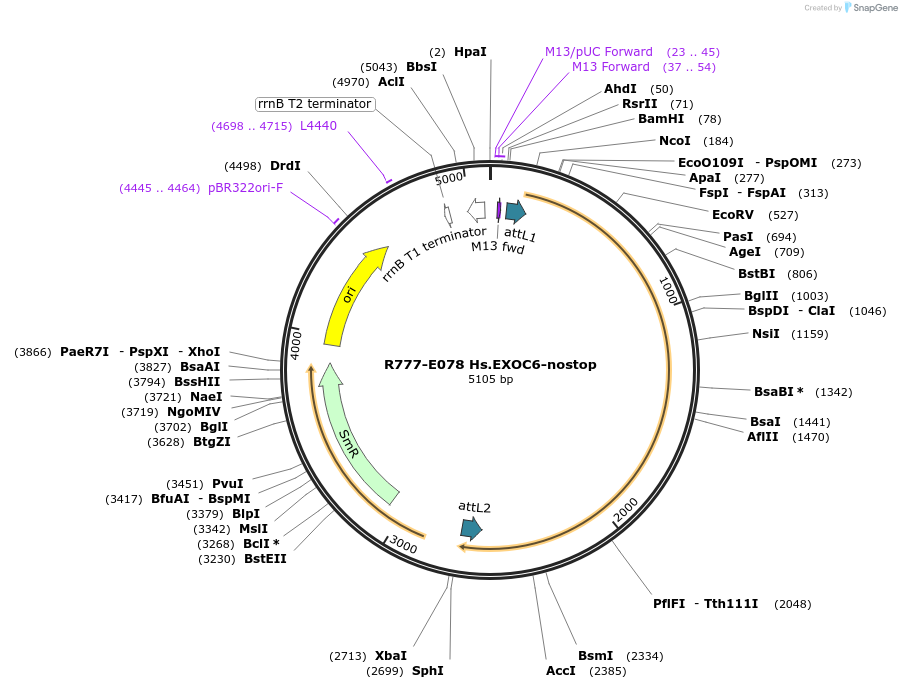
R777-E078 Hs.EXOC6-nostop
Plasmid#70362PurposeGateway ORF clone of human EXOC6 [NM_019053.4] without stop codon (for C-terminal fusions)DepositorInsertEXOC6 (EXOC6 Human)
UseGateway entry cloneAvailable SinceMarch 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
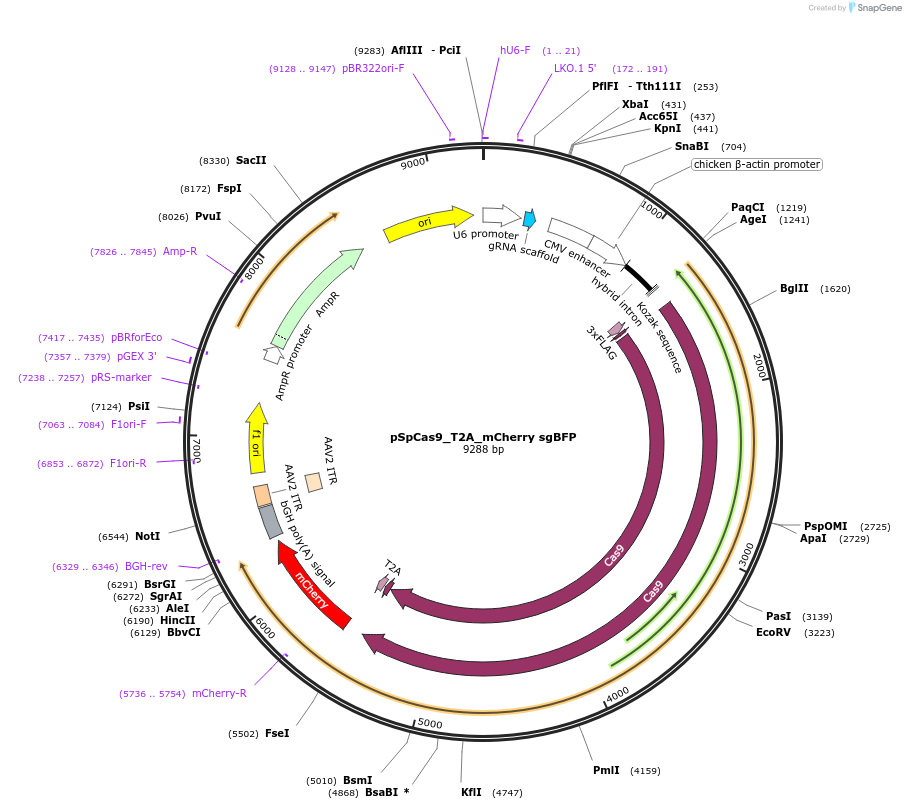
pSpCas9_T2A_mCherry sgBFP
Plasmid#192929PurposeAll-in-one plasmid for cutting BFP region in DSB Spectrum V2DepositorInsertSpCas9
TagsT2A mCherryExpressionMammalianAvailable SinceDec. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
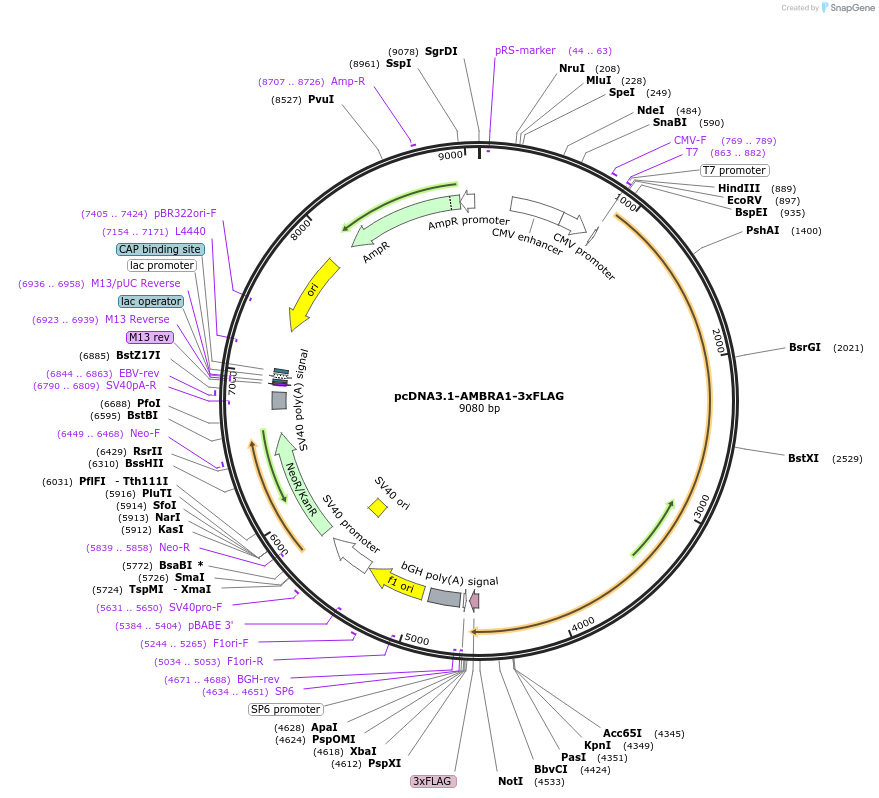
pcDNA3.1-AMBRA1-3xFLAG
Plasmid#172605PurposeExpresses 3xFLAG-tagged AMBRA1 in mammalian cellsDepositorAvailable SinceAug. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
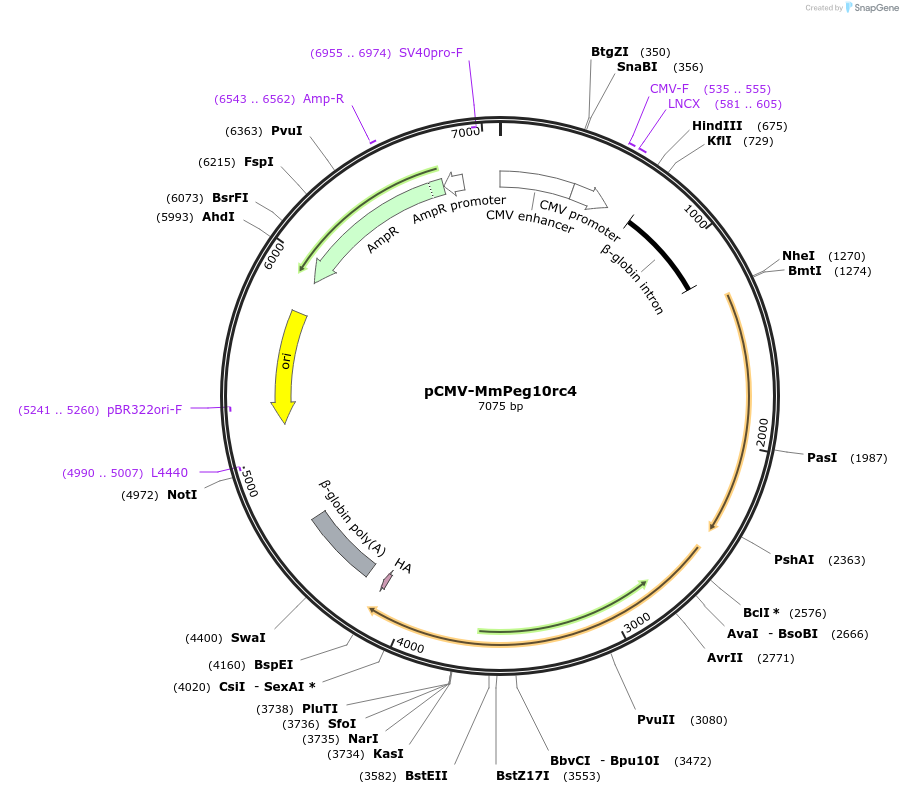
pCMV-MmPeg10rc4
Plasmid#174858PurposeExpresses recoded mouse Peg10 in mammalian cellsDepositorAvailable SinceSept. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
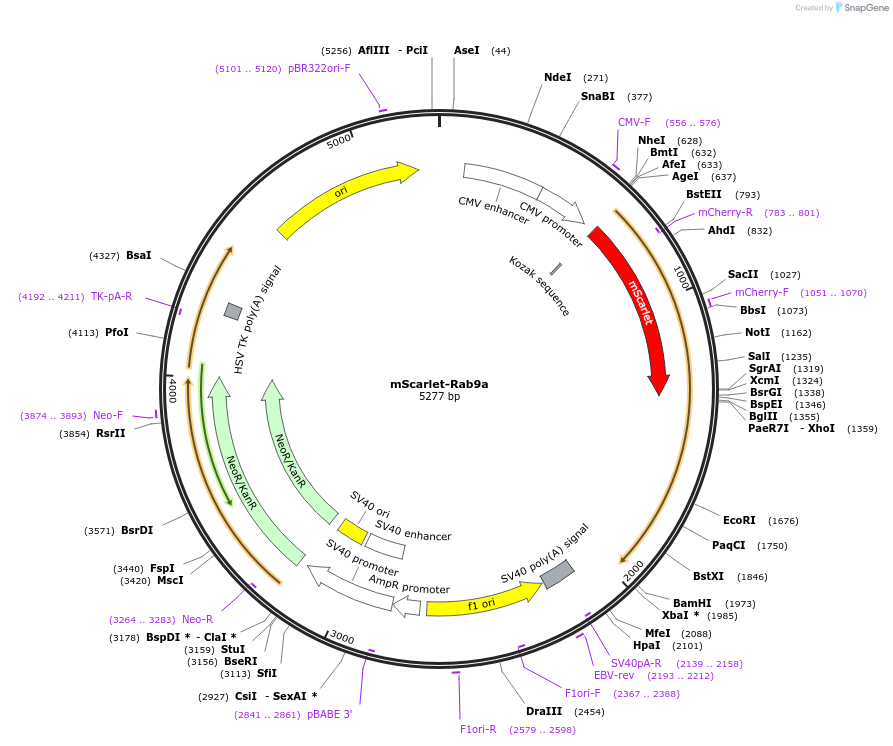
mScarlet-Rab9a
Plasmid#169074PurposeExpress human Rab9a in mammalian cellsDepositorAvailable SinceJan. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
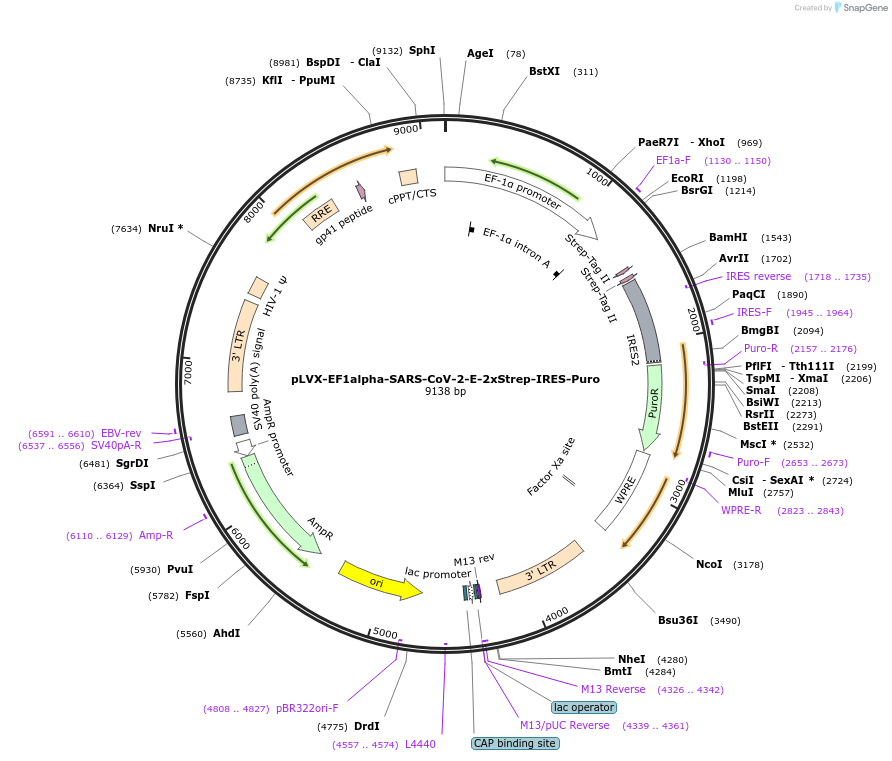
pLVX-EF1alpha-SARS-CoV-2-E-2xStrep-IRES-Puro
Plasmid#141385PurposeLentiviral expression of SARS-CoV-2 envelope (E) protein; transient expression and generate lentivirusDepositorInsertSARS-CoV-2-E (E SARS-CoV-2)
UseLentiviralTags2xStrepExpressionMammalianMutationhuman codon optimizedPromoterEF1alphaAvailable SinceApril 8, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
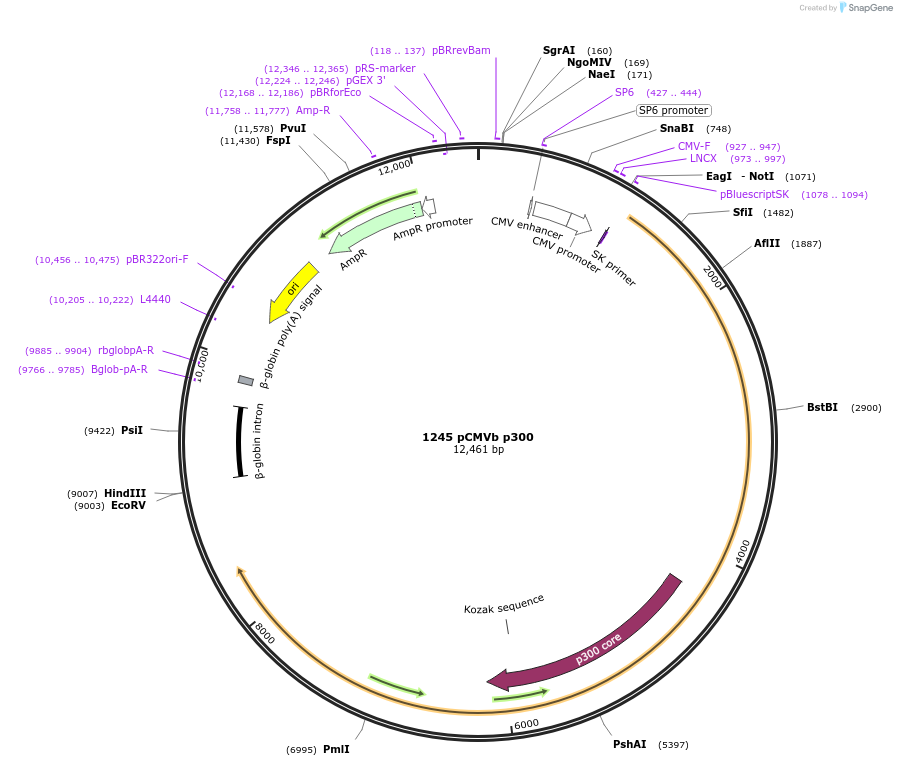
1245 pCMVb p300
Plasmid#10717DepositorInsertp300 (EP300 Human)
ExpressionMammalianAvailable SinceDec. 7, 2005AvailabilityAcademic Institutions and Nonprofits only -
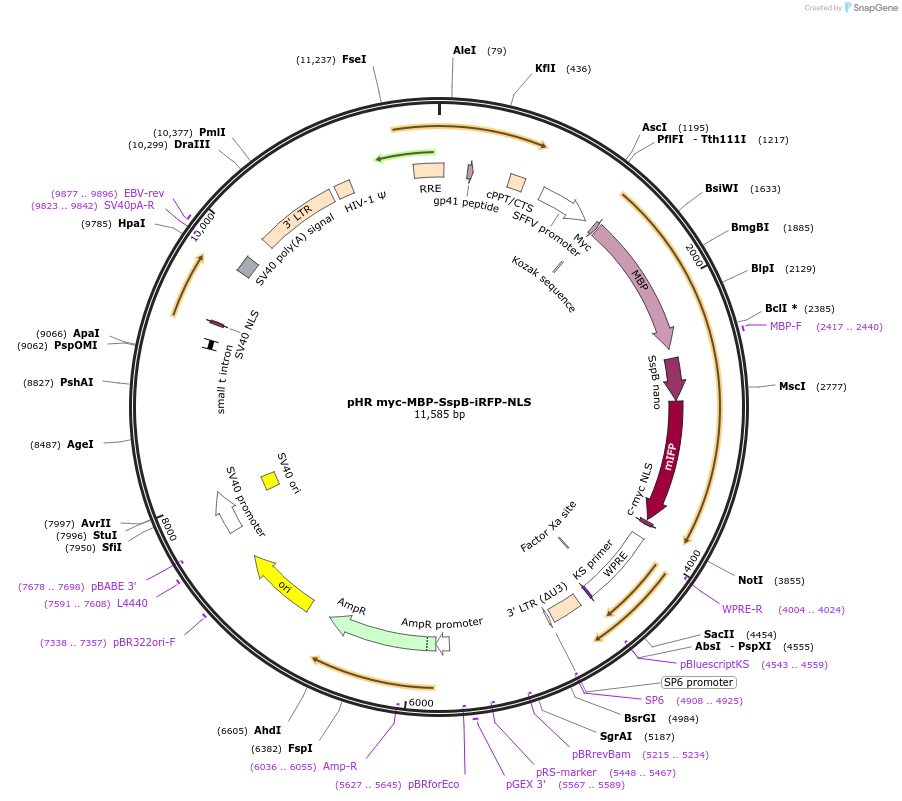
pHR myc-MBP-SspB-iRFP-NLS
Plasmid#223286PurposeAn SspB-tagged maltose binding protein (MBP) with an infrared fluorescent protein for visualization. Used to demonstrate condensate dissolution using the OptoMBP system.DepositorInsertMyc-MBP-SspB-irFP-NLS
UseLentiviralExpressionMammalianAvailable SinceAug. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
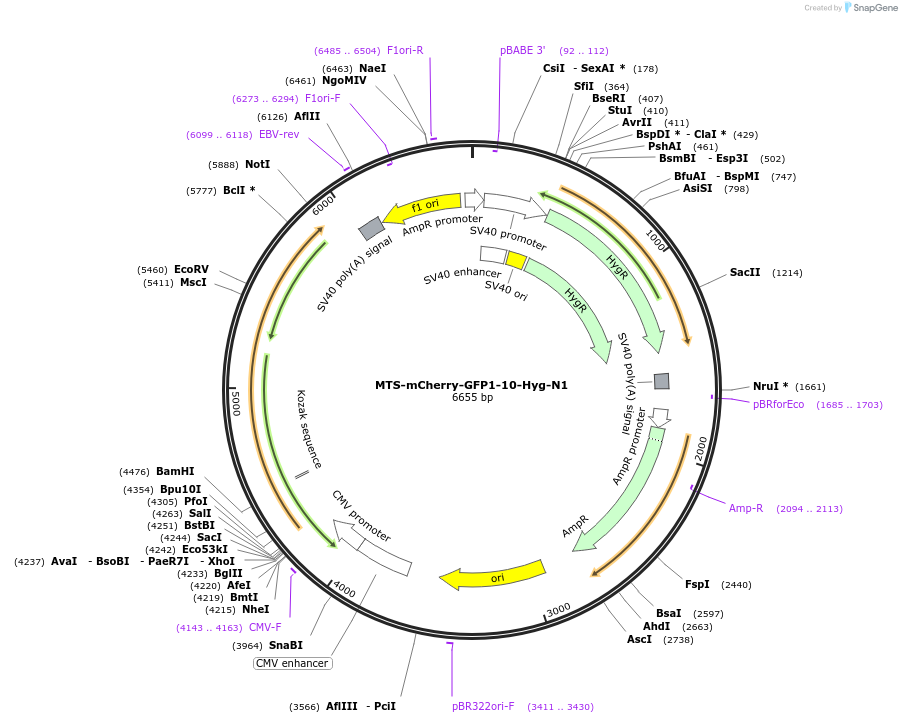
MTS-mCherry-GFP1-10-Hyg-N1
Plasmid#91957PurposeExpresses mCherry-GFP1-10 in mammalian cell mitochondriaDepositorInsertMTS-mCherry-GFP1-10
ExpressionMammalianPromoterCMVAvailable SinceJune 6, 2017AvailabilityAcademic Institutions and Nonprofits only -
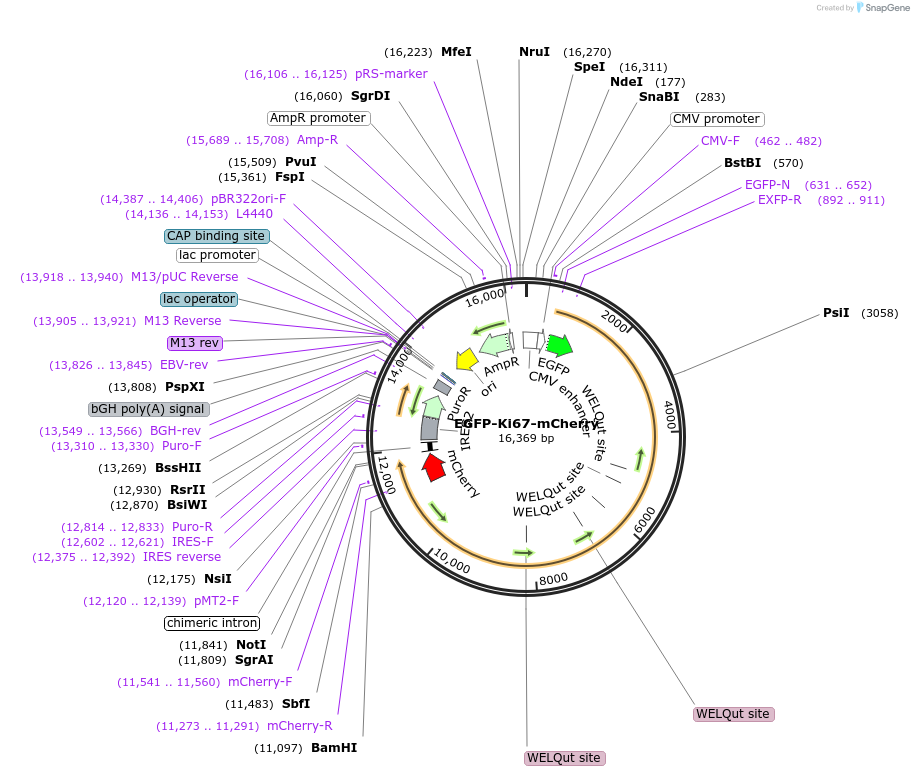
EGFP-Ki67-mCherry
Plasmid#183748PurposeExpresses EGFP-Ki67-mCherryDepositorInsertMKI67 (MKI67 Human)
TagsKi-67 with N-terminal EGFP tag and Ki-67 with C-…ExpressionBacterial and MammalianPromoterCMVAvailable SinceSept. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
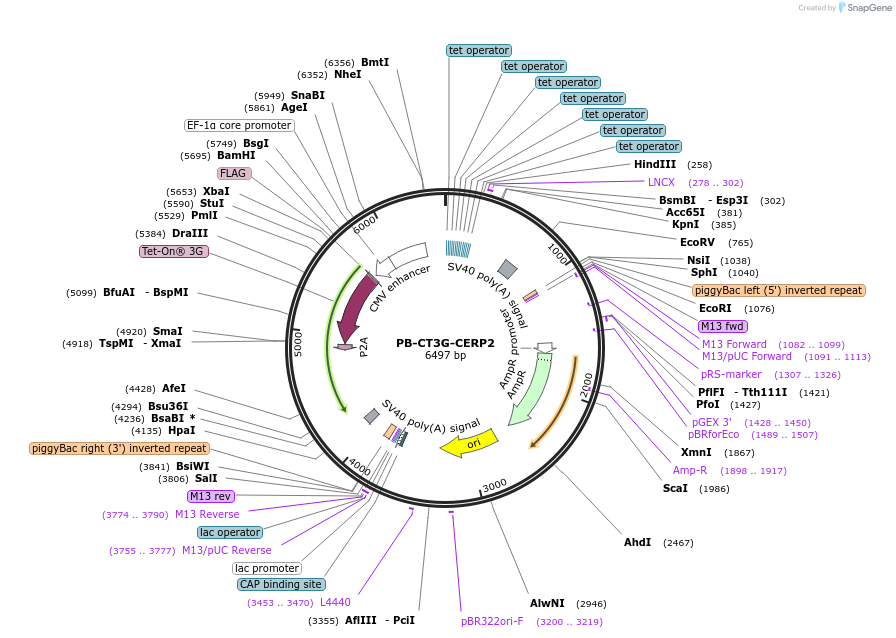
PB-CT3G-CERP2
Plasmid#175499PurposeAll-in-one piggyBac transposon vector for mCMV+dox-inducible expression of Golden Gate cloned elements (CMVe-hEF1a-driven rtTA and puromycin resistance)DepositorTypeEmpty backboneUseSynthetic BiologyExpressionMammalianPromoterTRE3G-mCMVAvailable SinceOct. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
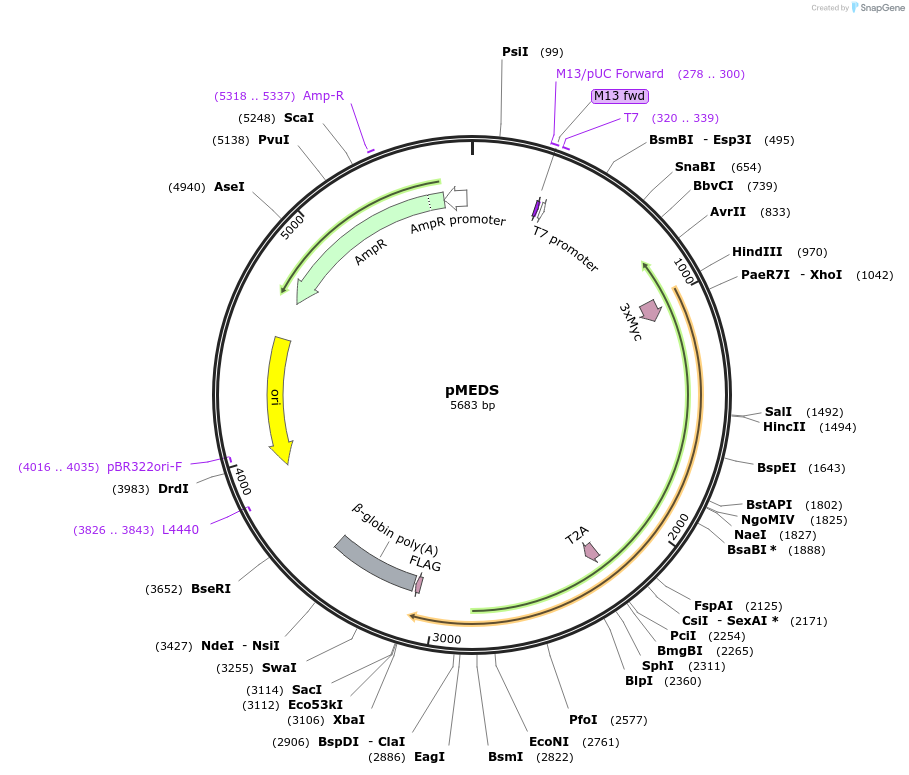
pMEDS
Plasmid#236568PurposeIntroduces a multiple expression and dual selection (MEDS) system to use alongside CRISPR for applications such as gene editing.DepositorInsertsCD/UPRT (negative selection)
NPT II (positive selection)
Tags3x Myc and FLAGExpressionMammalianAvailable SinceJune 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
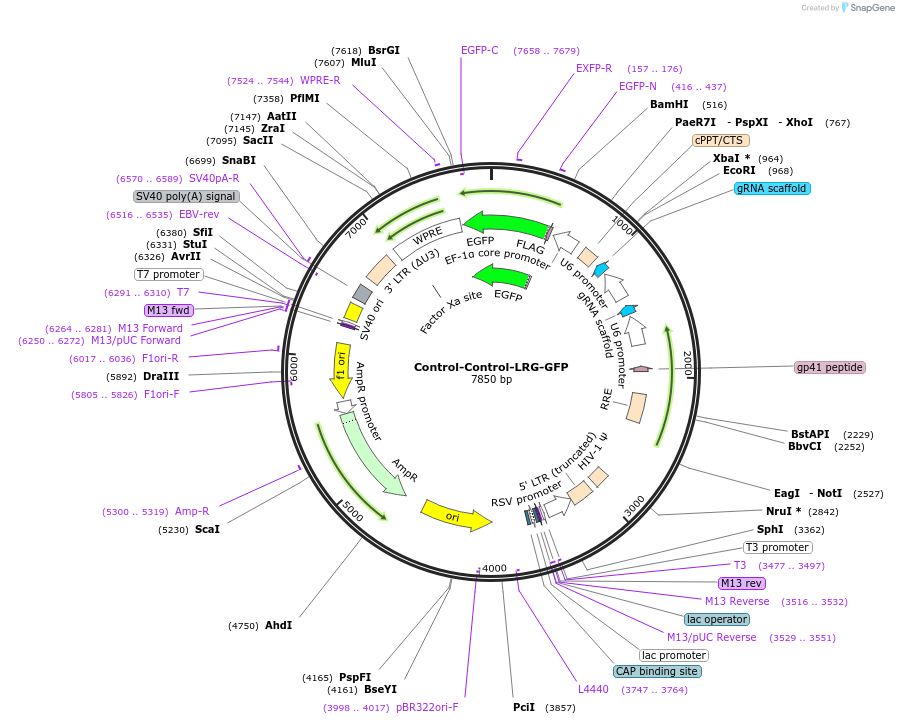
Control-Control-LRG-GFP
Plasmid#225873PurposeTwo copies of guide RNA targeting a control sequenceDepositorInsertControl
UseCRISPR and LentiviralAvailable SinceOct. 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
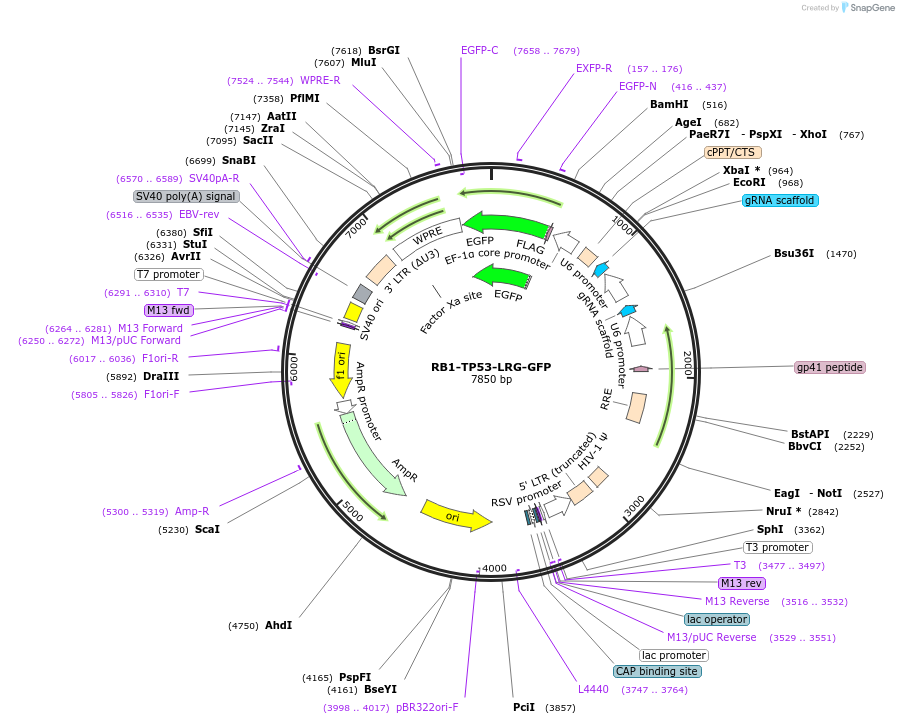
RB1-TP53-LRG-GFP
Plasmid#225876PurposeGuides targeting TP53 and RB1DepositorAvailable SinceOct. 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
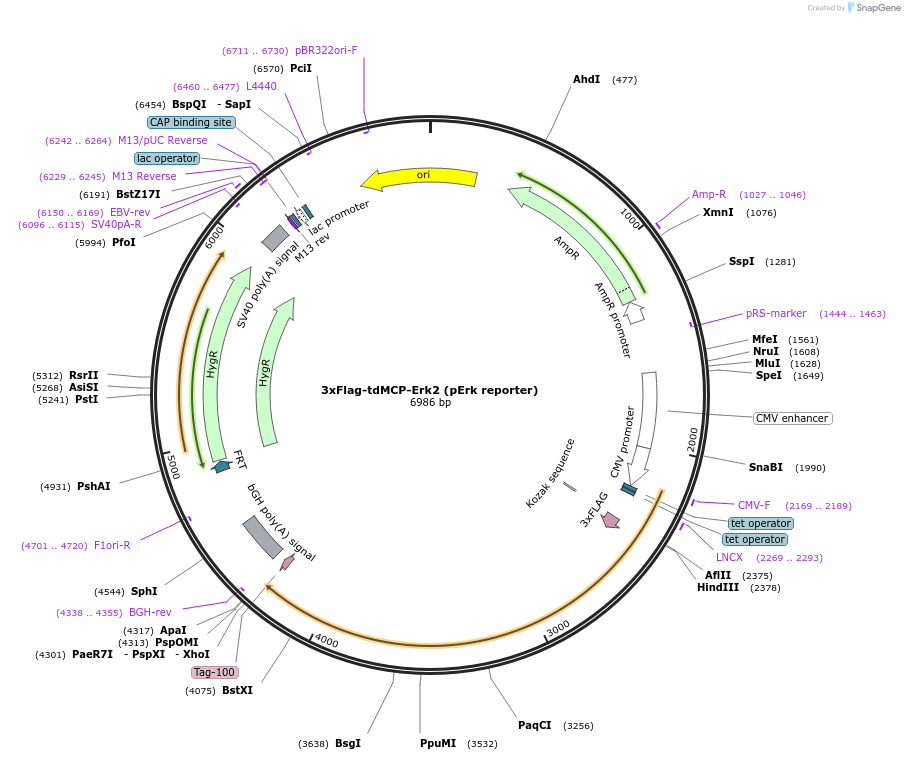
3xFlag-tdMCP-Erk2 (pErk reporter)
Plasmid#226167PurposeUsed to transiently express the pErk reporter in LABEL-seq MAPK activity assays.DepositorAvailable SinceNov. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
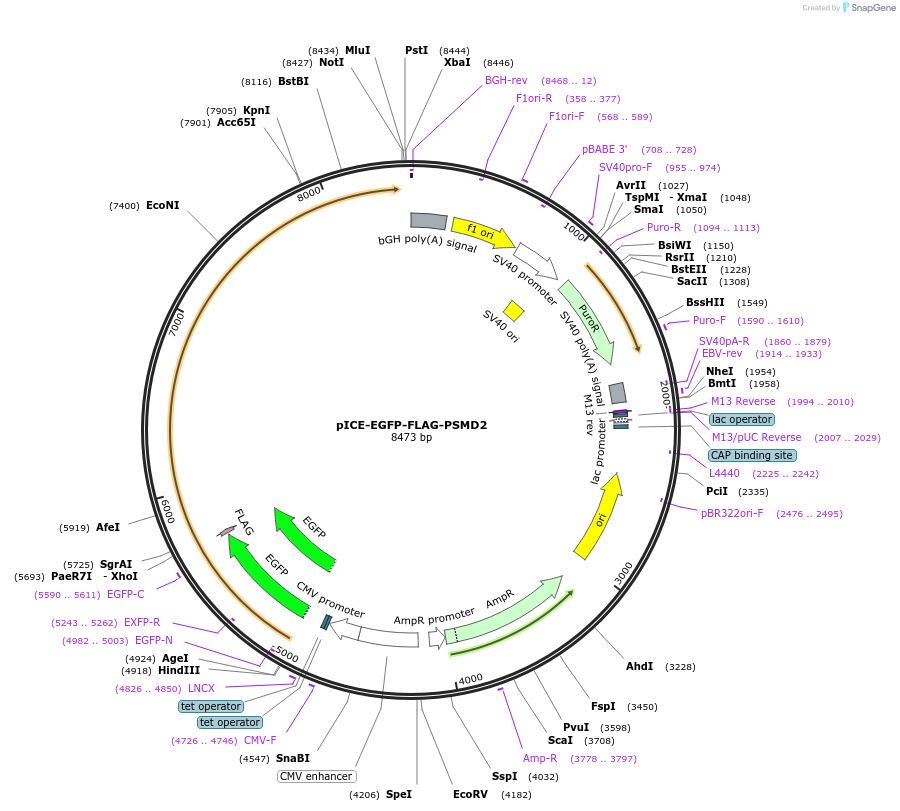
pICE-EGFP-FLAG-PSMD2
Plasmid#161917PurposeExpresses human PSMD2 with a N-terminal GFP-FLAG tag. Confers Puromycin resistance. Inducible in T-REx cells.DepositorInsert26S proteasome non-ATPase regulatory subunit 2 (PSMD2 Human)
TagsGFP-FLAGExpressionMammalianPromoterCMVAvailable SinceMay 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
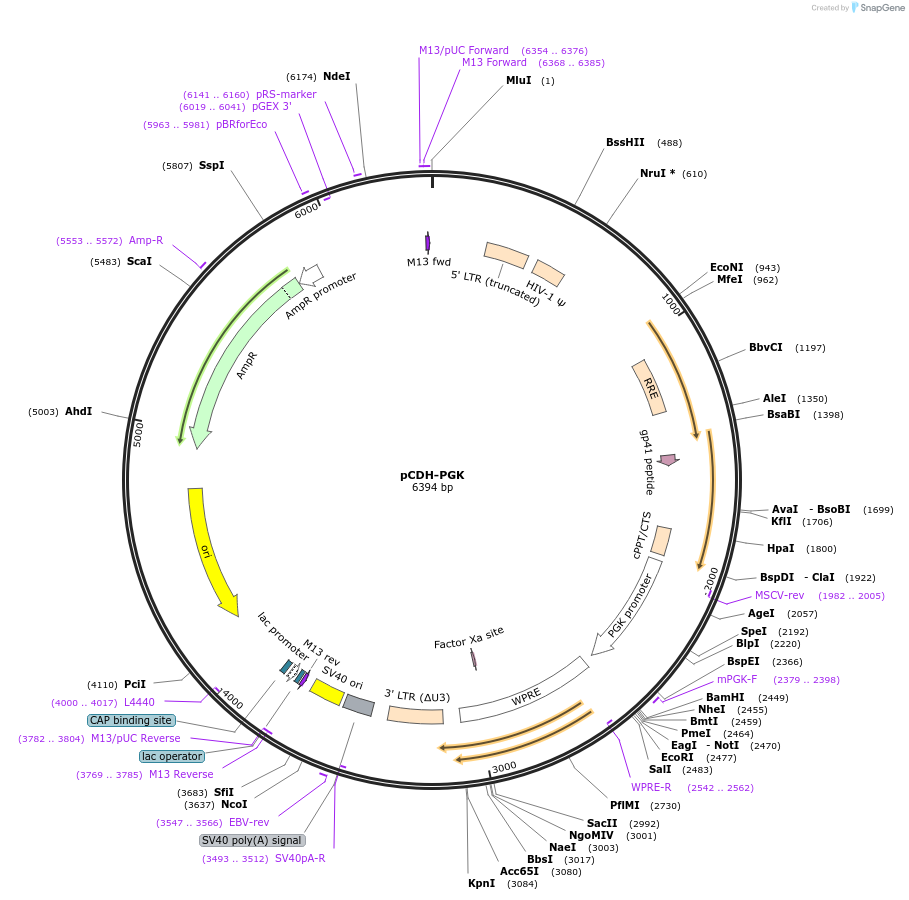
pCDH-PGK
Plasmid#72268PurposeExpress gene of interest under PGK (phosphoglycerate kinase I) promoterDepositorTypeEmpty backboneUseLentiviralExpressionMammalianPromoterPGKAvailable SinceJan. 11, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
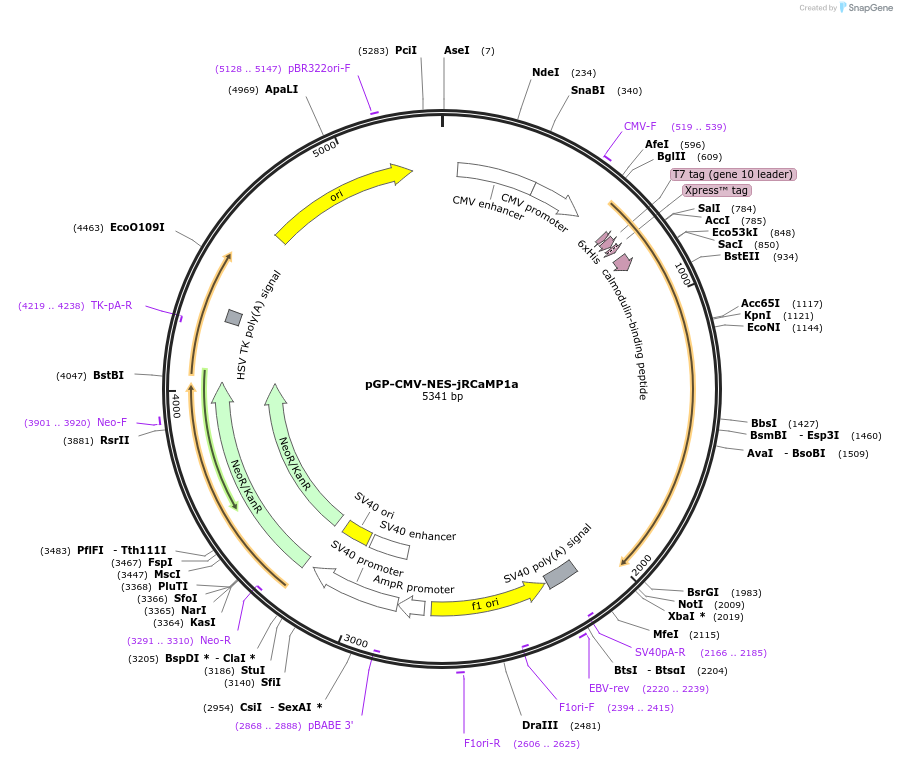
pGP-CMV-NES-jRCaMP1a
Plasmid#61562PurposeRed fluorescent calcium sensor proteinDepositorInsertNES-jRCaMP1a
Tagsnuclear export signalExpressionMammalianPromoterCMVAvailable SinceJan. 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
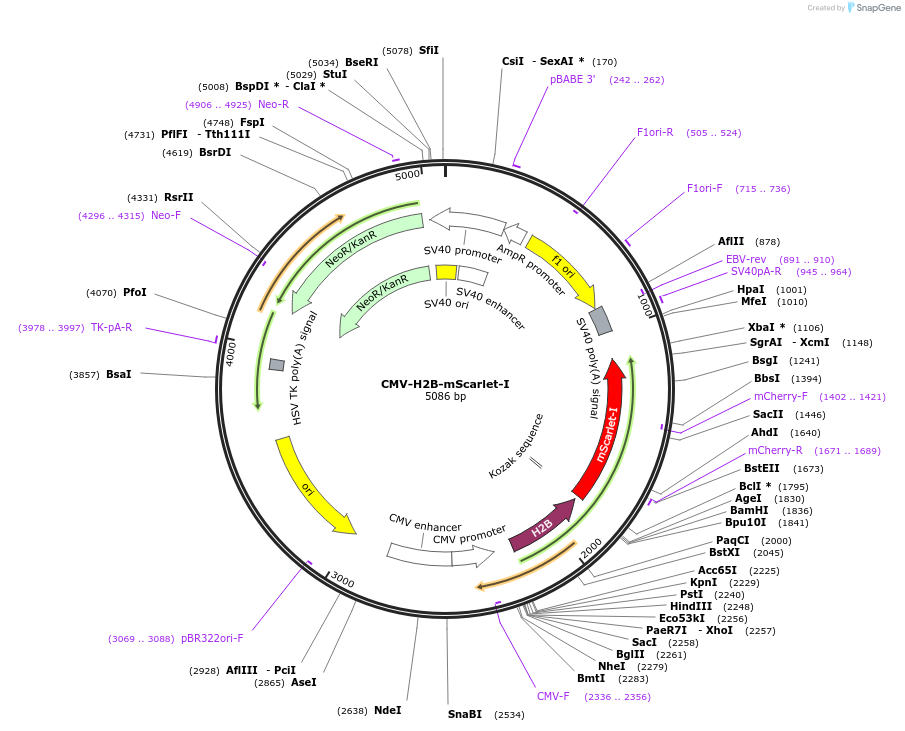
CMV-H2B-mScarlet-I
Plasmid#182297PurposeLocalization of mScarlet-I fluorescent protein to histone 2BDepositorInsertmScarlet-I
TagsmScarlet-IExpressionMammalianAvailable SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
-
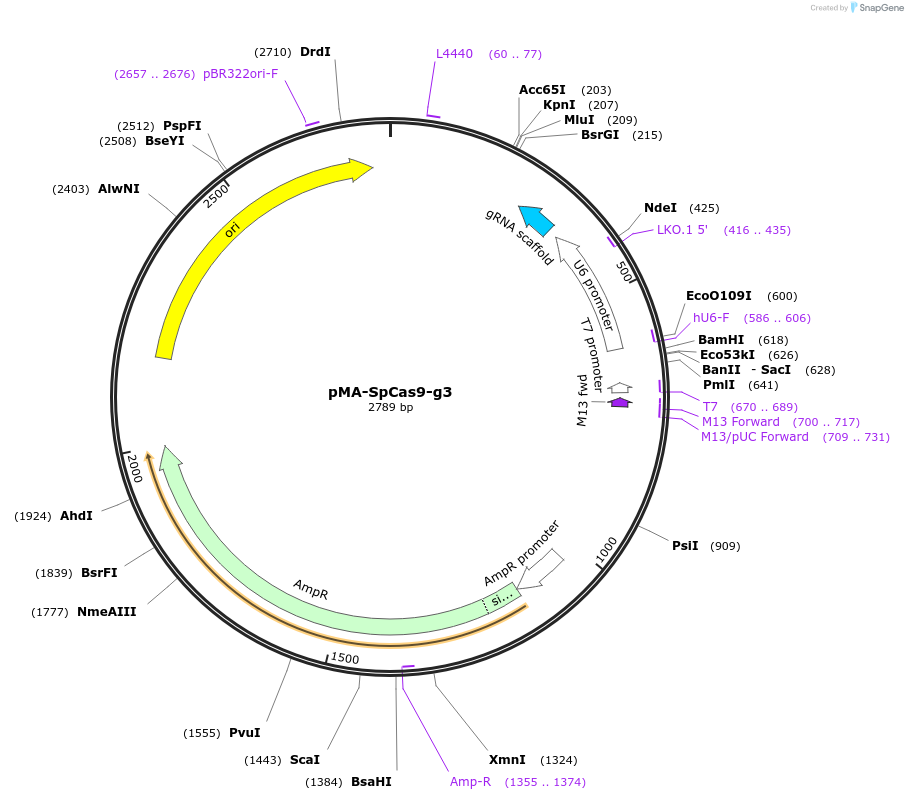
pMA-SpCas9-g3
Plasmid#80786PurposeBsaI-based cloning of SpCas9 gRNA guide sequence, position 3/13/23 in the arrayDepositorInsertCRISPR gRNA expression cassette (for SpCas9)
UseCRISPRTagsnaExpressionMammalianPromoterU6Available SinceSept. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
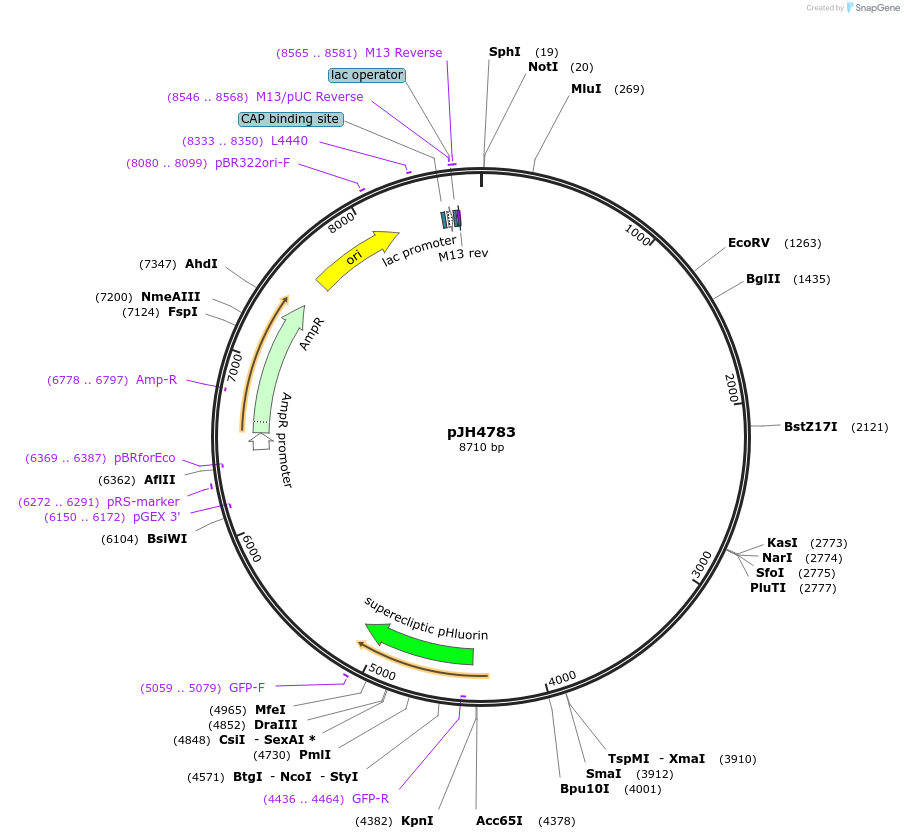
pJH4783
Plasmid#200785PurposePnpr-4 snb-1::pHluorin unc-54 3' UTR C.elegans AVA and other neurons expression of snb pHluorinDepositorAvailable SinceMay 7, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
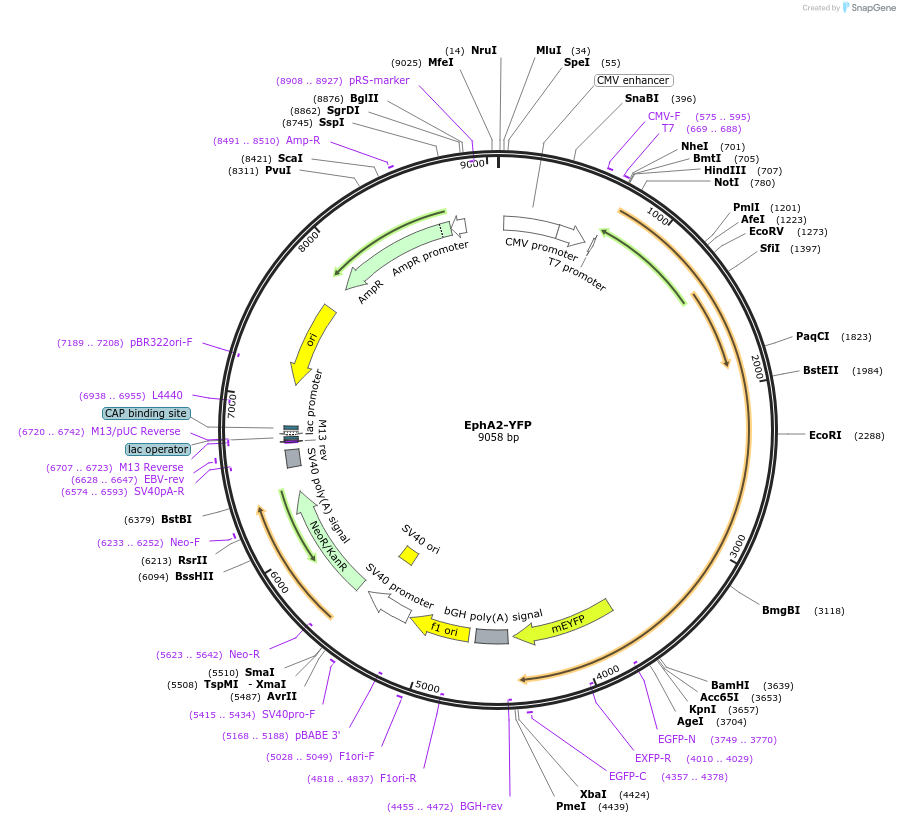
EphA2-YFP
Plasmid#108852PurposeEncodes for human EphA2 fluorescently labeled with eYFP on the C-Terminus via a 15 amino acid (GGS)5 flexible linkerDepositorInsertEPHA2 (EPHA2 Human)
TagsLabeled with eYFP on the C-Terminus via a 15 amin…ExpressionMammalianAvailable SinceApril 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
Cultivarium POSSUM Toolkit
Plasmid Kit#1000000234PurposeGolden Gate genetic toolkit developed to help researchers identify functional plasmids and establish genetic tractability in non-model bacteria.DepositorApplicationCloning and Synthetic BiologyVector TypeBacterial ExpressionCloning TypeGolden Gate (Loop)Available SinceFeb. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
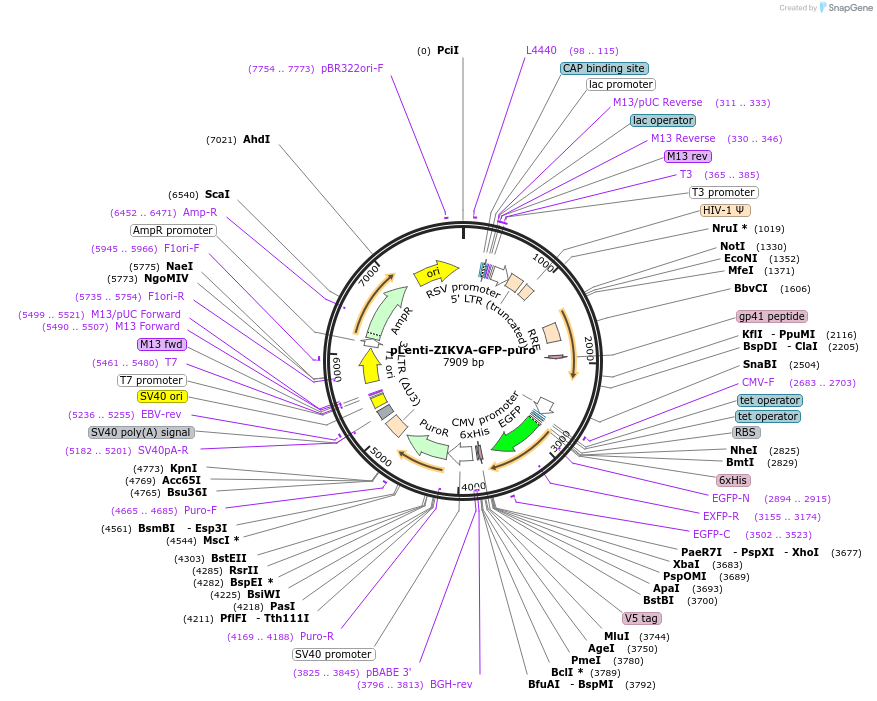
pLenti-ZIKVA-GFP-puro
Plasmid#140090PurposeFluorescence activatable reporter of ZIKV NS2B-NS3 protease activityDepositorInsertZIKVA-GFP
UseLentiviral; Low expressionTags6XHis tag and GFPExpressionMammalianPromoterCMVAvailable SinceJune 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
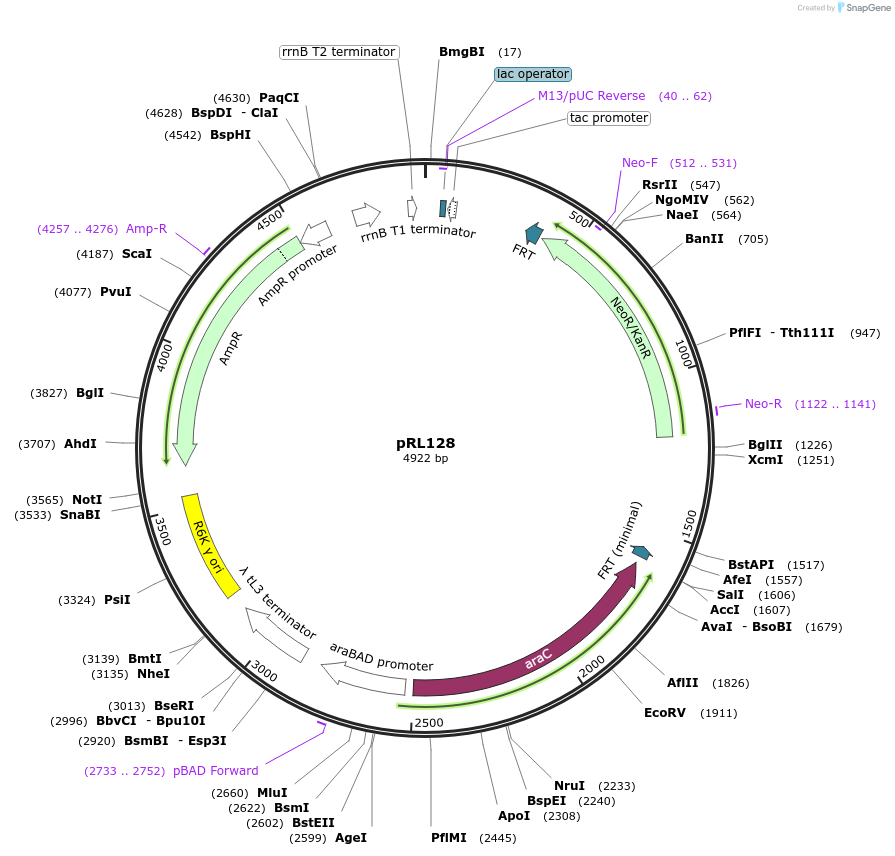
pRL128
Plasmid#40180DepositorInsertsPtac-lacO
araC-pBAD
PromoterBAD and tacAvailable SinceJan. 25, 2013AvailabilityAcademic Institutions and Nonprofits only -
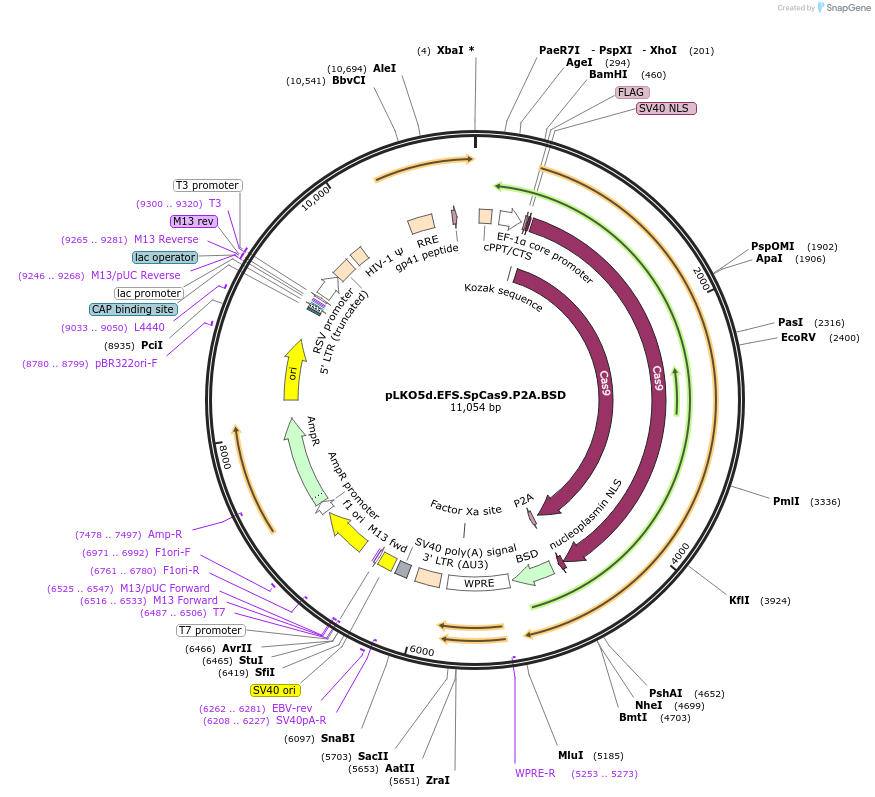
pLKO5d.EFS.SpCas9.P2A.BSD
Plasmid#57821PurposeLentiviral Vector for SpCas9 Expression without sgRNA, Blasticidin resistance, EFS Promoter drivenDepositorAvailable SinceAug. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
mCherry-MyosinIIB-N-18
Plasmid#55107PurposeLocalization: Cytoskeleton, Excitation: 587, Emission: 610DepositorAvailable SinceSept. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
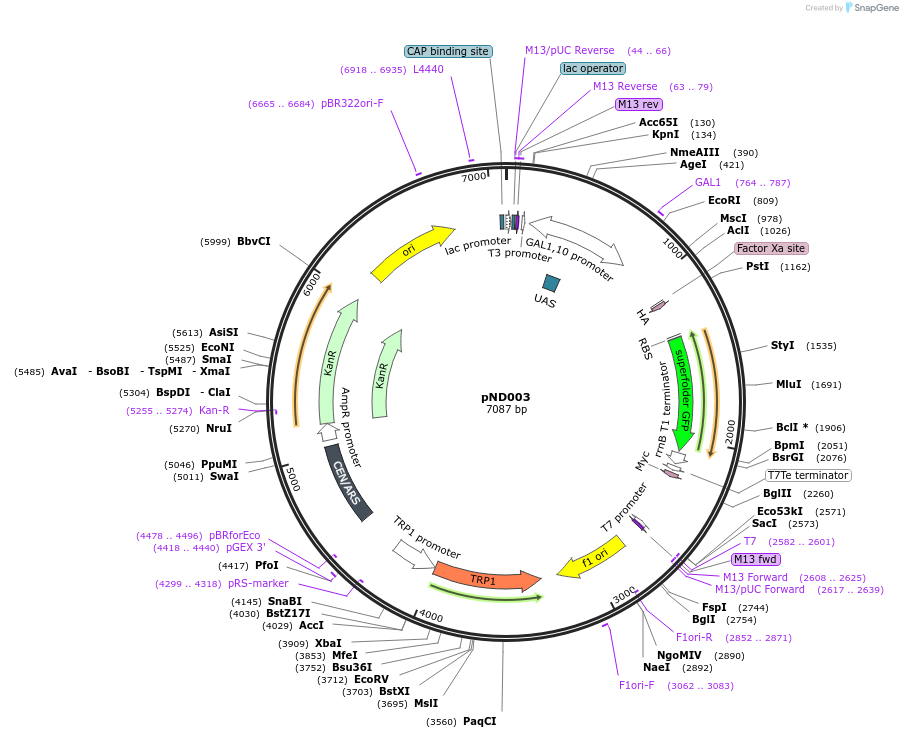
pND003
Plasmid#200949PurposeYeast surface display destination vector for Golden Gate cloningDepositorTypeEmpty backboneUseYeast surface displayTagsAGA2, HA, and MycExpressionYeastPromoterGAL1Available SinceJune 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
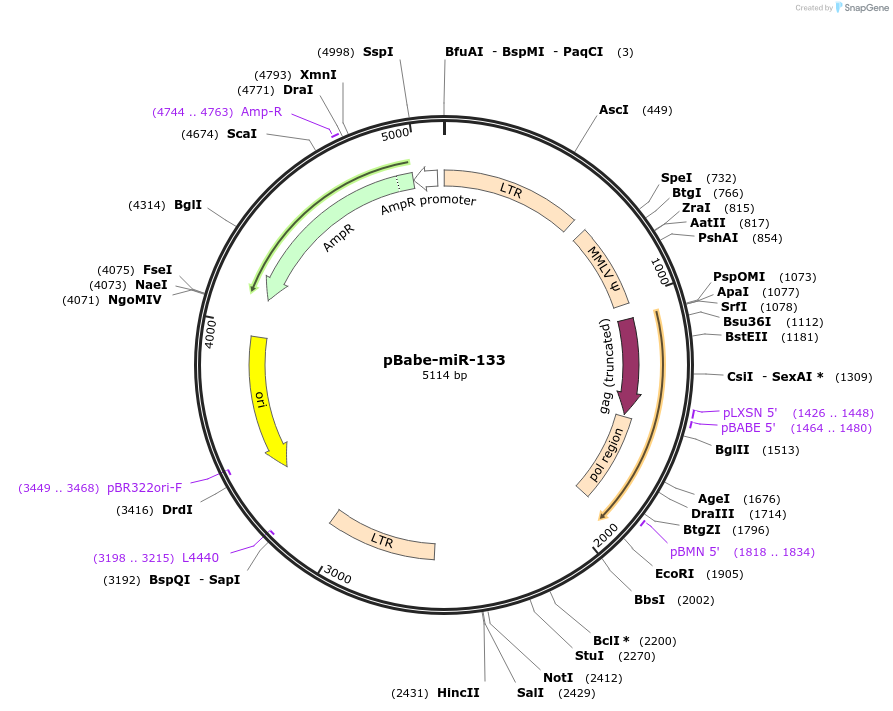
pBabe-miR-133
Plasmid#172392PurposeRetroviral expression vector for direct cardiac reprogrammingDepositorInsertmiR-133
UseRetroviralAvailable SinceAug. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
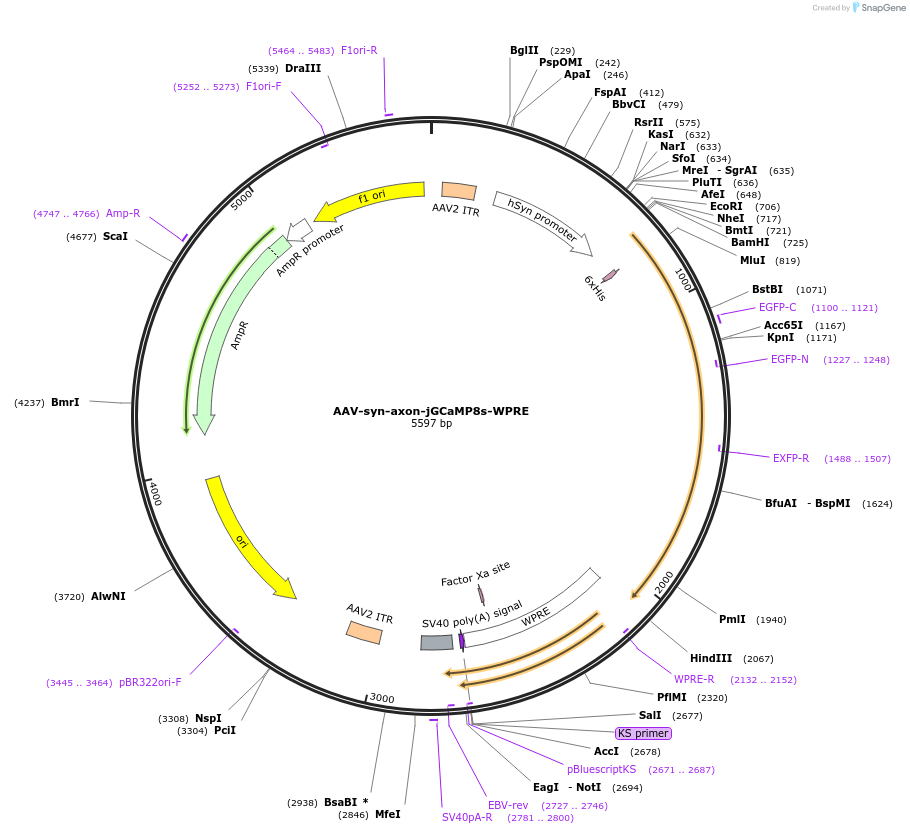
AAV-syn-axon-jGCaMP8s-WPRE
Plasmid#186043PurposejGCaMP8s expression in axons under the control of the synapsin promoterDepositorInsertaxon-jGCaMP8s
UseAAVTagsGAP43-6xHisExpressionMammalianPromotersynapsinAvailable SinceAug. 24, 2022AvailabilityAcademic Institutions and Nonprofits only